Abstract
Octylphenol (OP) is an estrogenic detergent breakdown product. Structurally similar nonylphenols are transformed via type II ispo substitution, resulting in the production of hydroquinone and removal of the branched side chain. Nothing is known, however, about the gene(s) encoding this activity. We report here on our efforts to clone the gene(s) encoding OP degradation activity from Sphingomonas sp. strain PWE1, which we isolated for its ability to grow on OP. A fosmid library of PWE1 DNA yielded a single clone, aew4H12, which accumulated a brown polymerization product in the presence of OP. Sequence analysis of loss-of-function transposon mutants of aew4H12 revealed a single open reading frame, opdA, that conferred OP degradation activity. Escherichia coli subclones expressing opdA caused OP disappearance, with the concomitant production of hydroquinone and 2,4,4-trimethyl-1-pentene as well as small amounts of 2,4,4-trimethyl-2-pentanol. These metabolites are consistent with a type II ipso substitution reaction, the same mechanism described for nonylphenol biodegradation in other sphingomonads. Based on opdA's sequence homology to a unique group of putative flavin monooxygenases and the recovery of hydroxylated OP intermediates from E. coli expressing opdA, we conclude that this gene encodes the observed type II ipso substitution activity responsible for the initial step in OP biodegradation.
Nonylphenol (NP) and octylphenol (OP), collectively referred to as alkylphenols, are detergent breakdown products with highly branched side chains that act as endocrine disrupters and are known to exhibit weak estrogenic activity (17). Tabira et al. (33) have shown that alkylphenols bind to recombinant human estrogen receptors in a dose-dependent fashion. Alkylphenols induce feminization in male amago salmon (26), cause changes in the sex ratio toward females and increase the frequency of intersexuality in Pacific oysters (27), and inhibit testicular growth in male rainbow trout during maturation (20).
Given the potential of alkylphenols to cause harm to fish and other sensitive aquatic organisms, much interest has been focused on understanding the potential of microorganisms to degrade alkylphenols. Several organisms have been reported to degrade NP or OP when oxygen is available, including several fungal species (4, 22), psychrotrophic and psychrophilic Pseudomonas species (31), and the sewage sludge isolates Sphingobium xenophagum Bayram (15), Sphingomonas cloacae (12), Sphingomonas sp. strain TTNP3 (35), and Sphingobium amiense strain YTT (36).
The degradation mechanism for specific NP isomers has been examined biochemically for both Sphingobium xenophagum Bayram and Sphingomonas strain TTNP3. Both strains metabolize various NP isomers by type II ipso substitution. Growth on NP appears to be limited to those isomers that contain fully substituted alpha carbons on the alkyl side chain (5, 14).
There have been examples of ipso substitution as a mechanism for degradation of halogenated phenols by cytochrome P450s (2, 29), although there is no direct evidence that similar enzymes act upon alkyl-substituted substrates. Kolvenbach et al. recently reported that Sphingomonas sp. strain TTNP3 appears to use a monooxygenase to transform NP, but no information regarding the gene coding for this activity was given (23). The present study describes the cloning of a putative flavin monooxygenase from Sphingomonas sp. strain PWE1 whose expression conferred on Escherichia coli the ability to transform OP to hydroquinone (HQ).
MATERIALS AND METHODS
Chemicals.
OP [4-(2′,4′,4′-trimethyl-pentyl)-phenol] and 1,2,4-benzenetriol were purchased through Sigma-Aldrich (St. Louis, MO). Ascorbate, HQ, NP, and 2,4,4-trimethyl-1-pentene were purchased from Acros (Morris Plains, NJ). All solvents were high-performance liquid chromatography (HPLC) grade and were purchased through Fisher Scientific (Pittsburgh, PA).
Isolation.
Activated sludge from the municipal wastewater treatment plant in Ithaca, NY, was spiked with 1,000 mg liter−1 NP and incubated at room temperature while being shaken at 150 rpm. After 7 days, 1 ml of this enrichment was then transferred to 100 ml of minimal salts medium (MSM) (24) containing 1,000 mg liter−1 NP and allowed to grow for an additional 7 days. This process was repeated three more times. On the fourth transfer, OP (1,000 mg liter−1) was used as a growth substrate rather than NP since this single isomer is available commercially. This enrichment was subjected to three more transfers on OP and then plated onto MSM agar plates containing 1,000 mg liter−1 OP. A single strain able to use OP as the sole carbon and energy source was isolated from these OP minimal medium plates and designated PWE1. The phylogenetic relatedness of PWE1 to other bacteria was determined by analyzing a portion of the 16S rRNA gene which had been PCR amplified using universal primers 27F (5′ AGAGTTTGATCMTGGCTCAG 3′) and 1492R (5′ TACGGYTACCTTGTTACGACTT 3′) and then sequenced at the Cornell University BioResource Center.
Growth on OP.
Growth was monitored in triplicate flasks of 100 ml MSM with 1,000 μg ml−1 OP at 24°C. Samples were taken in triplicate from each flask, and the absorbance was measured at 600 nm by use of a MicroQuant spectrophotomer from BioTek Instruments (Winooksi, VT).
