Abstract
Edwardsiella ictaluri is the leading cause of mortality in channel catfish culture, but little is known about its pathogenesis. The use of signature-tagged mutagenesis in a waterborne infection model resulted in the identification of 50 mutants that were unable to infect/survive in catfish. Nineteen had minitransposon insertions in miscellaneous genes in the chromosome, 10 were in genes that matched to hypothetical proteins, and 13 were in genes that had no significant matches in the NCBI databases. Eight insertions were in genes encoding proteins associated with virulence in other pathogens, including three in genes involved in lipopolysaccharide biosynthesis, three in genes involved in type III secretion systems (TTSS), and two in genes involved in urease activity. With the use of a sequence from a lambda clone carrying several TTSS genes, Blastn analysis of the partially completed E. ictaluri genome identified a 26,135-bp pathogenicity island containing 33 genes of a TTSS with similarity to the Salmonella pathogenicity island 2 class of TTSS. The characterization of a TTSS apparatus mutant indicated that it retained its ability to invade catfish cell lines and macrophages but was defective in intracellular replication. The mutant also invaded catfish tissues in numbers equal to those of invading wild-type E. ictaluri bacteria but replicated poorly and was slowly cleared from the tissues, while the wild type increased in number.
The gram-negative enteric bacterium Edwardsiella ictaluri causes enteric septicemia of catfish (ESC), an economically significant disease of farm-raised channel catfish. Commercial catfish production accounts for 85 to 90% of the total finfish aquaculture production in the United States, with almost 300,000 tonnes produced annually (36), and significant losses due to ESC were reported on over 60% of all farms in operation (84). The clinical signs and pathogenesis of E. ictaluri infections have been reviewed previously (83). Briefly, ESC generally presents in two forms: a rapid-onset, acute septicemia with high mortality and a chronic form confined to the central nervous system and characterized by a distinct “hole-in-the-head” lesion (75). Subacute infections also occur, with lower mortality rates than in acute infections. Presentation in any given epizootic is dependent on fish condition and water quality, especially temperature. Generally, fish less than 1 year old are more susceptible than older fish, and disease is more acute within the optimal temperature range of 22°C to 28°C and more chronic outside of that range (29, 55). Chronic disease is also seen in populations that have survived epizootics and have developed some immunity, especially if environmental conditions are suboptimal.
Although there is substantial descriptive data relative to the invasion, spread, and persistence of E. ictaluri in channel catfish (6, 64, 65, 83), little is known about the virulence factors involved in the process. Chondroitinase activity has been correlated to virulence by several authors (19, 78, 85) and may mediate the cartilage degradation in the chronic “hole-in-the-head” lesion (85). Lipopolysaccharide (LPS) (49, 50) and fibrillar processes that are apparently involved in attachment (64, 79) have also been implicated in the pathogenesis of E. ictaluri. There is direct evidence that E. ictaluri can survive in catfish neutrophils (2, 64, 79), and several reports allude to intracellular replication and survival in catfish macrophages and neutrophils, based on microscopic observations (2, 6, 61, 64, 75, 79). Booth et al. (11) recently reported the entry, survival, and replication of E. ictaluri in vitro in head kidney-derived macrophages (HKDM), and there are also reports of invasion into fish cell lines (76, 77).
Signature-tagged mutagenesis (STM) is a mutagenesis system involving minitransposons that carry unique DNA tags, enabling the identification either by hybridization (39) or by PCR (51) of individual mutants in a mixture of mutants carrying different tags. The methodologies associated with STM were recently reviewed (15, 17). Briefly, a mixture of tagged mutants is used to establish an infection, and the mutants that are initially present are later compared to the mutants that remain at the death of the host. Those lost during the infection process are presumed to be attenuated as a result of the insertion of the transposon into a gene that is required for survival in the host. Because the tagged transposons carry an antibiotic resistance marker, the region flanking the transposon can be subcloned from a restriction digest using antibiotic selection. The subcloned gene carrying the insertion can be identified by using the tag as a primer to sequence the DNA flanking the minitransposon and comparing the sequence to known bacterial DNA databases. Numerous colonization/virulence factors from a variety of bacterial pathogens have been identified by using STM, and summaries of those studies are available (58, 73). Based on the hypothesis that a basic knowledge of ESC pathogenesis and virulence factors is important to the ultimate prevention and control of the disease, the main objective of this study was to identify virulence-associated genes of E. ictaluri by using the STM procedures of Lehoux (51) in our channel catfish waterborne-infection model (82). This is the first report of virulence factors identified for E. ictaluri using modern molecular screening methods.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, Escherichia coli was grown in Luria-Bertani broth (LB) at 37°C and E. ictaluri strains were grown in brain heart infusion (BHI) broth at 28°C. Strain CC118 λpir of E. coli was used to maintain the STM delivery plasmids and to isolate plasmid DNA prior to introduction into the conjugation strain, S17-1 λpir. Antibiotics were used in the following concentrations: kanamycin (Km) at 50 μg ml−1, colistin (Col) at 10 μg ml−1, and ampicillin (Amp) at 200 μg ml−1. Edwardsiella ictaluri transconjugants reisolated from fish were grown on Trypticase soy agar plates supplemented with 5% sheep blood (BA).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Escherichia coli | ||
| CC118 λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA λpir lysogen | 23 |
| S17-1 λpir | Tp Sm thi pro recA HsdR− HsdM+; RP4-2-Tc::Mu::Km Tn7 λpir | 23 |
| XL1-Blue MRF′ | (mcrA)183 (mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proABlacIqZ.M15 Tn5(Kan)] | Stratagene, La Jolla, CA |
| Edwardsiella ictaluri | ||
| 93-146 | Wild-type E. ictaluri isolated in 1993 from moribund channel catfish in a natural outbreak of ESC on a commercial farm | LSU aquatic animal diagnostic laboratory |
| Plasmids | ||
| pBluescript SK- | Cloning vector | Stratagene, La Jolla, CA |
| pUT-miniTn5Km2 | Delivery plasmid carrying mini-Tn5Km | 23 |
| pUT-miniTn5Km-MCS | pUT-miniTn5Km2 with multiple cloning site containing EcoRV, XbaI, and ApaI restriction enzyme sites | This study |
| pUT-Km-STM | pUT-miniTn5Km-MCS carrying a specific tag in the MCS | This study |
SPF channel catfish.
Channel catfish egg masses were obtained from a commercial producer with no history of ESC outbreaks. The eggs were disinfected with 100 ppm free iodine and hatched in closed recirculating systems in the specific-pathogen-free (SPF) laboratory at the LSU School of Veterinary Medicine. The fish were reared on commercial catfish diets fed at 2 to 3% of their body weight per day until used for the infection and coinfection experiments. The 1.5- to 2-year-old fish used to harvest HKDM were reared entirely within the SPF laboratory and weighed 1,000 to 1,500 g.
Production of an STM plasmid and validation of the STM procedures.
When using both a wild-type E. ictaluri and an attenuated, tagged aroA mutant (82), our attempts to validate the use of the tagged pUTmini-Tn5Km2 plasmid and PCR conditions reported by Lehoux et al. (51) proved to be inconsistent for both mutant production and tag identification in E. ictaluri. We therefore modified pUT-miniTn5Km2 (22) for STM by inserting a new multiple cloning site containing EcoRV, XbaI, and ApaI sites (5′GATATCTCTAGAGGGCCC3′). The new plasmid, pUT-miniTn5Km-MCS, was digested with EcoRV and treated with calf intestinal alkaline phosphatase. The 12-tag sequences reported by Lehoux et al. (51) were synthesized and designated STM-A/B to STM-Y/Z, excluding the letters I and Q. Annealed tags were phosphorylated using T4 polynucleotide kinase (New England Biolabs) and ligated to the plasmid using T4 DNA ligase (New England Biolabs). The new plasmids were designated pUT-Km-STM-A/B to pUT-Km-STM-Y/Z, depending on which tag was inserted, and the orientation and single-copy insertion of each tag were verified by PCR. A minitransposon carrying tag STM-G/H was also ligated into the aroA gene (82) of wild-type E. ictaluri and used to validate the STM procedure.
Tagged E. ictaluri transconjugant libraries were made by conjugation using procedures previously described (57). Briefly, E. ictaluri 93-146 was mated with E. coli S17-1 λpir containing the tagged pUT-Km-STM, after which the transconjugants were suspended in 10 mM MgSO4. The specificities of the tags in the E. ictaluri genomic background were confirmed by PCR on a mixture containing DNA from individual mutants carrying each of the 12 tags, as well as a mixture containing all of the tags except the one being used as the primer. The presence of individual tags was confirmed by the amplification of a predicted 643-bp product. Three of the original tags (51), E/F, J/K, and W/X, were not used further because they amplified nonspecific products from the tagged E. ictaluri chromosomal DNA, even when DNA carrying the specific tags was not included in the screening (data not shown).
Generation and identification of E. ictaluri attenuated mutants by STM.
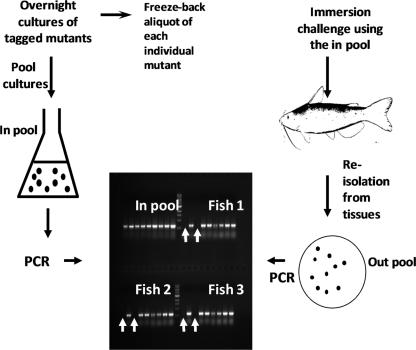
The procedures for STM used in this study are depicted in Fig. 1. Briefly, an aliquot of each tagged library was removed from the freezer, thawed, and spread on BHI plates with Km to select for transconjugants and Col to eliminate the E. coli donor (BHI-KmCol). Nine colonies, one from each plate of individual tags, were inoculated into individual tubes containing 6 ml of BHI-KmCol broth and incubated at 28°C for 16 h. After the incubation, 1-ml samples of individual transconjugants were frozen and stored in microfuge tubes at −80°C. The remaining 5 ml of culture for each transconjugant from each of the nine tagged libraries was combined to create 45-ml pools of transconjugants that were used in the immersion challenges (In pools). In addition, DNA was isolated from 500 μl of each In pool prior to challenge and frozen at −20°C to be used in PCR to verify that all nine mutants, each carrying an individual tag, were present.
