Abstract
The diversity of a collection of 102 lactococcus isolates including 91 Lactococcus lactis isolates of dairy and nondairy origin was explored using partial small subunit rRNA gene sequence analysis and limited phenotypic analyses. A subset of 89 strains of L. lactis subsp. cremoris and L. lactis subsp. lactis isolates was further analyzed by (GTG)5-PCR fingerprinting and a novel multilocus sequence analysis (MLSA) scheme. Two major genomic lineages within L. lactis were found. The L. lactis subsp. cremoris type-strain-like genotype lineage included both L. lactis subsp. cremoris and L. lactis subsp. lactis isolates. The other major lineage, with a L. lactis subsp. lactis type-strain-like genotype, comprised L. lactis subsp. lactis isolates only. A novel third genomic lineage represented two L. lactis subsp. lactis isolates of nondairy origin. The genomic lineages deviate from the subspecific classification of L. lactis that is based on a few phenotypic traits only. MLSA of six partial genes (atpA, encoding ATP synthase alpha subunit; pheS, encoding phenylalanine tRNA synthetase; rpoA, encoding RNA polymerase alpha chain; bcaT, encoding branched chain amino acid aminotransferase; pepN, encoding aminopeptidase N; and pepX, encoding X-prolyl dipeptidyl peptidase) revealed 363 polymorphic sites (total length, 1,970 bases) among 89 L. lactis subsp. cremoris and L. lactis subsp. lactis isolates with unique sequence types for most isolates. This allowed high-resolution cluster analysis in which dairy isolates form subclusters of limited diversity within the genomic lineages. The pheS DNA sequence analysis yielded two genetic groups dissimilar to the other genotyping analysis-based lineages, indicating a disparate acquisition route for this gene.
Lactococcus lactis is the primary constituent of many industrial and artisanal starter cultures used to ferment dairy products, especially hard and semihard cheeses. These starter cultures play a key role in determining shelf-life, preservation, and organoleptic quality and thus influence the quality and safety of these fermented products (35). As a result of the industrial importance of L. lactis, it has been the subject of numerous studies, which have resulted in detailed knowledge of its physiology and molecular biology. Moreover, due to the availability of a vast molecular toolbox and three whole genome sequences, of strains IL-1403, SK11, and MG1363 (3, 22, 40), L. lactis has gained a strong position as a model organism for low-GC gram-positive bacteria (20).
The model strains that are used in most studies almost exclusively originate from dairy fermentations. Isolates from (fermented) plant material have been only poorly characterized. However, in recent years there has been growing interest in plant isolates as several examples indicate phenotypes of industrial interest such as a unique flavor-forming potential or the production of bacteriocins with a broad mode of action (1, 17, 18).
Historically, three different industrial phenotypes have been recognized, those of L. lactis subsp. lactis, L. lactis subsp. cremoris, and L. lactis subsp. lactis biovar diacetylactis (34). These two subspecies and one biovar are taxonomically differentiated by only a few phenotypic characteristics (Table 1). L. lactis subsp. cremoris is characterized by the inability to produce ammonia from arginine and by low tolerance to elevated temperatures and salt concentrations. L. lactis subsp. lactis produces ammonia from arginine and is tolerant to 40°C and 4% NaCl (34). Some L. lactis subsp. lactis strains are able to ferment citrate and to produce the flavor compound diacetyl, and these are referred to as L. lactis subsp. lactis biovar diacetylactis strains (34).
TABLE 1.
Phenotypes and genotypes of Lactococcus species and subspecies and biovar diacetylactis
| Organism | Phenotypea
|
Genotype of clusterb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Growth at 40°C | Growth with 4% NaCl | Citrate utilization | Arginine dehydrolase | Maltosec | Lactose and galactosec | Ribosec | SSU rRNA gene and (GTG)5 fp | L. lactis subsp. lactis type- strain-like (n) | L. lactis subsp. cremoris type- strain-like (n) | |
| L. lactis subsp. lactis | + (−) | + | − | + | + | + | + | 1A, 1B, 1C | 1A (60), 1C (2) | 1B (9) |
| L. lactis subsp. lactis biovar diacetylactis | + (−) | + | + | + | + | + | + | 1A | 1A (7) | |
| L. lactis subsp. cremoris | − | − | − | − | − | + | − | 1A, 1B | 1B (11) | |
| L. lactis subsp. hordniae | − | − | − | + | − | − | − | 1A | NA | NA |
| L. raffinolactis | − | − | − | − (+) | + | + | − (+) | 3 | NA | NA |
| L. plantarum | V | + | − | − | + | − | − | 5 | NA | NA |
| L. garvieae | + | + | − | + | + (−) | + | + | 2 | NA | NA |
| L. piscium | − | NA | NA | − | + | + | − | 4 | NA | NA |
Adapted from Schleifer et al. and Holt et al. (13, 34). V, variable; NA, not available/applicable; +, positive; −, negative; +(−), most positive but occasional negative; −(+), most negative but occasional positive.
Data are from the present study. n, number of isolates in this study; NA, not available/applicable; fp, fingerprinting.
Results indicate whether acid is produce from this substance.
Numerous studies involving DNA-DNA hybridization, small subunit (SSU) rRNA gene sequences, and PCR fingerprint analyses show two genomic lineages (7, 23, 28, 31). Additionally, subspecific probes have been proposed and applied for the identification of environmental isolates (18, 32). This started an increasingly confusing period of subspecies classification and identification of L. lactis strains. For example, two different genotypes recognized by SSU rRNA sequence differences were referred to as L. lactis subsp. lactis and L. lactis subsp. cremoris (31), but several groups have recognized that phenotypic and genotypic groupings do not match (16, 27, 32, 38). An L. lactis subsp. cremoris-like genotype, i.e., a genotype similar to the genotype of the L. lactis subsp. cremoris type strain, may have the L. lactis subsp. lactis phenotype, with strain MG1363 a well-known representative of this group (38). Other investigators postulated that lactococci can be divided into three to five major groups with an unusual taxonomic structure (16, 36). In this structure, two genetically distinct groups of strains show indistinguishable phenotypes while, conversely, two distinct phenotypic groups are genetically homologous.
