Abstract
Bacteria were isolated from the rhizosphere and from inside the roots and stems of sugarcane plants grown in the field in Brazil. Endophytic bacteria were found in both the roots and the stems of sugarcane plants, with a significantly higher density in the roots. Many of the cultivated endophytic bacteria were shown to produce the plant growth hormone indoleacetic acid, and this trait was more frequently found among bacteria from the stem. 16S rRNA gene sequence analysis revealed that the selected isolates of the endophytic bacterial community of sugarcane belong to the genera of Burkholderia, Pantoea, Pseudomonas, and Microbacterium. Bacterial isolates belonging to the genus Burkholderia were the most predominant among the endophytic bacteria. Many of the Burkholderia isolates produced the antifungal metabolite pyrrolnitrin, and all were able to grow at 37°C. Phylogenetic analyses of the 16S rRNA gene and recA gene sequences indicated that the endophytic Burkholderia isolates from sugarcane are closely related to clinical isolates of the Burkholderia cepacia complex and clustered with B. cenocepacia (gv. III) isolates from cystic fibrosis patients. These results suggest that isolates of the B. cepacia complex are an integral part of the endophytic bacterial community of sugarcane in Brazil and reinforce the hypothesis that plant-associated environments may act as a niche for putative opportunistic human pathogenic bacteria.
Brazil is one of the world's largest sugarcane producers and has considerable influence over the international sugar market. Brazilian sugarcane is primarily used to produce sugar and alcohol. Production has increased over time to approximately 26 million tons of sugar and 16 million m3 of alcohol in 2006. Although ethanol has been used as an alternative source of fuel in Brazil since 1980, it is currently receiving worldwide interest as a biofuel to replace, at least in part, gasoline, thereby contributing to a reduction in carbon emissions (33). Consequently, sustaining and enhancing the growth and yield of sugarcane have become a major focus of research. The growth and performance of sugarcane in the field are adversely affected by a number of abiotic and biotic factors, including a wide range of fungal and bacterial diseases. Pokkah boeng, caused by the fungus Fusarium moniliforme, is one of the most widespread diseases and may cause serious yield losses in commercial sugarcane plantings (68). F. moniliforme can be disseminated horizontally by airborne spores or crop debris and vertically through seed pieces. Current control strategies involve the use of resistant varieties and fungicide applications. The efficacy of both control measures, however, is limited, and there is an increasing need for novel and environmentally sound strategies to control this and other diseases of sugarcane.
The overall goal of this study was to isolate and characterize beneficial bacteria that are intimately associated with sugarcane and have the potential to control pathogens and to promote the growth and yield of sugarcane. Among the plant-associated microorganisms, endophytic bacteria are regarded as a largely untapped resource for the discovery of isolates with novel antifungal and plant growth-promoting traits (52, 71, 77, 78). For several crops, endophytic bacteria have shown beneficial effects on plant growth and health, and the main modes of action described are nitrogen fixation, production of phytohormones and antifungal compounds, and induced systemic resistance (17, 38, 44, 52, 60, 71, 76). For sugarcane, studies on endophytic bacteria have focused on Gluconacetobacter diazotrophicus and its abilities to fix nitrogen (9, 24) and to inhibit Xanthomonas albilineans, the causal agent of leaf scald disease (8). However, information on the antifungal traits and different mechanisms involved in plant growth promotion, beyond nitrogen fixation, is limited. Moreover, no comprehensive analysis of the frequency, diversity, and activities of endophytic bacterial communities of sugarcane has, to our knowledge, been performed to date.
In this study, bacteria were isolated from the rhizosphere and from inside the roots and stems of sugarcane plants grown in the field in Brazil. We specifically focused on delineation of the cultivated endophytic bacterial isolates and characterization of their salient metabolic features. The diversity and putative identities of the cultivated endophytic bacteria were determined by genomic DNA fingerprinting by 16S rRNA gene and recA (13) sequence analyses. Antifungal activities toward F. moniliforme and specific metabolites produced by the endophytic bacteria were determined in bioassays, PCR-based analysis, and chromatography (thin-layer chromatography [TLC] and reverse-phase high-performance liquid chromatography [RP-HPLC]).
MATERIALS AND METHODS
Bacterial isolation, media, and growth conditions.