FIG. 1.
Overview of the signature-tagged mutagenesis procedures. The two PCR products missing for all three fish (white arrows) represent putative attenuated mutants for further characterization.
For the immersion challenge, experimental fish were placed into 20-liter tanks supplied with a continuous flow of dechlorinated municipal water maintained at 25°C ± 1°C at a flow rate of 500 to 600 ml per minute. Fish were stocked at a density of 25 per tank and fed commercial catfish feed ad libitum every other day during a 4-week acclimation period. One tank of 20 fish was infected with each In pool by adding enough bacterial culture to achieve a final concentration of approximately 1 × 108 CFU per ml of tank water. When fish began dying, generally 6 to 7 days postinfection, bacteria were isolated from three dead or moribund fish by spreading 200 μl of a liver tissue homogenate on BA plates. The resulting solid lawn of bacteria was washed off with 5 ml sterile saline and collected in sterile test tubes (Out pool). Each of these Out pools was processed by PCR, and transconjugants that were missing from the Out pools potentially carried mutations in virulence-related genes.
Cloning of flanking DNA and sequence analysis.
Each putative virulence-associated mutant was further evaluated to determine the gene of insertion. Briefly, chromosomal DNA was digested with ClaI, which does not cut the Km-STM minitransposon, ligated into the ClaI site of pBluescript SK− (Stratagene, La Jolla, CA), and electroporated into E. coli XL1-Blue MRF′. Km/Amp-resistant colonies containing plasmids carrying the mini-Tn5-Km-STM transposon and associated chromosomal DNA flanking the site of insertion were selected on LB-Km/Amp, and plasmid DNA was purified by using a QIAprep spin mini-prep kit (QIAGEN, Valencia, California). The region adjacent to the tagged end of the transposon was sequenced by using the tag-specific primer on an ABI prism 377 automated sequencer (PE-Applied Biosystems, Foster City, CA), and the resulting sequence was analyzed against the NCBI databases using Blastx to ascertain homologous sequence information.
CI assays.
For the competitive index (CI) assays, mutant strains and wild-type E. ictaluri bacteria were grown separately in BHI at 28°C for 18 h. All mutants tested grew well in broth culture, with final plate counts averaging 9.2 × 109 CFU/ml with a range of 5.8 × 109 to 1.1 × 1010 CFU/ml. Based on the values for optical density at 600 nm, cultures were diluted so that equal numbers of wild-type and mutant bacteria were mixed together and used in the immersion challenge at a final concentration of 1 × 108 CFU/ml of tank water. Liver samples were removed from at least three dead or moribund fish in each tank, homogenized, serially diluted, and drop plated on BHI to determine the total CFU recovered and on BHI-Km to determine the number of mutant CFU recovered. The CI was determined by dividing the recovery ratio of mutant CFU/wild-type CFU by the input ratio of mutant CFU/wild-type CFU. The values, ranging from 0 to 1, indicate the level of attenuation, with values closer to 0 indicating greater attenuation and values closer to 1 greater virulence. A CI of 0.0 indicates that no mutants were recovered.
LPS immunoblotting.
Putative LPS mutant and wild-type E. ictaluri cells were grown in BHI broth, harvested by centrifugation, and washed three times in phosphate-buffered saline, pH 7.3. The cells were resuspended in sterile, distilled, deionized water to a concentration of 0.1 ml of cell pellet/ml of water and sonicated until the cell suspensions cleared. The sonicated cells were centrifuged to remove particulate cell debris, and the supernatants were subjected to electrophoresis on a 12% polyacrylamide gel under denaturing conditions. Purified E. ictaluri LPS was run as a positive control. Lysates were transferred to 0.45 μM nitrocellulose membranes at a constant 100 V for 1 h in 25 mM Tris, 192 mM glycine, and 20% (vol/vol) methanol, pH 8.3. The blots were air dried and stored in the dark until the LPS was immunodetected by using mouse anti-LPS primary monoclonal antibody Ed9 (1) and goat anti-mouse immunoglobulin G heavy-plus-light chains-horseradish peroxidase-labeled secondary antibodies with an enhanced chemiluminescence (ECL) Western blotting analysis system (GE Healthcare, Piscataway, NJ).
Further sequencing of the E. ictaluri type III secretion system (TTSS).
A 259-bp PCR product was amplified from the pBluescript plasmid carrying the subcloned 65ST insertion by using primers identified from the E. ictaluri esaU sequence. The fragment was labeled by using the ECL DNA-labeling system (GE Healthcare) and was used to screen a previously described λ Zap Express E. ictaluri genomic library (82) according to the manufacturer's instructions (Stratagene, Inc., La Jolla, CA). The pBK-CMV phagemid was excised from hybridization-positive plaques according to the Zap Express protocol and was sequenced as described above. The sequence derived was subsequently used to search the partially completed E. ictaluri genome database (www.microgen.ouhsc.edu/cgi-bin/blast_form.cgi) to identify a 41,205-bp fragment carrying 33 genes (26,135 bp) in a pathogenicity island encoding a TTSS. Because of the similarity between the E. ictaluri and the Salmonella and E. tarda TTSS sequences, gene names were assigned based on homology. In order to evaluate similarity to putative Salmonella homologues when Blastx alignments were not identified, the L-ALIGN program at www.ch.embnet.org was used to directly compare sequences, and various proteomics programs at us.expasy.org were used to compare primary and secondary protein structures. Salmonella gene names were assigned to previously unnamed E. tarda open reading frames (ORFs) when L-ALIGN alignments and protein structure analysis indicated similarity (see Table 4).
TABLE 4.
Comparison of the E. ictaluri TTSS to the E. tarda and Salmonella pathogenicity island 2-encoded TTSS and putative functions
| E. ictaluri genea | Total no. of amino acids of product | % Identity/similarity at the amino acid level to gene product of:
|
Putative TTSS function | |
|---|---|---|---|---|
| E. tarda | S. enterica serovar Typhimurium | |||
| esaL | 341 | 88/92 | 28/47 | Regulation of translocon expression |
| esaK | 217 | 80/88 | 22/56 | Apparatus protein with glutamine-rich region |
| esaW (orf19) | 194 | 90/94 | 30/55 | Apparatus protein with leucine-rich region |
| esaJ | 243 | 88/94 | 41/59 | Outer membrane protein of the needle complex |
| esaI | 83 | 78/90 | 43/64 | Apparatus protein |
| esaH | 88 | 90/94 | 30/57 | Apparatus protein |
| esaG | 73 | 90/98 | 46/69 | Apparatus protein |
| esrC | 230 | 96/97 | AraC family regulatory protein | |
| esaE (orf13) | 74 | 88/94 | 25/45 | Hypothetical protein |
| esaD | 399 | 82/87 | 24/42 | Periplasmic channel |
| esaC | 494 | 93/95 | 46/67 | Oligomeric outer membrane secretin |
| esaB | 159 | 89/91 | 27/77 | Outer membrane component |
| eseGb | 301 | 70/79 | 26/44 | Effector protein |
| escB | 163 | 88/92 | 30/46 | sseF and sseG chaperone |
| eseE | 127 | 92/96 | 25/49 | Chaperone |
| eseD | 193 | 78/89 | 27/54 | Translocon protein |
| eseC | 508 | 78/88 | 27/48 | Translocon protein |
| escA | 155 | 92/96 | 30/53 | Chaperone |
| eseB | 198 | 77/87 | 33/52 | Translocon filament protein |
| escCc(orf2) | 140 | 80/86 | 25/58 | Unknown |
| esaQ | 304 | 86/90 | 39/57 | Apparatus protein that regulates translocation |
| esaP (orf1A) | 159 | 91/95 | 17/49 | Apparatus protein, integral membrane protein |
| esaO (orf1B) | 122 | 80/83 | 24/64 | Apparatus protein containing leucine zipper |
| esaN | 438 | 90/94 | 55/68 | Apparatus protein, ATP synthase |
| esaV | 685 | 93/96 | 52/69 | Apparatus membrane protein |
| esaM | 126 | 90/93 | 29/47 | Apparatus protein, secretion regulator |
| esaR | 250 | 96/99 | 58/76 | Integral apparatus membrane protein |
| esaS | 89 | 96/97 | 52/71 | Integral apparatus membrane protein |
| esaT | 260 | 93/96 | 43/64 | Integral apparatus membrane protein |
| esaU | 352 | 89/94 | 43/60 | Integral apparatus membrane protein |
| slt (orf26) | 220 | 81/85 | 30/65 | Soluble lytic murein transglycosylase |
| esrA | 937 | 90/93 | 32/48 | Regulatory sensor kinase |
| esrB | 214 | 96/98 | 48/66 | Response regulator |
Where named differently, comparable E. tarda genes are in parentheses. L-ALIGN and protein structure analysis of the E. ictaluri homologues of E. tarda orf2, orf1A, orf1B, orf13, orf19, and orf26 demonstrated significant similarity to the S. enterica serovar Typhimurium homologues, and the E. ictaluri homologues are named accordingly.
Based on analysis of sixfold coverage of the E. ictaluri genome in this region, E. ictaluri encodes an eseG gene that has a truncated amino terminus due to the loss of an A at position 11382 of the DQ233733 sequence that introduces a stop codon. The E. ictaluri ureG gene starts at bp 120 of the E. tarda ureG gene.
Formerly eseA (see reference 88).
Confirmation of a single transpositional event in the TTSS mutant 65ST.
Genomic DNA from 65ST was isolated using standard methods (5), and 10 μg was cut to completion with ClaI, which does not cut the mini-Tn5 Km-STM transposon. Cut genomic DNA was separated on a 1% agarose gel and transferred to an ECL Hybond N+ nylon membrane (GE Healthcare). A 373-bp Km-STM PCR product was amplified using primers Tn5kan+ (5′ACACGTAGAAAGCCAGTCCG3′) and Tn5kan− (5′CCCAGTCATAGCCGAATAG3′), labeled using the ECL nucleic acid-labeling system, hybridized to the 65ST genomic DNA on the membrane, and detected using the ECL detection reagents.
Intracellular replication of the TTSS mutant 65ST.
