Abstract
Vibrio furnissii M1 was recently reported to biosynthesize n-alkanes when grown on biopolymers, sugars, or organic acids (M. O. Park, J. Bacteriol. 187:1426-1429, 2005). In the present study, V. furnissii M1 was subjected to genomic analysis and studied biochemically. The sequence of the 16S rRNA gene and repetitive PCR showed that V. furnissii M1 was not identical to other V. furnissii strains tested, but the level of relatedness was consistent with its assignment as a V. furnissii strain. Pulsed-field gel electrophoresis showed chromosomal bands at approximately 3.2 and 1.8 Mb, similar to other Vibrio strains. Complete genomic DNA from V. furnissii M1 was sequenced with 21-fold coverage. Alkane biosynthetic and degradation genes could not be identified. Moreover, V. furnissii M1 did not produce demonstrable levels of n-alkanes in vivo or in vitro. In vivo experiments were conducted by growing V. furnissii M1 under different conditions, extracting with solvent, and analyzing extracts by gas chromatography-mass spectrometry. A highly sensitive assay was used for in vitro experiments with cell extracts and [14C]hexadecanol. The data are consistent with the present strain being a V. furnissii with properties similar to those previously described but lacking the alkane-producing phenotype. V. furnissii ATCC 35016, also reported to biosynthesize alkanes, was found in the present study not to produce alkanes.
The need for renewable energy sources will require the development of biofuel options other than ethanol. One excellent fuel option would be bio-alkanes. Alkanes comprise the major component of current petroleum-based fuels. A biological petroleum would be renewable and completely compatible with existing fuel infrastructure. Thus, considerable interest was generated by recent reports of high-level n-alkane formation by the bacterium Vibrio furnissii M1 (22, 23, 24).
V. furnissii strains were recognized as a distinct species in 1983 (6). Other Vibrio species, such as V. cholerae (8) and V. parahaemolyticus (30), have been more extensively studied because of their significant pathogenicity in humans. Both of the latter species (12), along with V. vulnificus (7) and V. fischeri (25), have been subjected to genomic sequencing that has been completed and published. V. furnissii has been most extensively studied with respect to its physiological and genetic mechanisms of chitin degradation (3, 17). Marine vibrios are prominent chitinolytic organisms (18).
Thus, the recent report of a V. furnissii strain biosynthesizing appreciable quantities of n-alkanes was unusual and interesting (22, 23, 24). The organism was isolated from activated sludge of a sewage disposal plant located in the Osaka prefecture of Japan (24). It was reported to produce a copious lipid layer that floated on top of liquid cultures. The culture lipids were found to consist of 48% alkanes (24), and the alkane-producing phenotype was the subject of three papers published between 2001 and 2005. The reports were significant for several reasons. First, n-alkanes were produced during growth on renewable carbon sources such as sugars and polysaccharides. The latter included starch, chitin, and xylan. This is of specific interest when considering substrates for large-scale growth for production of a usable biofuel. Second, the amount of alkanes was significant, accounting for as much as 35% of the carbon consumed (23). Third, the n-alkane backbone was proposed to derive largely from the reduction of fatty acids through a novel mechanism, the reduction of a fatty alcohol to an alkane (22). This mechanism would be both interesting and commercially appealing, as there would be no loss of carbon in a pathway condensing acetyl coenzyme A (acetyl-CoA) units into a long-chain acyl-CoA followed by six-electron reduction to yield an alkane.
The present study was conducted to investigate the alkane-producing phenotype of V. furnissii M1 using a combined approach of whole-genome sequencing and biochemical studies. The major findings were that alkane-producing genes could not be identified and alkane biosynthesis could not be demonstrated in vivo or in vitro.
MATERIALS AND METHODS
Microorganisms and cultivation.
V. furnissii M1 was kindly provided by Kazuya Watanabe of the Marine Biotechnology Institute, Japan. Other strains were obtained from the American Type Culture Collection (ATCC, Manassas, VA).
V. furnissii M1 was cultivated in three different media. Marine liquid medium contained 37.4 g marine broth 2216 (Becton Dickinson and Company, Franklin Lakes, NJ) per liter distilled water. Luria-Bertani broth was adjusted to 4% (wt/vol) NaCl. Medium 3 contained 2.0 mg EDTA·2Na, 2.8 mg H3BO3, 0.75 mg Na2MoO4·2H2O, 0.24 mg ZnSO4·7H2O, 1.6 mg MnSO4·H2O, 0.04 mg Cu(NO3)2·3H2O, 0.75 mg CaCl2·2H2O, 0.2 mg MgSO4·7H2O, 10 mg FeSO4·7H2O, 30 g NaCl, 1.32 g (NH4)2SO4, 0.2 g yeast extract, 8.7 g K2HPO4·3H2O, and 8.4 g KH2PO4 per liter distilled water. Carbon sources used were 1.64 g sodium acetate, 1.92 g sodium propionate, 2.70 g disodium succinate hexahydrate and 1.87 g disodium l-malate per liter. The pH was adjusted to 7.0 with KOH. After autoclaving, the following filter-sterilized reagents (Sigma-Aldrich, St. Louis, MO) were added: 0.025 mg thiamine-HCl, 0.025 mg d-biotin, 0.025 mg nicotinamide, and 0.025 mg p-amino benzoic acid per liter. Plates were prepared by the addition of 1.5% (wt/vol) granulated agar (Becton Dickinson and Company) to broth media.