Fosmid library.
PWE1 DNA was isolated via phenol-chloroform extraction and used to generate a fosmid library with a CopyControl fosmid library production kit (Epicentre Biotechnologies, Madison, WI) per the kit instructions. Fosmid clones were screened visually for the accumulation of putative ring-hydroxylated OP intermediates, as indicated by the production of a brown polymerization product (BPP) when grown with OP in the presence of p-toluidine and FeCl3 (30).
Transposon mutagenesis of fosmid clones.
A BPP-producing fosmid clone labeled aew4H12 was mutated with an EZ::TN5 <R6Kγori/Kan-2> transposon mutagenesis kit (Epicenter Biotechnology, Madison, WI) in order to obtain loss-of-function mutants. Briefly, the fosmid was extracted using a modified alkaline lysis method and then subjected to transposon mutagenesis according to the manufacturer's instructions. The reaction mixture was then transformed into TransforMax EPI300 electrocompetent E. coli (Epicenter Biotechnology, Madison, WI) and screened for loss of the BPP phenotype as described above. Fosmids from BPP− mutants were extracted by alkaline lysis and were then sequenced with outward-facing transposon primers to determine the site of transposon insertion. Sequences were aligned using the DNAStar program suite (DNAStar, Madison, WI) to identify open reading frames.
Further information was gathered by using PCR to amplify fragments of the fosmid that lay between the site of Tn5 insertion and the fosmid multicloning site. This was done using transposon-specific primers (R6KAN-2 RP-1 reverse primer 5′ CTACCCTGTGGAACACTACATCT 3′ and KAN-2 FP-1 5′ ACCTACAACAAAGCTCTCATCAACC 3′) and a fosmid-specific primer (pCC1/pEpiFOS reverse sequencing primer 5′ CTCGTATGTTGTGTGGAATTGTGAGC 3′). These additional amplicons were also sequenced.
In silico DNA analyses.
Sequence analysis of the mutant fosmids that had lost the ability to confer BPP production was done with DNAStar and Blast (1). A putative open reading frame which was common to all of the mutants was identified and named opdA. This open reading frame was PCR amplified using primers opdA forward (5′ TTC ATC CTG AAA GAC ACT GCC GGA 3′) and opdA reverse (5′ ACG CGC TTC CAG ACC AAC CTA TTT 3′) and subcloned into pGEM-T Easy (pGEM) (Promega, Madison, WI). The plasmid was designated pAW1 and transformed into E. coli JM109. Activity was assessed by monitoring formation of 2,4,4-trimethyl-1-pentene in the headspace of sealed cultures (see below).
Detecting HQ formation.
Overnight PWE1 cultures were diluted 1:1 with fresh medium and brought to a starting OP concentration of 480 μM. The fresh culture was incubated at room temperature while being shaken. After 1 h, the culture was filtered through glass wool to remove residual OP and then centrifuged to pellet the cells. The resulting supernatant was adjusted to pH 9 with 1.5% K2CO3. Acetic anhydride at 0.5% was then added to derivatize aromatic hydroxyls, and the supernatant was incubated while being shaken at room temperature for 1 h. The derivatized supernatant was then extracted with 30 ml of ethyl acetate. The extract was dried using anhydrous Na2SO4 and then evaporated under N2 at 40°C. The residue was redissolved in 1 ml of ethyl acetate for analysis via gas chromatography-mass spectrometry (GC-MS) with an HP 6890 GC equipped with an HP-5MS column (5% phenyl methyl siloxane; 30 m by 0.25 mm; 0.25-μm film thickness), using helium as the carrier gas with a flow rate of 1 ml/min. The temperature program included a hold at 40°C for 1 min, followed by an increase of 5°C/min to 150°C and a hold for 5 min. This was followed by an increase of 40°C/min to 300°C and a hold for 5 min. The detector was an HP 5973 MSD with the quadrapole and source set at 150°C and 230°C, respectively.
The accumulation of HQ in the supernatant of E. coli clones expressing opdA was also confirmed via HPLC using a mobile phase of 20% methanol and 80% of 40 mM acetic acid. The solvent was pumped at a rate of 1 ml min−1 using a Waters model 590 pump through a Varian Microsorb-MV C18 column (250 mm by 4.6 mm). Samples were injected by a Shimadzu SIL-10AD AP autoinjector and detected with a Shimadzu SPD-10A VP UV-Vis detector by monitoring absorbance at 290 nm. Quantitation was accomplished by comparison with a standard curve of authentic HQ.
Detecting side-chain metabolites.