A standard gentamicin survival assay (26) was used to evaluate the abilities of the wild-type and the TTSS mutant E. ictaluri bacteria to enter and to survive and replicate in channel catfish HKDM and the channel catfish ovary (CCO) cell line. Briefly, HKDM were isolated by the method of Booth et al. (11) and viable counts were determined by using trypan blue dye exclusion (70). Dissociated cells were suspended to a final concentration of 1 × 107 cells/ml in channel catfish macrophage medium (CCMM) consisting of RPMI 1640 medium (GIBCO, Invitrogen Corporation, Carlsbad, CA) diluted to a catfish tonicity of 243 mOsm/kg by adding 1 part sterile deionized/distilled water (RPMI 9:1) and containing 15 mM HEPES buffer solution (GIBCO), 0.18% sodium bicarbonate solution (GIBCO), 0.05 mM beta-mercaptoethanol (Sigma Chemicals Co., St. Louis, MO), and 5% heat-inactivated pooled channel catfish serum (27, 60). One ml of the cell suspension was added to each well of a 24-well plate and allowed to adhere for 16 h at 28°C with 5% CO2, after which the wells were washed three times with RPMI 9:1 to remove nonadherent cells and 1 ml of fresh CCMM was added per well. Following adherence and washing, the attached cells averaged 91.4% macrophages, 5.4% neutrophils, and 3.1% unidentified cells, as determined by nonspecific esterase and Sudan black B staining (25), and averaged 2.5 × 105 cells per well.
To evaluate the efficiency of entry and replication, 1 × 104 of either wild-type or TTSS mutant E. ictaluri cells that had been opsonized for 30 min in normal autologous serum were added to triplicate wells of the 16-h HKDM cultures, giving a multiplicity of infection (MOI) of 1 bacterium:10 HKDM. After infection, the plates were centrifuged at 200× gravity to synchronize contact of the bacteria with the adhered cell layer and allowed to incubate for 30 min. The medium was then removed from each well, and CCMM with 100 μg/ml gentamicin was added for 1 h to kill residual extracellular bacteria. The cells were then washed three times with RPMI 9:1, and CCMM containing a 0.35-μg/ml bacteriostatic dose of gentamicin was added to control the extracellular growth of bacteria released from the cells. At 0 (90 min postinfection), 5, and 10 h, the HKDM were lysed by the addition of 100 μl of a 1% solution of Triton X-100 (Fisher Scientific, Fair Lawn, NJ), and the numbers of surviving E. ictaluri cells were determined by spreading serial dilutions on BA. A similar assay was used for the CCO cells, except that L-15 medium (GIBCO) was used, the cells were split and passed using standard cell culture methods, a 100-μg/ml dose of gentamicin was used throughout the assay, and the MOI was 1:1, bacteria to cells. In addition to the enumeration of intracellular bacteria, coverslips with both uninfected and infected HKDM and CCO cells were removed from wells without being lysed, stained using a Hema 3 stain kit (Fisher Diagnostics, Middletown, VA), mounted on glass slides with Permount (Fisher Chemical, Fair Lawn, NJ), and observed by light microscopy.
Invasion and persistence of the TTSS mutant, 65ST, in vivo.
The comparative invasion and persistence of the wild-type E. ictaluri and the TTSS mutant, 65ST, were evaluated in vivo using SPF channel catfish fingerlings with an average weight of 5 g. Sixty fish were stocked in each of two tanks, and one of the groups was experimentally infected by immersion exposure with wild-type E. ictaluri while the other was experimentally infected with strain 65ST. Briefly, after the water levels were dropped to 4 liters per tank, overnight broth cultures of E. ictaluri 93-146 or strain 65ST were each added directly to the tanks at doses of 2.2 × 108 CFU/ml of tank water. The numbers of CFU/ml were estimated by using a standard curve for optical density at 600 nm and confirmed by drop plating serial dilutions onto BA. The water flow was stopped for 15 min following initial exposure and then resumed.
To assess the initial invasion, five fish were removed from each tank at 0.5, 2, 4, and 8 h postexposure and euthanized by being transferred to water containing 1 g/liter MS-222. In order to assess the persistence in the tissues, five fish were also collected at 24, 48, 72, 96, 120, 144, and 168 h. Five fish were also collected from each tank prior to the experimental infections for a preinfection sample. Using aseptic technique, samples of the head kidney were suspended in 0.5 ml sterile 0.9% saline solution, weighed, and homogenized. The resulting suspension was serially diluted in 0.9% saline solution in triplicate using 96-well plates, and 20-μl aliquots of each triplicate well were dropped onto BHI plates for quantification. Colonies were counted after incubation for 48 h, and the CFU/gm of tissue was calculated.
Statistical methods.
The percentages of uptake in the CCO and HKDM gentamicin survival assays were calculated by dividing the mean number of CFU/well at time zero by the number of CFU in the initial bacterial inoculum. Differences in percent uptake and growth between strains were analyzed by using the general linear model from Statistical Analysis Systems (SAS), version 9.1 (SAS Institute, Incorporated, Cary, North Carolina). In the in vivo invasion/persistence study, differences in CFU/g tissue for the mutant and the wild-type E. ictaluri bacteria were analyzed by using two-way analysis of variance following a log transformation of the CFU count data. When the overall model indicated significance at a P value of <0.05, Scheffe's test was used for pairwise comparison of the main effects, and a least square means procedure was used for pairwise comparison of the interaction effects.
RESULTS
Screening of the E. ictaluri transconjugant library.
A total of 119 pools, containing nine mutants per pool, were each used to infect one tank of 25 fish. Of the 1,071 mutants screened, 50, or about 4.7% of the total transconjugants, were missing from the Out pools, indicating that they carried an insertion in virulence-related genes. This corresponds to the upper end of the 1.5 to 6.5% range identified by using STM in other gram-negative pathogens (58, 73).
Identification of the gene of insertion.
Cloning and sequencing of the DNA flanking the inserted transposon of all 50 attenuated mutants indicated that none of the mutants contained vector plasmid integrations. This is consistent with our earlier results using a similar pGP704-based suicide vector, pLOF, in E. ictaluri, in which plasmid integrations occurred in only 0.6% of the mutants generated (57). Details of the matches found by analysis of the NCBI bacterial databases are presented in Table 2. In summary, 19 mutants had insertions in miscellaneous genes in the chromosome, 10 matched to various hypothetical proteins, and 8 had insertions that were in genes matching to virulence genes identified in other pathogens. Thirteen mutants had insertions in genes with no significant amino acid or nucleotide matches in the NCBI databases (Table 3).
TABLE 2.
E. ictaluri virulence-related genes identified by STMa
| Type and name of mutant | Homologous gene product (organism)b | Function of matching protein in NCBI databases (accession no.) | Length in amino acids | % Amino acid identity | % Amino acid similarity | % Nucleotide identity | CI |
|---|---|---|---|---|---|---|---|
| Miscellaneous chromosomal mutants | |||||||
| 50ST | NagA (YPK) | Glucosamine catabolism (NP_406152) | 134 | 71 | 85 | 71 | ND |
| 65NO | InsA (SHD) | Insertion element (AAA25031) | 50 | 80 | 82 | 86 | 0.0 |
| 69ST | MiaA (YPS) | Amino acyl tRNA synthesis (NP_404020) | 140 | 80 | 90 | 71 | 0.014 |
| 69YZ | GidA (SAT) | Glucose-inhibited division protein (NP_462773) | 212 | 91 | 96 | 78 | ND |
| 86LM | PyrD (SHF) | Pyrimidine biosynthesis (NP_706868) | 118 | 81 | 91 | 73 | ND |
| 95GH | LipA (SET) | Lipoic acid synthesis (NP_459625) | 71 | 95 | 97 | 81 | ND |
| 155LM | GuaB (SET) | Purine biosynthesis (NP_457045) | 205 | 84 | 88 | 81 | ND |
| 157AB | Ppc (SHF) | Anaerobic respiration (NP_709756) | 30 | 90 | 93 | 73 | ND |
| 172UV | FolD (SET) | Folic acid biosynthesis (NP_455132) | 16 | 87 | 93 | 66 | ND |
| 196GH | Fom1 (STW) | Glycolysis (BAA32495) | 223 | 53 | 69 | 51 | ND |
| 197GH | AdiA (SAT) | Biodegradation of arginine (NP_463161) | 91 | 66 | 82 | 64 | 0.16 |
| 199UV | NuoH (YPS) | Aerobic respiration, electron transport (NP_406080) | 193 | 90 | 96 | 79 | ND |
| 203AB | Hpt (SAT) | Purine metabolism (NP_459175) | 69 | 78 | 92 | 75 | ND |
| 207AB | 0304 (VCH) | Purine metabolism (NP_229959) | 211 | 51 | 73 | 59 | ND |
| 223UV | Ssb (SHF) | Single-stranded DNA binding protein (NP_709860) | 119 | 84 | 90 | 77 | ND |
| 225UV | HupA (YPS) | DNA binding protein HU-alpha (NP_407181) | 15 | 100 | 100 | 86 | ND |
| 231ST | Cls (PSS) | Phospholipid biosynthesis (ZP_00125405) | 113 | 32 | 56 | 46 | ND |
| 234AB | AdiA (SHF) | Biodegradation of arginine (NP_838857) | 192 | 61 | 74 | 67 | 0.00013 |
| 236UV | ECA | Universal stress protein (CAG74674.1) | 142 | 63 | 80 | 35 | 0.00000022 |
| Hypothetical proteins | |||||||
| 60AB | SET | Protein STY3950 (AL513382) | 162 | 46 | 57 | 62 | 0.000012 |
| 71AB | BRJ | Protein bll4065 (overlaps protein with no database match in 147AB) (NP_770705) | 194 | 45 | 65 | 44 | 0.00049 |
| 72NO | PST | Protein with pentapeptide repeats (Q52118) | 131 | 45 | 58 | 29 | 0.00000062 |
| 168AB | SAT | Methyl-accepting chemotaxis protein (NP_462053) | 162 | 33 | 55 | 44 | 0.0 |
| 180GH | YojN (SAT) | Putative signal transduction histidine kinase (NP_461211) | 157 | 41 | 53 | 62 | 0.00051 |
| 193UV | YPS | Putative fimbrial usher protein (YPO0302) | 73 | 29 | 52 | 40 | 0.00000044 |
| 194AB | SET | Putative exported protein (NP_457745) | 218 | 57 | 72 | 61 | 0.000011 |
| 196ST | OBI | Sigma-L-dependent transcriptional regulator (NP_691931) | 105 | 27 | 43 | 36 | 0.0000011 |
| 233PR | PHL | Putative adhesin (NP_927889) | 123 | 34 | 55 | 36 | 0.00089 |
| 245YZ | DSD | Putative Fic protein family; COG3177 (ZP_00129414) | 181 | 55 | 67 | 66 | 0.0 |
| Virulence-related mutants | |||||||
| 58PR | LsgA (HPD) | LPS biosynthesis protein (NP_873379) | 140 | 23 | 49 | 43 | 0.002 |
| 142YZ | WaaL (EDT) | LPS biosynthesis (AAL01247) | 22 | 86 | 95 | 95 | 0.0 |
| 173ST | WaaL (EDT) | LPS biosynthesis (AF326578) | 108 | 80 | 86 | 84 | 0.0000064 |
| 65ST | EsaU (CHV) | Type III secretion apparatus protein (NP_902280) | 266 | 43 | 57 | 59 | 0.0 |
| 84LM | UreG (YEN) | Urease accessory protein (P42871) | 35 | 86 | 100 | 79 | 0.000013 |
| 166ST | EIC | Repeat region, E. ictaluri plasmid pEI2, upstream from type III secretion chaperone/effector (AF244084) | 51 | 100 | 100 | 100 | 0.0 |
| 217UV | SspH1 (SAT) | Type III secretion effector protein (AAD40326) | 680 | 43 | 56 | 66 | 0.0000012 |
| 243NO | UreF (YPS) | Accessory for urease assembly (NP_406188) | 80 | 62 | 82 | 61 | 0.00000045 |
The length in amino acids measures the region sequenced from the tag. In some cases the insert was immediately upstream of the gene indicated but is predicted to affect expression. Amino acid identity and similarity values show the comparison of the amino acids adjacent to the tag with those in the most-similar homologue. In instances where nucleotide matches were not identified by Blast, nucleotide identity was determined by alignment in L-ALIGN at www.ch.embnet.org/software/LALIGN_form.html. CI is calculated relative to the wild-type strain 93-146 as described in Materials and Methods. CI values are the averages of the results for a minimum of three fish. In fish where no mutant bacteria were recovered, the CI is recorded as 0.0.