More recently, microbes have been characterized by DNA sequence analysis of several housekeeping genes or other protein-coding gene sequences in an approach called multilocus sequence analysis (MLSA) or typing (21). MLSA methods have brought a new dimension to the elucidation of genomic relatedness at inter- and intraspecific levels, providing microbiologists with tools to search for phylogenetic markers independent of ribosomal DNA genes, and are most promising in obtaining biologically meaningful and functional groupings (21). Several studies have been published describing the application of MLSA to estimate the relative contribution of recombination and (point) mutation to clonal diversification in order to study phylogeny and microbial diversity in specific ecological niches (4, 14, 21). So far, MLSA and multilocus sequence typing methods have been used primarily in epidemiology including the population analysis of food-related pathogens like Salmonella, Listeria monocytogenes, or Bacillus cereus (12, 33, 37). Most recently, a method was reported for Lactobacillus plantarum (6).
In the present study genotypic and the traditionally used differentiating phenotypic characteristics (Table 1) are determined to asses the diversity of 102 lactococcus isolates including a subset of 89 strains of L. lactis subsp. cremoris and L. lactis subsp. lactis isolates from dairy and nondairy (plant) origin. Additionally, the development and application of a novel MLSA scheme targeting housekeeping and flavor-related genes for diversity analysis within L. lactis is reported. The scheme shows the existence of three separate lineages. Additionally, diversity was studied using SSU rRNA gene sequence analysis and repetitive element-based PCR genomic fingerprint analysis (30).
MATERIALS AND METHODS
Bacterial isolates and medium.
A total of 103 isolates were subjected to genotypic analysis (Fig. 1). The collection included 102 lactococcus isolates representing L. garvieae, L. piscium, L. plantarum, L. raffinolactis, L. lactis subsp. hordniae, L. lactis subsp. cremoris, and L. lactis subsp. lactis. Isolates from the latter two subspecies were labeled dairy or nondairy according to their sources of isolation, and care was taken to maximally cover the diversity within the species. Strains were maintained in M17 broth (Oxoid Ltd., Basingstoke, Hampshire, England) with 0.5% glucose (wt/vol) as a carbon source (GM17). Serial transfer was minimized to prevent the occurrence of mutations as a result of adaptation to laboratory medium and conditions. Additionally, Enterococcus casseliflavus NIZO 2193 was used in this study as an outgroup in DNA sequence analysis.
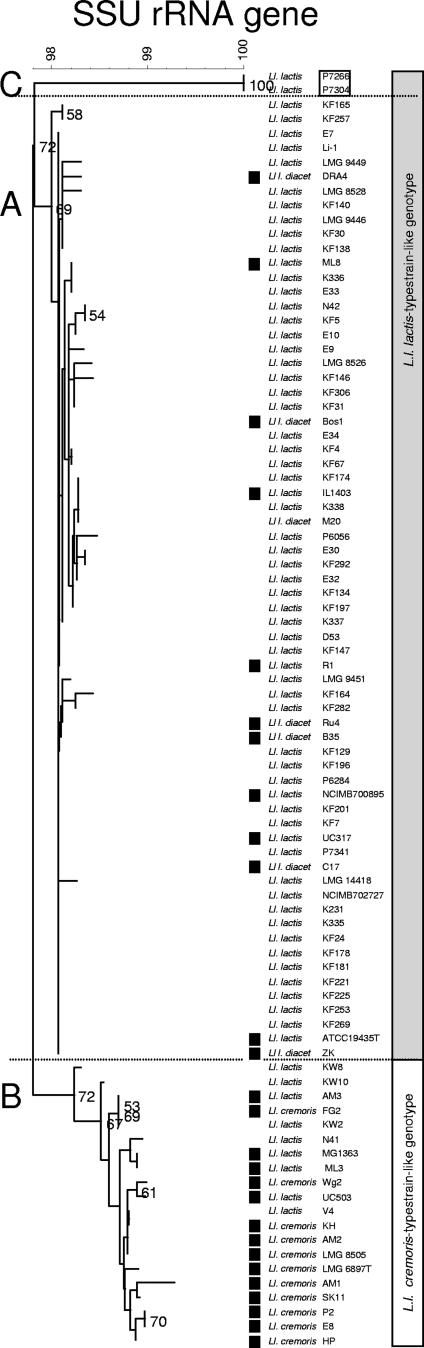
FIG. 1.
Neighbor-joining cluster analysis of the partial SSU rRNA gene sequence (Escherichia coli positions 41 to 705) of 102 Lactococcus isolates with E. casseliflavus as the outgroup. Bootstrap percentages (≥50) after 500 simulations are shown. All L. lactis subsp. lactis isolates and L. garvieae showed growth at 37°C; other isolates did not (data not shown). Further phenotypes characteristic for L. lactis subsp. cremoris and L. lactis subsp. lactis, according to Schleifer et al. (37), are indicated and include the production of ammonia from arginine, acid from maltose, and salt tolerance at 4% NaCl. Additionally, citrate production on WPCCA medium is specified for differentiation of biovar diacetylactis (data not shown). Isolates obtained from dairy substrates are indicated (▪). Five distinct species clusters (1 to 5) support the Lactococcus classification. The L. lactis isolate cluster 1 includes three subgroups; one major subgroup, 1B, comprises 11 L. lactis subsp. cremoris and 9 L. lactis subsp. lactis isolates, whereas the other major subgroup, 1A, includes 67 L. lactis subsp. lactis and 2 L. lactis subsp. hordniae isolates. Finally, two L. lactis subsp. lactis strains form a distinct minor subcluster (1C). Culture references: LMG, Laboratorium voor Microbiologie, University of Ghent, Belgium; ATCC: American Type Culture Collection, Manassas, VA; DSM(Z), Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; NCFB, NCDO National Collection of Food Bacteria; National Collections of Industrial and Marine Bacteria Ltd., Aberdeen, Aberdeen, Scotland, United Kingdom. KF and KW strains originate from Kelly's laboratory, Palmerston North, New Zealand; other strains are from the NIZO culture collection.