The bacterial strains used in this study were isolated from sugarcane plants (cv. SP80-1842) grown for 3 months in an experimental field located in Piracicaba, Brazil (22°41′S 47°33′W). Rhizosphere and root endophytic bacteria were isolated from root segments collected at a depth of 5 to 15 cm from the stem base. The root segments were washed in tap water to remove adhering soil particles. Rhizosphere bacteria were isolated by vigorously shaking 2 g of root segments in 200 ml of PBS buffer (140 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.4]) for 1 h, followed by dilution plating on tryptic soy agar (TSA; Difco, Le Pont de Claix, France) supplemented with 50 mg ml−1 of the fungicide imazalil (Agricur, Brazil) and incubation at 28°C. For the isolation of root endophytes, washed root segments were surface sterilized by sequential washing in 70% ethanol for 1 min, sodium hypochlorite (2%, vol/vol) for 3 min, and 70% ethanol for 30 s and two rinses with ample sterilized distilled water. Surface sterilization was verified by plating aliquots (100 μl) of the sterile distilled water used in the final rinse onto TSA plates. The surface-sterilized root segments were ground in a mortar with a pestle by using 2 ml of PBS buffer, and 100-μl aliquots were plated on TSA supplemented with 50 mg ml−1 imazalil. Plates were incubated at 28°C, and the colonies were counted after 10 days. Stem endophytic bacteria were isolated from stem sections sampled at 15 and 30 cm from the stem base. Segments of 5 cm were washed and surface sterilized as described for the roots. Afterwards, the epidermis was aseptically removed and plant extract was collected from the stem section by centrifugation at 3,000 × g for 10 min (46). Aliquots of 100 μl were plated onto TSA supplemented with imazalil and incubated at 28°C, and colonies were counted after 10 days. All bacterial isolates were stored at −80°C in 40% (vol/vol) glycerol.
Genotypic characterization of endophytic bacteria.
To determine the genotypic diversity of the isolated bacteria, BOX-PCR analysis with the BOX-1AR primer (42) was performed according to the protocol described in detail by Rademaker et al. (67). Amplifications were carried out with an MJ Research PTC-200 thermocycler. Six-microliter aliquots of the PCR products were loaded onto a 25-cm 1.5% agarose gel in 1× TBE buffer (90 mM Tris-borate, 2 mM EDTA [pH 8.0]) and run at 40 V for 16 h. The gel was stained with ethidium bromide and visualized under UV light. Image analysis was performed with GelCompar II software (Applied Maths, Sint-Martens-Latem, Belgium).
Bacterial isolates were sent to Macrogen Inc. (Seoul, Korea) to have the nearly full-length 16S rRNA gene (∼1,500 bp) sequenced according to company specifications. Sequencing of the gene for RecA recombinase (recA, 1,000 bp) was performed by BaseClear (Leiden, The Netherlands) with primers BCR1 and BCR2 (54). Sequences were examined and edited with the BioEdit Sequence Alignment Editor (35). The Basic Local Alignment Tool (BLAST) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and sequence match of the Ribosomal Database Project (http://rdp.cme.msu.edu) were used to search for similar known sequences. These sequences were aligned with CLUSTAL_X version 1.8 (80). Phylogenetic and molecular evolutionary analyses were conducted with MEGA version 3.1 (45). Neighbor-joining consensus trees were obtained with the Kimura two-parameter substitution model (41) and bootstrap test (28). In the 16S rRNA gene and recA gene analyses conducted, we focused primarily on comparisons and best hits with sequences of well-characterized (type) strains from the Burkholderia cepacia complex experimental strain panel (16, 56).
Phenotypic characterization of bacterial strains.
The activity of the bacterial strains toward hyphal growth of F. moniliforme strain CBS 218.76 was performed on one-fifth-strength potato dextrose agar medium (pH 7.0) (Oxoid, Hampshire, England) as described previously (22). Indoleacetic acid (IAA) production was determined by using a modification of the qualitative method developed by Bric et al. (11). Six strains were spot inoculated onto 1/10-strength TSA plates amended with 5 mM l-tryptophan (Sigma, Steinheim, Germany) and overlaid with a nitrocellulose membrane (Amersham, Buckinghamshire, England). After incubation for 24 h at 25°C, the membrane was removed from the plate and treated with Salkowski reagent (2% [wt/vol] 0.5 M FeCl3 in 35% perchloric acid) for 15 min at room temperature. IAA-producing bacteria were identified by a red halo on the membrane surrounding the bacterial colony.
Protease activity was evaluated on agar medium containing (per liter) 15 g of skim milk powder, 4 g of blood agar base, and 0.5 g of yeast extract (Oxoid, Hampshire, England). Inoculated plates were incubated at 27°C for 24 h, and extracellular protease activity was detected by the presence of a halo surrounding the bacterial colony.