Most-similar homologue of gene product, from organism indicated in parentheses. BRJ, Bradyrhizobium japonicum; CHV, Chromobacterium violaceum; DSD, Desulfovibrio desulfuricans; ECA, Erwinia carotovora; EPEC, enteropathogenic Escherichia coli; EIC, Edwardsiella ictaluri; EDT, Edwardsiella tarda; HPD, Haemophilus ducreyi; OBI, Oceanobacillus iheyensis; PHL, Photorhabdus luminescens; PSS, Pseudomonas syringae; PST, Pantoea stewartii subsp. stewartii; SAT, Salmonella enterica serovar Typhimurium; SET, Salmonella enterica subsp. enterica serovar Typhi; SHD, Shigella dysenteriae; SHF, Shigella flexneri; STW, Streptomyces wedmorensis; VCH, Vibrio cholerae; YEN, Yersinia enterocolitica; YPK, Yersinia pestis KIM; YPS, Yersinia pestis.
TABLE 3.
CIs for E. ictaluri mutants with transposon insertions in genes with no significant matches in the genetic databases at either the amino acid or nucleotide level
| Mutant | % G+C contenta | CI |
|---|---|---|
| 61ST | 40.7 | 0.000020 |
| 147ABb | 48.7 | 0.000001 |
| 184GH | 54.3 | 0.0000041 |
| 185GH | 48.8 | 0.0000022 |
| 195CD | 51.3 | 0.0 |
| 203UV | 46.0 | 0.000045 |
| 205NO | 46.0 | 0.0 |
| 219YZ | 48.2 | 0.00016 |
| 220PR | 43.4 | 0.000000053 |
| 222 CD | 59.8 | 0.079 |
| 242 PR | 58.8 | 0.00000070 |
| 242 YZ | 43.9 | 0.0 |
| 254 CD | 56.2 | 0.0000011 |
The E. ictaluri genome has a reported 53% G+C content (37). The percentages of G+C were calculated using 500 bp flanking both sides of the STM transposon insertion site.
Overlaps with hypothetical protein in 71AB.
Miscellaneous chromosomal insertions.
Although most of the transconjugants with an insertion in miscellaneous genes in the chromosome have obvious housekeeping roles (Table 2), several may have direct roles in virulence. Mutant 65NO has an insertion in a gene with high similarity to insA, the transposase of the insertion element, IS1, a common insertion sequence in the Enterobacteriaceae that is often present in multiple copies in a genome. The capacity of insertion sequences to modify gene expression, sequester genes, and promote genome rearrangements (31) makes 65NO an interesting mutant, and the severe level of attenuation, with a CI of 0.0, indicates that an important gene has been affected. Mutant 69ST carries an insertion in the miaA gene, which is involved in the posttranscriptional modification of tRNA. Transconjugants 197GH and 234AB both carry the transposon insertion in the arginine decarboxylase (adiA) gene.
Insertions in hypothetical genes in other pathogens.
Of the 10 mutants with insertions in hypothetical genes, 2 have insertions in genes encoding proteins that have similarity to putative adhesion-related proteins. The insertion in 193UV is in a gene possibly encoding a fimbrial assembly/usher protein that has a CI of 0.00000044, indicating an important role in virulence. Further sequence analysis revealed homologues to proteins identified as a putative fimbrial chaperone and putative outer membrane proteins. These putative genes are flanked by transposon insertions, and the region has a 43.8% G+C content, compared to the 53% G+C content of the E. ictaluri chromosome (37), suggesting the horizontal transfer of a pathogenicity island (34).
The insertion in 233PR is in a gene with similarity to hypothetical adhesion-related genes but is more moderately attenuated, with a CI of 0.00089. Analysis of surrounding sequences revealed several genes with similarity to putative hemolysin/adhesion genes associated with a genomic island in some of the verotoxin-producing strains of E. coli CL3 (74) and Yersinia pestis (68) and a region that encodes sequences homologous to CdiA and CdiI, which are associated with contact-dependent inhibition of growth in E. coli (3). This region, containing about 10 ORFs, is also bordered by TnpA transposon sequences, again indicating possible horizontal transfer of this region, but has a percent G+C content similar to that of the overall E. ictaluri genome.
The mutation in 72NO, with a CI of 0.00000062, is in a gene whose product is related to a variety of hypothetical proteins with similarity to a family of gene products that carry pentapeptide repeats. These repeats are described by the motif A(D/N)*XX, where the asterisk denotes a polar amino acid and the X is any amino acid (7). The first 75 amino acids following the transposon insertion in 72NO comprise a pentapeptide-repeat domain with a very similar repeat motif, except that asparagine is not as common at position two.
Two other mutants had insertions in genes with hypothetical roles in signal recognition. The insertion in 180GH is in a gene encoding a putative signal transduction histidine kinase (HK). Further sequence analysis immediately downstream from the HK revealed a positive response regulator containing a CheY-like receiver domain and a helix-turn-helix domain. There is a 7-bp overlap of the two ORFS, suggesting that they comprise an operon encoding a two-component signal transduction system (54). Two-component systems respond to environmental cues by transferring a phosphate group from a conserved histidine residue on the HK to a conserved aspartate residue on the response regulator, causing a transformational change in the protein structure, resulting in either activation or repression of transcription. Because several of these two-component systems were identified using in vivo expression technologies (44, 86), a role in host environment recognition is postulated, although the relatively high CI of 0.0005 would indicate a more minor role. The role in virulence of the putative methyl-accepting chemotaxis protein (MCP) mutated in 168AB was confirmed by the fact that no 168AB mutants were recovered from any of the three fish evaluated in the competitive assay.
Insertions in known virulence-related genes in other pathogens.
In total, eight insertions were in genes encoding E. ictaluri proteins with similarity to proteins known to be directly involved in the virulence of other bacterial pathogens (Table 2). Three of those, 58PR, 142YZ, and 173ST, encode proteins involved in the synthesis of LPS. Immunodetection of Western-blotted whole-cell lysates with monoclonal antibody to E. ictaluri O-antigen (monoclonal antibody Ed9) (1) determined that all three are negative for the production of O-antigen side chains (data not shown). The severe level of attenuation of the LPS mutants is consistent with the high level of attenuation previously reported for E. ictaluri LPS mutants (50).
Two of the mutants, 84LM and 243NO, carried insertions in genes involved in urease activity, and both were confirmed in the competitive assay to be severely attenuated, with CI values of 0.000013 and 0.00000045, respectively. This is interesting in light of the fact that E. ictaluri is urease negative in standardized biochemical tests. The insert in 84LM is in a gene similar to ureG, which encodes a GTP-binding protein that is thought to function in energy-dependent urease assembly (63). The insert in 243NO is in a gene similar to ureF, encoding a protein thought to function in the generation or delivery of carbon dioxide to the metallocenter of the enzyme to facilitate interaction between the urease apoprotein and the UreE holoprotein (63). Further analysis of the E. ictaluri genome has identified a urease operon containing seven genes with a gene arrangement similar to those in Escherichia coli O157:H7, Morganella morganii, and Yersinia enterocolitica, as well as putative urea and ammonia transporters (10).
The three remaining virulence-related mutants, 65ST, 166ST, and 217UV, had insertions in genes encoding proteins with similarity to proteins from TTSS, and all were confirmed to be severely attenuated in the competitive assays (Table 2). The insertion in 65ST is in a gene encoding a protein with homology to EsaU/SsaU and to the homologous proteins in a number of other gram-negative bacteria carrying a TTSS. In other gram-negative pathogens, EsaU/SsaU homologues are associated with the inner-membrane structural component of the type III secretion apparatus, which is essential for type III secretion and virulence (20, 41). The CI value of 0.0 for 65ST suggests an essential role in virulence for EsaU and the TTSS in E. ictaluri. Consequently, the EsaU mutant is characterized further below.