Genotypic diversity. (i) DNA extraction.
Bacterial DNA was extracted using an InstaGene Matrix (Bio-Rad, Hercules, CA) according to the manufacturer's instructions.
(ii) Genomic fingerprinting and computer-assisted analysis.
(GTG)5-PCR genomic fingerprints were obtained as described previously (11, 30). Computer-assisted analysis of the genomic fingerprints was performed using BioNumerics software, version 4.50 (Applied Maths, Saint-Martens-Latem, Belgium), as described previously (29). Briefly, the similarity between pairs of genomic fingerprints was calculated using the product-moment correlation coefficient (r value), applied to the whole densitometric curve of the gel tracks. Cluster analysis of the pairwise similarity values was performed using the unweighted-pair group method using arithmetic averages (UPGMA) algorithm.
(iii) Selection of loci for sequence analysis and primer design.
Multiple protein-coding loci, both housekeeping genes and functional genes, were evaluated for their applicability in assessing the diversity and relationships of different isolates in an MLSA approach. Criteria used were the following: (a) broad distribution among whole genome sequences of lactic acid bacteria ([LAB] 14 completed genomes and 10 draft genome sequences were included), (b) presence in only one copy per organism, and (c) mutually unlinked in location. The final selection included three housekeeping genes: atpA, encoding ATP synthase alpha subunit; pheS, encoding phenylalanine tRNA synthetase; and rpoA, encoding RNA polymerase alpha chain. Three functional genes involved in production of flavor compounds during fermentation were also included: bcaT, encoding branched chain amino acid aminotransferase; pepN, encoding aminopeptidase N; and pepX, encoding X-prolyl dipeptidyl peptidase. Multiple sequence alignments of these genes from all LAB and phylogenetic analyses showed that conserved regions, suitable for primer design, were not always present among all included LAB sequences. This rendered the construction of universal LAB primers unfeasible. Therefore, primers were designed on the conserved regions within the lactococcal genomes and extended with related genomes where possible, as long as two suitable conserved regions remained. With respect to the housekeeping genes, sequences from the following genomes were included: Enterococcus faecalis (V583), Lactobacillus plantarum (WCFS1), L. lactis subsp. lactis (IL-1403), L. lactis subsp. cremoris (SK11), Streptococcus pneumoniae (TIGR4 and R6), Streptococcus agalactiae (NEM316 and 2603 V/R), Streptococcus pyogenes (MGAS8232, SSI-1, MGAS315, and SF370), and Streptococcus mutans (UA159). For bcaT, sequences from E. faecalis (V583), L. lactis subsp. lactis (IL-1403), L. lactis subsp. cremoris (SK11), S. pneumoniae (TIGR4 and R6), S. agalactiae (NEM316 and 2603 V/R), S. pyogenes (MGAS8232, SSI-1, MGAS315, and SF370), and S. mutans (UA159) were included; for pepN both lactococcus genomes, Lactobacillus johnsonii (NCC 533), and Lactobacillus gasseri were included, whereas for pepX only the lactococcus genomes were used. Primer design was performed by standard procedures using Kodon, version 2.04, software (Applied Maths).
(iv) Multilocus and SSU rRNA gene sequence analysis.
DNA sequence analysis was performed of approximately 400-bp intragenic regions of the atpA, pheS, rpoA, bcaT, pepN, and pepXP genes and the SSU rRNA gene. The primer combinations (Proligo, Paris, France) for the primary PCR and cycle sequencing and the annealing temperatures used are listed in Table 2. The amplification of the gene fragments was carried out for 30 cycles in a 50-μl mixture containing each deoxynucleoside triphosphate at a concentration of 2.5 mM, 250 ng of each primer, 1 U of Taq polymerase (Applied Biosystems, Nieuwerkerk aan de IJssel, The Netherlands), and 5.0 μl of genomic DNA. After incubation for 3 min at 95°C and for 15 s at 94°C, samples were subjected to 30 cycles of 30 s at the annealing temperature (Table 2), followed by 1 min 45 s at 72°C; the reaction was completed by 4 min at 72°C and kept at 4°C using a GeneAmp PCR System 9600 thermocycler (Applied Biosystems). Presence, size, and approximate quantity of the resulting PCR product were analyzed using a 1% agarose gel with 0.5 μg of ethidium bromide per ml and a low DNA mass ladder (E-Gel; Invitrogen). The obtained PCR products were purified using a GFX PCR kit (Amersham Biosciences, Roosendaal, The Netherlands) according to the manufacturer's instructions. The forward primer of the primary PCR was also applied in cycle sequencing using an ABI PRISM BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems) according to the manufacturer's protocol. Briefly, 2.5 μl of the primary PCR and 3 pmol of the forward primer were used in a 20-μl total sequencing reaction mixture. Cycle sequencing started with 1 min at 96°C followed by 25 cycles of 96°C for 10 s and 50°C for 5 s, with a final extension for 4 min at 60°C. Alternatively, 46°C for 5 s was used for annealing when no PCR product was obtained at 50°C, but still no specific amplicons were obtained for limited genes and strains, as reported below. Reaction products were purified using Autoseq G-50 columns (Amersham Biosciences) according to the manufacturer's instructions. After denaturation, 10 μl of the purified template was mixed with 30 μl of Hi-Di (Applied Biosystems) and incubated for 5 min at 95°C. Electrophoresis was performed using an ABI Prism 310 Genetic Analyser, and the sequences were captured using GeneScan analysis, version 6.7 (Applied Biosystems).
TABLE 2.
