Abstract
A 16S rRNA-targeted, Cy3-labeled oligonucleotide probe was designed to detect members of the genus Bdellovibrio by fluorescence in situ hybridization. Specific hybridization conditions were established; however, the detection of bdellovibrios in environmental samples required enrichment, confirming that Bdellovibrio spp. are not present in large numbers in the environment.
Bdellovibrio and like organisms (BALOs) are predatory gram-negative bacteria that are ubiquitous in terrestrial and aquatic environments. Prey include free-living bacteria as well as plant, animal, and human pathogens (12, 13, 17). The life cycle of Bdellovibrio has two major stages: a motile, nonreplicative stage spent searching for prey (the attack phase) and a stage spent inside the periplasm of the gram-negative prey cell after the formation of an osmotically stable body termed the bdelloplast (the growth phase) (15, 27). Until recently, all bacteria that exhibited these life cycle traits were included in the genus Bdellovibrio. Phylogenetic studies based on 16S rRNA sequences have demonstrated extensive diversity among the organisms in this genus, and two new genera have been established: Bacteriovorax (4) and Peredibacter (8). To date, the genus Bdellovibrio comprises only one species, Bdellovibrio bacteriovorus. However, a Bdellovibrio-like organism (BLO) with a different life cycle was isolated from sewage on lawns of Caulobacter crescentus cells. This epibiotic strain, JSS, does not enter the periplasmic space of the prey cell (20, 26). Phylogenetic studies show that it belongs to the genus Bdellovibrio (8) and is a new species (S. F. Koval, unpublished data). Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes has become a commonly used technique for the direct identification of prokaryotes and is widely used to determine bacterial community composition (2, 10). Previously designed BALO-targeted oligonucleotides for use in FISH were not specific enough for the identification of the genus Bdellovibrio, as the targets detected included non-BALOs (8, 16). However, Bdellovibrio-specific primers were used to detect Bdellovibrio sequences in soil (9). In this study, an oligonucleotide probe designated BDE525, targeting a region of the 16S rRNA gene sequence of Bdellovibrio, was designed. The probe was used in a FISH procedure with B. bacteriovorus strains, other BLO isolates, and environmental samples.
Cultures.
B. bacteriovorus strain 109J was cultured on Escherichia coli ML35 according to the method of Flannagan et al. (11). B. bacteriovorus 6-5-S was cultured on Aquaspirillum serpens VHL and BLO strains JSS and KL1 were cultured on C. crescentus CB2A as described previously by Koval and Hynes (20). BLO strains DM7C, DM8A, and DM11A were cultured on Burkholderia cenocepacia strains grown in Lennox Luria-Bertani medium (Difco) for 24 h at 30°C. For the maintenance of predators, 3 ml of prey cells and 1 ml of predators were mixed in 20 ml of HM buffer (3 mM HEPES [Sigma], pH 7.6, with 1 mM CaCl2 and 0.1 mM MgSO4). Cocultures were incubated at 30°C for 18 to 24 h with vigorous aeration until prey cells were lysed. The prey-independent strain B. bacteriovorus 109JA and the facultative predator Bacteriovorax stolpii UKi2 were grown on PY medium (peptone [Bacto] at 10 g liter−1, yeast extract at 3 g liter−1, pH 6.8 to 7.2) (27) for 24 h at 30°C. Shewanella baltica 63 (NCTC 10735; obtained from T. J. Beveridge, University of Guelph) was grown aerobically in Trypticase soy medium for 24 h at 30°C.
Cells were harvested for fixation when the coculture was composed of a mixture of bdelloplasts and attack-phase cells of predators. B. bacteriovorus 109JA, Bacteriovorax stolpii UKi2, and S. baltica 63 were harvested in log phase after 24 h of incubation. Cells were concentrated by centrifugation of 20 ml of each culture at 10,000 × g for 15 min at 4°C. Each pellet was resuspended in 2 ml of phosphate-buffered saline (PBS; pH 7.4) and fixed in a 1:3 dilution of neutral 10% buffered formalin (pH 7.0; 10 ml of 36 to 38% formaldehyde [Sigma], 0.65 g of sodium phosphate dibasic [anhydrous], 0.4 g of sodium phosphate monobasic, 90 ml of double-distilled water) on ice at 4°C for 3 h. Cells were then centrifuged, washed twice in 20 ml of PBS, resuspended in 2 ml of PBS-96% ethanol solution (1:1), and stored at −20°C prior to hybridization.
