Abstract
Little is known about bacterial communities that colonize mucosal surfaces in the human gastrointestinal tract, but they are believed to play an important role in host physiology. The objectives of this study were to investigate the compositions of these populations in the distal small bowel and colon. Healthy mucosal tissue from either the terminal ileum (n = 6) or ascending (n = 8), transverse (n = 8), or descending colon (n = 4) of 26 patients (age, 68.5 ± 1.2 years [mean ± standard deviation]) undergoing emergency resection of the large bowel was used to study these communities. Mucosa-associated eubacteria were characterized by using PCR-denaturing gradient gel electrophoresis (DGGE), while real-time PCR was employed for quantitative analysis. Mucosal communities were also visualized in situ using confocal laser scanning microscopy. DGGE banding profiles from all the gut regions exhibited at least 45% homology, with five descending colon profiles clustering at ca. 75% concordance. Real-time PCR showed that mucosal bacterial population densities were highest in the terminal ileum and that there were no significant differences in overall bacterial numbers in different parts of the colon. Bifidobacterial numbers were significantly higher in the large bowel than in the terminal ileum (P = 0.006), whereas lactobacilli were more prominent in the distal large intestine (P = 0.019). Eubacterium rectale (P = 0.0004) and Faecalibacterium prausnitzii (P = 0.001) were dominant in the ascending and descending colon. Site-specific colonization in the gastrointestinal tract may be contributory in the etiology of some diseases of the large intestine.
The human colonic microbiota is part of a highly complex bacterial ecosystem (26), and several hundred different types of bacteria are known to colonize the gut (8, 9, 43). Evidence implicates the microbiota in the pathophysiology of several diseases of the large intestine, including colorectal cancer, Crohn's disease, and ulcerative colitis (9, 16, 18, 25, 28). These diseases usually show distinct patterns of incidence in the large bowel. For example, 50% of colorectal cancers occur in the distal large gut (4), while in contrast, the transverse colon is a relatively rare site of tumor formation, with the incidence increasing in the ascending colon and cecum (4). Similarly, ulcerative colitis principally affects the distal large bowel and most frequently involves the rectum (34), whereas Crohn's disease is localized in the ileocecal junction in 60% of patients, in the small intestine alone in 20%, and in the colon alone in a further 20% (12).
Much of our knowledge of the diversity and distribution of the intestinal microbiota derives from bacteriologic studies on fecal material, employing traditional techniques of culture, microscopy, and determinations of the fermentative and other biochemical capabilities of the isolates (27, 30). This work, while still important in many types of study, is labor intensive and requires the use of specialized apparatus that allows extremely oxygen-sensitive bacteria to be cultivated (23, 30, 36). Moreover, it has been reported that about two-thirds of the total microscopic count in the gut is not recovered, even using strict anaerobic culture methods (44, 48).
Molecular techniques based on the analysis of 16S rRNA gene sequences, however, allow detailed studies to be made on intestinal bacterial communities (30). In particular, PCR coupled with denaturing gradient gel electrophoresis (DGGE) has been a useful tool in providing visual fingerprints of the gut microbiota (1, 38, 49), while the more recent development of real-time PCR provides a sensitive and quantitative device for population analysis (5, 10, 15). Recent metagenomic studies, based on high-throughput PCR, cloning, and sequencing, have enabled phylogenetic information about both culturable and nonculturable species colonizing the human digestive tract to be obtained (8, 13). Such investigations have been used, for example, to demonstrate that changes in the relative abundance of the Bacteroidetes and the Firmicutes biotas in the mouse gut resulted in significant, transmissible changes in host adiposity (24) and to provide a comprehensive indication of the species present and their relative abundances (8, 13, 49).
Because they grow in close juxtaposition to host cells, mucosal bacteria are thought to play a particularly important role in gut health, and distinct bacterial populations have been observed to occur on the gut epithelium in both health and disease (29, 31). Molecular studies have been done previously on mucosal communities in the gastrointestinal tract. Using DGGE, Zoetendal et al. (49) reported that mucosa-associated bacteria were similar along the length of the gastrointestinal tract, and more recently, comparative qualitative analysis of bacterial 16S rRNA gene sequences on the colonic mucosa of three healthy individuals indicated the occurrence of a large number of uncultured bacteria (8). The objective of the present investigation was to use a combination of molecular techniques (real-time PCR, DGGE, and fluorescence in situ hybridization [FISH]) to quantitate and visualize the bacterial diversity of the predominant bacteria colonizing the bowel wall in the terminal ileum, as well as in the ascending, transverse, and descending colon.
MATERIALS AND METHODS
Patients and specimens.
Mucosal biopsies were taken from 26 patients who underwent emergency surgery requiring colonic resection. None of the patients had bowel preparation before surgery. For each patient, a biopsy sample of healthy tissue was taken from one of the following areas: the terminal ileum (n = 6), the ascending or transverse colon (n = 8 each), or the descending colon (n = 4). Fourteen patients were male, with a mean age of 68 years (range, 59 to 81), and 12 were female, with a mean age of 65 (range, 61 to 76). Pathologists took mucosal sections at points furthest away from diseased areas, and histology later confirmed that there was a clear margin between healthy and diseased tissue. Diagnoses of colorectal cancer were confirmed in all but two cases, which were diagnosed as localized perforated diverticulitis. The biopsy samples, with associated mucus, were collected aseptically, rinsed gently with sterile anaerobic phosphate-buffered saline, and frozen immediately at −80°C for subsequent PCR analysis; biopsy samples for FISH were covered in Tissue-Tek freezing medium (Sakura Finetek Europe B.V., The Netherlands) to preserve the mucus layer and stored at −80°C in sterile Eppendorf tubes. Ethical approval was obtained from the Tayside committee on medical research ethics and the MRC/CRUK tissue bank committee, Ninewells Hospital, Dundee.
DNA extraction and purification.