Interestingly, two of the TTSS-related insertions, in transconjugants 166ST and 217UV, are located either in or immediately upstream of putative TTSS effector proteins previously reported on the two native plasmids of E. ictaluri, pEI1 and pEI2 (28). The transposon insertion in 166ST is in repeat 1 of pEI2, 120 bp upstream from the start codon for orf1 (28). This region does not encode any recognizable ORFs, but the mutant has a CI of 0.0 and the insertion may be in a region with a regulatory role in the expression of orf1. The similarity in size, pI, and sequence to the spa15-encoded chaperone of Shigella flexneri (67) suggests that orf1 of pEI2 is a TTSS chaperone. orf1 is in an apparent operon with a putative TTSS effector that has similarity to OspB, a TTSS effector in several gram-negative pathogens. The insertion in 217UV is in orf1 of pEI1 (28), which has similarity to leucine-rich-repeat (LRR) effector proteins in a variety of pathogens, including several Salmonella-translocated effectors (STE), several IpaH proteins in Shigella, YPO1007 and YopM in Yersinia, Y4FR in Rhizobium, and ID431 and blr1676 in Bradyrhizobium.
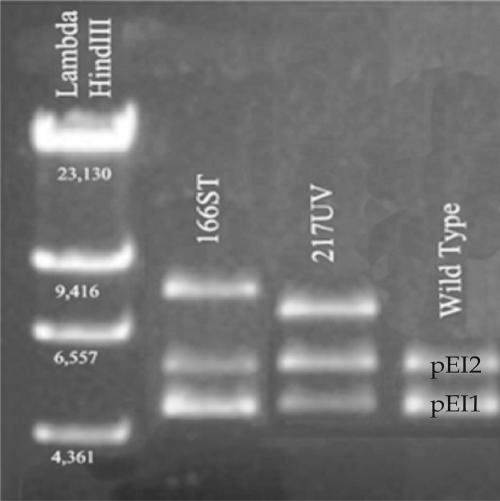
The identification of an attenuated phenotype as a result of insertions in the plasmids by STM is somewhat surprising. Both pEI1 and pEI2 are multicopy plasmids (data not shown), and not all of the copies would be expected to carry the transposon if an insertion occurs in one of the plasmids. The examination of plasmid profiles for 166ST and 217UV confirms that both wild-type and mutant plasmids persist, although differences in band intensity suggest a reduction in copy number for the wild-type plasmid that corresponds to the plasmid in which the insertion occurred (Fig. 2). The intact genes and associated promoters in the remaining wild-type plasmids, however, would be expected to complement the activity lost in association with either mutation. The low CI values of both 166ST and 217UV, however, suggest that a reduction in the number of copies of the genes in question results in attenuation, possibly due to competitive binding by nonfunctional mutant proteins or to a need for a threshold number of protein molecules to mediate function.
FIG. 2.
Agarose gel showing plasmid preparations from wild-type Edwardsiella ictaluri and two mutant strains with transposon insertions in plasmids pEI1 and pEI2. Note the mutant pEI2 plasmid in 166ST carrying the 2,378-bp transposon insertion and the lower relative brightness of the wild-type pEI2 plasmid, suggesting an apparent reduction in copy number. Also note similar results for pEI1 in 217UV. The left lane is a size ladder with numbers of base pairs indicated.
Insertions in genes with no known genetic matches.
The most intriguing mutants uncovered in this study are those that carry insertions in genes with no matches in the bacterial databases at NCBI (Table 3). Most of the mutants with insertions in genes with no matches have very low CI values, and several have very low G+C content, suggesting that they are located in E. ictaluri pathogenicity islands. Further work to identify the roles of these unknown proteins may lead to the identification of factors unique to the pathogenesis of E. ictaluri.
CI assays.
The completion of the CI assays resulted in CI values ranging from 0.0 to 0.079 (Tables 2 and 3). Of the mutants with insertions in known virulence genes, three had CI values of 0.0, including one in a putative LPS biosynthesis gene, one immediately upstream of a putative TTSS chaperone and effector, and one in a TTSS apparatus protein. All of the mutants with insertions in genes encoding hypothetical proteins or proteins with no matches in the databases had CI values of less than 0.079, with six values of 0.0.
E. ictaluri TTSS.
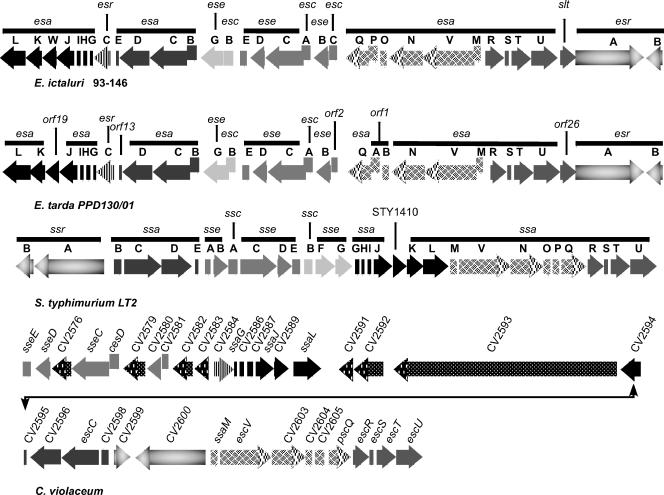
The hybridization of an esaU probe to the E. ictaluri λ library resulted in the identification of a 4,662-bp E. ictaluri genomic fragment carrying six complete ORFs encoding proteins with homology to the inner membrane components of the needle complex of TTSS of a number of gram-negative bacteria. Using this sequence, subsequent Blastx analysis of the partial E. ictaluri genome identified a 41,205-bp fragment containing a 26,135-bp pathogenicity island encoding 33 genes of a TTSS (GenBank accession no. DQ233733). The E. ictaluri TTSS is highly similar to the TTSS described for Edwardsiella tarda (81), and both are similar to the SPI-2 class of TTSS carried by Salmonella enterica serovar Typhimurium, Escherichia coli, and Chromobacterium violaceum (8). Gene alignment and comparisons are presented in Fig. 3 and Table 4.
FIG. 3.
Genetic organization of the type III secretion system genes of E. ictaluri compared to homologous regions of other TTSS. Arrows with the same pattern indicate homologous proteins. Note that E. ictaluri gene escC (88) was formerly known as eseA.
Characterization of the TTSS mutant 65ST.
The transposon insertion in esaU follows the sequence encoding amino acid residue 61 of 351 total amino acids. esaU is oriented in the same transcriptional direction as esaR, esaS, esaT, and slt, and these genes may be in the same transcriptional unit, because no obvious transcriptional termination sequences are found between them. There is an apparent sigma-70 transcription initiation site upstream of esaR, and esaR appears to be translationally coupled to esaS, esaT, esaU, and slt by hairpins associated with their translation initiation regions. Sequence analysis indicates that the transposon insertion in 65ST is after bp 184 of esaU, resulting in a frame shift that generates a stop codon 9 bp downstream. Consequently, it is unlikely that the slt gene, which is translationally linked, is expressed in 65ST.
The pathogenicity of E. ictaluri is dependent on its ability to infect and multiply within host cells (11). The genetic similarity to the SPI-2 class of TTSS, which are involved in intracellular survival and replication in Salmonella (18, 38-40) and E. tarda (81), suggests a similar role for the E. ictaluri TTSS. Consequently, the esaU insertion mutant, 65ST, was evaluated for its ability to enter the host following immersion challenge and to enter and to survive and replicate in the CCO cell line and in HKDM. Before these studies were initiated, Southern blot analysis of 65ST genomic DNA confirmed that the attenuated phenotype was due to a single transposon insertion (data not shown). As indicated in Tables 5 and 6, the insertion in esaU resulted in markedly reduced replication but did not affect uptake by HKDM. The mutation in E. ictaluri esaU also did not affect entry into the nonphagocytic CCO cells (Table 5), but the mutant's replication was poor compared to that of the wild-type strain (Fig. 4; Table 6). Although the initial uptake into both HKDM and CCO cells was not significantly different between the TTSS mutant and the wild-type strain, uptake in the CCO cells was poor relative to that in the HKDM, with less than 1% of the cells recovered after the gentamicin exposure compared to about 50% in the HKDM (Table 5).
TABLE 5.
Percentage of bacteria initially added to wells that were recovered from CCO cells or HKDM
| Strain | MOI (CFU/cell) | % CFU recovered froma:
|
|
|---|---|---|---|
| CCO cells | HKDM | ||
| Wild type | 1:10 | 0.37 ± 0.12 | 56.0 ± 15.95 |
| 1:1 | 0.41 ± 0.15 | ND | |
| 65ST | 1:10 | 0.42 ± 0.05 | 37.3 ± 5.49 |
| 1:1 | 0.55 ± 0.03 | ND | |
Percentage of bacteria recovered following a 30-min exposure to either wild-type E. ictaluri or the EsaU mutant 65ST and a 60-min exposure to a killing dose of gentamicin. Values are the means ± standard errors of the results of three experiments. There were no significant differences between the results for the mutant and wild-type strains. ND, not determined.
TABLE 6.
Increase in numbers of CFU over time for wild-type Edwardsiella ictaluri and the EsaU mutant, 65ST, following infection of HKDM and CCO cellsa
| Cell type and time (h) postinfection | No. of CFU of wild type/well | Fold increase from time zerob | No. of CFU of 65ST/well | Fold increase from time zerob |
|---|---|---|---|---|
| HKDM | ||||
| 0 | 1,483 ± 33.3 | 616 ± 83.3 | ||
| 5 | 6,500 ± 2,179.4 | 4.3 | 517 ± 120.2** | 0.85 |
| 10 | 53,333 ± 23,154.1 | 35.3 | 817 ± 183.3*** | 1.3 |
| CCO cells | ||||
| 0 | 413 ± 48.1 | 477 ± 8.8 | ||
| 4 | 1,793 ± 218.3 | 4.3 | 283 ± 18.6*** | 0.59 |
| 8 | 5,233 ± 611.9 | 12.7 | 226 ± 21.9*** | 0.47 |
CCO and HKCM cells were infected in 24-well plates at MOIs of 1 CFU:10 HKDM cells and 1 CFU:1 CCO cell. CFU data are the means ± standard errors of the results for triplicate wells and are representative of one of three experiments. A significant difference for mutant compared to wild type following a log transformation of the CFU data are indicated by asterisks. **, P < 0.01; ***, P = 0.001.