All strains were maintained as frozen stocks and grown at 37°C with agitation at 210 rpm, unless otherwise noted. Strains were initially transferred from frozen stocks onto marine agar plates. After 18 to 24 h, isolated colonies were used to inoculate 3 ml of medium 3. After approximately 18 h, measurements of optical density at 600 nm (OD600) (Beckman DU 7400) ranged from 1.3 to 2.0. Two hundred microliters of each preculture was used to inoculate 20 ml of marine broth or medium 3 in 25-ml Erlenmeyer flasks. Cultures were grown under numerous conditions to test for alkane formation. Among the factors tested were degree of aeration, alternate carbon sources, different medium types, and harvesting media and cultures at various growth stages over 1 to 7 days. The conditions used in different experiments are provided with the relevant results below.
Chemicals.
Chromasolv chloroform (Sigma-Aldrich, St. Louis, MO), spectrophotometric-grade methanol (Sigma-Aldrich), hexanes (Mallinckrodt, Hazelwood, MO), diethyl ether (Fisher Scientific, Hampton, NH), heptane (Sigma-Aldrich), octacosane (Acros Organics, Geel, Belgium), and hexadecane (Sigma-Aldrich) were obtained from the sources indicated.
[1-14C]hexadecanol (Moravek Biochemicals Inc., Brea, CA) was 56 mCi/mmol and had a radiochemical purity of 99.3%.
Analytical methods.
Analytical methods largely followed those described by Park et al. (23, 24). Cultures were extracted following the Bligh and Dyer protocol (4). The concentrated extract was developed with 80:20:1 hexanes-diethyl ether-water on silica gel thin-layer chromatography plates. Spots were eluted with chloroform, concentrated to 100 μl, and analyzed by gas chromatography-mass spectrometry (GC-MS). To ensure that alkanes were not lost during initial chromatography, extracts were also directly analyzed by GC-MS. GC-MS analysis was conducted with an HP6890 gas chromatograph connected to an HP5973 mass spectrometer (Hewlett Packard, Palo Alto, CA). GC was conducted under the following conditions: helium gas, 1 ml/min; HP-5 column (5% phenylmethyl siloxane capillary; 30 m by 250 μm by 0.25 μm); temperature ramp, 100 to 300°C; 10°C/min. The mass spectrometer was run under the following conditions: electron impact at 70 eV and 35 μA.
In experiments where analytical standards were spiked into growth medium, 125 nmol hexadecane was added at the end of the growth phase to a 20-ml culture grown on medium 3 for 11 days. Octacosane (0.25 μmol) was used to spike a 50-ml culture grown on medium 3 containing 10 mM d-glucose for 24 h. The cultures were then extracted and handled as described previously.
Methods relating to alkane contamination reduction.
The following methods were instituted to reduce alkane contamination during the course of the present study. The chloroform solvent used for extractions was switched from Chromasolv to Chromasolv Plus (both from Sigma-Aldrich, St. Louis, MO). All glassware was rinsed twice with Chromasolv Plus solvent prior to use. Stoppers were neoprene rubber, and Teflon stopcocks were used in separatory funnels. Contact of any solvents or other components with plasticware was avoided. In total, these methods greatly reduced the introduction of contaminating alkanes.
In vitro experiments.
Cell-free enzyme fractions were obtained and assays were conducted as described previously by Park (22). Minor modifications were as follows: dispersal was accomplished by shaking rather than sonication to prevent generating radioactive aerosols; 10-mg protein aliquots were used in the assay for greater sensitivity.
REP-PCR and pulsed-field gel electrophoresis (PFGE).
Genomic relatedness of Vibrio strains was investigated utilizing repetitive extragenic palindromic PCR (REP-PCR) DNA fingerprinting with the primers ERIC 1R (3′-CACTTAGGGGTCCTCGAATGTA-5′) and ERIC 2 (5′-AAGTAAGTGACTGGGGTCAGCG-3′) (9). PCR amplification was performed using the following protocol: 95°C for 2 min; 30 cycles of 94°C for 3 min, 92°C for 30 s, 50°C for 1 min, and 65°C for 8 min; final extension at 65°C for 8 min. Samples were separated on a 1.5% SeaKem LE agarose (Cambrex Bioscience, Rockland, ME) gel in 1× Tris-acetate-EDTA (TAE) at 4°C for 16 h at 68 mV and stained for 20 min with a solution containing 0.5 μg of ethidium bromide per ml. Gel images were analyzed by BioNumerics v.2.5 software (Applied-Maths, Sint-Martens-Latem, Belgium) and normalized to an external 1-kb reference ladder. DNA fragments less than 300 bp long were not used in analyses. DNA fingerprint similarities were calculated using Pearson's product-moment correlation coefficient with 1% optimization. Dendrograms were generated using the unweighted pair-group method using arithmetic averages (9, 13, 14).