DNA was extracted from cryopreserved tissue sections using a QIAamp DNA spin column (QIAGEN Ltd., West Sussex, United Kingdom). Briefly, biopsy specimens (2 to 5 mg) were suspended in lysis buffer and lysozyme (50 mg ml−1) solutions before incubation at 55°C. Proteinase K, buffer ATL (QIAGEN), and ethanol were sequentially added. The mixture was bead beaten for 2 min before and after incubation at 70°C using a MiniBeadbeater-8 (Biospec Products, Bartlesville, OK). Bacterial cell lysates were obtained by centrifugation at 5,000 × g (3 min). The resultant DNA was adsorbed onto minicolumn membranes, washed, and eluted according to the QIAGEN DNeasy tissue kit procedure. The purified DNA was eluted in 200 μl of elution buffer and stored at −80°C until analysis.
Amplification in conventional PCR.
A Techne genius PCR machine (Techne Ltd., Duxford, United Kingdom) was used for conventional PCR. The V2-V3 region of the 16S rRNA gene (corresponding to positions 339 to 539 of the Escherichia coli gene) was then amplified with (GC-rich) eubacterium-specific primers HDA1 (5′-GC CGC CCG GGG CGC GCC CCG GGC GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) (45). The reactions were done in 0.2-ml reaction tubes. In all cases, the reaction was carried out using 25 μl Red Taq DNA polymerase ready mix (Sigma, Poole, Dorset, United Kingdom), HDA primers (2 μl of each; 5 mM), nanopure water (16 μl), and extracted community DNA (5 μl). Previous optimization studies (21, 38) have shown that extracted DNA requires a minimum of a 1:10 dilution to ensure reliable PCRs. The thermal program was as follow: 94°C (4 min) followed by 30 thermal cycles of 94°C (30 s), 56°C (30 s), and 68°C (60 s). The procedure was completed with a final elongation step at 68°C (7 min). PCR products were run on an agarose electrophoresis gel for confirmation of product concentration, quality, and size (ca. 200 bp) against a DNA molecular weight marker (2-log DNA ladder N3200L; New England Biolabs Ltd., Herts, United Kingdom). Primers were purchased from Invitrogen Life Technologies (Paisley, United Kingdom).
DGGE analysis.
A D-Code universal mutation detection system (Bio-Rad, Hemel Hempstead, United Kingdom) with 16-cm by 16-cm (1-mm-deep) polyacrylamide gels (10%), run with 1× TAE buffer diluted from 50× TAE buffer (40 mM Tris base, 20 mM acetic acid, and 40 mM EDTA), was used for these analyses. Initially, the separation parameters were optimized by running PCR products from selected pure cultures of bacteria and PCR amplicons from extracted community DNA on gels with a 0%-to-100% denaturing gradient perpendicular to the direction of electrophoresis (a 100% denaturing solution contained 40% [vol/vol] formamide and 7.0 mM urea). Denaturing gradients were formed with two 10% acrylamide (acrylamide-bisacrylamide, 37.5:1) stock solutions (Sigma, Poole, Dorset, United Kingdom). On this basis, a denaturation gradient for parallel DGGE analysis ranging from 30 to 60% was selected. PCR amplicons from the following bacteria were run on a parallel gel in order to confirm these separation conditions: Clostridium clostridioforme DUN-120, Escherichia coli ATCC 11775, Enterococcus faecalis ATCC 51299, Eubacterium limosum DUN-112, Lactobacillus plantarum DUN-145, Collinsella aerofaciens DUN-207, Bacteroides fragilis NCTC 9343, Desulfococcus multivorans NCIMB 12965, Bifidobacterium bifidum NCIMB 702715, and Lactobacillus paracasei DUN-213. The bacteria were obtained from the National Collection of Industrial and Marine Bacteria (Aberdeen, United Kingdom), the National Collection of Type Cultures (London, United Kingdom), the American Type Culture Collection (Manassas, VA) and laboratory stock cultures at the University of Dundee (DUN-). Electrophoresis was done at 150 V at 60°C for approximately 5 h (electrophoresis was terminated when the xylene cyanol dye reached the bottom of the gels). All gels were stained using SYBR gold stain diluted to 10−4 in 1× TAE (Molecular Probes Europe, Leiden, The Netherlands) for 30 min. The gels were viewed and images documented by using a BioDocit system (UVP, Upland, CA).
Sequencing of DGGE bands.
For analysis of the major resolved DGGE amplicons, selected bands were cut out of the polyacrylamide gels using a sterile scalpel under UV illumination and incubated at 4°C for 20 h, together with 20 μl nanopure water in nuclease-free universal bottles. Portions (5 μl) were removed and used as a template for a PCR identical to that outlined in the DGGE analysis described above. PCR products were purified using QIAquick PCR purification kits (QIAGEN) and sequenced using the reverse (non-GC clamp) primer (HDA2). The sequencing cycles were 94°C (4 min) followed by 25 cycles of 96°C (30 s), 50°C (15 s), and 60°C (4 min). Once chain termination was complete, sequencing was done using BigDye terminator sequencing on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA). For excised DGGE bands, the presence of a GC clamp on sequence analyses confirmed that the correct target, rather than a contaminant, had been reamplified.
Sequencing of DGGE amplicons.
BLAST searches on each sequence were done against those in the EMBL nucleotide sequence database (2).
Dendrogram construction from DGGE gels.
The gel images were analyzed using the BioNumerics Fingerprint package (Applied Maths, Sint-Martens-Latem, Belgium). The bands in each lane were detected automatically. Each lane on the gel was then compared, allowing matching profiles to be generated. Matching profiles for each lane were used to produce a dendrogram by using the unweighted pair group method with arithmetic mean so that potential clustering patterns could be observed. For each band represented in the synthetic reference lane, corresponding bands that had been excised were selected for sequencing.
Real-time PCR analysis.
Quantitation of bacterial 16S rRNA genes in mucosal tissue was done using an iCycler real-time PCR detection system (Bio-Rad, Hemel Hempstead, United Kingdom), using the experimental protocols described by Fite et al. (10). Briefly, DNA melting curves were used to monitor product specificities; detection was based on fluorescence energy transfer, with SYBR green 490 as the fluorophore. The cycle number at which the signal was first detected corresponded to the original concentration of DNA template, and the starting copy number of amplicons was inversely proportional to the real-time threshold cycle. Analyses of PCR amplification and melting curves were made using iCycler Optical System software, version 3.0 (Bio-Rad). The conditions included one cycle at 95°C for 3 min, followed by 38 cycles of denaturation at 95°C for 30 s, primer annealing at the optimum temperature for 30 s (optimal temperature and primers used in the study are shown in Table 1), and one final cycle at 95°C for 30 s. The melt curve analysis was run for 43 repeats, increasing concordantly by 1°C. Plasmid standards and samples were assayed simultaneously in duplicate. A range of concentrations from 101 to 108 of plasmid DNA was used in each real-time PCR assay to generate standard curves for quantitation of target DNA in test samples. Primers were purchased from Invitrogen.