Time zero is at 90 min postinfection (30 min of exposure and 60 min of gentamicin killing).
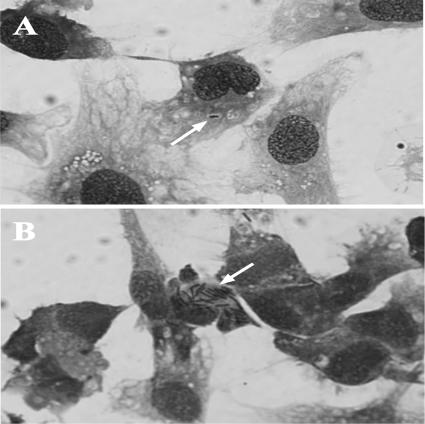
FIG. 4.
CCO cells infected with E. ictaluri at 8 h postinfection in a gentamicin survival assay. (A) Cells infected with an E. ictaluri esaU mutant, 65ST, showing typical single bacterium per cell. (B) Cells infected with wild-type E. ictaluri strain 93-146, showing typical large number of bacterial cells per infected CCO cell. Both the mutant and the wild type typically had 1 to 2 bacterial cells per CCO cell immediately following infection.
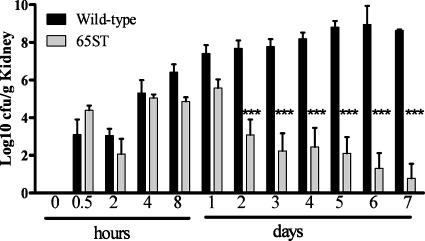
As indicated in Fig. 5, there was no significant difference in vivo between the uptake or initial growth of the TTSS mutant in comparison to the uptake and initial growth of the wild-type E. ictaluri bacteria following immersion challenge. The tissue levels of the TTSS mutant strain, however, were significantly lower and declined for all times sampled after 24 h out to day 7, indicating slow clearance from the tissue, compared to increasing numbers of the wild type. Three fish challenged with the mutant had minimal bacteria remaining on day 8 (data not shown), but could not be compared to the fish challenged with the wild-type bacteria because five of the wild-type-challenged fish succumbed to the disease prior to sampling. No deaths occurred in fish challenged with the TTSS mutant. These results suggest that the E. ictaluri TTSS is not involved in the initial penetration of tissues from the environment but is required for survival and replication in the host. Fish mortality following challenge was 56.7% with the wild-type bacteria, with no deaths in fish challenged with 65ST.
FIG. 5.
Comparison of the early invasion and persistence of wild-type Edwardsiella ictaluri and an esaU mutant, 65ST, in channel catfish. Head kidney samples were taken from each of five fish at each time point, and the data represent the means of the results, with bars representing the standard errors of the means. ***, significant difference from the corresponding wild type (P > 0.001).
DISCUSSION
The STM procedure provided a powerful tool for the identification of genes required for the survival of E. ictaluri in channel catfish. Every strain identified by STM is, by definition, attenuated in the catfish host. Transconjugants that can be transcomplemented by the production of a soluble virulence factor produced by another transconjugant in the pool or that carry the transposon in a gene required to produce disease but not required for survival and growth in the host will not be detected.
Miscellaneous chromosomal insertions.
The miaA gene has been implicated in the regulation of the virulence gene virF in Shigella flexneri and of the vir regulon in Agrobacterium tumefaciens (24, 33). In both of these organisms, the expression of a variety of virulence-related genes is downregulated from 3- to 10-fold in miaA mutants, depending on the gene in question. Mutations in miaA in A. tumefaciens, however, had minimal effects on virulence in a potato challenge model and had no effects in tobacco, kalanchoe, or tomato (33). The relatively low level of attenuation in 69ST, with a CI of 0.014, is consistent with these results but does not rule out an effect on the expression of an unknown virulence gene.
Although the CI for 197GH is only 0.16, indicating a negligible effect on virulence, the 197GH insertion is only 51 bp from the stop codon of adiA, which could result in the production of a truncated, partially functional enzyme. The insertion in 234AB, however, is in the amino terminus, which would result in the production of a severely truncated product that would be unlikely to have enzymatic activity. The lower CI for 234AB, at 0.00013, indicates that the mutation in 234AB has a significant attenuating effect and suggests that AdiA plays a role in E. ictaluri pathogenesis. The metabolism of arginine in eukaryotic cells is a complex process involving inducible nitric oxide synthase, arginases, and AdiA (9). Significant interregulation of these pathways is involved in the control of a range of metabolic functions, including inducible nitric oxide synthase activity and programmed cell death, which are also important in relation to intracellular bacterial infections (42). Although bacterial arginase activity is known to be involved in the inhibition of host cell nitric oxide production by Helicobacter pylori (32), there are no reports connecting bacterial AdiA and bacterial virulence, except for several reports that evaluate the role of AdiA in environmental acid resistance (16, 52, 53). There is a report, however, that elevated dietary arginine significantly reduces catfish mortality following an E. ictaluri immersion challenge. The dietary arginine levels for the maximum growth of catfish were determined to be 0.8 to 0.9% of a 24%-protein diet (12). When evaluated relative to exposure to E. ictaluri, however, providing arginine at 0.5, 1, 2, and 4% of the diet resulted in cumulative percent mortalities of 16, 15, 7, and 7%, respectively, after 28 days (12). The fact that doubling dietary arginine increased the resistance of catfish to E. ictaluri infection further indicates that arginine metabolism, possibly involving AdiA, is important in E. ictaluri pathogenesis. Booth (10) reported that E. ictaluri produces an acid-inducible urease enzyme that is required for replication in catfish macrophages and speculated that AdiA could be involved in the de novo synthesis of urea, which could be metabolized to ammonia by the urease enzyme, resulting in an increase in environmental pH.
Insertions in hypothetical genes in other pathogens.
A role for MCP in Vibrio cholerae and Vibrio vulnificus virulence is suspected because MCP expression was induced in vivo following the infection of mice with V. cholerae (14) and in humans with V. vulnificus (45). In Vibrio anguillarum, the chemotactic gene cheR was important to virulence in fish following immersion challenge, but not following intraperitoneal injection (66). Vibrio cholerae encodes TcpI, a 620-amino-acid protein with similarity to the highly conserved regulatory domain of MCPs. TcpI is thought to regulate the major pilin subunit of the toxin-coregulated pilus while at the same time reducing chemotactic motility, possibly to facilitate V. cholerae microcolony formation (35). Motility and pilus formation are also coregulated by an MCP in Pseudomonas aeruginosa (35). All of these studies indicate an important role for MCP in the initiation of infection, and the high degree of attenuation of 168AB in catfish may indicate a similar role for MCP in the initiation of an E. ictaluri infection in catfish, especially when exposure to the host is via a waterborne, immersion route.
The presence of pentapeptide repeats in 72NO places the putative protein in a family of proteins that have predicted β-helical structures. The pentapeptide-repeat family of proteins are both cytoplasm and membrane bound. They are most common in cyanobacteria but have also been identified in Bacillus subtilis, E. coli, and Erwinia stewartii, as well as plant proteins (43). The function of the repeats is uncertain, as is the molecular activity of the proteins, but the predicted structure suggests a targeting or structural function (7). Analysis in the PSORT II server indicates a predicted cytoplasmic or nuclear location for the E. ictaluri protein. In any case, the extremely low CI of 0.00000062 suggests an important role in E. ictaluri virulence.
Insertions in known virulence-related genes in other pathogens.
Interestingly, two of the mutants found in this study had insertions in genes encoding a urease complex, even though E. ictaluri is reported as urease negative in standard biochemical testing (85). The urease enzyme is a multisubunit molecule that catalyzes the breakdown of urea to form carbonate and ammonia, resulting in a net increase in pH. Urease is known to play a significant role in the virulence of several pathogens, including H. pylori, Klebsiella pneumoniae, Proteus mirabilis, and E. coli O157:H7 (13, 30, 56, 62). Sequence analysis, however, links the nine genes of the E. ictaluri urease enzyme complex most closely to the complexes of Y. enterocolitica, M. morganii, and Lactococcus fermentum, with the ureABCEFGD gene sequence maintained and 67.1 to 91.2% amino acid identity to the Y. enterocolitica urease proteins. These three ureases are distinct among bacterial ureases because they are acid activated, with a pH optimum of around 3 to 4, and they encode a conserved acid activation motif in the catalytic site of the urease α subunit (UreC) (80, 87). This acid activation motif is conserved in E. ictaluri, and a requirement for acidic conditions may explain the fact that E. ictaluri is urease negative in standard biochemical tests (85), which are done at neutral pH. For a facultative intracellular pathogen capable of surviving and replicating in catfish HKDM (11), the ability to increase the environmental pH could be important in passage through the gastrointestinal tract or in survival in the acid phagosomes of macrophages.
The strong similarity in structure and amino acid sequence of Orf1 of pEI1 and the STEs suggests a similarity in function. In general, LRR proteins are structural in nature and are involved in diverse protein-protein interactions, including signal transduction, cell adhesion, cell development, DNA repair, and RNA processing (46, 47). Of the three STEs with the greatest similarity to pEI1 Orf1, the most studied are SspH1 and SspH2, both of which are involved in S. enterica serovar Typhimurium virulence in cattle (59). In calves infected with ΔsspH1 or ΔsspH2 mutants, severe diarrhea and death occurred 1 to 4 days postinfection, similar to those infected with the wild type. An ΔsspH1-ΔsspH2 double mutant, however, did not cause diarrhea or death (59). Although Blastn analysis of the E. ictaluri sspH1-sspH2 gene homologue and the E. ictaluri genome database indicates the presence of at least three additional effectors with SspH1/2 homology that vary in the number of LRR and vary in amino acid homology with SspH1/2 from 51 to 71%, the high level of attenuation associated with the single mutation in orf1 of pEI1 (CI = 0.0000062) indicates that the level of attenuation is similar to that of the S. enterica serovar Typhimurium double mutants in calves. This suggests either a more integral role in pathogenesis for orf1 in E. ictaluri than for either sspH1 or sspH2 in Salmonella or that the TTSS effectors of E. ictaluri are simpler and less redundant than those of Salmonella.