For pGEM subclones in E. coli, 500 μl of an overnight culture was added to 4.5-ml aliquots of 1/10 LB in 25-ml Balch tubes. The medium was supplemented with 150 μg ml−1 ampicillin, and the cultures were incubated with shaking at 37°C for 2 h, at which time opdA expression was induced by the addition of 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Uninduced controls did not receive IPTG but rather had 50 mM glucose added upon inoculation in order to repress expression from the lac promoter of pGEM. After 2 more hours of incubation, OP in a methanol solution was spiked into cultures and the tubes were immediately sealed with rubber stoppers and crimped. Given the apparent toxicity of HQ to E. coli, only 1/4 the amount of OP (120 μM) added to PWE1 was added to these E. coli cultures. Headspace samples of 0.25 ml were periodically removed and analyzed by GC-MS as follows: 40°C hold for 1 min, increased by 5°C/min to 100°C and held for 3 min. The temperature was then increased by 10°C/min to 165°C and finally increased at 60°C/min to 240°C. All other GC-MS conditions were as described above. The appearance of 2,4,4-trimethyl-1-pentene was quantified by comparison with dilutions of an authentic standard made in similar Balch tubes.
Quantifying OP disappearance.
After 70 h, the above-described Balch tube cultures were sacrificed for further chemical analysis. The tubes were unsealed, and 500 μl of culture was removed and diluted with an equal volume of methanol. The methanol-amended culture was centrifuged to remove cellular debris, and the resulting supernatant was filtered through a 4-mm, 0.2-μm regenerated cellulose syringe filter (Corning, Corning, NY). The filtrate was analyzed for both HQ and OP via HPLC. OP was resolved with a mobile phase of 80% methanol and 20% 80 mM acetic acid and detected at 220 nm, whereas HQ was resolved using the method described above.
Nucleotide sequence accession numbers.
The full sequence of the putative OP monooxygenase gene (opdA) and a partial sequence of the Sphingomonas sp. strain PWE1 16S rRNA gene have been deposited in GenBank under accession numbers EU002557 and EU004850, respectively.
RESULTS
PWE1 growth on OP.
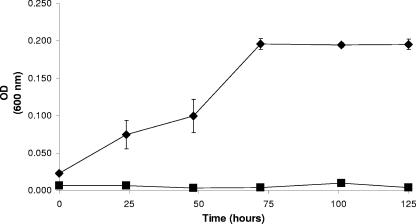
Sphingomonas sp. strain PWE1 was isolated based on its ability to grow with OP as a sole carbon and energy source. Nucleotide sequence analysis of the 16S rRNA gene in PWE1 showed 99% homology with Sphingomonas cloacae, a known NP-degrading microorganism (12). In minimal medium, stationary phase was reached by 72 h and the optical density at 600 nm did not exceed 0.2, as determined in a 96-well plate spectrophotometer (Fig. 1). TTNP3 showed a similar growth pattern in minimal medium with OP over a similar time span, reaching an optical density at 550 nm of only 0.23 (34). PWE1 grew to a much higher optical density in a complex rich medium (data not shown), just as TTNP3 growth increased with the addition of sodium acetate to the OP-degrading cultures (34).
FIG. 1.
Growth of Sphingomonas strain PWE1 on OP. Triplicate values of a representative culture flask of PWE1 grown in MSM with OP (⧫) compared to PWE1 grown in MSM without OP (▪). OD (600 nm), optical density at 600 nm.
Identification of opdA.
The reported production of ring-hydroxylated intermediates by other Sphingomonas strains led us to hypothesize that a screen dependent upon the polymerization of these intermediates would yield a diagnostic BPP in the presence of OP. BPP production could then be used to identify PWE1 fosmid clones harboring the gene(s) which encoded this activity. More than 900 fosmid library clones were screened on 1/10 LB with OP. p-Toluidine and FeCl3 were also added to enhance polymerization (30). One clone accumulated BPP when incubated with OP. It was designated aew4H12. No BPP was observed in the supernatant of E. coli harboring aew4H12 in the absence of OP or in the supernatant of any other fosmids in the presence of OP. Sequence analysis of BPP− transposon mutants of aew4H12 was used to target a putative open reading frame that appeared to encode OP degradation and was labeled opdA. When opdA was subcloned from aew4H12 into pGEM-T Easy to give pAW1 and expressed in E. coli JM109, it conferred BPP production but the vector control did not.
In silico analyses of opdA performed using Psi-Blast (1) identified a conserved monooxygenase domain and a flavin adenine dinucleotide (FAD)-binding domain that shared homology with those found in UbiH from E. coli (Fig. 2). Blastx analysis of opdA showed it to have weak predicted amino acid sequence similarity (32% identity, 48% similarity over 535 amino acids) to a putative polyketide hydroxylase from Stigmatella aurantiaca DW4/3-1 (RefSeq accession number ZP_01459560.1). Other putative genes that showed some predicted amino acid similarity with opdA included those encoding FAD-binding monooyxgenases, such as the PheA/TfdB family FAD monooxygenase from Myxococcus xanthus DK1622 (RefSeq accession number YP_635433), 2-polyprenyl-6-methoxyphenol hydroxylase from Burkholderia cenocepacia PC184 (GenBank accession number EAY67308), and 4-methyl-5-nitrocatechol monooxygenase from Burkholderia sp. strain DNT (GenBank accession number ABC00744). The closest related gene encoding a protein of known function was mhpA, 3-hydroxyphenylpropionate 2-monooxygenase, from E. coli W3110 (GenBank accession number BAA13052). A multisequence alignment of OpdA from PWE1, MhpA, UbiH, and close Blastx matches showed regions of conserved residues (Fig. 2). Specifically, two motifs recognized for ADP binding and flavin binding that had been identified in MhpA (10) were also found to be in OpdA.