Probe design.
An oligonucleotide probe was designed using the ARB probe tool (http://www.arb-home.de). The design of the oligonucleotide probe was based on the phylogeny of Bdellovibrio inferred by 16S rRNA sequence analysis. A search for a region that would allow the recognition of all nine clusters of the Bdellovibrio clade established by Davidov and Jurkevitch (8) did not reveal a suitable probe, which indicates the broad phylogenetic diversity of BALOs. The sequence of an oligonucleotide probe (E. coli positions 525 to 542) designated BDE525 (Table 1), or S-G-Bde-0525-a-A-18 according to the Oligonucleotide Database Project (1), was chosen, as it perfectly matched all of the 16S rRNA sequences of the Bdellovibrio clade except for those of the unidentified bacterial isolates (clone 6i39 in cluster 2 and clones SM1G01 and SM1H08 in cluster 8) (Table 1). Clone 6i39 exhibited four mismatches relative to the probe. Clones SM1G01 and SM1H08 each exhibited one mismatch at the 3′ end. The sequence of BDE525 was also determined to provide a good site for probe-conferred fluorescence. According to Behrens et al., the site spanning positions 525 to 542 on the predicted secondary structure of E. coli gives a brightness of class III with a relative FISH intensity of 0.6 to 0.41 when hybridized with an 18-mer oligonucleotide probe labeled with the fluorescent dye Cy3 (5). Probe specificity was tested against target and nontarget organisms based on a comparative analysis of recently available aligned 16S rRNA gene sequences from the BLAST database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) with the assistance of the Probe Match function of Ribosome Database Project II version 9 (6). Some of these sequences are shown in Table 1. The probe was synthesized and labeled at the 5′ end with Cy3 by Sigma-Aldrich Canada Ltd. (Oakville, Ontario, Canada). Target and nontarget organisms were examined with probe BDE525 in a FISH protocol to confirm the specificity of the probe. Control cells and BLO isolates were also examined using a Cy3-labeled universal eubacterial probe, EUB338 (S-D-Bact-0338-a-A-18; 5′ GCTGCCTCCCGTAGGAGT 3′).
TABLE 1.
Comparison of 16S rRNA sequences of target and nontarget organisms with the sequence of probe BDE525
| Probe or organism | GenBank accession no. | Sequencea |
|---|---|---|
| Probe BDE525 | 5′ GATCCCTCGTCTTACCGC 3′ | |
| B. bacteriovorus 109J | AY094125 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| B. bacteriovirus 6-5-S | AY094106 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| B. bacteriovirus 100 HD | AF084850 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| B. bacteriovirus BEP2 | AF148938 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| B. bacteriovirus SRA9 | AF263833 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| B. bacteriovirus E | AF084851 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| B. bacteriovirus 2484Se2 | AY094126 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| B. bacteriovirus BEP2 | AF148938 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| Bdellovibrio sp. strain W ATCC 27047 | AJ292518 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| Bdellovibrio sp. strain JSS | EF687743 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| Bdellovibrio sp. strain Hefner | AY094114 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| Bdellovibrio sp. strain Vietnam | AY094123 | 3′ CTAGGGAGCAGAATGGCG 5′ |
| Bacteriovorax stolpii UKi2 | M34125 | 3′ GTGGGAAGCAGTATGGCG 5′ |
| Bacteriovorax marinus JS | AF084854 | 3′ GTGGGAAGCAGTATGGCG 5′ |
| Bacteriovorax litoralis JS5 | AF084859 | 3′ GTGGGAAGCAGTATGGCG 5′ |
| Peredibacter starrii A3.12 | AF084852 | 3′ CTGGGAAGCATAATGGCG 5′ |
| Shewanella sp. clone D1Mn | AF271657 | 3′ CGAGGGAGCAGTATGGCG 5′ |
| Shewanella baltica 63 NCTC 10735 | AJ000214 | 3′ CTGGGAGGCAGAATGGCG 5′ |
| Uncultured bacterium clone 6i39 | AY177783 | 3′ CTAGGGACAGCAATGGCG 5′ |
| Uncultured deltaproteobacterium clone SM1G01 | AF445695 | 3′ CCAGGGAGCAGAATGGCG 5′ |
| Uncultured deltaproteobacterium clone SM1H08 | AF445705 | 3′ CCAGGGAGCAGAATGGCG 5′ |
| Klebsormidium flaccidum plastid (eukaryote) | X75522 | 3′ GTAGGGGGCAGAATGGCG 5′ |
Mismatching nucleotides are underlined.