DNA for PFGE was prepared from cells lysed in plugs (11). Cells were grown on marine agar plates overnight at 37°C, washed once in 1 ml resuspension buffer (100 mM Tris [pH 8.0], 100 μM EDTA), and then resuspended to an absorbance at 600 nm of 2.1. One part cell suspension was mixed with 20 μl of 20-mg/ml protein kinase A (VWR International) and 1 part 2% SeaPlaque GTG agarose (Cambrex BioScience, Rockland, ME) in 1× TAE and molded into plugs. Plugs were lysed at 55°C in 10 ml lysing solution (50 mM Tris [pH 8.0], 50 μM EDTA, 1% sodium dodecyl sulfate, 1% N-laurylsarcosine, 0.1 mg/ml protein kinase A) for 2 h. Plugs were washed four times in Tris-EDTA and twice in 0.5× TAE. Plugs were immediately used in PFGE.
PFGE was performed with a CHEF DRII system (Bio-Rad, Richmond, CA). Agarose-imbedded DNA was run on 0.8% Megabase agarose (Bio-Rad) in 0.5× TAE at 14°C. Conditions were 72 h, initial pulse time of 1,200 s, final pulse time of 1,800 s, at 2 mV/cm (11). The gel was stained with a solution containing 0.5 μg of ethidium bromide per ml. Chromosome sizes were estimated based on the mobility of unknowns in comparison with DNA samples of known base pair composition (Bio-Rad).
Genome sequencing and annotation.
DNA for sequencing was collected as previously described (26). V. furnissii M1 was grown overnight in marine broth. Cells were centrifuged at 10,000 rpm for 10 min at 4°C, washed once in TEN buffer (50 mM Tris, 20 μM disodium EDTA, and 50 mM NaCl, pH 8.0), and resuspended in 8 ml TEN buffer. Cells were lysed by adding 1 ml of a 5-mg/ml lysozyme solution (Promega, St. Louis, MO) at 37°C for 30 min followed by 1 ml of 5 mg/ml predigested pronase solution (Promega) at 37°C for 30 min, and then 1 ml 20% N-laurylsarcosine (Promega) at 37°C for 1 h. Eleven grams of cesium chloride was added, followed by 1 ml of 10-mg/ml ethidium bromide. The solution was centrifuged at 40,000 rpm at 20°C for 40 h. DNA bands were isolated using a syringe, ethidium bromide was extracted using salt-saturated butanol, and the DNA was dialyzed overnight against four washes of 0.01 M Tris-HCl, pH 8.0, containing 0.1 mM EDTA. The DNA was confirmed to be from V. furnissii by 16S rRNA sequencing. 16S rRNA was amplified by PCR using the primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 785r (5′-GGACTACCIGGGTATCTAATCC-3′), 530f (5′-GTGCCAGCMGCCGCGG-3′) and 1100r (5′-GGGTTGCGCTCGTTG-3′), and 926f (5′-AAACTYAAAKGAATTGACGG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′), using the following conditions: 95°C for 5 min; 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 90 s; 72°C for 5 min. PCRs were performed in duplicate and sequenced at the Biomedical Genomics Center at the University of Minnesota. The DNA fragments used for genomic sequencing were determined to be longer than 15 kb via electrophoresis on a 1% agarose gel.
Genome sequencing and contig assembly were performed by the Center for Genomic Sciences at the Allegheny-Singer Research Institute, using a 454 sequencer (454 Life Sciences, Branford, CT). The Newbler assembly program was used to order the sequences into 121 contigs. In order to annotate the contigs, a single pseudochromosome was constructed using a linker sequence which allows identification of partial genes at contig margins, as described by Tettelin et al. (29).
The resulting pseudochromosome was subjected to automated annotation via GenDB (19). Pfam (27) and hidden Markov models (HMM) (10) for local and global alignments were used to search the M1 genome for specific protein targets. Additional functionality was screened by searching the M1 genome using BLASTP (1).
Functional analysis of ORF 275.
V. furnissii M1 genomic DNA and the primers GGATTATGGCATATGATGTTAGAT and TCTTTTCGAAACTTAACGCA were used to amplify open reading frame (ORF) 275 using the PCR. Primers contained the NdeI and HindIII restriction sites, respectively. The gene was cloned separately into either pET28b+ or pET30a+ vectors (Novagen, San Diego, CA) and transformed into Escherichia coli BL-21 cells. Starter cultures of the recombinant E. coli strain, grown at room temperature to an OD600 of 0.5 to 0.6, were used to inoculate 100-ml cultures. These cultures were grown at 15°C to an OD600 of 0.4 to 0.5 and induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside for 19.5 h before harvesting. Crude extracts were prepared from both pET vectors by sonication on a Biosonik sonicator (Bronwill Scientific, Rochester, NY) at 80% intensity. Purified His-tagged protein was prepared from the pET28b+ construct. Crude extracts were passed over a nickel column (Novagen) and eluted with a solution of 1 M imidazole, 20 mM Tris [pH 7.9], 50 mM NaCl.