Primers and annealing temperatures used for gene-specific PCR amplification and sequencing of L. lactis isolates
| Gene | Forward primera | Reverse primera | Annealing temp (°C) |
|---|---|---|---|
| rpoA | 5′-ATGATYGARTTTGAAAAACC-3′ | 5′-ACHGTRTTRATDCCDGCRCG-3′ | 46 |
| atpA | 5′-TAYRTYGGKGAYGGDATYGC-3′ | 5′-CCRCGRTTHARYTTHGCYTG-3′ | 50 |
| bcaT | 5′-TTTKSHRTGCCDGTWGG-3′ | 5′-GGWCCHACTTCYGTYTC-3′ | 46 |
| pepN | 5′-ATKTCTTAYGCWGAYRTYGT-3′ | 5′-TTKCTTCAAGSMAWGSCC-3′ | 50 |
| pepX | 5′-TTTGGGTTGAAAGTCCAGT-3′ | 5′-CCAAGAAGAAATTCCAGC-3′ | 46 |
| pheS | 5′-CAYCCNGCHCGYGAYATGC-3′ | 5′-CCWARVCCRAARGCAAARCC-3′ | 50 |
| SSU rRNA | 5′-GCGGCGTGCCTAATACATGC-3′ | 5′-ATCTACGCATTTCACCGCTAC-3′ | 50 |
Y, R, H, D, K, Y, W, M, and N are degenerate nucleotides according to the degenerate nucleotide alphabet: R for A or G; M for A or C; Y for C or T; K for G or T; W for A or T; N for A, G, C or T; S for G or C; H for A, C or T; and D for A or G or T.
(v) Computer-assisted analysis of DNA sequence data.
Single forward sequences were trimmed, aligned, and analyzed using BioNumerics, version 4.50, software. For each gene fragment, sequences were compared using neighbor-joining cluster analysis based on global alignment similarities. E. casseliflavus NIZO 2193 was used as outgroup in SSU rRNA gene sequence cluster analysis. Bootstrap analysis was applied with 500 simulations with single-sequence cluster analyses. Additionally, a composite data set was defined averaging the similarity matrices of five gene sequence analyses.
Phenotypic diversity. (i) Growth tests.
For screening purposes isolates were grown at 30°C in 96-well microplates using 250 μl of GM17 medium. Overnight cultures of isolates were incubated in quadruplicate, and each plate contained strain ML3 (NIZO 643) as a reference and no inoculum as a negative control. The following test media and conditions were evaluated: GM17 medium at 37 and 45°C; GM17 medium containing 4% NaCl at 30°C. When the test medium was not GM17, strains were subcultured twice to prevent false positives due to carryover from GM17 medium. Growth was analyzed by determining the turbidity at 600 nm and scored as negative (0), weak (1), or positive (2). Growth was scored after 24 h.
(ii) Fermentation tests.
For the determination of arginine hydrolase activity, isolates were grown in 96-well microplates in arginine broth as described by Niven et al. (26) but supplemented with 5g/liter peptone, 250 mg/liter MgSO4·7H2O, and 0.5 g/liter ascorbic acid. Growth was scored after 48 h by determining the turbidity at 600 nm. Cells were removed by centrifugation, and 10 μl of the resulting supernatant was mixed with reagent according to Niven et al. (5 g of KI, 5 g of HgCl2, 4 g of NaOH, and 100 ml of H2O, filtered) (26), and arginine hydrolyzing activity was scored as negative (0), weak (1), or strong (2), judged by the intensity of the orange color. Strain LMG 8505 (NIZO 2419) did not grow on the medium used by Niven et al. (26), and consequently no arginine hydrolase activity was determined.
The ability to ferment citrate was measured with whey permeate calcium citrate Casitone sgar (WPCCA) as described by Galesloot et al. (8) but with milk replaced by whey permeate.
The fermentation of maltose, lactose, galactose, and ribose was determined with the API 50 CHL assay (BioMérieux, l'Etoile, France). Strains were cultivated overnight in GM17 medium at 30°C, after which the culture was washed and resuspended in M17 with 1/4 sodium β-glycerol phosphate buffer (4.75 g/liter). Fermentation activity was determined after 48 h of incubation at 30°C using the recommendations of the manufacturer. The activity was scored using a seven-step incremental scale from negative (0) to positive (6).
Nucleotide sequence accession numbers.
The sequences generated in this study are available from GenBank/EMBL databases under the accession numbers EU091374 to EU091475 (SSU rRNA partial gene sequences), EU111131 to EU111178 (rpoA partial gene sequences), EU111220 to EU111267 (pheS partial gene sequences), EU111568 to EU111615 (atpA partial gene sequences), EU111309 to EU111354 (pepX partial gene sequences), EU111396 to EU111442 (pepN partial gene sequences), and EU111483 to EU111529 (bcaT partial gene sequences).
RESULTS
Taxonomic framework based on SSU rRNA gene sequence and phenotypic analysis.
Cluster analysis of partial SSU rRNA gene sequences of all isolates revealed five distinct species clusters (Fig. 1, clusters 1 to 5) that support the Lactococcus species classification of L. lactis, L. garvieae, L. raffinolactis, L. piscium, and L. plantarum. Additionally, all strains were screened using the phenotypic tests as proposed by Schleifer et al. (34) for the subspecific classification of L. lactis subsp. cremoris and L. lactis subsp. lactis. These tests include the production of ammonia from arginine and acid from maltose as well as salt tolerance at 4% NaCl (Table 1), and the results are depicted in Fig. 1. Eleven L. lactis isolates showed the inability to grow at NaCl concentrations of more than 4% at 37°C and did not form acid from maltose and ammonia from arginine, the discriminating phenotypic traits for L. lactis subsp. cremoris (Fig. 1). Furthermore, citrate production is specified for differentiation of biovar diacetylactis, as indicated by the WPCCA column in Fig. 1. The L. lactis cluster (Fig. 1, cluster 1) includes two major subgroups: subgroup B comprises 11 L. lactis subsp. cremoris isolates (unable to grow at 37°C) and in addition 9 L. lactis. subsp. lactis isolates (able to grow at 37°C), whereas subgroup A includes the other 67 L. lactis subsp. lactis isolates (able to grow at 37°C) and 2 L. lactis subsp. hordniae isolates (unable to grow at 37°C). The L. lactis subsp. hordniae sequences differ by one nucleotide (underlined) in the V1 region, having a T in AGTGGGGGA instead of the C in AGCGGGGGA of the other L. lactis subspecies and Lactococcus species. A third minor subgroup C is composed of L. lactis subsp. lactis strains P7266 NIZO 2206) and P7304 (NIZO 2207); both were able to grow at 37°C. Biovar diacetylactis strains were not differentiated from other L. lactis subsp. lactis cultures using partial SSU rRNA gene sequence analysis. Note that MG1363, which is commonly referred to as L. lactis subsp. cremoris, is actually L. lactis subsp. lactis, with a genotype like that of the L. lactis subsp. cremoris type strain (19, 38).