FISH of pure cultures.
The FISH procedure was performed according to the method of Mahmoud et al. (22) with CLEARCELL slides ER-203B-2 (Erie Scientific, Portsmouth, NH) and a 10-μl sample size. For hybridization, a master mix containing 50 ng of labeled probe in 100 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.4], 0.1% sodium dodecyl sulfate, and different concentrations of formamide; final probe concentration, 0.5 ng/μl) was applied to each well on a multiwell slide, and the wells were covered. Slides were incubated for 2 h at 46°C and then placed in a wash buffer (20 mM Tris-HCl [pH 7.4], 0.1% sodium dodecyl sulfate, 5 mM EDTA, and the appropriate concentration of NaCl according to the table by Pernthaler et al. [25]) and incubated at 48°C for 20 min. Slides were rinsed in sterile distilled water and air dried in the dark at room temperature. Each treatment was replicated once within an experiment, and each experiment was performed twice. The whole-cell hybridizations were recorded with a QImaging Retiga 1300 cooled monochrome 12-bit camera connected to an Axioskop II epifluorescence microscope (Zeiss, Hamburg, Germany) equipped with an HBO100/2 mercury lamp for epifluorescence illumination. The camera was controlled with Northern Eclipse software version 6.0 (Empix Imaging Inc., Mississauga, Ontario, Canada). Hybridizations with the Cy3 probes were visualized with the Zeiss filter set 15 (red excitation light around 546 nm). Cells stained with DAPI (4′,6′-diamidino-2-phenylindole) were visualized with the Zeiss filter set 2 (blue excitation light around 365 nm). Target and nontarget cells showed no autofluorescence at the excitation wavelength for the Cy3 labeling.
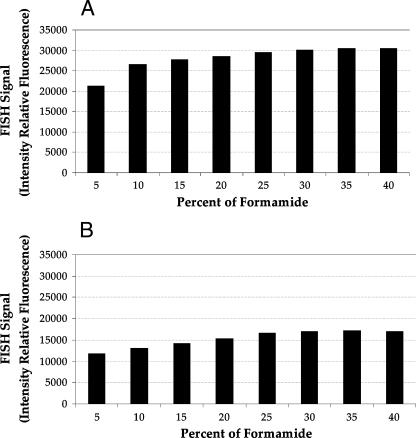
To determine the optimum concentration of formamide for the FISH protocol with Bdellovibrio, the stringency of hybridization was adjusted by gradually increasing the formamide concentration in the hybridization buffer (in 5% intervals). We used a coculture of B. bacteriovorus 109J and E. coli prey cells (Fig. 1A), as the bdelloplasts of E. coli cells are uniformly of the same shape and size. The probe was accessible to cells growing in bdelloplasts as well as free-swimming attack cells. This result is important as the predator Bdellovibrio is often found in bdelloplasts in the environment. The single-cell fluorescence intensities were determined by converting wavelength-specific cell brightness into a grayscale pixel value that ranged from 0 to 64,500 per cell. By manual comparison of images taken during stages of the life cycle, the lower and upper limits of the fluorescence range were determined. These limits excluded faint and very bright cells. For attack-phase cells and cells growing in bdelloplasts, the ranges were 5,000 to 35,000 and 14,000 to 40,000 U, respectively. A minimum of 500 cells were counted at each concentration of formamide for attack-phase cells, and a minimum of 200 were counted for bdelloplasts. The variations in FISH signal intensities at formamide concentrations of 30 to 40% were almost identical for cells growing in bdelloplasts and attack-phase cells of the predator (Fig. 2). Therefore, 35% formamide was chosen for hybridization events.
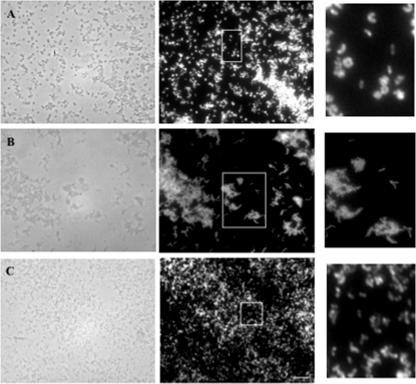
FIG. 1.