Acetaldehyde dehydrogenase (CoA-acetylating) activity was measured by monitoring NADH production or consumption at 340 nm on a Beckman DU 640 spectrophotometer (Beckman Coulter, Fullerton, CA). All solutions were prepared in 20 mM Tris, pH 7.5. Conversion of acetaldehyde and CoA to acetyl-CoA was measured with both crude extracts and purified protein. The final assay buffer contained 2.5 mM NAD, 50 mM acetaldehyde, and 1 mM coenzyme A. The reverse reaction was monitored with purified protein in an assay buffer containing 100 μM acetyl-CoA and 250 μM NADH2.
Nucleotide sequence accession number.
The V. furnissii M1 16S rRNA sequence obtained in this study has been deposited in GenBank under accession number EU204961.
RESULTS
V. furnissii M1 general characteristics and comparison to other vibrios.
The V. furnissii M1 culture showed a thick, floating, waxy layer as described by Park et al. (24). The previous study also reported that the 16S rRNA sequence was consistent with identification as a V. furnissii strain, but the sequence itself was not reported. The V. furnissii M1 16S rRNA sequence obtained in this study also showed the highest similarity, >99%, to two V. furnissii 16S rRNA sequences in GenBank.
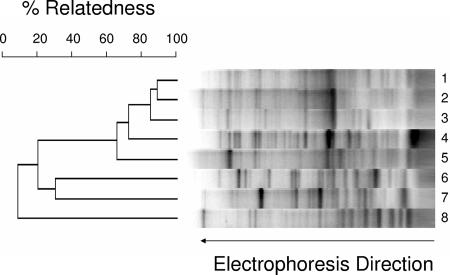
Subsequently, a more powerful method for comparing V. furnissii M1 to other Vibrio strains was used: REP-PCR. In this technique, PCR amplification of closely spaced repetitive elements throughout the genome can be used as a genomic signature. REP-PCR was performed with primers designed to amplify repetitive elements found in diverse microbial genomes. A representative gel is shown in Fig. 1. A dendrogram was constructed as described in Materials and Methods. V. furnissii M1 was not identical to other V. furnissii strains tested here. However, the relatedness observed, on the order of 60 to 90%, is consistent with comparisons in the literature between organisms of the same species, confirming that V. furnissii M1 is indeed a V. furnissii strain.
FIG. 1.
REP-PCR of genomic DNA from V. furnissii M1 and control strains. Percent relatedness was determined as described in Materials and Methods. The lanes contained DNA from the following strains: 1, V. furnissii ATCC 33841; 2, V. furnissii ATCC 35627; 3, V. furnissii ATCC 35016; 4, V. furnissii M1; 5, V. furnissii ATCC 35628; 6, V. parahaemolyticus LM5312; 7, V. harveyi B392; 8, E. coli K-12.
PFGE.
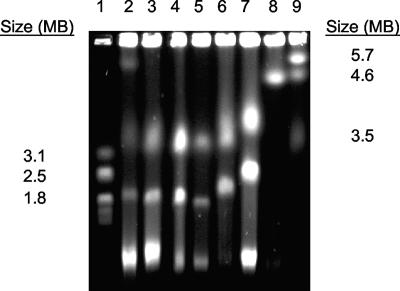
Genomic DNA of V. furnissii M1, other V. furnissii strains, and standard strains were analyzed by PFGE (Fig. 2) to determine the size of the genome and its organization. The data suggested that V. furnissii M1 has a bipartite genome consisting of two chromosomal elements with a total size of approximately 5 Mb. This is similar to characteristics of other Vibrio species, as reported in the literature and shown in Fig. 2 (7, 21).
FIG. 2.
Comparative PFGE of genomic DNA from V. furnissii M1 (lane 2), V. furnissii 35016 (lane 3), V. furnissii 35627 (lane 4), V. furnissii 35628 (lane 5), V. parahaemolyticus LM5312 (lane 6), V. harveyi B392 (lane 7), and E. coli DH5α (lane 8). Hansenula wingei (lane 1) and Schizosaccharomyces pombe (lane 9) chromosomes were used as size markers.
Genome annotation.
The genome of V. furnissii M1 was sequenced, computationally assembled and the genes annotated. The raw sequencing data consisted of 106 Mb, which represented an estimated 21-fold coverage of the total genome. The base reads were assembled into 121 contigs that converged at 4.95 Mb of genome data, consistent with the genome size estimated by PFGE. The contigs were randomly ordered and assembled into a pseudochromosome for annotation purposes.
A major goal of the genome annotation was to identify putative genes that might be involved in alkane biosynthesis. Among the candidate alkane-biosynthetic genes examined specifically were those encoding acyl-CoA reductase, aldehyde reductase, and alcohol reductase and genes that might encode an enzyme catalyzing the reduction of an alcohol to an alkane. HMMs for the local and global Pfam alignments for acyl-CoA reductase (PF05893) were used to search the M1 genome. No sequence was found with an expectation value (e-value) less than 1.0. Dozens of putative aldehyde and alcohol reductases were identified. The genome was examined for aldehyde and alcohol dehydrogenase genes that were clustered with genes encoding enzymes resembling ribonucleotide reductase or other radical-dependent oxidoreductases, an anticipated gene constellation that might encode a carboxylic acid-to-alkane biosynthetic pathway, as proposed by Park (22).