FIG. 2.
Alignment of OpdA with single-component monooxygenases. The predicted amino acid sequence of OpdA is compared with those of Blastx matches Escherichia coli W3110 MhpA (BAA13052.1) (GenBank or RefSeq accession numbers are in parentheses), Polaromonas naphthalenivorans CJ2 putative FAD-binding monooxygenase (YP_980856.1), Myxococcus xanthus DK1622 PheA/TfdB family putative FAD-binding monooxygenase (YP_635433.1), Stigmatella aurantiaca DW4/3-1 putative polyketide hydroxylase (ZP_01459560.1), Stigmatella aurantiaca putative cytochrome P450 dependent monooxygenase (CAD19095.1), and Burkholderia sp. strain DNT 4-methyl-5-nitrocatechol monooxygenase (ABC00744.1). Escherichia coli K-12 UbiH (NP_417383.1) is also part of the alignment, based upon identification of a conserved region in OpdA that was shared with UbiH by use of Psi-Blast. Highlighted residues are those shared in common with OpdA. Underlined regions designate the ADP-binding and flavin-binding motifs, with the boldface residues in the consensus line being the specific conserved residues. Residues 26 to 65 in the consensus correspond with an ADP-binding motif (10, 37). Residues 295 to 335 in the consensus correspond with a flavin-binding motif (8, 10).
Testing opdA activity.
Based on similarities with other sphingomonads that can grow on alkylphenols (11), we expected that JM109 pAW1would metabolize OP to HQ. The complete disappearance of OP (0.61 μmol) added to 5-ml cultures of JM109 pAW1 (final concentration of 120 μM OP) after 70 h of incubation was accompanied by the production of HQ, which reached a maximal detectable amount of 0.15 μmol (final concentration of 30 μM HQ) (Table 1). HQ was not produced in cultures that lacked OP or by either JM109 pAW1 without IPTG or the vector control. Levels of OP disappearance in media inoculated with JM109 pAW1 without polymerizing agents were highly variable. Further analysis of cell viability suggested that this was likely due to the toxicity of the accumulating HQ, as 90 μM HQ was sufficient to reduce E. coli CFU by a factor of 1,000 (data not shown). Addition of FeCl3 and toluidine, which facilitated HQ polymerization, alleviated this toxicity somewhat (data not shown). As further evidence of toxicity, when pAW1 was maintained in E. coli DH5α (which lacks the lac repressor), the BPP phenotype was hypervariable and rapidly lost during subculturing. Interestingly, sequence analysis of opdA amplified from BPP− DH5α pAW1 revealed the presence of one silent and three coding mutations. The latter resulted in the following substitutions: N163S, Q205E, and A241C.
TABLE 1.
OP conversion to HQ and 2,4,4-trimethyl-1-pentene in an E. coli subclone harboring opdA
| Substrate | Starting mass (μmol) | Expected mass (μmol) | Observed mass (μmol) | % Expected |
|---|---|---|---|---|
| OP | 0.61 | 0 | NDa | |
| HQ | 0 | 0.61 | 0.15 | 25 |
| 2,4,4-Trimethyl-1-pentene | 0 | 0.61 | 0.35 | 57 |
ND, OP was not detectable in the 70-h culture samples that were used for HQ quantification.
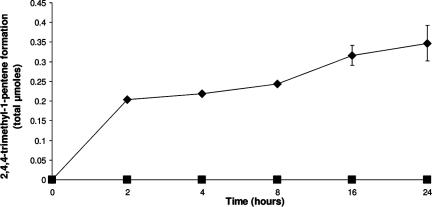
The long-chain alcohol expected to form as a result of OP metabolism (13, 34) was not detectable under the extraction conditions used to identify metabolites in wild-type PWE1 or in JM109 pAW1. GC-MS analysis of the headspace of JM109 pAW1 cultures was therefore used to capture any volatile metabolites and surprisingly revealed two distinct peaks. One peak corresponded in retention time (1.8 min) and mass spectrum to a commercial standard of 2,4,4-trimethyl-1-pentene, with a base peak m/z (relative abundance) of 57 (100), and yielded additional fragments at m/z 55 (28.7), 69 (8.64), 97 (17.6), and 112 (18.6). The other peak had a retention time of 3.8 min and its mass spectrum revealed a base peak m/z of 59 (100), as well as additional fragments at m/z 55 (34.9), 57 (93.3), 97 (21.0), and 115 (7.49). The mass spectrum of this second peak was identical to the mass spectrum Fujita and Reinhard (13) described for 2,4,4-trimethyl-2-pentanol. The peak area of the pentene was approximately 20 times greater than that of the pentanol, although we could not quantify the pentanol since no commercial standards were available. Over the course of 24 h, there was increased formation of both side-chain products in the headspace of E. coli pAW1 cultures, although results for only 2,4,4-trimethyl-1-pentene are shown in Fig. 3. There was very little change in the headspace concentration past 24 h. The final amount of 2,4,4-trimethyl-1-pentene at 70 h was 0.35 μmol (Table 1).