FISH of pure cultures with probe BDE525. Phase-contrast (left) and epifluorescence (right) images of the same fields are shown. (A) B. bacteriovorus 109J; (B) B. bacteriovorus 109JA; (C) BLO strain JSS. White boxes indicate the sections shown at greater magnification on the right. The bar in panel C is 10 μm and applies to all panels.
FIG. 2.
Average intensities of the BDE525 FISH signal for B. bacteriovorus 109J at different formamide concentrations. (A) Cells growing inside bdelloplasts; (B) attack-phase cells.
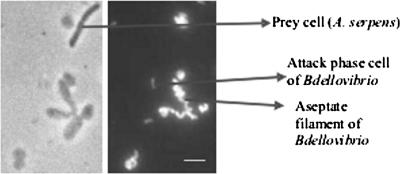
As seen in Fig. 2, the fluorescence signal from cells growing in bdelloplasts was more intense than the signal from released progeny cells. This result was effectively illustrated in a coculture of B. bacteriovorus 6-5-S and A. serpens VHL, as the bdelloplasts were larger than those derived from E. coli, though often not of uniform shape or round. Long, aseptate, helical cells of the predator were clearly seen inside bdelloplasts (Fig. 3). The few Aquaspirillum prey cells left in the coculture did not hybridize with probe BDE525. The BDE525 probe also hybridized with prey-independent B. bacteriovorus 109JA harvested during the log growth phase (Fig. 1B).
FIG. 3.
FISH of cells in a coculture of B. bacteriovorus 6-5-S and A. serpens VHL with probe BDE525. Phase-contrast (left) and epifluorescence (right) images of the same field are shown. The bar is 5 μm.
This probe was tested against Bacteriovorax stolpii as the most phylogenetically related, culturable, nontarget organism. Bacteriovorax stolpii UKi2 exhibited four mismatches compared to probe BDE525 (Table 1) and did not hybridize with this probe at the different formamide concentrations tried (data not shown). S. baltica 63, with three mismatches relative to the probe (Table 1), also did not hybridize with probe BDE525 at any of the formamide concentrations (data not shown). Probe EUB338 (used to assess the performance of FISH) hybridized with B. bacteriovorus strain 109J, E. coli ML35, A. serpens VHL, and Bacteriovorax stolpii UKi2 (data not shown). We therefore judged the BDE525 probe to be specific for the intended genus.
Because FISH requires bacterial ribosomes to which the oligomer probe binds, the concentration of RNA in the cell of interest is important. The signal conferred by fluorescently labeled rRNA-targeted probes on fixed cells can be correlated to cellular rRNA content and growth rates (2). The ribosome content of a cell is positively correlated with the growth rate, and thus cells for FISH are usually harvested during the log phase of growth. However, in most environmental samples, the cellular RNA content is low, as the bacteria are slow growing (21). The growth rate of BALOs is not easy to define as BALOs do not grow by the doubling of cell numbers. In the growth phase, the cell in the bdelloplast increases in size as a single, elongating, spiral cell with unseptated cytoplasm. Therefore, the concept of the doubling time of a cell does not apply to BALOs. The strong FISH signals of cells in bdelloplasts may reflect their active metabolism. Alternatively, the location of these cells may give rise to higher local concentrations of the probe, which may affect hybridization kinetics, leading to intense hybridizations within the 2-h hybridization period.
To further test the specificity of the BDE525 probe, the hybridization of other BALO isolates was performed using the conditions established for B. bacteriovorus 109J. Strain KL1, like strain JSS, was isolated from sewage on lawns of C. crescentus CB2A cells but in a different year. Both strains are epibiotic predators (26) and both hybridized with probe BDE525 (Fig. 1C). Strain JSS has been identified as a Bdellovibrio sp. (8). BLO strains DM7C, DM8A, and DM11A were isolated on B. cenocepacia strains J2315 (DM7C) and K56-2 (DM8A and DM11A). Strains DM7C and DM8A were isolated from compost; strain DM11A was isolated from sewage. These isolates have a periplasmic life cycle characteristic of B. bacteriovorus (23). The three strains hybridized with probe BDE525 (data not shown), which suggests that they are Bdellovibrio spp.