Further sequence analysis identified a gene immediately downstream from the spa15 homologue encoded by orf1 of pEI2 that encodes a protein with high homology to OspB. OspB is a secreted effector of Shigella with unknown function (55, 71), although a role in invasion, intracellular replication, induction of macrophage apoptosis, or decreased intracellular ATP could not be demonstrated (71). The operon structure of orf1 and the OspB homologue is typical of chaperone/effector gene arrangements, suggesting that orf1 is a chaperone for the OspB homologue.
The E. ictaluri TTSS.
Both Edwardsiella TTSS differ from the other SPI-2 class of TTSS in that they encode an AraC-type regulator, EsrC. In E. tarda, the expression of EsrC is under the control of the EsrA/EsrB two-component regulators, both of which then act in concert to regulate the expression of different components of the TTSS (89). Both systems also encode a soluble lytic murein transglycosylase (slt) that may be cotranscribed and translationally coupled to the esaR-U inner membrane components of the TTSS apparatus and is presumably involved in local rearrangement of the peptidoglycan to allow needle complex formation in the periplasmic space (48, 69). Although peptidoglycan modulation is important in the formation of the Salmonella SPI-1 needle complex (48, 69), the slt genes of S. enterica serovar Typhimurium and E. coli are located at centisomes 99.621 and 99.802, substantially removed from the SPI-1-type TTSS at centisomes 61.892 to 62.6759 for Salmonella and 84.26 to 84.767 for E. coli.
The comparison of nucleotide content relates the E. ictaluri TTSS more closely to C. violaceum, with 60.4 and 56.4% G+C content, respectively, than to the S. enterica serovar Typhimurium and E. coli systems, with 46.6 and 40.4% G+C content, respectively. This high G+C content is unusual for TTSS clusters of animal pathogens, which are generally lower in G+C content than the surrounding genome at 40 to 48% (4). At 60.4% G+C content, the E. ictaluri TTSS G+C content is higher than that of the surrounding genome (4, 37) and is more similar to the TTSS G+C content of plant pathogens (4), despite its obvious similarity to the SPI-2 class of TTSS. The E. tarda TTSS has an even higher percent G+C content, at 63.8%. The Edwardsiella TTSS genes should provide interesting new information in the evolutionary analysis of TTSS.
Further analysis indicated that E. ictaluri also carries sequences similar to orf29 and orf30 of E. tarda (89). Because E. tarda orf29 and orf30 are only 591 bp upstream from esrB and are reportedly under the regulatory control of both EsrA-EsrB and EsrC, Zheng et al. (89) considered them part of the TTSS island of E. tarda. In addition, based on the presence of a predicted coiled-coil secondary structure that is common in TTSS effectors (21), Zheng et al. (89) suggested that Orf29 and Orf30 are E. tarda effectors. In comparison, orf29 and orf30 of the E. ictaluri TTSS are located 1,667 bp upstream from the start of esrB, and several ORFs whose products have similarity to a variety of transposases of gram-negative transposons are encoded in the intervening sequence. In addition, based on sixfold coverage of this region in the E. ictaluri genome project, the region carrying orf29 and orf30 appears to encode a single protein, Orf29/30. Putative transcription and translation initiation regions are found upstream from the start of orf29-30, but a putative translation initiation region was not apparent immediately upstream of the putative orf30 start codon of E. ictaluri. Although Orf29/30 of E. ictaluri has extensive coiled domains, a role as an effector remains to be determined. Additional work is required to evaluate the relationship of Orf29/30 to the E. ictaluri TTSS and its role in pathogenesis.
The severe attenuation of the E. ictaluri TTSS mutants is consistent with data for S. enterica serovar Typhimurium showing that SPI-2-encoded TTSS mutants have at least a fivefold increase in 50% lethal dose values in mice (72). The ability of the E. ictaluri TTSS mutants to cross the epithelial barrier is also consistent with S. enterica serovar Typhimurium SPI-2 TTSS mutants, which are equally attenuated by either oral or intraperitoneal exposure (72), indicating that SPI-2 is required for events in the infectious process that occur after penetration. Mutations in the TTSS of E. tarda resulted in a 1-log increase in the 50% lethal dose value, but neither efficiency of entry nor persistence was evaluated (81).
In conclusion, the identification of 50 mutants that are important for the survival of E. ictaluri in channel catfish offers significant insight into the pathogenesis of this pathogen, although a number of them are in housekeeping genes that have predictable growth- or replication-related phenotypes. The identification of effectors with close homology to the SspH1 and SspH2 effectors of the Salmonella SPI-2-encoded TTSS on plasmid pEI1 and in the chromosome implies a role in intracellular replication. Characterization of the TTSS esaU apparatus mutant demonstrates the importance of the secretion system in the pathogenesis of E. ictaluri in catfish and in intracellular survival. The variation in effectors compared to those of the Salmonella SPI-2 TTSS, in combination with the identification of a TTSS effector and its putative chaperone on plasmid pEI2 that have homology to an uncharacterized effector from Shigella, suggests that the E. ictaluri TTSS operates with some variation on the SPI-2 TTSS model.
Acknowledgments
This research was supported by the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service with grant number 2002-35204-12605 to Ronald L. Thune.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Ainsworth, A. J., G. Capley, P. Waterstreet, and D. Munson. 1986. Use of monoclonal antibodies in the indirect fluorescent antibody technique for the diagnosis of Edwardsiella ictaluri. J. Fish Dis. 9:439-444. [Google Scholar]
- 2.Ainsworth, A. J., and D. X. Chen. 1990. Differences in the phagocytosis of four bacteria by channel catfish neutrophils. Dev. Comp. Immunol. 14:201-209. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245-1248. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, D. L., A. Pitman, and R. W. Jackson. 2003. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4:407-420. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. E. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 6.Baldwin, T. J., and J. C. Newton. 1993. Pathogenesis of enteric septicemia of channel catfish, caused by Edwardsiella ictaluri: bacteriologic and light and electron microscopic findings. J. Aquat. Anim. Health 5:189-198. [Google Scholar]
- 7.Bateman, A., A. G. Murzin, and S. A. Teichmann. 1998. Structure and distribution of pentapeptide repeats in bacteria. Protein Sci. 7:1477-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betts, H. J., R. R. Chaudhuri, and M. J. Pallen. 2004. An analysis of type-III secretion gene clusters in Chromobacterium violaceum. Trends Microbiol. 12:476-482. [DOI] [PubMed] [Google Scholar]
- 9.Blantz, R. C., J. Satriano, F. Gabbai, and C. Kelly. 2000. Biological effects of arginine metabolites. Acta Physiol. Scand. 168:21-25. [DOI] [PubMed] [Google Scholar]
- 10.Booth, N. J. 2005. The role of urease in the pathogenesis of Edwardsiella ictaluri. Ph.D dissertation. Louisiana State University, Department of Pathobiological Sciences, Baton Rouge.
- 11.Booth, N. J., A. ElKamel, and R. L. Thune. 2006. Intracellular replication of Edwardsiella ictaluri in channel catfish macrophages. J. Aquat. Anim. Health 18:101-108. [Google Scholar]
- 12.Buentello, J. A., and D. M. Gatlin. 2001. Effects of elevated dietary arginine on resistance of channel catfish to exposure to Edwardsiella ictaluri. J. Aquat. Anim. Health 13:194-201. [Google Scholar]
- 13.Burall, L. S., J. M. Harro, X. Li, C. V. Lockatell, S. D. Himpsl, J. R. Hebel, D. E. Johnson, and H. L. T. Mobley. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72:2922-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camilli, A., D. S. Merrell, and J. J. Mekalanos. 2001. Strategies to identify bacterial pathogenicity factors, p. 133-175. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, CA.
- 16.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. [DOI] [PubMed] [Google Scholar]
- 18.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 19.Cooper, R. K. 1991. Molecular studies of the virulence factors associated with Edwardsiella ictaluri. Ph.D. thesis. University of Georgia, Athens.
- 20.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 21.Delahay, R. M., and G. Frankel. 2002. Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol. Microbiol. 45:905-916. [DOI] [PubMed] [Google Scholar]
- 22.Delorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative Eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5-derived and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 24.Durand, J. M., G. R. Bjork, A. Kuwae, M. Yoshikawa, and C. Sasakawa. 1997. The modified nucleoside 2-methylthio-N6-isopentenyladenosine in tRNA of Shigella flexneri is required for expression of virulence genes. J. Bacteriol. 179:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellsaesser, C. F., N. W. Miller, C. J. Lobb, and L. W. Clem. 1984. A new method for the cytochemical staining of cells immobilized in agarose. Histochemistry 80:559-562. [PubMed] [Google Scholar]
- 26.Elsinghorst, E. A. 1997. Measurement of invasion by gentamycin resistance, p. 667-681. In V. L. Clark and P. M. Bavoil (ed.), Bacterial pathogenesis. Academic Press, San Diego, CA.
- 27.Faulmann, E., M. A. Cuchens, C. J. Lobb, N. W. Miller, and L. W. Clem. 1983. An effective culture system for studying in vitro mitogenic responses to channel catfish lymphocytes. Trans. Amer. Fish. Soc. 112:673-679. [Google Scholar]
- 28.Fernandez, D. H., L. Pittman-Cooley, and R. L. Thune. 2001. Sequencing and analysis of the Edwardsiella ictaluri plasmids. Plasmid 45:52-56. [DOI] [PubMed] [Google Scholar]
- 29.Francis-Floyd, R., M. H. Beleau, P. R. Waterstrat, and P. R. Bowser. 1987. Effect of water temperature on the clinical outcome of infection with Edwardsiella ictaluri in channel catfish. J. Am. Vet. Med. Assoc. 191:1413-1416. [PubMed] [Google Scholar]
- 30.Friedrich, A. W., R. Kock, M. Bielaszewska, W. Zhang, H. Karch, and W. Mathys. 2005. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J. Clin. Microbiol. 43:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, DC.