Detection and analysis of antibiotic genes and metabolites.
PCR-based screening for genes involved in the biosynthesis of pyrrolnitrin, 2,4-diacetylphloroglucinol, phenazines, and pyoluteorin was performed as described by De Souza and Raaijmakers (21). Special attention was given to the prnD gene, which is involved in catalyzing the oxidation of the amino group of aminopyrrolnitrin to a nitro group to form pyrrolnitrin (reviewed in reference 21). The 1,092-bp prnD gene was partially (666 bp) sequenced by BaseClear (Leiden, The Netherlands) with primers PRND1 and PRND2 (21). Pyrrolnitrin detection by TLC was performed as described by De Souza and Raaijmakers (19). Pure pyrrolnitrin was used as a reference and was kindly provided by M. A. de Waard of the Laboratory of Phytopathology, Wageningen University, Wageningen, The Netherlands. For detection of the antibiotics 2,4-diacetylphloroglucinol, phenazines, and pyoluteorin, RP-HPLC linked to a photodiode array spectrophotometer was used by following protocols described previously (10).
To determine the activity of metabolites in cell extracts of cultures of the bacterial isolates, TLC plates were sprayed with a spore suspension (106 spores ml−1) of Cladosporium cucumerinum in potato dextrose broth (Difco, Le Pont de Claix, France) and incubated at 25°C for 5 days.
Site-directed mutagenesis.
To assess the role of pyrrolnitrin in fungal growth inhibition, a pyrrolnitrin-deficient mutant of endophytic strain ESS4 was obtained by site-directed mutagenesis of the prnD gene. A 396-bp fragment of the prnD gene of strain ESS4 was amplified with primers PRNhF (5′-TTTTTAAGCTTTGCACTTCGCGTTCGAGAC-3′) and PRNxR (5′-TTTTTTCTAGACGAGATGAGCATGTGCATG-3′), which contain restriction sites for HindIII and XbaI (underlined), respectively. These primers were designed on the basis of an alignment of the GenBank sequences AF161183, AF161186, CP000150, CP000125, CP000441, PFU74493, and CP000076. After digestion with HindIII and XbaI, the 396-bp fragment was cloned into plasmid pMP5285 (43) and transferred into a spontaneous rifampin-resistant derivative of strain ESS4 by triparental mating. Putative prnD mutants were selected on Pseudomonas Agar F medium (Difco, Le Pont de Claix, France) containing 200 μg ml−1 rifampin and 1,200 μg ml−1 kanamycin. The disruption of the prnD gene was confirmed by PCR amplification with primers PRND2 (21) and P5285R (5′-CCAGATAGCCCAGTAGCTG-3′). Primer PRND2 targets a specific sequence in the prnD gene, whereas primer P5285R anneals to the plasmid flanking region cointegrated into the prnD gene. The amplified 600-bp PCR product was checked for the presence of the HindIII restriction site. The deficiency in pyrrolnitrin production was determined by TLC analysis of cell extracts of the putative prnD mutant culture.
Nucleotide sequence accession numbers.
The sequences obtained in this study were deposited in GenBank (for the accession numbers, see Table 2).
TABLE 2.