To estimate a limit of detection of Bdellovibrio by FISH, a 48-h coculture of B. bacteriovorus 109J and E. coli prey cells was diluted in a 10-fold series. Each dilution was plated to enumerate the PFU per milliliter (the yield was 2 × 109 PFU/ml), and at the same time, a 100-μl volume of cells was fixed for FISH (for which a 10-μl sample size was used). At least 20 to 50 Bdellovibrio cells per field of view were detected by the BDE525 probe in dilutions containing 2 × 109 to 2 × 106 PFU/ml. Fewer Bdellovibrio cells were detected in the 2 × 105-PFU/ml sample, which would contain 2 × 103 Bdellovibrio cells per 10 μl of sample. Therefore, any culture or environmental sample to be tested by FISH needs to contain at least 105 PFU/ml.
FISH analysis of environmental samples.
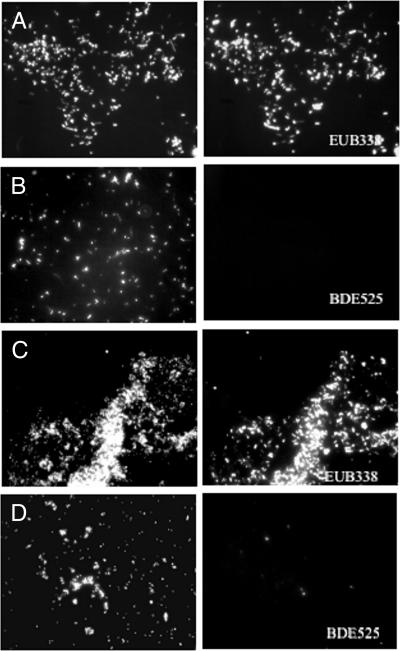
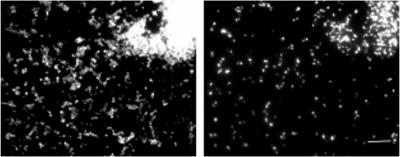
The ability of the BDE525 probe to detect Bdellovibrio in environmental samples was tested with samples collected in the autumn from the following habitats: runoff from a cow-milking facility, cornfield soil, residential compost and soil, and raw sewage and activated sludge from the Adelaide Street pollution control plant in London, ON. Bdellovibrios have been isolated from soil (9, 12, 17), compost (this study), and sewage (12, 20). For the fixation of solid environmental samples, approximately 1.0 g of soil (18, 29) was fixed in 30 ml of 3.6 to 3.8% formaldehyde (prepared as described for the fixation of pure cultures) and placed on ice at the site of collection. For the fixation of liquid environmental samples, 10 ml of liquid was fixed in 30 ml of neutral 10% buffered formalin on ice at the site of collection. All samples were then fixed overnight on ice in a refrigerator (4°C). After fixation, samples were centrifuged at 12,000 × g, washed twice in PBS, resuspended in a final volume of 2 ml of a 1:1 PBS-100% ethanol solution, and stored at −20°C. To test for the number of bacterial cells in fixed samples, prior to FISH analysis, 200 μl of each fixed sample was stained with DAPI (final concentration, 1 μg ml−1) for 15 min. Between 30 and 50 cells per field of view were required for further FISH analysis and were found in the milking facility runoff, the cornfield soil, and the compost samples. The raw-sewage and activated-sludge samples were concentrated five times by centrifugation (10 ml resuspended in 2 ml) prior to staining. Bacterial cells were identified in all samples by hybridization with the EUB338 probe (Fig. 4A), but none of the cells in the activated-sludge sample (Fig. 4B) or any other samples hybridized with the BDE525 probe. The analysis of the composition of microbial communities in bulk soil samples by FISH often leads to a low detection yield (14).
FIG. 4.
FISH analysis of environmental samples. DAPI (left) and FISH (right) images of the same fields are shown. (A and B) Activated-sludge samples were subjected to FISH with EUB338 and BDE525. (C and D) Enrichment cultures of activated sludge with E. coli ML35 prey cells hybridized with EUB338 and BDE525.