TABLE 1.
16S rRNA PCR primer sets used for real-time PCR in this study
| Bacterium | Forward sequence (5′-3′) | Position | Reverse sequence (5′-3′) | Position | Product length (bp) | ToCa | Reference |
|---|---|---|---|---|---|---|---|
| Desulfovibrio genus | CCGTAGATATCTGGAGGAACATCAG | 691 | ACATCTAGCATCCATCGTTTACAGC | 826 | 135 | 62 | 10 |
| Faecalibacterium prausnitzii | GATGGCCTCGCGTCCGATTAGb | 223 | CCGAAGACCTTCTTCCTCC | 420 | 199 | 58 | 47 |
| Clostridium clostridioforme | AATCTTGATTGACTGAGTGGCGGAC | 99 | CCATCTCACACTACCGGAGTTTTTC | 247 | 148 | 62 | 5 |
| Peptostreptococcus anaerobius | GCTCGGTGCCTTCACTAACGb | 1310 | AGCCCCGAAGGGAAGGTGTG | 1498 | 188 | 64 | 41 |
| Lactobacillic | GATAGAGGTAGTAACTGGCCTTTA | 478 | GCGGAAACCTCCCAACA | 853 | 390 | 55 | 32 |
| Bifidobacteria | AGGGTTCGATTCTGGCTCAG | 8 | CATCCGGCATTACCACCCb | 164 | 156 | 62 | 19 |
| Bacteroides | GTCAGTTGTGAAAGTTTGCb | 605 | CAATCGGAGTTCTTCGTG | 732 | 127 | 56 | 6 |
| Enterobacteria | CATTGACGTTACCCGCAGAAGAAGC | 457 | CTCTACGAGACTCAAGCTTGC | 652 | 195 | 63 | 5 |
| Eubacterium rectale | CGGTACCTGACTAAGAAGC | 477 | CCTAGTATTCATCGTTTACGGCGTGb | 825 | 347 | 60 | 42 |
| Enterococcus faecalis | GCTTTCGGGTGTCGCTGATG | 199 | CGTCCTTGTTCTTCTCTAAC | 452 | 253 | 58 | 5 |
| All eubacteria | ACTCCTACGGGAGGCAGCAGT | 330 | GTATTACCGCGGCTGCTGGCAC | 530 | 200 | 54 | 39 |
ToC indicates the optimum melting temperature determined by conventional temperature gradient PCR.
Primer modified from primer in reference.
Lactobacillus primers were determined by sequencing and experimental tests to detect bacterial subgroups associated with L. delbrueckii and L. plantarum, which include L. acidophilus, L. crispatus, L. johnsonii, L. gasseri, and L. brevis.
FISH and confocal laser scanning microscopy (CLSM).
Biopsy samples were cut into 10-μm sections using a cryomicrotome and placed on Teflon-coated 10-well glass slides (VWR, Merck Eurolab Ltd., Poole, Dorset, United Kingdom). A range of fluorescent 16S rRNA oligonucleotide probes was used to visualize bacteria present on the mucosal surface (7) as described previously (29). Probe Bif 164 (5′-CATCCGGCATTACCACCC-3′) was used to identify bifidobacteria (20), and Bac 303 (5′-CCAATGTGGGGGACCTT-3′) was used for bacteroides and prevotella (33). Probe Ato 291 (5′-GGTCGGTCTCTCAACCC-3′) was used for the atopobium cluster (14), and probe Erec 482 (5′-GCTTCTTAGTCAGGTACCG-3′) was used for the Eubacterium rectale/Clostridium coccoides group (11). The nucleic acid stain DAPI (4′,6′-diamidino-2-phenylindole) was added to the final wash buffer after hybridization (29). Intestinal isolates and a range of culture collection type strains were used as controls in testing the specificities of the oligonucleotide probes (5). Organisms were cultured in Wilkins-Chalgren broth (Oxoid, Basingstoke, Hamps, United Kingdom) in an anaerobic chamber (atmosphere: 80% N2, 10% CO2, 10% H2) at 37°C for 24 h. The bacteria were then fixed in fresh paraformaldehyde for 4 h at room temperature, washed in phosphate-buffered saline, and stored at −20°C (3). Probes were purchased from Thermohybaid, Interactiva Division (Ulm, Germany), and 5′-labeled with fluorochrome Cy3, fluorescein isothiocyanate (FITC), or Cy5. Slides were visualized using a Nikon Eclipse E800 upright microscope attached to a Nikon PCM 2000 confocal system, with a 488-nm argon laser (green fluorescence) and a 543-nm helium-neon laser (red fluorescence). A 60× Plan Apo immersion lens (numerical aperture, 1.4) was used for all images, which were captured and overlaid using C-Imaging software (Compix, Inc., Cranberry Township, PA).
Statistical analysis.
Data analysis was done using GraphPad Prism version 4.0 (GraphPad Software, Inc., San Diego, CA). The copy numbers of the 16S rRNA gene per gram of sample were transformed into logarithms, and the normally distributed data was subjected to statistical analysis. One-way analysis (Tukey's multiple comparison) of variance was used for comparison of bacterial densities between different specimens. Probability values of <0.05 were taken as statistically significant.
Chemicals.
Unless stated otherwise, all chemicals were obtained from Sigma.
Nucleotide sequence accession numbers.
The 35 species-specific sequences identified from the DGGE amplicons have been placed in the EMBL/GenBank/DDBJ database under accession numbers AM850147 to AM850181.
RESULTS
DGGE analysis.