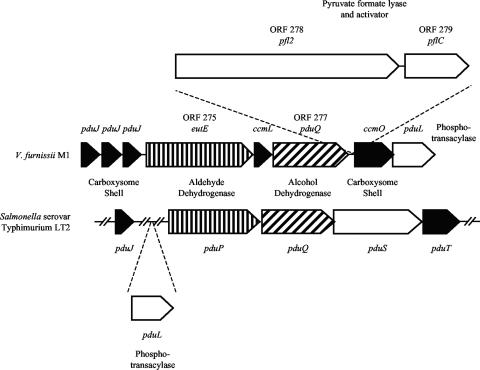
In this context, one interesting gene cluster was annotated as consisting of genes for an aldehyde dehydrogenase (ORF 275), an iron-dependent alcohol dehydrogenase (ORF 277), a pyruvate formate lyase homolog (ORF 278), and an accompanying pyruvate formate lyase activator (ORF 279) (Fig. 3). These functional genes were flanked by genes that were annotated as metabolosome (carboxysome) shell proteins (Fig. 3). The flanking gene structure is consistent with the function of pduJ and ccmO genes in generating a carboxysome or metabolosome structure (5, 15), which is a bacterial intracellular protein shell involved in compartmentalizing metabolites or a set of reactions.
FIG. 3.
Genome region from V. furnissii M1 showing similarities to a gene region in Salmonella. Identified protein types are highlighted: carboxysome shell proteins (black), aldehyde dehydrogenases (vertical lines), and alcohol dehydrogenases (diagonal lines). The ORF numbers (275, 277, 278, and 279) are given for proteins discussed in the text.
Further bioinformatics analysis of other genomes revealed a very similar cluster of genes in E. coli strain F11 (accession number NZ_AAJU00000000), a pathogen not expected to produce alkanes. Moreover, the metabolosome-like structure may replace the multidomain protein particle catalyzing the conversion of pyruvate to ethanol in E. coli K-12 (16). This was tested by cloning and expressing ORF 275 in E. coli. The recombinant E. coli strain was assayed for different activities as described in Materials and Methods. ORF 275 was shown with purified enzyme to encode a bidirectional acetaldehyde dehydrogenase (CoA-acetylating) enzyme. Formation of acetaldehyde occurred at a rate of 1.5 μmol/min/mg, while the reverse reaction had a rate of 2.7 μmol/min/mg. It was thus functional as an acetyl-CoA reductase, an activity complementary with a pyruvate formate lyase and an aldehyde-oxidizing alcohol dehydrogenase. These activities in total are consistent with a metabolosome that compartmentalizes a fermentative pathway metabolizing pyruvate to ethanol.
Whole-cell studies attempting to detect alkanes.
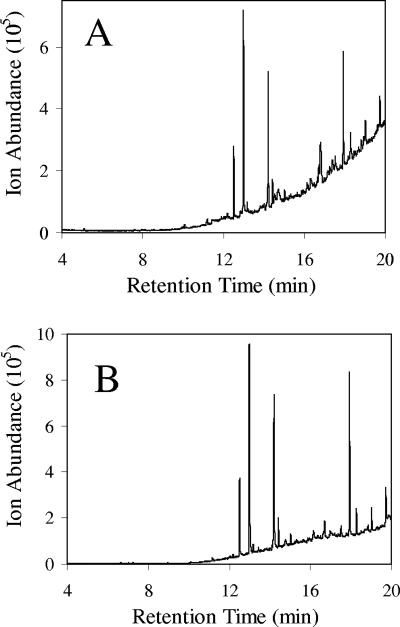
V. furnissii M1 cells and media were extracted by methods described by Park et al. (23, 24). In preliminary experiments, alkanes were detected by GC and confirmed by MS (Fig. 4A). Subsequent analysis of media, solvents, and glassware revealed that they were contaminated with alkanes and other materials. The solvents used in extractions were the most significant source of contamination (Fig. 4B). A series of methodological alterations were made to eliminate contamination, as described in Materials and Methods, which led to greatly diminished peaks via GC.
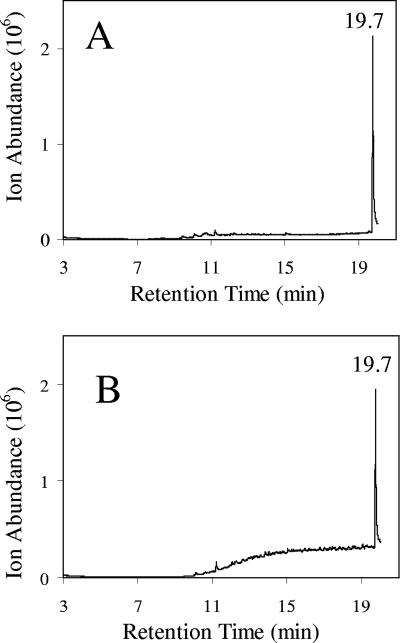
FIG. 4.
Initial gas chromatograms of V. furnissii M1 (A) and a chloroform blank showing the extent of contamination (B). The prominent peaks in both were identified by MS as methyl palmitate (12.5 min), dibutyl phthalate (13.0), octadecenoic acid methyl ester (14.2 min), diethylhexyl phthalate (17.9 min), and octacosane (19.7 min).