Genetic characteristics of endophytic bacteria isolated from sugarcane plantsa
| Strain | Plant tissue | 16S rRNA gene
|
recA gene
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. | Hit in NCBI database | Genomovar | % Identity | Reference strain | Accession no. | Hit in NCBI database | Genomovar | % Identity | Reference strain | ||
| ESS21 | Stem | EF602568 | Microbacterium testaceum | 99 | SE017 | ||||||
| ESR94 | Root | EF602564 | P. fluorescens | 99 | ATCC 13525 | ||||||
| ESS10 | Stem | EF602554 | Pantoea stewartii | 99 | LMG2715 | ||||||
| ESS12 | Stem | EF602555 | Pantoea ananatis | 99 | LMG20106 | ||||||
| ESS29 | Stem | EF602556 | Pantoea ananatis | 99 | LMG20106 | ||||||
| ESS2 | Stem | EF602551 | B. cenocepacia | III | 100 | AU1054 | EF602569 | B. cenocepacia | III | 100 | CEP511 |
| ESS9 | Stem | EF602553 | B. cenocepacia | III | 100 | AU1054 | EF602573 | B. cenocepacia | III | 100 | CEP511 |
| ESR100 | Root | EF602566 | B. cenocepacia | III | 100 | HI2424 | EF602581 | B. cenocepacia | III | 99 | HI2424 |
| ESR99 | Root | EF602565 | B. cenocepacia | III | 100 | HI2424 | EF602571 | B. cenocepacia | III | 99 | HI2424 |
| ESR60 | Root | EF602557 | B. cenocepacia | III | 99 | HI2424 | EF602574 | B. cenocepacia | III | 99 | CEP511 |
| ESR108 | Root | EF602567 | B. cenocepacia | III | 99 | LGM12615 | EF602572 | B. cenocepacia | III | 99 | MRL10 |
| ESS4 | Stem | EF602552 | B. cenocepacia | III | 99 | LGM12615 | EF602576 | B. cenocepacia | III | 98 | MRL10 |
| ESR90 | Root | EF602562 | B. cepacia | I | 99 | ATCC 25416 | EF602580 | B. cenocepacia | III | 98 | MRL10 |
| ESR85 | Root | EF602560 | B. cepacia | I | 99 | ATCC 25416 | EF602579 | B. cenocepacia | III | 98 | MRL10 |
| ESR92 | Root | EF602563 | B. cepacia | I | 99 | ATCC 25416 | EF602570 | B. cenocepacia | III | 98 | MRL10 |
| ESR63 | Root | EF602558 | B. cepacia | I | 99 | ATCC 25416 | EF602578 | B. cepacia | I | 98 | ATCC 17759 |
| ESR73 | Root | EF602559 | B. cenocepacia | III | 99 | LGM12615 | EF602577 | B. cepacia | K | 99 | R9929 |
| ESR87 | Root | EF602561 | B. cepacia | I | 99 | ATCC 25416 | EF602575 | B. cepacia | - | 97 | J503 |
The isolates represent 18 genotypic groups based on BOX-PCR analysis.
RESULTS AND DISCUSSION
Frequency, diversity, and identity of endophytic bacteria from sugarcane.
The population density of culturable bacteria in the rhizosphere of sugarcane was approximately 20-fold higher than the density of bacterial endophytes in the roots (Table 1). In the stem, bacterial density was several orders of magnitude lower than in the roots and rhizosphere of sugarcane plants (Table 1). This distribution pattern, in which lower plant parts harbor higher frequencies of endophytes, confirms and extends results reported previously for corn (31), pea (25), and soybean (44) plants. A total of 154 bacterial isolates was randomly selected and subjected to BOX-PCR analysis to assess the genotypic diversity. On the basis of a cutoff similarity value of 70% (58, 59), 24 distinct genotypic groups were defined among the stem endophytes, 23 groups were defined among the root endophytes, and 25 groups were defined among the rhizosphere isolates (Table 1). Dual-culture plate assays revealed that 20, 39, and 25% of the bacterial isolates selected from the stem, root, and rhizosphere, respectively, significantly inhibited the hyphal growth of F. moniliforme, the causal agent of Pokkah boeng disease of sugarcane (Table 1).
TABLE 1.
Frequency, diversity, and activity of bacteria isolated from the rhizosphere, roots, and stems of sugarcane plants grown in the field in Brazil
| Isolate source | Mean density (CFU g−1 [fresh wt]) ± SEMa | No. of randomly selected isolates | No. of BOX-PCR groups | No. (%) of isolates showing:
|
|
|---|---|---|---|---|---|
| Inhibition of F. moniliforme growth | IAA production | ||||
| Stem endophytes | (2.87 ± 1.33) × 102 | 49 | 24 | 10 (20) | 36 (74) |
| Root endophytes | (3.27 ± 2.01) × 106 | 44 | 23 | 17 (39) | 20 (46) |
| Rhizosphere | (7.90 ± 2.43) × 107 | 61 | 25 | 15 (25) | 8 (13) |
Mean values of 18 replicates are shown.
On the basis of BOX-PCR and antagonistic activity against F. moniliforme, 18 endophytic bacterial isolates, consisting of 11 isolates from the root and 7 from the stem, were selected for 16S rRNA gene sequencing. Analysis of the 16S rRNA gene sequences revealed that most (13 out of 18) of the endophytic isolates belong to the genus Burkholderia, whereas the other 5 isolates were classified as Pantoea, Pseudomonas, and Microbacterium isolates (Table 2). Previous studies have shown that isolates belonging to these bacterial genera promote the growth of different crops and are able to control specific plant diseases. For example, the rice endophyte Pantoea agglomerans YS19 showed nitrogen-fixing activity in vitro, produced four different phytohormones, including IAA, and promoted plant growth (29). Recently, a new endophytic nitrogen-fixing Pantoea sp. was isolated from sugarcane plants in Cuba (53), although its role in plant growth promotion has not been established. Endophytic Pseudomonas fluorescens strains Endo2 and Endo35 induced systemic disease protection against Macrophomina phaseolina, the causal agent of dry root rot of black gram (40). Also, endophytic Burkholderia species have received increased interest in the last years because of their capacity to fix nitrogen and their potential to promote plant growth. Burkholderia species have been isolated from several crops (5, 79), including sugarcane (61), rice (23), wine plants (18), onion (75), maize, and coffee (26, 27).