FIG. 3.
2,4,4-Trimethyl-1-pentene formation in the headspace of E. coli expressing opdA. Sealed culture tubes with 0.61 μmol OP in the aqueous phase were monitored by headspace sampling for the evolution of 2,4,4-trimethyl-1-pentene in the gas phase over time. E. coli pAW1 (▪) and the vector control (⧫) are shown.
Metabolites from wild-type PWE1.
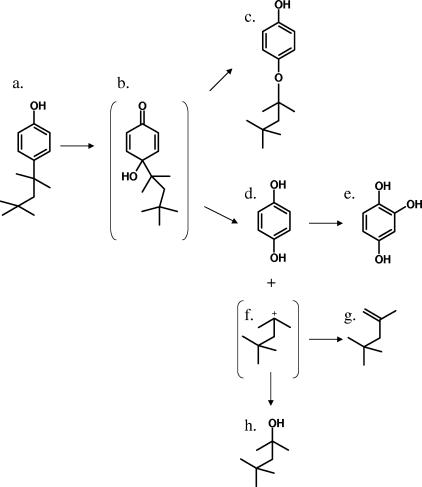
GC-MS analysis of acetylated extracts from PWE1 cultures revealed the presence of a metabolite with the same mass spectrum and retention time (20.2 min) as a similarly derivatized sample of authentic HQ. A putative trihydroxylated intermediate was also detected and found to have the same mass spectrum and retention time (28.7 min) as authentic 1,2,4-benzenetriol when derivatized by acetylation. Another putative metabolite was also detected in ethyl acetate extracts from both the acetylated (30.1 min) and underivatized (29.5 min) supernatants of PWE1 cultures. Without acetylation the parent ion had an m/z of 222 (23.1), with a base peak of m/z 151 (100), and an additional fragment at an m/z of 123 (37.8). After aqueous acetylation the metabolite yielded a weak molecular ion with an m/z of 264 (10.0) and showed a loss of m/z 42, yielding a fragment with an m/z of 222 (28.0), a base peak with an m/z of 151 (100), and another fragment with an m/z of 123 (26.5). The underivatized spectrum was consistent with either hydroxyoctylphenol (1,2-dihydroxy-4-octylbenzene) or octyloxyphenol (Fig. 4, compound c), although the addition of only one acetyl group during derivatization strongly suggests a single free hydroxyl group on the ring and is therefore more consistent with octyloxyphenol (Fig. 4, compound c).
FIG. 4.
Proposed pathway for OP monooxygenation encoded by opdA in Sphingomonas sp. strain PWE1. (a) OP; (b) putative ipso hydroxylation intermediate; (c) tentatively identified octyloxyphenol; (d) HQ; (e) 1,2,4-trihydroxybenzene; (f) putative alkyl side-chain carbocation; (g) 2,4,4-trimethyl-1-pentene; (h) 2,4,4-trimethyl-2-pentanol.
DISCUSSION
We report here on the successful cloning and expression of a gene (opdA) whose product is capable of transforming OP to HQ. Although several thorough recent reports have described in detail the mechanism whereby specific NP isomers are degraded to HQ by sphingomonads closely related to PWE1 (5, 14, 16, 23), no genes that code for such ring hydroxylating and dealkylating activity have previously been identified. It appears that PWE1 utilizes the same enzyme to accomplish both of these activities, as expression of OpdA was both necessary and sufficient for the transformation of OP to HQ and for the production of the side-chain products in E. coli. However, we cannot yet rule out the possibility that an endogenous E. coli protein catalyzes the dealkylating activity.
While other aromatic monooxygenases, such as phenol monooxygenase in Pseudomonas putida CF600, contain multicomponent monooxygenases with different genes encoding the FAD-binding domain and the catalytic monooxygenase subunit, opdA likely encodes a single-component enzyme containing both domains. This was suggested from Psi-Blast analyses indicating homology with the respective conserved regions of an E. coli ubiquinone synthesis enzyme, 2-octaprenyl-6-methoxyphenol hydroxylase (UbiH). Interestingly, as with OP, the ubiquinone alkyl side chain is highly branched; however, unlike OpdA, UbiH acts on an unsubstituted carbon para to an existing hydroxyl group (38). By contrast, the apparent site of hydroxylation for OpdA is the para carbon already occupied by the octyl side chain. OpdA also shared homology with MhpA, which catalyzes the hydroxylation of 3-(3-hydroxyphenyl)propionate to 3-(2,3-dihydroxyphenyl)propionate. However, MhpA-associated hydroxylation results in a catecholic intermediate (10), not HQ. Importantly, neither UbiH nor MhpA activities result in removal or rearrangement of the alkyl side chains.