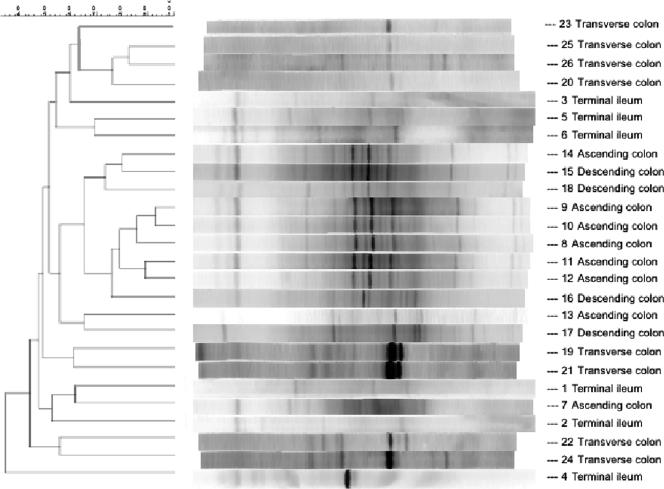
Parallel DGGE using PCR products from pure cultures of C. clostridioforme, E. coli, E. faecalis, E. limosum, L. plantarum, C. aerofaciens, B. fragilis, D. multivorans, B. bifidum, and L. paracasei demonstrated that each species formed a single major band on the gels (data not shown). Figure 1 shows a dendrogram generated from DGGE fingerprints of mucosa-associated eubacteria. In general, most DGGE fingerprints were characterized by the presence of 1 to 3 dominant bands, with microbial complexity varying markedly between different regions of the gut. Thus, some ascending colon community lanes exhibited up to 20 discrete bands of various density. Intraindividual variation was generally low, as demonstrated by the homology which was apparent between samples from each region in cluster analysis, particularly for ascending colon samples (maximum sample concordance, 90%) and transverse colon samples (maximum concordance, 80%). DGGE banding profiles from all gut regions exhibited at least 45% homology, with five descending colon profiles clustering at ca. 75% concordance.
FIG. 1.
Dendrogram produced with the unweighted pair group method with arithmetic mean showing the percent matching of DGGE fingerprints derived from terminal ileum, ascending colon, transverse colon and descending colon tissue samples. The dendrogram was produced using the BioNumerics Fingerprint software package.
Phylogenetic analysis of selected DGGE amplicons.
In the 69 bands selected, excised, and sequenced from the gels, 35 species-specific sequences were identified (Table 2). In the terminal ileum, the most prominent bands corresponded to EMBL accession number EF067826 of B. fragilis, AF418173 of D. multivorans, and AM042696 of Bacteroides vulgatus. A high degree of interindividual similarity was evident in DGGE band patterns from the ascending colon samples. The three main bands corresponded to X83935 of B. fragilis, AY804151 of E. rectale, and AB021418 of Comamonas terrigena. The major bands from the transverse colon samples corresponded to X83935 of B. fragilis, AY804152 of E. rectale, and AM042696 of B. vulgatus, while AB186304 of Bifidobacterium pseudolongum, AY373589 of Lactobacillus fermentum, and M59089 of C. clostridioforme were the sequences corresponding to the prominent bands on DGGE gels run with bacterial DNA amplified from the distal colon (Fig. 1).
TABLE 2.
Sequence identities of PCR amplicons derived from DGGE gels
| Sequencea | Sequence length in bp (% ambiguity) | Organism with highest sequence homology, sequence accession no. (% homology)b |
|---|---|---|
| TI1 | 193 (3.1) | Bacteroides fragilis, EF067826 (96) |
| TI2 | 182 (0) | Desulfococcus multivorans, AF418173 (99) |
| TI3 | 180 (1.6) | Bacteroides vulgatus, AM042696 (100) |
| TI4 | 185 (0) | Intestinal bacterial clone, DQ441271 (99) |
| TI5 | 178 (0) | Intestinal bacterial clone, EF404525 (98) |
| TI6 | 235 (2.4) | Mycoplasma faecium, AF125590 (97) |
| TI7 | 215 (3.3) | Clostridium fallax, M59088 (92) |
| TI8 | 199 (5.4) | Vibrio campbellii, X74692 |
| TI9 | 230 (0) | Clostridium acetobutylicum, AM231182 (99) |
| AC1 | 215 (3.2) | Eubacterium rectale, AY804151 (97) |
| AC2 | 179 (3.4) | Comamonas terrigena, AB021418 (98) |
| AC3 | 217 (5.1) | Environmental bacterial clone, AY879308 (94) |
| AC4 | 194 (3.9) | Lactobacillus plantarum, DQ239695 (97) |
| AC5 | 183 (0) | Enterococcus faecalis, AM157433 (98) |
| AC6 | 191 (1.0) | Bacteroides fragilis, X83935 (96) |
| AC7 | 210 (0) | Ruminococcus sp., AY442822 (100) |
| AC8 | 202 (3.0) | Intestinal bacterial clone, DQ100449 (96) |
| AC9 | 236 (3.9) | Clostridium innocuum, DQ440561 (97) |
| TC1 | 186 (0) | Lactobacillus rhamnosus, DQ010418 (93) |
| TC2 | 167 (0) | Bifidobacterium breve, AY267190 (98) |
| TC3 | 191 (0) | Bacteroides fragilis, X83935 (96) |
| TC4 | 180 (1.1) | Bacteroides vulgatus, AM042696 (98) |
| TC5 | 200 (1.5) | Eubacterium rectale, AY804152 (98) |
| TC6 | 184 (0) | Enterococcus faecalis, AY692453 |
| DC1 | 179 (2.2) | Bifidobacterium pseudolongum, AB186304 (97) |
| DC2 | 183 (0) | Lactobacillus acidophilus, M58802 99 |
| DC3 | 198 (0) | Lactobacillus fermentum, AY373589 (97) |
| DC4 | 183 (0) | Lactobacillus salivarius, AY389804 (98) |
| DC5 | 176 (0) | Clostridium clostridioforme, M59089 (100) |
| DC6 | 198 (1.5) | Lactobacillus fermentum, AY373589 (97) |
| DC7 | 226 (0) | Intestinal bacterial clone, EF403114 (100) |
| DC8 | 289 (0) | Clostridium fallax, AY208919 (100) |
| DC9 | 177 (1.7) | Enterococcus faecalis, AJ301831 |
| DC10 | 163 (0) | Bifidobacterium infantis, AY151398 |
| DC11 | 240 (0) | Bifidobacterium breve, M84776 (100) |
TI, terminal ileum; AC, ascending colon; TC, transverse colon; DC, descending colon.