MLSA of L. lactis strains.
Partial DNA sequences of atpA, rpoA, pheS, bcaT, pepN, and pepXP were obtained for 89 L. lactis subsp. lactis and L. lactis subsp. cremoris isolates using the primers and temperatures listed in Table 2. No amplification products were obtained with L. lactis subsp. hordniae and other Lactococcus species. Decreasing the annealing temperature by 4°C resulted in additional, presumably nonspecific, amplicons in some of the isolates, while others still did not yield a PCR product and, therefore, were not used for further analysis. As a consequence, the following results are limited to L. lactis subsp. lactis and L. lactis subsp. cremoris diversity only.
A limited number of sequences with nucleotide ambiguities were observed for bcaT (five sequences with 1 or 2 ambiguities), pepN (seven sequences with up to 15 ambiguities), and pepXP (seven sequences with 1 or 2 single nucleotide deletions). All positions containing ambiguities were excluded from further analysis. Moreover, no sequences were obtained of pepN in strains KF7 (NIZO 2219) and KF 178 (NIZO 2234), of pepXP in strains P7266 and P7304, and of bcaT in strains P7266, KF 178, KW2 (NIZO 2247), and AM3 (NIZO 2251). Sequences of these isolates were omitted from the composite MLSA cluster analysis presented in Fig. 2 and Table 3. Cluster analysis of the sequences of atpA, rpoA, bcaT, pepN, and pepXP individually (85 to 89 isolates) and as a composite data set (five loci for MLSA; 83 isolates) (Fig. 2) revealed two distinct clusters corresponding to subgroups A and B in the cluster analysis based on partial SSU rRNA gene sequences (Fig. 1 and 2). Isolates of SSU rRNA gene sequence subgroup C yielded deviating five-locus MLSA sequences, each forming a single strain cluster, designated C1 and C2, supporting a more distant relation to the other isolates in this study. In the five-locus MLSA cluster analysis, subgroup A includes all isolates showing a biovar diacetylactis phenotype cluster together at high similarity (>99%) with isolates from dairy substrates (Fig. 2, 5 loci MLSA). This is in contrast with the partial SSU rRNA gene sequence cluster analysis, where the L. lactis subsp. lactis dairy isolates were scattered throughout cluster A (Fig. 2, SSU rRNA gene). Only ATCC 19435 clusters at a greater distance from the other isolates from dairy substrates. The single biovar diacetylactis isolate M20 not of dairy origin also clusters at some distance from most of the isolates from dairy substrates.
FIG. 2.
Comparative diversity analysis of 89 L. lactis isolates. Neighbor-joining cluster analysis of the partial SSU rRNA gene sequence (Escherichia coli positions 41 to 705), a five-locus MLSA based on a composite data set of DNA sequences of ∼0.4 kb of the intragenic regions of housekeeping genes atpA and rpoA and flavor-related genes bcaT, pepN, and pepXP and the single partial pheS sequence. Bootstrap percentages (≥50) after 500 simulations are shown for the single sequence analyses. Strains with the L. lactis subsp. lactis type-strain-like genotype [SSU rRNA gene sequence and (GTG)5-PCR fingerprint type] are represented with gray bars. Strains with an L. lactis subsp. cremoris type-strain-like genotype [SSU rRNA gene sequence and (GTG)5 PCR fingerprint type] are represented with white bars. Isolates obtained from dairy substrates are indicated (▪). The results of the partial SSU rRNA gene sequence and five-locus MLSA cluster analysis allow the discrimination of two major genomic lineages designated the L. lactis subsp. cremoris and L. lactis subsp. lactis type-strain-like genotypes, respectively. Strains P7304 and P7266 (boxed) grouped separately from the two major lineages. No sequences of pepN for isolates KF7 and KF 178, of pepXP for strains P7266 and P7304, and of bcaT for isolates P7266, KF 178, KW2, and AM3 are included in the composite five-locus MLSA cluster analysis presented since they were not obtained experimentally (see explanation in text).
TABLE 3.
Genetic diversity at seven loci based on 76 L. lactis isolates included in Fig. 2 and 3 for which sequences are available for all locia
| Locus | Gene length (bp) | Amplified fragment length (bp) | Analyzed fragment length (bp) | No. of polymorphic sites | % of variable nucleotide sites | No. of alleles | Avg G+C content (%) | dN/dS ratiob |
|---|---|---|---|---|---|---|---|---|
| atpA | 1,503 | 1,141 | 409 | 55 | 13.5 | 31 | 42 | 0.1408 |
| rpoA | 939 | 814 | 438 | 29 | 6.6 | 22 | 42 | ND |
| pheS | 2,533 | 618 | 397 | 46 | 11.6 | 30 | ND | ND |
| pepN | 1,023 | 482 | 411 | 65 | 15.8 | 38 | 37 | 0.0741 |
| bcaT | 1,047 | 493 | 315 | 87 | 27.6 | 39 | 39 | 0.0738 |
| pepX | 2,269 | 602 | 422 | 81 | 19.3 | 26 | 39 | 0.0806 |
| SSU rRNA | 4,050 | 650 | 420 | 14 | 3.3 | 15 | ND | ND |
This subset, apart from pheS, does not comprise any ambiguous nucleotides.