NP and OP degradation were initially thought to occur through ring hydroxylation adjacent to the phenolic hydroxyl group (7, 35), as was shown for the degradation of 3- and 4-n-alkylphenols, yielding catecholic intermediates with subsequent meta cleavage (19). More recently, Corvini et al. (6) showed that degradation of alkylphenols with branched side chains occurs via oxidation at the quaternary alpha carbon in NP isomers p353NP and p262NP in Sphingomonas sp. strain TTNP3. Based on the formation of HQ and the detection of side-chain alcohol products (5), it has been determined that strain TTNP3 transforms NP via type II ipso substitution (5). ipso substitution in general is characterized by an intermediate whose leaving group is not hydrogen and in which both groups temporarily share the same position during electrophilic substitution of an aromatic ring. For alkylphenols, type I and II ispo substitutions differ in the charges of the leaving groups and the natures of the resulting ring products: OP degradation via type I ipso substitution would result in an anionic leaving group and the formation of p-benzoquinone, whereas type II ipso substitution of OP would result in the formation of HQ (29). In the case of NP degradation by Bayram and TTNP3, a putative bisubstituted intermediate is thought to decompose and ultimately result in the formation of HQ and a 9-carbon carbocation that then undergoes an Sn1 reaction with water to produce the observed alcohol (15, 25). Kolvenbach et al. (23) have shown, using 18O oxygen, that the new hydroxyl group of HQ is derived from molecular oxygen. The HQ is then further metabolized and serves as the true growth substrate.
The branching pattern of the alkyl side chain of NP isomers that serve as growth substrates for TTNP3 and Bayram is different than that of OP; however, those isomers and OP share in common a fully substituted alpha carbon on the alkyl side chain. This feature seems to be a prerequisite for side-chain removal (5, 14). Based on the similarity of the intermediates detected in the supernatants of PWE1 and of E. coli expressing opdA to those produced by Bayram and TTNP3, we propose that PWE1 uses a similar type II ipso substitution mechanism to degrade OP. However, in PWE1 we found that a large portion of the putative carbocation was converted to 2,4,4-trimethyl-1-pentene (Fig. 4). This is likely the result of an E1 elimination reaction and actually strengthens the case for a carbocation intermediate, since carbocations are known to undergo both Sn1 and E1 reactions (9). Although we did not confirm that similar alkenes were produce during NP degradation, any that were produced would likely have escaped detection previously, as none of the earlier work describing NP degradation employed headspace analysis (6, 14).
Despite OP disappearing to levels below detection in induced cultures of E. coli pAW1, we could account for only 57% of the side chain as the pentene and 25% as HQ (Table 1). This may have been due to the formation of the octyloxyphenol, a tentatively identified product we previously detected via GC-MS in samples from wild-type cultures incubated with OP. However, octyloxyphenol was not detectable in E. coli pAW1 cultures. This may have been due to the significantly lower concentration of OP used in those assays than in the wild-type-PWE1 experiments. Others have shown that similar NP metabolites accumulate in culture supernatants and do not undergo further metabolism (6, 7, 16). It is also possible that some of the HQ produced in E. coli was further transformed to alleviate toxicity or may have polymerized and was therefore not detected using our methods.
HQ is a metabolic intermediate in a variety of aromatic catabolic pathways, including those of p-nitrophenol (32), 4-chlorophenol (28), pentachlorophenol (3), γ-hexachlorocyclohexane (25), and 4-ethylphenol (21). Unlike TTNP3, PWE1 was unable to grow on HQ even when ascorbate was added to prevent HQ polymerization. However, resting cells of PWE1 incubated with HQ produced a yellow color that disappeared upon acidification and had a maximum absorbance at 320 nm, which is characteristic of HQ meta ring fission product formation (18). HQ has been reported to be directly cleaved via meta cleavage in some instances (25), but in other instances HQ was found to be transformed further to 1,2,4-trihydroxybenzene, which then served as a substrate for ring fission (21, 28). In contrast to the results reported for TTNP3, we were able to detect 1,2,4-trihydroxybenzene in ethyl acetate extracts of PWE1 supernatant, but only when the supernatant was first derivatized with acetic anhydride. 1,2,4-Trihydroxybenzene did not serve as a growth substrate when supplied exogenously and was not readily cleaved via meta cleavage by PWE1 resting cells, so it is unclear if this is a true metabolic intermediate or a dead-end product. Approximately 3.5 kb of DNA on either side of opdA was sequenced but did not appear to encode any genes for putative ring cleavage enzymes, as has often been found for other ring-hydroxylating enzymes.
Although the additional steps whereby HQ is degraded by PWE1 require further investigation, we have presented evidence that the degradation of OP to HQ is mediated by a putative flavin monooxygenase encoded by opdA from Sphingomonas sp. strain PWE1. This is the first example of a gene associated with the ring oxidation and side-chain removal of branched-chain alkylphenols.
Acknowledgments
A.W.P. was supported by a USDA multidisciplinary graduate education traineeship and an EPA STAR fellowship. This work was partially funded by USDA 189412.