Homology is to sequences in EMBL database. Percent similarity values are based on pair-wise alignments.
Real-time PCR analysis.
Bacteria from colonic tissue samples were quantitated by real-time PCR using eubacterium-, genus-, and species-specific primer sets (Table 1). The major groups of bacteria found on the DGGE gels were quantified, and the counts were converted to the log number of 16S rRNA genes per milligram of mucosal tissue. The numbers of individual bacteria measured by real-time PCR are shown in Table 3. Enterobacteria were not identified in the DGGE gels; however, real-time PCR analysis using a genus-specific primer demonstrated that these organisms were present in high levels in all parts of the colon. Statistical analysis showed that bacterial colonization of the mucosa in the terminal ileum, measured using real-time PCR and the universal primer set, was significantly greater than that of all parts of the colon (P < 0.001). However, there were no significant differences in total bacterial counts in the ascending, transverse, and descending colon. Bifidobacteria and F. prausnitzii were significantly higher in the proximal colon than in the terminal ileum (P = 0.006 and P = 0.003, respectively), but there were no significant differences in the numbers of these organisms in different parts of the large bowel. The numbers of Eubacterium rectale, C. clostridioforme, and F. prausnitzii were significantly higher in the ascending and descending colon than in the transverse colon and terminal ileum (P = 0.0004, P = 0.004, and P = 0.001, respectively), while significantly higher numbers of lactobacilli were found in the descending and transverse colon than in the terminal ileum and proximal colon (P = 0.019). Significantly higher numbers of E. faecalis were found in the proximal and transverse colon than in the terminal ileum (P = 0 003). No significant differences were observed in the distributions of Peptostreptococcus anaerobius (P = 0.378), desulfovibrios (P = 0.428), enterobacteria (P = 0.0884), Clostridium butyricum (P = 0.122), or bacteroides (P = 0.55) throughout the large gut.
TABLE 3.
Quantitation of bacterial populations in the terminal ileum and different regions of the large intestine using real-time PCR
| Organism | Log10 16S rRNA gene copies/mg mucosal tissuee
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Terminal ileum (n = 6)
|
Ascending colon (n = 8)
|
Transverse colon (n = 8)
|
Descending colon (n = 4)
|
|||||||||
| Range | Mean | SD | Range | Mean | SD | Range | Mean | SD | Range | Mean | SD | |
| Total eubacteria | 7.9-9.5 | 9.1a,b,c | 0.6 | 6.8-7.9 | 7.4a | 0.4 | 6.9-8.0 | 7.3b | 0.4 | 6.9-8.1 | 7.5c | 0.5 |
| Bacteroides | 5.8-6.4 | 6.0 | 0.2 | 6.1-6.4 | 6.3 | 0.1 | 6.1-6.7 | 6.3 | 0.2 | 6.1-6.4 | 6.3 | 0.1 |
| Bifidobacteria | 4.4-4.8 | 4.7a | 0.2 | 4.9-5.9 | 5.3a | 0.4 | 4.4-5.4 | 4.9 | 0.3 | 4.7-6.2 | 5.2 | 0.7 |
| Desulfovibrios | 4.0-4.6 | 4.3 | 0.2 | 3.8-4.6 | 4.2 | 0.3 | 3.8-6.1 | 4.3 | 0.8 | 3.6-5.1 | 4.3 | 0.6 |
| Lactobacilli | 2.7-6.4 | 5.2a | 1.5 | 2.8-5.7 | 4.5a,d | 1.2 | 5.0-6.2 | 5.7 | 0.4 | 5.7-6.2 | 6.0d | 0.2 |
| Enterobacteria | 4.1-7.4 | 5.7 | 1.3 | 5.1-6.6 | 5.5 | 0.5 | 4.3-6.5 | 5.7 | 0.7 | 4.5-6.2 | 5.4 | 0.8 |
| Peptostreptococcus anaerobius | 3.8-6.4 | 4.8 | 1.0 | 3.0-6.5 | 4.0 | 1.1 | 2.9-5.8 | 3.9 | 0.9 | 3.4-4.2 | 3.7 | 0.4 |
| Clostridium clostridioforme | 6.1-6.5 | 6.2 | 0.2 | 6.1-6.7 | 6.4 | 0.2 | 6.0-6.5 | 6.2 | 0.2 | 6.0-7.2 | 6.5 | 0.5 |
| Clostridium butyricum | 0-3.6 | 2.2 | 1.7 | 2.7-3.2 | 2.9 | 0.2 | 0-3.4 | 2.0 | 1.6 | 0-3.3 | 1.5 | 1.7 |
| Eubacterium rectale | 4.2-7.1 | 5.7 | 1.1 | 5.8-7.1 | 6.5d | 0.4 | 4.2-6.4 | 5.4 | 0.8 | 5.0-7.4 | 6.0d | 1.1 |
| Faecalibacterium prausnitzii | 3.7-5.6 | 4.7a | 0.7 | 5.2-6.6 | 6.0a | 0.6 | 3.9-6.4 | 5.2 | 0.9 | 4.3-7.4 | 6.0 | 1.3 |
| Enterococcus faecalis | 0-4.2 | 2.6a,b | 2.0 | 4.0-4.7 | 4.3a | 0.2 | 4.1-5.1 | 4.5b | 0.3 | 0-4.5 | 3.2 | 2.2 |
A significant difference (P < 0.5) was found between populations in the terminal ileum and proximal colon.
A significant difference (P < 0.5) was found between populations in the terminal ileum and transverse colon.
A significant difference (P < 0.5) was found between populations in the terminal ileum and distal colon.
A significant difference (P < 0.5) was found between populations in the proximal and distal colon.
No significant differences were found between populations in the proximal and transverse colon or between populations in the transverse and distal colon.
CLSM of tissue samples.