To determine whether the extraction and new workup procedures were appropriate for isolating and concentrating alkanes for detection, an internal standard was added to cultures of V. furnissii M1. The internal standards hexadecane and octacosane were used in independent experiments. The choice of using a C16 and C28 alkane, respectively, was made to largely bracket the entire range of n-alkane chain lengths previously reported to be produced by V. furnissii M1 (23, 24). Solvent extracts from these spiked cultures were processed and subjected to GC-MS using procedures that minimized alkane contamination. A parallel experiment was conducted using an E. coli culture containing the same internal standard alkanes. Figure 5 shows a representative chromatogram of the resultant extracts analyzed by GC-MS with octacosane-spiked medium. The large octacosane peak was clearly identifiable by both retention time and the characteristic mass spectrum. The level of octacosane added (0.25 μmol) matched the level of individual alkanes reported to be present in GC analyses by Park et al. (24). Clearly, no peaks comparable to the added standard were discernible. Very minor peaks were observed above the baseline, but the same minor peaks were found in V. furnissii M1 (Fig. 5A) and E. coli (Fig. 5B) extracts, suggesting that they are derived from a common source and are not made biosynthetically. E. coli and V. furnissii M1 cultures were grown in the same growth medium and were extracted in parallel. Results with hexadecane-spiked cultures produced similar results.
FIG. 5.
Gas chromatograms of extracts of V. furnissii M1 (A) and E. coli K-12 (B) using cleaner solvents and methods. Cultures were spiked with octacosane prior to extraction and workup. The 19.7-min peak was confirmed by MS to be octacosane.
Since alkane formation could be dependent on growth conditions, analysis for alkanes was conducted with V. furnissii M1 cultures grown in different media, under aerobic and microaerophilic (nonshaking) conditions, and over a period of 1 to 7 days. Alkanes were not detected above background levels under any of these conditions. The media were quite different. Medium 3 is completely defined, and marine broth is a standard commercial culture medium for V. furnissii. Under microaerophilic conditions, the culture was observed to have a floating pellicle, and birefringence was seen in marine broth cultures on the top of the culture. While these observations were initially thought to be potential indicators of hydrocarbon formation, no alkanes attributable to V. furnissii M1 were obtained from extracts of these cultures.
Cell-free enzyme assays for alkane formation.
Cell-free enzyme preparations from V. furnissii M1, prepared as described by Park (22), were tested for hexadecanol reductase activity (Table 1). No significant radioactivity was detected in the spot on a thin-layer chromatography plate corresponding to the Rf value of authentic hexadecane. The percentage of the starting radioactivity in the hexadecane fraction in all cases was less than 0.1%, a level significantly below that of the radiochemical impurities of the starting material, or 0.9%. Total recovery of radioactivity in substrate (hexadecanol) and putative product (hexadecane) fractions was 57% in the no-enzyme control and 25 to 47% in the enzyme treatments. This is similar to the total recovery reported by Park (22). A level of activity several percent of that reported by Park (22) would have been detected in this experiment. In separate experiments, V. furnissii ATCC 35028 was tested for reductase activity with hexadecanol, but no activity was detected.
TABLE 1.
In vitro assay for hexadecanol reduction to hexadecane using [14C]hexadecanol
| Enzyme fraction | Radioactivity (dpm)
|
|
|---|---|---|
| Hexadecanol | Hexadecane | |
| No enzyme | 1,267,002 ± 65,067 | 563 ± 38 |
| V. furnissii M1, soluble | 674,895 ± 23,202 | 303 ± 32 |
| V. furnissii M1 membranes | 799,030 ± 143,600 | 812 ± 162 |
| E. coli, soluble | 797,954 ± 246,564 | 642 ± 185 |
| E. coli membranes | 795,810 ± 127,375 | 474 ± 152 |
No evidence for hydrocarbon oxidation.
It was considered that cells producing alkanes may also have the capability to oxidize hydrocarbons, thus recapturing carbon and energy. Experiments were conducted to determine the potential growth of V. furnissii M1 in the presence of alkanes as the sole carbon source or in admixture with limiting alternative carbon sources such as glucose. No evidence for growth was observed using dodecane (C12), hexadecane (C16), octadecane (C18), eicosane (C20), docosane (C22), or tetracosane (C24), alkanes that Park reported to be produced by V. furnissii M1. In addition, bioinformatics tools were used to search for genes encoding proteins homologous to AlkA, AlkB, and cytochrome P450 monooxygenases. These genes, established to encode alkane-oxidizing enzymes in other bacteria, could not be discerned in the genome of V. furnissii M1. Using the HMMER 2 tool (10) in conjunction with the Pfam HMMs for cytochrome P450 (accession number PF00067), in local and global alignments against the V. furnissii M1 genome sequences, no match with an e-value lower than 4.0 was found. For AlkB, no HMMs were available, so the Pseudomonas oleovorans AlkB sequence (gi 113639) was used with the BLAST tool. The best match found in the V. furnissii M1 genome was 0.02. While this could indicate weak homology, there was no characteristic clustering of genes, as is found in alkane degraders. Specifically, we could not find evidence for the presence of the gene cluster alkFGHJKL or the regulatory elements alkST.