Pairwise ratios of nonsynonymous to synonymous substitutions (dN/dS) in sequences were calculated by using the method of Nei and Gojobori (25). ND, not determined.
The genes involved in flavor formation show a larger sequence variation than the housekeeping genes (Table 3). In particular, sequence variation in rpoA is limited and close to the SSU rRNA gene.
The observed sequence variation in the partial pheS gene sequence (Fig. 2) is substantial, and neighbor-joining cluster analysis resulted in groupings different from those described above. Again, two subgroups were found, one referred to as B+ comprising the members of cluster B (Fig. 1 and 2) with 26 additional L. lactis subsp. lactis isolates; the other, A−, comprises 43 L. lactis subsp. lactis isolates including all biovar diacetylactis isolates (Fig. 2, PheS). Limited unexplained ambiguous nucleotides were found in the pheS gene sequences in a pattern shared among (most of) the isolates. These nucleotides were not included in the cluster analysis (Fig. 2, PheS).
(GTG)5-PCR genomic fingerprint analysis.
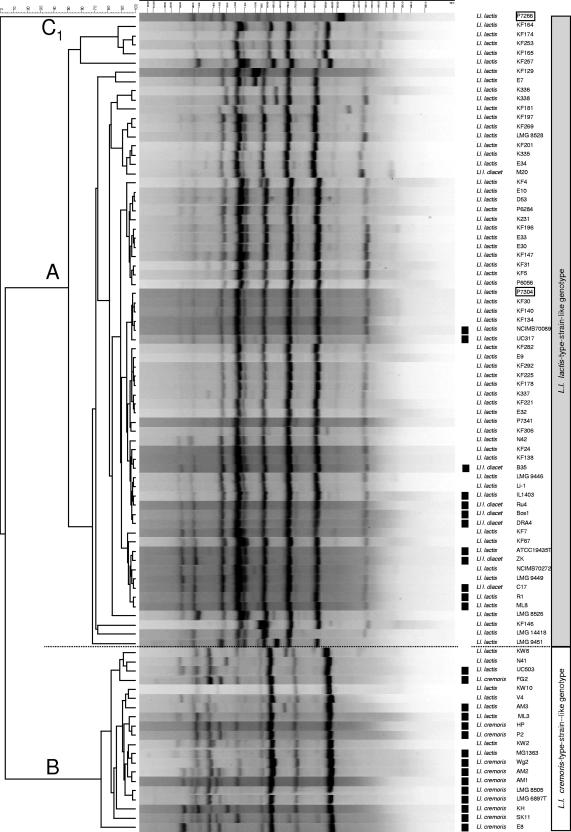
In addition to the genetic approaches described above, the subspecific diversity of L. lactis was analyzed with (GTG)5-PCR genomic fingerprinting. Product moment-UPGMA cluster analysis revealed two distinct clusters (Fig. 3) highly similar to the subgroups A and B observed with SSU rRNA gene sequence analysis and the composite five-locus MLSA clustering (Fig. 2, SSU rRNA gene sequence and five-locus MLSA). From the minor subgroup C, isolate P7304 is found in cluster A. Within the subgroups little diversity was observed. The results confirm the close relationship of L. lactis subsp. lactis and L. lactis subsp. lactis biovar diacetylactis since these strains remained indistinguishable using (GTG)5-PCR genomic fingerprinting analysis. The phenotypic differences between isolates are apparently not reflected in the genotype as sampled in the current collection by this method. The (GTG)5-PCR genomic fingerprint profiles of the two L. lactis subsp. hordniae strains show similarity with group B (data not shown) while partial SSU rRNA gene sequences cluster with group A (Fig. 1).
FIG. 3.
Product-moment and UPGMA cluster analysis of (GTG)5-PCR genomic fingerprint profiles of L. lactis isolates. The isolates cluster in two subgroups, cluster A comprising 11 L. lactis subsp. cremoris and 9 L. lactis subsp. lactis isolates and cluster B including the other 67 L. lactis subsp. lactis isolates. Isolates obtained from dairy substrates are indicated (▪).
DISCUSSION
Analysis of a collection of strains from various dairy and plant fermentations and a wide range of geographic locations, including Europe, Asia, New Zealand, and diverse environmental niches, provides a comprehensive view of the molecular and phenotypic diversity of the species L. lactis.
Three molecular typing methods were applied for analysis of L. lactis diversity, which allowed high-resolution analysis of the molecular diversity within the species. This newly developed MLSA typing scheme for L. lactis revealed unique sequence types for most strains, allowing high-resolution diversity analysis of the current strain collection. Partial SSU rRNA gene sequence analysis, (GTG)5-PCR genomic fingerprinting analysis, and a novel MLSA scheme all revealed two major, distinct genomic lineages within the species that are dissimilar to groups defined on the basis on extensive phenotypic analysis, including specific assays applied in the validly published classification of the species (15, 34).
Two of the three genomic lineages found are already supported with early work by Garvie and Farrow (9), DNA-DNA homology studies by Jarvis and Jarvis (15), and more recent genotypic analyses including gene sequence analyses (2, 24, 27, 38). One genomic lineage consists almost exclusively of strains of the L. lactis subsp. lactis including biovar diacetylactis. The second lineage comprised 10 out of the 11 analyzed L. lactis subsp. cremoris isolates and 10 isolates with an L. lactis subsp. lactis phenotype. In the current study 9 L. lactis subsp. lactis isolates with an L. lactis subsp. cremoris type-strain-like genotype were identified based on SSU rRNA gene sequence, five-locus MLSA, and (GTG)5 genomic fingerprinting. A reciprocal condition was not reported by Jarvis and Jarvis (15) and Garvie (10), but it was described by Kelly and Ward (16). In our analysis a clear third genomic lineage can be recognized that, to our knowledge, has not been described before. It consists of two isolates with an L. lactis subsp. lactis phenotype (P7266 and P7304) isolated from litter on pastures, and their partial SSU rRNA gene sequences (Fig. 1 and 2) and five-locus MLSA sequences (Fig. 2) group separately from the other L. lactis isolates. Phenotypic differences, however, are less distinct. The SSU rRNA gene sequences of the strains showed high similarity to Ribosomal Database Project II-derived lactococcal sequences of isolates from freshwater prawn (5), an organism that may be found in litter on pastures.