- 32.Gobert, A. P., D. J. McGee, M. Akhtar, G. L. Mendz, J. C. Newton, Y. Cheng, H. L. Mobley, and K. T. Wilson. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98:13844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray, J., J. Wang, and S. B. Gelvin. 1992. Mutation of the miaA gene of Agrobacterium tumefaciens results in reduced vir gene expression. J. Bacteriol. 174:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hacker, J., and J. B. Kaper. 1999. The concept of pathogenicity islands, p. 1-11. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, DC.
- 35.Harkey, C. W., K. D. Everiss, and K. M. Peterson. 1994. The Vibrio cholerae toxin-coregulated pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect. Immun. 62:2669-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey, D. J. 2006. Aquaculture outlook. Electronic outlook report. U.S. Department of Agriculture yearbook of agriculture. U.S. Department of Agriculture, Washington, DC. http://www.ers.usda.gov/Publications/Outlook/.
- 37.Hawke, J. P., A. C. McWhorter, A. G. Steigerwalt, and D. J. Brenner. 1981. Edwardsiella ictaluri sp. nov., the causative agent of enteric septicemia of catfish. Int. J. Syst. Bacteriol. 31:396-400. [Google Scholar]
- 38.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 39.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 40.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 41.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaufmann, S. H. E. 1993. Immunity to extracellular bacteria, p. 1251-1286. In W. E. Paul (ed.), Fundamental immunology. Raven Press, Ltd., New York, NY.
- 43.Kieselbach, T., A. Mant, C. Robinson, and W. P. Schroder. 1998. Characterization of an Arabidopsis cDNA encoding a thylakoid lumen protein related to a novel “pentapeptide repeat” family of proteins. FEBS Lett. 428:241-244. [DOI] [PubMed] [Google Scholar]
- 44.Kim, S. Y., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 45.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobe, B., and J. Deisenhofer. 1994. The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19:415-421. [DOI] [PubMed] [Google Scholar]
- 47.Kobe, B., and J. Deisenhofer. 1995. Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 5:409-416. [DOI] [PubMed] [Google Scholar]
- 48.Koraimann, G. 2003. Lytic transglycosylases in macromolecular transport systems of gram-negative bacteria. Cell. Mol. Life Sci. 60:2371-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence, M. L., M. M. Banes, P. Azadi, and B. Y. Reeks. 2003. The Edwardsiella ictaluri O polysaccharide biosynthesis gene cluster and the role of O polysaccharide in resistance to normal catfish serum and catfish neutrophils. Microbiology 149:1409-1421. [DOI] [PubMed] [Google Scholar]
- 50.Lawrence, M. L., M. M. Banes, and M. L. Williams. 2001. Phenotype and virulence of a transposon-derived lipopolysaccharide O side-chain mutant strain of Edwardsiella icataluri. J. Aquat. Anim. Health 13:291-299. [Google Scholar]
- 51.Lehoux, D. E., F. Sanschagrin, and R. C. Levesque. 1999. Defined oligonucleotide tag pools and PCR screening in signature-tagged mutagenesis of essential genes from bacteria. BioTechniques 26:473. [DOI] [PubMed] [Google Scholar]
- 52.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lubetsky, J. B., and A. M. Stock. 2005. Two-component signal transduction and chemotaxis, p. 17-36. In G. Waksman, M. Caparon, and S. Hultgren (ed.), Structural biology of bacterial pathogenesis. ASM Press, Washington, DC.
- 55.MacMillan, J. R. 1985. Infectious diseases, p. 405-496. In C. S. Tucker (ed.), Channel catfish culture. Elsevier, New York, NY.
- 56.Maroncle, N., D. Balestrino, C. Rich, and C. Forestier. 2002. Identification of Klebsiella pneumoniae genes involved in intestinal colonization and adhesion using signature-tagged mutagenesis. Infect. Immun. 70:4729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maurer, K. J., M. L. Lawrence, D. H. Fernandez, and R. L. Thune. 2001. Evaluation and optimization of a DNA transfer system for Edwardsiella ictaluri. J. Aquat. Anim. Health 13:163-167. [Google Scholar]
- 58.Mecsas, J. 2002. Use of signature-tagged mutagenesis in pathogenesis studies. Curr. Opin. Microbiol. 5:33-37. [DOI] [PubMed] [Google Scholar]
- 59.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 60.Miller, N. W., and E. C. Mckinney. 1994. In vitro culture of fish leukocytes, p. 339-351. In P. W. Hochachka (ed.), Biochemistry and molecular biology of fishes. Elsevier, London, United Kingdom.
- 61.Miyazaki, T., and J. A. Plumb. 1985. Histopathology of Edwardsiella ictaluri in channel catfish Ictalurus punctatus. J. Fish Dis. 8:389-392. [Google Scholar]
- 62.Mobley, H. L., M. J. Cortesia, L. E. Rosenthal, and B. D. Jones. 1988. Characterization of urease from Campylobacter pylori. J. Clin. Microbiol. 26:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrison, E. E., and J. A. Plumb. 1994. Olfactory organ of channel catfish as a site of experimental Edwardsiella ictaluri infection. J. Aquat. Anim. Health 6:101-109. [Google Scholar]
- 65.Nusbaum, K. E., and E. E. Morrison. 1996. Entry of 35S-labelled Edwardsiella ictaluri into channel catfish. J. Aquat. Anim. Health 8:146-149. [Google Scholar]
- 66.O'Toole, R., D. L. Milton, and H. Wolf-Watz. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625-637. [DOI] [PubMed] [Google Scholar]
- 67.Page, A. L., P. Sansonetti, and C. Parsot. 2002. Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43:1533-1542. [DOI] [PubMed] [Google Scholar]
- 68.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 69.Pucciarelli, M. G., and F. Garcia-del Portillo. 2003. Protein-peptidoglycan interactions modulate the assembly of the needle complex in the Salmonella invasion-associated type III secretion system. Mol. Microbiol. 48:573-585. [DOI] [PubMed] [Google Scholar]
- 70.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 71.Santapaola, D., C. F. Del, A. Petrucca, S. Uzzau, M. Casalino, B. Colonna, R. Sessa, F. Berlutti, and M. Nicoletti. 2006. Apyrase, the product of the virulence plasmid-encoded phoN2 (apy) gene of Shigella flexneri, is necessary for proper unipolar IcsA localization and for efficient intercellular spread. J. Bacteriol. 188:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shea, J. E., J. D. Santangelo, and R. G. Feldman. 2000. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr. Opin. Microbiol. 3:451-458. [DOI] [PubMed] [Google Scholar]
- 74.Shen, S., M. Mascarenhas, K. Rahn, J. B. Kaper, and M. A. Karmali. 2004. Evidence for a hybrid genomic island in verocytotoxin-producing Escherichia coli CL3 (serotype O113:H21) containing segments of EDL933 (serotype O157:H7) O islands 122 and 48. Infect. Immun. 72:1496-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shotts, E. B., V. S. Blazer, and W. D. Waltman. 1986. Pathogenesis of experimental Edwardsiella ictaluri infections in channel catfish (Ictalurus punctatus). Can. J. Fish. Aquat. Sci. 43:36-42. [Google Scholar]
- 76.Silva, R. C. 1998. Effects of sodium chloride on Edwardsiella ictaluri: studies in channel catfish, fish cell lines, and bacterial cultures. Ph.D. thesis. Auburn University, Auburn, AL.
- 77.Skirpstunas, R. T., and T. J. Baldwin. 2002. Edwardsiella ictaluri invasion of IEC-6, Henle 407, fathead minnow and channel catfish enteric epithelial cells. Dis. Aquat. Organ. 51:161-167. [DOI] [PubMed] [Google Scholar]
- 78.Stanley, L. A. 1991. Characteristics of pathogenic mechanisms of Edwardsiella ictaluri in the channel catfish, Ictalurus punctatus. Ph.D. thesis. Clemson University, Clemson, SC.
- 79.Stanley, L. A., J. S. Hudson, T. E. Schwedler, and S. S. Hayasaka. 1994. Extracellular products associated with virulent and avirulent strains of Edwardsiella ictaluri from channel catfish. J. Aquat. Anim. Health 6:36-43. [Google Scholar]
- 80.Suzuki, K., Y. Benno, T. Mitsuoka, S. Takebe, K. Kobashi, and J. Hase. 1979. Urease-producing species of intestinal anaerobes and their activities. Appl. Environ. Microbiol. 37:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan, Y. P., J. Zheng, S. L. Tung, I. Rosenshine, and K. Y. Leung. 2005. Role of type III secretion in Edwardsiella tarda virulence. Microbiology 151:2301-2313. [DOI] [PubMed] [Google Scholar]
- 82.Thune, R. L., D. H. Fernandez, and J. R. Battista. 1999. An aroA mutant of Edwardsiella ictaluri is safe and efficacious as a live, attenuated vaccine. J. Aquat. Anim. Health 11:358-372. [Google Scholar]
- 83.Thune, R. L., L. A. Stanley, and R. K. Cooper. 1993. Pathogenesis of gram-negative bacterial infections in warm water fish. Annu. Rev. Fish Dis. 3:37-68. [Google Scholar]
- 84.U.S. Department of Agriculture. 2003. National animal health monitoring system reports-catfish 2003. U.S. Department of Agriculture yearbook of agriculture. U.S. Department of Agriculture, Washington, DC. http://www.aphis.usda.gov/animal_health/.
- 85.Waltman, W. D., E. B. Shotts, and T. C. Hsu. 1986. Biochemical characteristics of Edwardsiella ictaluri. Appl. Environ. Microbiol. 51:101-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang, J., A. Mushegian, S. Lory, and S. Jin. 1996. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 93:10434-10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Young, G. M., D. Amid, and V. L. Miller. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng, J., N. Li, Y. P. Tan, J. Sivaraman, Y. K. Mok, Z. L. Mo, and K. Y. Leung. 2007. EscC is a chaperone for the Edwardsiella tarda type III secretion system putative translocon components EseB and EseD. Microbiology 153:1953-1962. [DOI] [PubMed] [Google Scholar]
- 89.Zheng, J., S. L. Tung, and K. Y. Leung. 2005. Regulation of a type III and a putative secretion system in Edwardsiella tarda by EsrC is under the control of a two-component system, EsrA-EsrB. Infect. Immun. 73:4127-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]