Other Vibrio strains.
A patent filed on V. furnissii M1 in Japan claimed that other Vibrio strains also produce alkanes, albeit in smaller amounts than V. furnissii M1 (20). In the present study, V. furnissii ATCC 35628 was tested for alkane formation in vivo, and no levels above background were detected. Additionally, membrane and soluble enzyme fractions were prepared from V. furnissii ATCC 35628 and tested in vitro with [14C]hexadecanol. The radioactivity (in dpm) in the region of a hexadecane standard was on the order of 0.1% of the initial radioactivity, a level consistent with background radiation in negative controls. In other experiments, a strain reported in the patent to make alkanes, V. furnissii ATCC 35016, was obtained from the ATCC and tested. No alkanes were detected.
DISCUSSION
The V. furnissii M1 strain used in this study strongly resembles the strain described previously (22, 23, 24), except that no alkane formation was observed here. It is not possible to use DNA sequence data to rigorously ascertain the relationship to the previously described V. furnissii M1 strain, because no DNA sequences had been reported in the literature or deposited in GenBank. However, in this study, 16S rRNA sequence data and REP-PCR data support the idea that the organism used here was a V. furnissii strain and that it differed from V. furnissii ATCC cultures that were tested.
In this study, V. furnissii M1 did not make alkanes under any in vivo growth condition tested, and protein extracts did not catalyze alkane formation in vitro. Some in vivo studies used standards carried throughout the extraction and purification protocols to show that the methods employed would have detected alkanes, with significant sensitivity, had they been present. The conditions of growth and analysis used here followed the procedures of Park et al. (22, 23, 24) closely. Both cells and media were extracted to ensure that any alkane present would not be missed. In vitro assays were also very sensitive. Picomole levels of alkane would have been detectable, but nothing above background could be discerned. Levels that were orders of magnitude lower than those reported by Park (22) could have been detected in the assays conducted here.
The lack of alkane biosynthetic activity in strain M1 is consistent with the lack of activity in V. furnissii strains ATCC 35627, 35628, and 33841 (H. R. Beller, personal communication), which were assayed under a range of conditions comparable to those described by Park and coworkers; these in vivo assays involving GC-MS entailed high extraction efficiency (typically >99% based on recoveries of the surrogate compound decane-d22) and would have been able to detect 0.001% of the alkane concentrations that were reported by Park and coworkers (H. R. Beller, personal communication).
In the present study, V. furnissii ATCC 35016 was shown not to produce alkanes under the conditions tested. A patent filed in Japan by Miyamato (20) reported that V. furnissii ATCC 35016 produced alkanes, albeit at lower levels than V. furnissii M1. This could not be reproduced in the present study.
Several observations reported by Park et al. were unexpected and unexplained. Different papers reported different hydrocarbons being produced that would derive from divergent mechanisms: even-chain alkanes, odd-chain alkenes, branched-chain alkanes, and alkenes. Differences were reported with different growth substrates, but some differences were surprising. For example, growth with acetate resulted in only a C18 alkene being formed, but butyl acetate, which would almost surely be metabolized via ester hydrolysis to yield acetate, gave rise to C18, C21, C24, and C27 branched-chain alkanes. Butyric acid, another likely metabolite from butyl acetate, was reported to give rise to linear C16 to C18 alkanes.
No obvious genes that might be related to alkane biosynthesis were identified in this study. It must be acknowledged that alkane biosynthesis is currently poorly understood, and hence the genes may not be obvious. However, nothing resembling putative plant decarbonylases was detected. Fatty acid aldehydes and alcohols derive from acyl-CoA reductases. Only one acyl-CoA reductase homolog was identified that clustered with other genes that might be involved in alkane production. That gene was cloned, expressed, and found likely to carry out a different function (see below).
The genome sequence was also annotated to search for alkane degradation genes. The logic behind this was that bacteria producing other energy-rich, carbon-rich molecules (polyhydroxyalkanoates, triacylglycerides, and glycogen) typically oxidize these carbon storage molecules (2, 28). Thus, we looked for the readily identifiable enzymes involved in alkane oxidation: cytochrome P450 monooxygenases and Alk proteins. None of these enzyme systems were identified. In BLAST searches, expectation values for homologs were generally greater than 1.0. Moreover, these systems are multicomponent and thus encoded by gene clusters. These gene clusters should be readily identifiable, if present, even if the sequences were fairly divergent.