One of the six MLSA gene targets, pheS, showed two major groups, different from those revealed by the other genotyping methods and not reported before. One group is formed by the L. lactis subsp. cremoris type-strain-like genotype (based on SSU rRNA gene sequence and five-locus MLSA) amended with a large portion of L. lactis subsp. lactis isolates. The other group comprised the remaining L. lactis subsp. lactis isolates. Congruence of a pheS genotype with another genotype or phenotype was not observed. This phenomenon cannot be explained by a single horizontal gene transfer event, as has been identified with other genes from comparative genome analysis of L. lactis (2), nor can it be explained by gene duplication. Gene duplication of pheS was not found in strains IL-1403, SK11, and MG1363, for which full genome sequences have been published (3, 22, 40).
L. lactis subsp. lactis and L. lactis subsp. cremoris are defined based on their phenotypes (34). In general, the isolates with an L. lactis. subsp. lactis phenotype showed a partial SSU rRNA gene sequence similar to the L. lactis subsp. lactis type strain while a limited number of isolates showed an L. lactis subsp. cremoris-(type strain)-like genotype. Unfortunately, this has been turned around by several groups using a subspecific classification based on genotype, causing confusion in the taxonomy of L. lactis. This started with the application of supposedly subspecies-specific probes (18, 31, 32) and continued with additional methods for genetic differentiation such as SSU rRNA gene sequencing (39) and other genotyping methods (see reference 38 and references therein). However, based on our findings, there seems to be no basis for redefinition of the L. lactis subspecies. The phenotypic differences listed in the current classification are sufficient to distinguish the subspecies (34). However, one should be aware that the subspecific identification of L. lactis in the literature from the 1990s to the present is often not based on the phenotype and may therefore be confusing. This confusion can be avoided by using the current phenotypic classification amended with a “type strain-like-genotype” classification, which will provide a direct subspecific phylogenetic reference.
The remarkable taxonomic structure of L. lactis is also evident from the genome sequences of three different strains that have been published (3, 40). These strains include well-known model strains of dairy origin such as strain IL-1403 (L. lactis subsp. lactis type-strain-like genotype and L. lactis subsp. lactis phenotype), strain MG363 (L. lactis subsp. cremoris type-strain-like genotype and L. lactis subsp. lactis phenotype), and strain SK11 (L. lactis subsp. cremoris type-strain-like genotype and L. lactis subsp. cremoris phenotype). Strain IL-1403 is actually a plasmid-cured derivative of the biovar diacetylactis strain CNRZ157 rendering it cit negative. This analysis and the reports of other investigators (27, 38) show that these three strains clearly represent only a small part of the diversity present within this subspecies. It can be expected that the adaptation to diverse plant substrates of the nondairy strains has resulted in the development of unique traits and phenotypes that can be utilized in dairy and other food fermenting applications. Recently, we have sequenced the genomes of two plant isolates (38). Further analysis of these data and genomotyping with DNA microarrays in our laboratory will provide insight into the lack of congruence of the pheS genotype with other features and will allow a deeper understanding of the genomic and functional diversity within the species.
Acknowledgments
We acknowledge Jan Hoolwerf and Arjen Wagendorp for their technical assistance. The following people are acknowledged for kindly providing strains used in this study: Andreas Ulrich and Thomas Müller, Centre for Agricultural Landscape and Land Use Research Münchenberg Institute of Microbial Ecology and Soil Biology Münchenberg, Germany; Sang-Hee Park, Laboratory of Veterinary Public Health Graduate School of Agricultural and Life Sciences, the University of Tokyo, Japan; and U. Shillinger, Federal Research Centre for Nutrition Institute of Hygiene and Toxiology Karlsruhe, Germany. Meike te Giffel and Giovanna Felis are acknowledged for valuable comments on the manuscript and Bart Hoste and Marc Vancanneyt for useful discussions including Lactococcus taxonomy.
D.G. and J.S. are indebted to the Fund for Scientific Research, Flanders (Belgium), for a position as a postdoctoral fellow and research grants, respectively.
Footnotes
Published ahead of print on 21 September 2007.
REFERENCES
- 1.Ayad, E. H., A. Verheul, W. J. Engels, J. T. Wouters, and G. Smit. 2001. Enhanced flavour formation by combination of selected lactococci from industrial and artisanal origin with focus on completion of a metabolic pathway. J. Appl. Microbiol. 90:59-67. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin, A., B. Quinquis, A. Sorokin, and D. S. Ehrlich. 2004. Recent genetic transfer between Lactococcus lactis and enterobacteria. J. Bacteriol. 186:6671-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bougnoux, M. E., D. Diogo, N. Francois, B. Sendid, S. Veirmeire, J. F. Colombel, C. Bouchier, H. Van Kruiningen, C. d'Enfert, and D. Poulain. 2006. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J. Clin. Microbiol. 44:1810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Las Rivas, B., A. Marcobal, and R. Munoz. 2006. Development of a multilocus sequence typing method for analysis of Lactobacillus plantarum strains. Microbiology 152:85-93. [DOI] [PubMed] [Google Scholar]
- 7.Erlandson, K., and C. A. Batt. 1997. Strain-specific differentiation of lactococci in mixed starter culture populations using randomly amplified polymorphic DNA-derived probes. Appl. Environ. Microbiol. 63:2702-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galesloot, T. E., I. Hassing, and J. Stadhouders. 1961. Agar media for the isolation and enumeration of aroma bacteria in starters. Neth. Milk Dairy J. 15:145-150. [Google Scholar]
- 9.Garvie, E., and J. Farrow. 1982. Streptococcus lactis subsp. cremoris (Orla-Jensen) comb. nov. and Streptococcus lactis subsp. diacetilactis (Matuszewski et al.) nom. rev., comb. nov. Int. J. Syst. Bacteriol. 32:453-455. [Google Scholar]
- 10.Garvie, E. I. 1978. Streptococcus raffinolactis (Orla-Jensen and Hansen): a group N streptococcus found in raw milk. Int. J. Syst. Bacteriol. 28:190-193. [Google Scholar]
- 11.Gevers, D., G. Huys, and J. Swings. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31-36. [DOI] [PubMed] [Google Scholar]
- 12.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A. B. Kolsto. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams and Wilkins Co., Baltimore, MD.