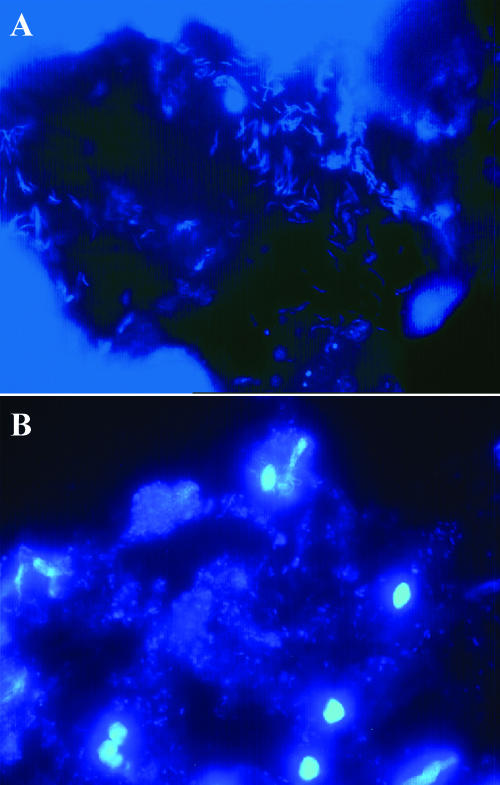
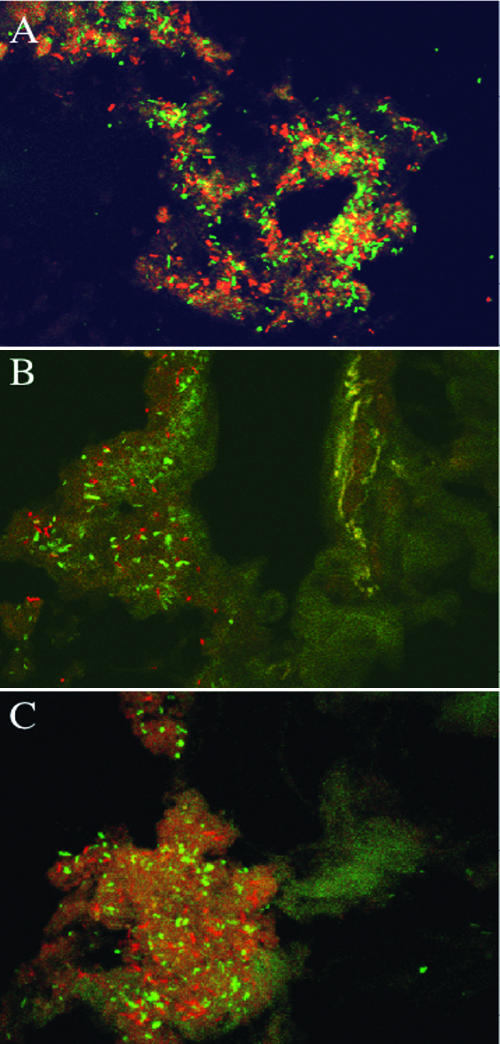
CLSM, in combination with 16S rRNA oligonucleotide probes, was used to visualize the spatial organization of communities on the tissue samples. The majority of the bacteria found by using real-time PCR and DGGE were visualized with the range of fluorescent probes used. Figure 2 shows transverse sections of mucosal tissue from the terminal ileum (Fig. 2A) and ascending colon (Fig. 2B) treated with the DNA stain DAPI for visualization of the total mucosal communities. In Fig. 2A, helical bacteria can be seen attached in large numbers to the mucus layer lining a crypt in the ileum. In contrast, in Fig. 2B, the presence of a substantial, heterogeneous mixture of organisms can be seen in the mucin layer of the descending colon. Figure 3 shows bacteria in mucin layers of transverse sections of biopsy samples from the ascending, transverse, and descending colon, respectively. Figure 3A was stained with a bacteroides-specific, fluorescently labeled (Cy3) 16S rRNA oligonucleotide probe (Bac 303) and with a bifidobacterium-specific 16S rRNA oligonucleotide probe with an FITC fluorophore (Bif 164). Figure 3B and C were both stained in combination with a Cy3-labeled probe (Erec 482) specific for the E. rectale/C. coccoides group and an FITC-labeled atopobium cluster probe (Ato 291). In Fig. 3A, large numbers of bacteroides and bifidobacteria can be seen attached to the mucus layer in the ascending colon, while in Fig. 3B and C, members of both the E. rectale group and the atopobium cluster were found to be present in the loosely attached mucin layer of the transverse and descending colon.
FIG. 2.
CLSM micrographs (magnification, ×60) of transverse sections of healthy gut mucosa, stained with DAPI, showing the occurrence of helical bacteria in the mucus layer lining a crypt and individual bacterial cells in the mucus layer of an ileum sample (A) and a heterogeneous assembly of microorganisms in the mucus layer of a tissue sample from the ascending colon (B).
FIG. 3.
CLSM fluorescent light micrographs (magnification, ×60). (A) Healthy rectal mucosa from the ascending colon stained with a Bacteroides genus-specific 16S rRNA probe labeled with Cy3 and with a Bifidobacterium genus-specific 16S rRNA oligonucleotide probe labeled with FITC. (B and C) Transverse sections of biopsy samples from the transverse colon (B) and descending colon (C), stained with the E. rectale/C. coccoides group probe (Cy3) and an atopobium cluster probe labeled with FITC. The bacteria can be seen to be present throughout the mucosae in the samples, growing in discrete colonies and in assemblages.
DISCUSSION
In the majority of previous studies on mucosal communities in humans, tissues were taken from colons that had been cleaned out with strong bowel cleansers or laxatives prior to sampling (8, 22, 40, 46, 49), which may change the composition of mucosal populations. In the present study, samples were taken from patients undergoing emergency surgery required for resection of the colon, which involved no prophylactic antibiotic treatment or bowel evacuation before the operation. The samples were obtained from parts of the gut remote from any diseased areas, and in all cases, histopathology reports confirmed the presence of localized disease and clear margins between healthy and diseased tissue. Two patients were diagnosed with colonic carcinoma and two with localized diverticulitis. The mean age of the patients in the study was 68 years in males and 65 in females, reflecting an aging cohort. This is because colonic pathologies requiring bowel resection are more common in this age group and are relatively rare in the population below age 50.
Although DGGE analysis has previously been used in studies to produce fingerprints of colonic mucosal samples, to date, few investigations have combined DGGE, cluster analysis, and sequencing with quantitative PCR and FISH in a relatively large subject group.