To rule out the possibility that something in the environmental samples was blocking the FISH detection, either by preventing proper fixation or by interfering with hybridization, the raw-sewage sample was spiked with B. bacteriovorus 109J cells, both fixed and unfixed, at a ratio of 1:1. These spiked samples showed a strong FISH signal with both probes (data not shown). Therefore, the fixation process was suitable for environmental samples and the environment tested did not interfere with FISH. It was concluded that the numbers of Bdellovibrio cells in the environmental samples were too low for the cells to be detected directly. For this study, samples were collected at one time point (in the autumn); however, for an ecological study, samples would need to be taken at several times during the year.
To detect Bdellovibrio in the environment, it was decided that, due to low cell numbers, enrichment of samples was required. Previous work in our laboratory had elucidated an appropriate method for the enrichment of both raw sewage and activated sludge with BALOs (19, 20). Therefore, these two environmental samples were selected for further analysis and were enriched using both A. serpens and E. coli as prey cells and dilute nutrient broth (0.08% nutrient broth [Difco] with 1 mM CaCl2 and 0.1 mM MgSO4) (7). When sufficient numbers of bdelloplasts were visible by light microscopy, samples were fixed and FISH was performed. At the same time, plaque assays were done to determine the numbers of BALOs present in the enrichment cultures and to determine a detection limit for environmental sample analysis. The yield of BALOs (2 × 102 PFU ml−1) in the A. serpens enrichment culture was too low for FISH analysis, as an average of two BALOs per well would be present. However, bdellovibrios were detected in both activated sludge (Fig. 4D) and raw sewage enriched with E. coli prey cells, in which the yield was much higher (2 × 106 PFU ml−1) and corresponded to an average of 2 × 104 BALOs per well. This result confirms our detection limit established previously.
To test the sensitivity of the FISH procedure, the hybridization of cells directly from a single plaque of the activated-sludge enrichment culture was tried. An individual plaque was picked and transferred to a microcentrifuge tube, and cells were fixed by the addition of formaldehyde, as described above. Only prey cells were detected by DAPI, as there was no signal with the BDE525 probe (data not shown). However, subsequent coculture of the BALOs in the plaque did provide enough cells for the identification of the predators as Bdellovibrio (Fig. 5). BLOs in individual plaques were grown by picking plaques with a Pasteur pipette and mixing cells in 5 ml of dilute nutrient broth. E. coli ML35 (0.5 ml of a 24-h culture) was added, and the coculture was incubated at 30°C until lysis of the prey cells had occurred. These results support the observations of Davidov et al. (9) that BALOs do not form numerically dominant groups within microbial communities. It also demonstrates that in order for microscopic analysis of FISH to be an effective tool for the identification of Bdellovibrio in the environment, enrichment of the Bdellovibrio subpopulation may be required. Alternatively, flow cytometry could be applied to samples previously stained with 16S rRNA probes (3, 28) or with DNA molecular beacons (21). These flow cytometry techniques may allow the detection of Bdellovibrio in environmental samples without the expense associated with peptide nucleic acid probes or multilaser flow cytometry or the technical demands of catalyzed reporter deposition FISH (24). If successful, these techniques would allow the identification and enumeration of Bdellovibrio cells without the prey bias associated with enrichment cultures (19). Despite the difficulties, the identification of Bdellovibrio in environmental samples, although not directly, has been made possible through the use of FISH.
FIG. 5.
FISH of a coculture from a single plaque with BDE525. DAPI (left) and FISH (right) images of the same field are shown. The bar is 10 μm.
Acknowledgments
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant to S. F. Koval.