- 14.Hommais, F., S. Pereira, C. Acquaviva, P. Escobar-Paramo, and E. Denamur. 2005. Single-nucleotide polymorphism phylotyping of Escherichia coli. Appl. Environ. Microbiol. 71:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis, A. W., and B. D. Jarvis. 1981. Deoxyribonucleic acid homology among lactic streptococci. Appl. Environ. Microbiol. 41:77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly, W., and L. Ward. 2002. Genotypic vs. phenotypic biodiversity in Lactococcus lactis. Microbiology 148:3332-3333. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, W. J., G. P. Davey, and L. J. Ward. 1998. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int. J. Food Microbiol. 45:85-92. [DOI] [PubMed] [Google Scholar]
- 18.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1995. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl. Environ. Microbiol. 61:788-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kok, J., G. Buist, A. L. Zomer, S. A. van Hijum, and O. P. Kuipers. 2005. Comparative and functional genomics of lactococci. FEMS Microbiol. Rev. 29:411-433. [DOI] [PubMed] [Google Scholar]
- 20.Liu, M., F. H. van Enckevort, and R. J. Siezen. 2005. Genome update: lactic acid bacteria genome sequencing is booming. Microbiology 151:3811-3814. [DOI] [PubMed] [Google Scholar]
- 21.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkinse, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Díaz-Muñiz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimerd, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangin, I., D. Corroler, A. Reinhardt, and M. Gueguen. 1999. Genetic diversity among dairy lactococcal strains investigated by polymerase chain reaction with three arbitrary primers. J. Appl. Microbiol. 86:514-520. [DOI] [PubMed] [Google Scholar]
- 24.Mori, S., K. Mori, I. Suzuki, and T. Kasumi. 2004. Phylogenetic analysis of Lactococcus lactis subspecies based on decoding the sequence of the pepT tripeptidase gene, the pepV dipeptidase gene and 16S rRNA. Syst. Appl. Microbiol. 27:414-422. [DOI] [PubMed] [Google Scholar]
- 25.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 26.Niven, C. F., K. L. Smiley, and J. M. Sherman. 1942. The hydrolysis of arginine by streptococci. J. Bacteriol. 43:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura, M., M. Kobayashi, T. Narita, H. Kimoto-Nira, and T. Okamoto. 2006. Phenotypic and molecular characterization of Lactococcus lactis from milk and plants. J. Appl. Microbiol. 101:396-405. [DOI] [PubMed] [Google Scholar]
- 28.Pu, Z. Y., M. Dobos, G. K. Limsowtin, and I. B. Powell. 2002. Integrated polymerase chain reaction-based procedures for the detection and identification of species and subspecies of the gram-positive bacterial genus Lactococcus. J. Appl. Microbiol. 93:353-361. [DOI] [PubMed] [Google Scholar]
- 29.Rademaker, J. L. W., and F. J. deBruijn. 2004. Computer-assisted pattern analysis of electrophoretic fingerprints and database construction. Chapter 1.7.5, p. 1-50. In G. Kowalchuk, F. deBruijn, I. Head, A. Akkermans, and J. van. Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 30.Rademaker, J. L. W., F. J. Louws, J. Versalovic, and F. J. deBruijn. 2004. Characterization of the diversity of ecological important microbes by rep-PCR genomic fingerprinting. Chapter 5.3.2, p. 1-33. In G. Kowalchuk, F. deBruijn, I. Head, A. Akkermans, and J. van. Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Salama, M., W. Sandine, and S. Giovannoni. 1991. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl. Environ. Microbiol. 57:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salama, M. S., W. E. Sandine, and S. J. Giovannoni. 1993. Isolation of Lactococcus lactis subsp. cremoris from nature by colony hybridization with rRNA probes. Appl. Environ. Microbiol. 59:3941-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salcedo, C., L. Arreaza, B. Alcala, L. de la Fuente, and J. A. Vazquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schleifer, K. H., J. Kraus, C. Dvorak, R. Kilpper-Balz, M. D. Collins, and W. Fisher. 1985. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst. Appl. Microbiol. 6:183-195. [Google Scholar]
- 35.Smit, G., B. A. Smit, and W. J. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 36.Tailliez, P., J. Tremblay, S. D. Ehrlich, and A. Chopin. 1998. Molecular diversity and relationship within Lactococcus lactis, as revealed by randomly amplified polymorphic DNA (RAPD). Syst. Appl. Microbiol. 21:530-538. [DOI] [PubMed] [Google Scholar]
- 37.Torpdahl, M., M. N. Skov, D. Sandvang, and D. L. Baggesen. 2005. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J Microbiol. Methods 63:173-184. [DOI] [PubMed] [Google Scholar]
- 38.van Hylckama Vlieg, J. E., J. L. Rademaker, H. Bachmann, D. Molenaar, W. J. Kelly, and R. J. Siezen. 2006. Natural diversity and adaptive responses of Lactococcus lactis. Curr. Opin. Biotechnol. 17:183-190. [DOI] [PubMed] [Google Scholar]
- 39.Ward, L. J., J. C. Brown, and G. P. Davey. 1998. Two methods for the genetic differentiation of Lactococcus lactis ssp. lactis and cremoris based on differences in the 16S rRNA gene sequence. FEMS Microbiol. Lett. 166:15-20. [DOI] [PubMed] [Google Scholar]
- 40.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. Gasson, O. Kuipers, D. v. Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]