Footnotes
Published ahead of print on 21 September 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, H. S., J. M. Lee, and S.-T. Lee. 1996. Biodegradation of 4-chlorophenol via a hydroquinone pathway by Arthrobacter ureafaciens CPR706. FEMS Microbiol. Lett. 145:125-129. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy, M. B., H. Lee, J. T. Trevors, and R. B. Zablotozicz. 1999. Chlorophenol and nitrophenol metabolism by Sphingomonas sp. UG30. J. Ind. Microbiol. Biotechnol. 23:232-241. [DOI] [PubMed] [Google Scholar]
- 4.Corti, A., S. Frassinetti, G. Vallini, S. D'Antone, C. Fichi, and R. Solaro. 1995. Biodegradation of nonionic surfactants. I. Biotransformation of 4-(1-nonyl)phenol by a Candida maltosa isolate. Environ. Pollut. 90:83-87. [DOI] [PubMed] [Google Scholar]
- 5.Corvini, P. F. X., J. Hollender, R. Ji, S. Schumacher, J. Prell, G. Hommes, U. Priefer, R. Vinken, and A. Schaffer. 2006. The degradation of α-quaternary nonylphenol isomers by Sphingomonas sp. strain TTNP3 involves a type II ipso-substitution mechanism. Appl. Microbiol. Biotechnol. 70:114-122. [DOI] [PubMed] [Google Scholar]
- 6.Corvini, P. F. X., R. Vinken, G. Hommes, M. Mundt, J. Hollender, R. J. W. Meesters, H. F. Schroder, and B. Schmidt. 2004. Microbial degradation of a single branched isomer of nonylphenol by Sphingomonas TTNP3. Water Sci. Technol. 50:189-194. [PubMed] [Google Scholar]
- 7.de Vries, Y. P., Y. Takahara, Y. Ikunaga, Y. Ushiba, M. Hasegawa, Y. Kasahara, H. Shimomura, S. Hayashi, Y. Hirai, and H. Ohta. 2001. Organic nutrient-dependent degradation of branched nonylphenol by Sphingomonas sp. YT isolated from a river sediment sample. Microbes Environ. 16:240-249. [Google Scholar]
- 8.DiMarco, A. A., B. A. Averhoff, E. E. Kim, and L. N. Ornston. 1993. Evolutionary divergence of pobA, the structural gene encoding p-hydroxybenzoate hydroxylase, in an Acinetobacter calcoaceticus strain well-suited for genetic analysis. Gene 125:25-33. [DOI] [PubMed] [Google Scholar]
- 9.Ege, S. 1999. Organic chemistry: structure and reactivity, 4th ed. Houghton Mifflin Co., Boston, MA.
- 10.Ferrandez, A., J. L. Garcia, and E. Diaz. 1997. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway in Escherichia coli K-12. J. Bacteriol. 179:2573-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii, K., N. Urano, H. Ushio, M. Satomi, H. Iida, N. Ushio-Sata, and S. Kimura. 2000. Profile of a nonylphenol-degrading microflora and its potential for bioremedial applications. J. Biochem. 128:909-916. [DOI] [PubMed] [Google Scholar]
- 12.Fujii, K., N. Urano, H. Ushio, M. Satomi, and S. Kimura. 2001. Sphingomonas cloacae sp. nov., a nonylphenol-degrading bacterium isolated from wastewater of a sewage-treatment plant in Tokyo. Int. J. Syst. Evol. Microbiol. 51:603-610. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, Y., and M. Reinhard. 1997. Identification of metabolites from the biological transformation of the nonionic surfactant residue octylphenoxyacetic acid and its brominated analog. Environ. Sci. Technol. 31:1518-1524. [Google Scholar]
- 14.Gabriel, F. L. P., M. Cyris, N. Jonkers, W. Giger, K. Guenther, and H.-P. E. Kohler. 2007. Elucidation of the ipso-substitution mechanism for side-chain cleavage of α-quaternary 4-nonylphenols and 4-t-butoxyphenol in Sphingobium xenophagum Bayram. Appl. Environ. Microbiol. 73:3320-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriel, F. L. P., W. Giger, K. Guenther, and H.-P. E. Kohler. 2005. Differential degradation of nonylphenol isomers by Sphingomonas xenophaga Bayram. Appl. Environ. Microbiol. 71:1123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabriel, F. L. P., A. Heidlberger, D. Rentsch, W. Giger, K. Guenther, and H.-P. E. Kohler. 2005. A novel metabolic pathway for degradation of 4-nonylphenol environmental contaminants by Sphingomonas xenophaga Bayram. ipso-Hydroxylation and intramolecular rearrangement. J. Biol. Chem. 280:15526-15533. [DOI] [PubMed] [Google Scholar]
- 17.Giger, W., P. Brunner, and C. Schafner. 1984. 4-Nonylphenol in sewage sludge: accumulation of toxic metabolites from nonionic surfactants. Science 225:623-625. [DOI] [PubMed] [Google Scholar]
- 18.Haigler, B. E., G. R. Johnson, W.-C. Suen, and J. C. Spain. 1999. Biochemical and genetic evidence for meta-ring cleavage of 2,4,5,-trihydroxytoluene in Burkholderia sp. strain DNT. J. Bacteriol. 181:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong, J. J., J. H. Kim, C.-K. Kim, I. Hwang, and K. Lee. 2003. 3- and 4-Alkylphenol degradation pathway in Pseudomonas sp. strain KL28: genetic organization of the lap gene cluster and substrate specificities of phenol hydroxylase and catechol 2,3-dioxygenase. Microbiology 149:3265-3277. [DOI] [PubMed] [Google Scholar]