The DGGE profiles of bacterial communities in the ascending, transverse, and descending colon of the different individuals were shown to be similar to each other, which contrasts with previous studies reporting a high degree of consortial conservation along the length of the colon but marked interindividual variation (21, 22, 46, 49).
Quantitative analysis with real-time PCR using universal primers indicated that the terminal ileum had higher bacterial cell densities than the colon and that overall bacterial numbers were generally similar within the ascending, transverse, and descending colon. This may be due to higher numbers of unidentified bacteria in the terminal ileum, since no differences were found with genus- and species-specific primers. At the genus level, bifidobacteria were more prominent in the colon as a whole than in the terminal ileum, whereas lactobacilli were more dominant further down the large intestine (transverse and descending colon). However, bacteroides, enterobacteria, and dissimilatory sulfate-reducing bacteria were distributed uniformly in the large bowel and terminal ileum. This observation is in agreement with the results of work published by Zoetendal et al. (49). At the species level, variations in the numbers of some organisms in different parts of the large gut could be seen. Enterococcus faecalis occurred in higher numbers in the colon than in the ileum, while E. rectale, F. prausnitzii, and C. clostridioforme were more important in the proximal bowel (ascending and descending colon). However, no significant differences were seen for P. anaerobius or C. butyricum numbers in different parts of the large intestine. In summary, therefore, at both the genus and species level, changing bacterial cell densities in mucosa-associated biofilms were seen at various sites in the large gut. This may be of some health significance, because differences in these communities could contribute to the pathogenesis of some large intestine diseases. For example, over half of large bowel cancers occur in the descending colon and distal large gut (4), while the rectum is always involved in ulcerative colitis (34).
Although a number of molecularly based techniques are now available for studying bacterial populations colonizing the bowel wall, PCR-DGGE, real-time PCR, and FISH in combination provide a useful assessment of microbiological diversity in qualitative and quantitative terms. An advantage of DGGE over other qualitative techniques is that, in principle, any amplifiable target sequence that is above the detection threshold can be identified (37). DGGE is therefore not biased towards those phyla for which probes can be designed, which by definition will normally be limited to those expected to be important in the target community. However, PCR-DGGE has a number of limitations in that bias can be introduced during PCR amplification: DNA fragments with similar electrophoretic mobilities may migrate to the same position, while the relative concentrations of PCR products do not necessarily reflect the composition of the community under study (17, 30, 37). Real-time PCR provides a more specific, sensitive, and reproducible measure of the initial amount of template sample and is preferable to other forms of quantitative PCR, which only detect the final amount of amplified product. With real-time PCR, fluorescence emission is monitored during the reaction as an indicator of amplicon production during each PCR cycle, as opposed to endpoint detection by conventional quantitative PCR methods (10). A drawback of the technique is that its usefulness is dependent on the availability of specific PCR primer sets, so that investigators can only find what they are looking for in a sample.
Due to the presence of microcolonies in mucosal biofilms, many of the bacteria cannot be reliably quantitated in situ using microscopic techniques. However, FISH methods have the advantage of showing cell localization and providing an identification of groups of bacteria directly on the mucosal surface. Hence, a combination of techniques that allows fingerprinting, quantitation, and visualization enhances studies of mucosa-associated bacterial populations in the gut.
Microscopy using CLSM and DGGE using a range of 16S rRNA probes which covered the major groups of bacteria found on the gut mucosa showed that the organisms were present as heterogeneous assemblages in the mucosal layer, being present as individual colonies or dispersed in the biofilms. Many of the bacteria quantitated by using real-time PCR were visualized with the fluorescent probes. Members of the group of organisms in the atopobium cluster, which includes C. aerofaciens, were also visualized in tissues; however, they were not detected by DGGE or the real-time PCR primer sets used. Using FISH, these organisms were previously shown to be a major constituent of the fecal microbiota of formula-fed infants (15), and primers have now been developed allowing their quantification in fecal material (35).
It was of note that some of the ileal biopsies harbored large numbers of helical bacteria (Fig. 2A). These organisms were not detected in colonic tissues and were not seen by using the range of FISH probes or detected by DGGE. The presence of these helical organisms and of members of the atopobium cluster warrants further study, and these organisms may account for some of the differences in bacterial numbers detected in the ileum.
In conclusion, it has been demonstrated that using a combination of culture-independent techniques, i.e., PCR-DGGE, real-time PCR, and FISH, allows the identification and quantitation of different populations of bacteria in mucosal biofilms in the human digestive tract. Whether differences in mucosal communities can be related to the etiology of gut disease now needs to be investigated. Further work is also needed to determine how epithelial biofilm communities interact with host immunity and metabolic processes in the mucosa.
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council.
Footnotes
Published ahead of print on 21 September 2007.
REFERENCES
- 1.Akkermans, A. D. L., E. G. Zoetendal, C. F. Favier, H. G. H. J. Heilig, W. M. Akkermans-van Vliet, and W. M. de Vos. 2000. Temperature and denaturing gradient gel electrophoresis analysis of 16S rRNA from human fecal samples. Biosci. Microflora 19:93-98. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chrisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 2004. Cancer Statistics registrations; registrations of cancer diagnosed in 2001, England. Series MB1, no. 32. Office for National Statistics, London, United Kingdom.