Most of the genes examined have homologs in other Vibrio species that are not known to produce alkanes. One gene region that differed from other Vibrio species sequenced to date contained structural genes with highly significant sequence identity to metabolosomes, or carboxysomes. Metabolosomes are intracellular, multiprotein structures consisting of shell proteins harboring metabolic proteins (15). They were initially known as carboxysomes because carbon dioxide-fixing enzymes were found associated with the first metabolosomes identified. More recently, other types of metabolism have been found to be harbored by shell proteins homologous to carboxysome shell proteins. The carboxysome gene region in V. furnissii M1 was initially considered intriguing. It included genes encoding oxidoreductases and a pyruvate formate lyase homolog; the latter activity could conceivably be involved in an alcohol-to-alkane reduction reaction. However, bioinformatics analysis and comparison to a similar region in E. coli F11 led us to the tentative conclusion that the gene cluster and metabolosome in V. furnissii M1 likely function in the fermentation of ethanol. This hypothesis was tested by cloning V. furnissii M1 ORF 275 in E. coli and assaying the protein extract from recombinant cells. The data indicated that ORF 275 encoded a bidirectional acetaldehyde dehydrogenase (CoA-acetylating) enzyme, consistent with its hypothetical role in an ethanol fermentation. While E. coli strain F11 has a homologous metabolosome-like structure, E. coli K-12 produces a multifunctional, spiral-shaped polypeptide that is thought to channel pyruvate to ethanol (16). The channeling multidomain protein and a metabolosome may represent different biological mechanisms for channeling metabolic flux through a potentially toxic aldehyde intermediate.
Conclusions.
V. furnissii strains, including strain M1, were observed to have chromosomes of approximately 3.2 and 1.8 Mb. No apparent alkane-producing genes or phenotypes were observed. The latter was checked in vivo and in vitro with V. furnissii M1 and ATCC 35628 and in vivo with V. furnissii ATCC 35016.
Acknowledgments
We acknowledge the help of Satoshi Ishii with the REP-PCR and Tao Yan with the PFGE. We thank Jack Richman for helpful advice on chemistry and Harry Beller of Lawrence Livermore National Laboratory for providing unpublished data.
This research was partly funded by a grant from the Institute for Renewable Energy and the Environment, grant LG-B13-IREE.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, H. M., and A. Steinbuchel. 2002. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60:367-376. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., C. Yu, Y. C. Lee, and S. Roseman. 1991. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276-24286. [PubMed] [Google Scholar]
- 4.Bligh, E. G., and W. J. Dyer. 1959. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 5.Bobik, T. A. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 70:517-525. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, D. J., F. W. Hickman-Brenner, J. V. Lee, A. G. Steigerwalt, G. R. Fanning, D. G. Hollis, J. J. Farmer, R. E. Weaver, S. W. Joseph, and R. J. Seidler. 1983. Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J. Clin. Microbiol. 18:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colwell, R. R. 2004. Infectious disease and environment: cholera as a paradigm for waterborne disease. Int. Microbiol. 7:285-289. [PubMed] [Google Scholar]
- 9.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 11.Fakhr, M. K., L. K. Nolan, and C. M. Logue. 2005. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 43:2215-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishii, S., T. Yan, D. A. Shively, M. N. Byappanahalli, R. L. Whitman, and M. J. Sadowsky. 2006. Cladophora (Chlorophyta) spp. harbor human bacterial pathogens in nearshore water of Lake Michigan. App. Environ. Microbiol. 72:4545-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerfeld, C. A., M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby, and T. O. Yeates. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936-938. [DOI] [PubMed] [Google Scholar]
- 16.Kessler, D., I. Leibrecht, and J. Knappe. 1991. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 281:59-63. [DOI] [PubMed] [Google Scholar]
- 17.Keyhani, N. O., and S. Roseman. 1996. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J. Biol. Chem. 271:33414-33424. [DOI] [PubMed] [Google Scholar]
- 18.Li, X., and S. Roseman. 2004. The chitinolytic cascade in vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. USA 101:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB—an open source genome annotation system for prokaryotic genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamato, K. 2001. Microorganisms and process for producing petroleum substitute oil by using these microorganisms. Japanese patent P2001-190323.
- 21.Okada, K., T. Iida, K. Kita-Tsukamoto, and T. Honda. 2005. Vibrios commonly possess two chromosomes. J. Bacteriol. 187:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, M. O. 2005. New pathway for long-chain n-alkane synthesis via 1-alcohol in Vibrio furnissii M1. J. Bacteriol. 187:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, M. O., K. Heguri, K. Hirata, and K. Miyamoto. 2005. Production of alternatives to fuel oil from organic waste by the alkane-producing bacterium, Vibrio furnissii M1. J. Appl. Microbiol. 98:324-331. [DOI] [PubMed] [Google Scholar]
- 24.Park, M. O., M. Tanabe, K. Hirata, and K. Miyamoto. 2001. Isolation and characterization of a bacterium that produces hydrocarbons extracellularly which are equivalent to light oil. Appl. Microbiol. Biotechnol. 56:448-452. [DOI] [PubMed] [Google Scholar]
- 25.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadowsky, M. J., R. E. Tully, P. B. Cregan, and H. H. Keyser. 1987. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybeans. Appl. Environ. Microbiol. 53:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnhammer, E. L. L., S. R. Eddy, and R. Durbin. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405-420. [DOI] [PubMed] [Google Scholar]
- 28.Steinbuchel, A., and S. Hein. 2001. Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. Adv. Biochem. Eng. Biotechnol. 71:81-123. [DOI] [PubMed] [Google Scholar]
- 29.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. DeBoy, T. M. Davidsen, M. Mora, M. Scarselli, I. M. Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. B. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genomic analysis of multiple pathogenic islands of Streptococcus agalactiae: Implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeung, P. S., and K. J. Boor. 2004. Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog. Dis. 1:74-88. [DOI] [PubMed] [Google Scholar]