We thank W. Montgomery McKillop for his assistance in installing the ARB software and the reviewers for their helpful comments.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer, M. L., J. Ravel, J. Chun, R. T. Hill, and H. N. Williams. 2000. A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 50:219-224. [DOI] [PubMed] [Google Scholar]
- 5.Behrens, S., B. M. Fuchs, F. Mueller, and R. Amann. 2003. Is the in situ accessibility of the 16S rRNA of Escherichia coli for Cy3-labeled oligonucleotide probes predicted by a three-dimensional structure model of the 30S ribosomal subunit? Appl. Environ. Microbiol. 69:4935-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 1:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, T. W., and M. F. Thomashow. 1992. A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J. Bacteriol. 174:689-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidov, Y., and E. Jurkevitch. 2004. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov. comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoraceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:1439-1452. [DOI] [PubMed] [Google Scholar]
- 9.Davidov, Y., A. Friedjung, and E. Jurkevitch. 2006. Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like-organisms. Environ. Microbiol. 8:1667-1673. [DOI] [PubMed] [Google Scholar]
- 10.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single microbial cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 11.Flannagan, R. S., M. A. Valvano, and S. F. Koval. 2004. Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109J from bdelloplasts of bacterial prey cells. Microbiology 150:649-656. [DOI] [PubMed] [Google Scholar]
- 12.Fratamico, P. M., and P. H. Cooke. 1996. Isolation of bdellovibrios that prey on Escherichia coli O157:H7 and Salmonella species and application for removal of prey from stainless steel surfaces. J. Food Safety 16:161-173. [Google Scholar]
- 13.Fratamico, P. M., and R. C. Whiting. 1995. Ability of Bdellovibrio bacteriovorus 109J to lyse gram-negative food-borne pathogenic and spoilage bacteria. J. Food Prot. 58:160-164. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, D., R. I. Amann, W. Ludwig, A. D. L. Akkermans, and K. H. Schleifer. 1992. Detection of microorganisms in soil after in situ hybridization with rRNA-targeted, fluorescently labeled oligonucleotides. J. Gen. Microbiol. 138:879-887. [DOI] [PubMed] [Google Scholar]
- 15.Jurkevitch, E. 2006. The genus Bdellovibrio, p. 12-30. In M. Dworkin et al. (ed.), The prokaryotes: a handbook on the biology of bacteria, 3rd ed. Springer-Verlag, New York, NY.
- 16.Jurkevitch, E., and B. Ramati. 2000. Design and uses of Bdellovibrio 16S rRNA-targeted oligonucleotides. FEMS Microbiol. Lett. 184:265-271. [DOI] [PubMed] [Google Scholar]
- 17.Jurkevitch, E., D. Minz, B. Ramati, and G. Barel. 2000. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microbiol. 66:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobabe, S., D. Wagner, and E.-V. Pfeiffer. 2004. Characterization of microbial community composition of a Siberian tundra soil by fluorescence in situ hybridization. FEMS Microbiol. Ecol. 50:13-23. [DOI] [PubMed] [Google Scholar]
- 19.Koval, S. F. 2006. The search for hunters: culture-dependent and -independent methods for analysis of Bdellovibrio and like organisms, p. 191-211. In E. Jurkevitch (ed.), Predatory prokaryotes: biology, ecology and evolution. Springer-Verlag, Berlin, Germany.
- 20.Koval, S. F., and S. H. Hynes. 1991. Effect of paracrystalline protein surface layers on predation by Bdellovibrio bacteriovorus. J. Bacteriol. 173:2244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenaerts, J., H. M. Lappin-Scott, and J. Porter. 2007. Improved fluorescent in situ hybridization method for detection of bacteria from activated sludge and river water by using DNA molecular beacons and flow cytometry. Appl. Environ. Microbiol. 73:2020-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahmoud, K. K., L. G. Leduc, and G. D. Ferroni. 2005. Detection of Acidithiobacillus ferrooxidans in acid mine drainage environments using fluorescent in situ hybridization (FISH). J. Microbiol. Methods 61:33-45. [DOI] [PubMed] [Google Scholar]
- 23.McNeely, D. 2004. Biocontrol of multidrug resistant Burkholderia cepacia. Ph.D. thesis. University of Ulster, Ulster, Northern Ireland.
- 24.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes, p. 207-226. In J. Paul (ed.), Methods in microbiology: marine microbiology, vol. 30. Academic Press Ltd., London, United Kingdom. [Google Scholar]
- 26.Shemesh, Y., Y. Davidov, S. F. Koval, and E. Jurkevitch. 2003. Small eats big: ecology and diversity of Bdellovibrio and like organisms, and their dynamics in predator-prey interactions. Agronomie 23:433-439. [Google Scholar]
- 27.Stolp, H. 1981. The genus Bdellovibrio, p. 618-629. In M. P. Starr, H. Stolp, H. G. Truper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Springer-Verlag, New York, NY.
- 28.Wallner, G., R. Amann, and W. Beisker. 1995. Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl. Environ. Microbiol. 61:1859-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarda, B., D. Hahn, A. Chatzinotas, W. Schonhuber, A. Neef, R.I. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]