- 20.Jobling, S., D. Sheahan, J. Osborne, P. Matthiessen, and J. Sumpter. 1996. Inhibition of testicular growth in rainbow trout (Oncorhynchus mykiss) exposed to estrogenic alkylphenolic chemicals. Envion. Toxicol. Chem. 15:194-202. [Google Scholar]
- 21.Jones, K. H., P. W. Trudgill, and D. J. Hopper. 1994. 4-Ethylphenol metabolism by Aspergillus fumigatus. Appl. Environ. Microbiol. 60:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junghanns, C., M. Moeder, G. Krauss, C. Martin, and D. Schlosser. 2005. Degradation of the xenoestrogen nonylphenol by aquatic fungi and their laccases. Microbiology 151:45-57. [DOI] [PubMed] [Google Scholar]
- 23.Kolvenbach, B., N. Schlaich, Z. Raoui, J. Prell, S. Zuhlke, A. Schaffer, F. P. Guengerich, and P. F. X. Corvini. 2007. Degradation pathway of bisphenol A: does ipso substitution apply to phenols containing a quaternary α-carbon structure in the para position? Appl. Environ. Microbiol. 73:4776-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCullar, M. V., V. Brenner, R. H. Adams, and D. D. Focht. 1994. Construction of a novel polychlorinated biphenyl-degrading bacterium: utilization of 3,4′-dichlorobiphenyl by Pseudomonas acidovorans M3GY. Appl. Environ. Microbiol. 60:3833-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyauchi, K., Y. Adachi, Y. Nagata, and M. Takagi. 1999. Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of γ-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 181:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura, M., H. Nagoya, and T. Hirai. 2002. Nonylphenol induces complete feminization of the gonad in genetically controlled all-male amago salmon. Fish. Sci. 68:1387-1389. [Google Scholar]
- 27.Nice, N. E., D. Morritt, M. Crane, and M. Thorndyke. 2003. Long-term and transgenerational effects of nonylphenol exposure at a key stage in the development of Crassostrea gigas. Possible endocrine disruption? Mar. Ecol. Prog. Ser. 256:293-300. [Google Scholar]
- 28.Nordin, K., M. Unell, and J. K. Jansson. 2005. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 71:6538-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohe, T., T. Mashino, and M. Hirobe. 1997. Substituent elimination from p-substituted phenols by cytochrome P450 ipso-substitution by the oxygen atom of the active species. Drug Metab. Dispos. 25:116-122. [PubMed] [Google Scholar]
- 30.Parke, D. 1992. Application of p-toluidine in chromogenic detection of catechol and protocatechuate, diphenolic intermediates in catabolism of aromatic compounds. Appl. Environ. Microbiol. 58:2694-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soares, A., B. Guieysse, O. Delgado, and B. Mattiasson. 2003. Aerobic biodegradation of nonylphenol by cold-adapted bacteria. Biotechnol. Lett. 25:731-738. [DOI] [PubMed] [Google Scholar]
- 32.Spain, J. C., and D. T. Gibson. 1991. Pathway for the biodegradation of p-nitrophenol in a Moraxella sp. Appl. Environ. Microbiol. 57:812-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabira, Y., M. Nakai, D. Asai, Y. Yakabe, Y. Tahara, T. Shinmyozu, M. Noguchi, M. Takatsuki, and Y. Shimohigashi. 1999. Structural requirements of para-alkylphenols to bind to estrogen receptor. Eur. J. Biochem. 262:240-245. [DOI] [PubMed] [Google Scholar]
- 34.Tanghe, T., W. Dhooge, and W. Verstraete. 2000. Formation of the metabolic intermediate 2,4,4-trimethyl-2-pentanol during incubation of a Sphingomonas sp. strain with the xeno-estrogenic octylphenol. Biodegradation 11:11-19. [DOI] [PubMed] [Google Scholar]
- 35.Tanghe, T., W. Dhooge, and W. Verstraete. 1999. Isolation of a bacterial strain able to degrade branched nonylphenol. Appl. Environ. Microbiol. 65:746-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ushiba, Y., Y. Takahara, and H. Ohta. 2003. Sphingobium amiense sp. nov., a novel nonylphenol-degrading bacterium isolated from a river sediment. Int. J. Syst. Evol. Microbiol. 53:2045-2048. [DOI] [PubMed] [Google Scholar]
- 37.Wierenga, R. K., P. Tepstra, and W. G. J. Hol. 1986. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]
- 38.Young, I. G., P. Stroobant, C. G. Macdonald, and F. Gibson. 1973. Pathway for ubiquinone biosynthesis in Escherichia coli K-12: gene-enzyme relationships and intermediates. J. Bacteriol. 114:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]