- 5.Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. T. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal water by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Child, M. W., A. Kennedy, A. W. Walker, B. Bahrami, S. Macfarlane, and G. T. Macfarlane. 2006. Studies on the effect of system retention time on bacterial populations colonizing a three-stage continuous culture model of the human large gut using FISH techniques. FEMS Microbiol. Ecol. 55:299-310. [DOI] [PubMed] [Google Scholar]
- 8.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2000. Diversity of the human intestinal microbial flora. Science 10:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finegold, S. M., H. R. Attebery, and V. L. Sutter. 1974. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am. J. Clin. Nutr. 27:1456-1469. [DOI] [PubMed] [Google Scholar]
- 10.Fite, A., G. T. Macfarlane, J. H. Cummings, M. J. Hopkins, S. C. Kong, E. Furrie, and S. Macfarlane. 2004. Identification and quantitation of mucosal and faecal desulfovibrios using real-time PCR. Gut 53:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces quantified by fluorescence in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasche, C., J. Scholmerich, J. Brynskov, G. d'Haens, S. B. Hanauer, E. J. Irvine, D. P. Jewell, D. Rachmilewitz, D. B. Sachar, W. J. Sandborn, and L. R. Sutherland. 2000. A simple classification of Crohn's disease: report of the working party for the World Congress of Gastroenterology, Vienna 1998. Inflamm. Bowel Dis. 6:8-15. [DOI] [PubMed] [Google Scholar]
- 13.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmsen, H. J. M., A. C. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins, M. J., G. T. Macfarlane, E. Furrie, A. Fite, and S. Macfarlane. 2005. Characterisation of intestinal bacteria in infant stools using real-time PCR and Northern hybridisation analyses. FEMS Microbiol. Ecol. 54:77-85. [DOI] [PubMed] [Google Scholar]
- 16.Horie, H., K. Kanazawa, M. Okada, S. Narushima, K. Itoh, and A. Terada. 1999. Effects of intestinal bacteria on the development of colonic neoplasm: an experimental study. Eur. J. Cancer Prev. 8:237-245. [DOI] [PubMed] [Google Scholar]
- 17.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes, R., and I. R. Rowland. 2003. Nutritional and microbial modulation of carcinogenesis, p. 208-220. In R. Fuller and G. Perdigon (ed.), Gut flora, nutrition, immunity and health. Blackwell Publishing, Oxford, United Kingdom.
- 19.Kok, R. G., A. de Waal, F. Schut, G. W. Welling, G. Weenk, and K. J. Hellingwerf. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledder, R. G., P. Gilbert, S. A. Huws, L. Aarons, M. P. Ashley, P. S. Hull, and A. J. McBain. 2007. Molecular analysis of the subgingival microbiota in health and disease. Appl. Environ. Microbiol. 73:516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepage, P., P. Seksik, M. Sutren, M. F. de la Cochetiere, R. Jian, P. Marteau, and J. Dore. 2005. Biodiversity of the mucosa-associated microbiota is stable along the digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11:473-480. [DOI] [PubMed] [Google Scholar]
- 23.Levett, P. N. 1991. Anaerobic microbiology. Oxford University Press, Oxford, United Kingdom.
- 24.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 25.Linskens, R. K., X. W. Huijsdens, P. H. Savelkoul, C. M. Vandenbroucke-Grauls, and S. G. Meuwissen. 2001. The bacterial flora in inflammatory bowel disease: current insights in pathogenesis and the influence of antibiotics and probiotics. Scand. J. Gastroenterol. 234:29-40. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane, G. T., and A. J. McBain. 1999. The human colonic microbiota, p. 1-25. In G. R. Gibson and M. B. Roberfroid (ed.), Colonic microbiota, nutrition and health. Kluwer Academic Publishers, Doordrecht, The Netherlands.
- 27.Macfarlane, G. T., G. R. Gibson, and J. H. Cummings. 1992. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72:57-64. [DOI] [PubMed] [Google Scholar]
- 28.Macfarlane, S., E. Furrie, A. Kennedy, J. H. Cummings, and G. T. Macfarlane. 2005. Mucosal bacteria in ulcerative colitis. Br. J. Nutr. 93:S67-S72. [DOI] [PubMed] [Google Scholar]
- 29.Macfarlane, S., E. Furrie, J. H. Cummings, and G. T. Macfarlane. 2004. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 38:1690-1699. [DOI] [PubMed] [Google Scholar]
- 30.Macfarlane, S., and G. T. Macfarlane. 2004. Bacterial diversity in the human gut. Adv. Appl. Microbiol. 54:261-289. [DOI] [PubMed] [Google Scholar]
- 31.Macfarlane, S., M. J. Hopkins, and G. T. Macfarlane. 2000. Bacterial growth and metabolism on surfaces in the large intestine. Microb. Ecol. Health Dis. 2:64-72. [Google Scholar]
- 32.Malinen, E., A. Kassinen, T. Rinttila, and A. Palva. 2003. Comparison of real-time PCR with SYBR green 1 or 5′-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology 149:269-277. [DOI] [PubMed] [Google Scholar]
- 33.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 34.Maratka, Z. 1986. Pathogenesis and aetiology of inflammatory bowel disease. p. 29-65. In F. T. de Dombal, J. Myrer, I. A. D. Bouchier, and G. Watkinson (ed.), Inflammatory bowel disease. Some international data and reflections. Oxford Medical Publications, Oxford, United Kingdom.
- 35.Matsuki, T., K. Wartanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, W. E., E. P. Cato, and L. V. Holdeman. 1978. Special problems associated with the isolation and identification of intestinal bacteria in fecal flora studies. Am. J. Clin. Nutr. 31:1450-1455. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer, G., C. Brinkhoff, U. Nübel, C. Santegoeds, H. Schafer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis, (DGGE) in microbial ecology, p. 1-23. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3. Kluwer Academic Publishers, Doordrecht, The Netherlands. [Google Scholar]
- 38.Muyzer, G., and K. Smalla. 1998. Application of denaturant gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 39.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primer set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 40.Poxton, I. R., R. Brown, A. Sawyer, and A. Ferguson. 1997. Mucosa-associated flora of the human colon. J. Med. Microbiol. 46:85-91. [DOI] [PubMed] [Google Scholar]
- 41.Riggio, M. P., and A. Lennon. 2002. Development of a PCR assay specific for Peptostreptococcus anaerobius. J. Med. Microbiol. 51:1097-1101. [DOI] [PubMed] [Google Scholar]
- 42.Schwiertz, A., U. Lehmann, G. Jacobisch, and M. Blaut. 2002. Influence of resistant starch on the SCFA production and cell counts of butyrate-producing Eubacterium spp. in the human intestine. J. Appl. Microbiol. 93:157-162. [DOI] [PubMed] [Google Scholar]
- 43.Simon, G. L., and S. L. Gorbach. 1984. Intestinal flora in health and disease. Gastroenterology 86:174-193. [PubMed] [Google Scholar]
- 44.Tannock, G. W., K. Munro, K. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturant gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, M., S. Ahrne, B. Jeppsson, and G. Molin. 2005. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54:219-231. [DOI] [PubMed] [Google Scholar]
- 47.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]