Genotypic characterization of endophytic Burkholderia isolates.
The high frequency of Burkholderia species among the endophytic bacteria from sugarcane plants and their strong growth-inhibitory activity against F. moniliforme (Table 3) make these isolates potential candidates for the control of Pokkah boeng disease. Previous studies have shown that strains belonging to the genus Burkholderia are effective biocontrol agents (6) and represent the active ingredient in several commercially available biocontrol products, including Deny (Helena Chemicals, Memphis, TN) and Intercept (Soil Technologies Corp., Fairfield, IA). However, many strains of different species belonging to the so-called B. cepacia complex may act as opportunistic pathogens of humans, especially of cystic fibrosis (CF) patients and immunocompromised individuals (34). Strains belonging to the B. cepacia complex are resistant to a wide range of therapeutic antibiotics, and in spite of stringent control policies, the number of infections in hospitals caused by B. cepacia complex strains has not decreased (15, 72). The B. cepacia complex consists of at least nine discrete genomic species (described previously as genomovars I through IX) (16, 56), and all of these genomovars contain strains that are able to infect humans or to colonize plants (3). Therefore, accurate identification of the Burkholderia isolates from sugarcane plants is a crucial step toward further development of these isolates for biological control of Pokkah boeng and other sugarcane diseases.
TABLE 3.
Phenotypic characteristics of endophytic bacteria isolated from sugarcane plants
| Strain | Antagonism toward F. moniliformea | IAA | Protease | Pyrrolnitrin
|
Growth at 37°C | |
|---|---|---|---|---|---|---|
| PCR | TLC | |||||
| ESS21 | − | +b | + | − | − | + |
| ESR94 | ++ | + | + | + | + | − |
| ESS10 | + | + | − | − | − | + |
| ESS12 | + | + | − | − | − | + |
| ESS29 | + | + | − | − | − | + |
| ESS2 | +++ | + | + | − | − | + |
| ESS9 | +++ | + | + | − | − | + |
| ESR100 | ++ | −b | + | − | − | + |
| ESR99 | +++ | + | + | − | − | + |
| ESR60 | +++ | + | + | − | − | + |
| ESR108 | +++ | − | + | + | + | + |
| ESS4 | +++ | + | + | + | + | + |
| ESR90 | +++ | − | + | + | + | + |
| ESR85 | +++ | − | + | + | + | + |
| ESR92 | +++ | − | + | + | + | + |
| ESR63 | +++ | − | + | + | + | + |
| ESR73 | ++ | − | + | + | + | + |
| ESR87 | +++ | + | + | + | + | + |
−, no inhibition; +, zone of inhibition of 1 to 3 mm; ++, zone of inhibition of 4 to 8 mm; +++, zone of inhibition of 9 to 13 mm.
+, positive; −, negative.
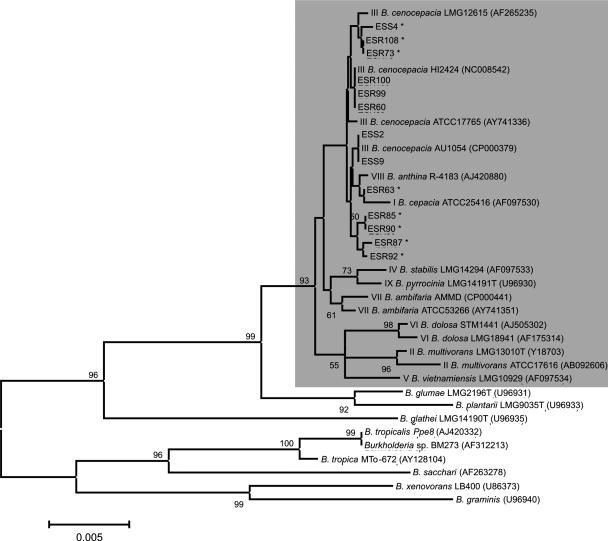
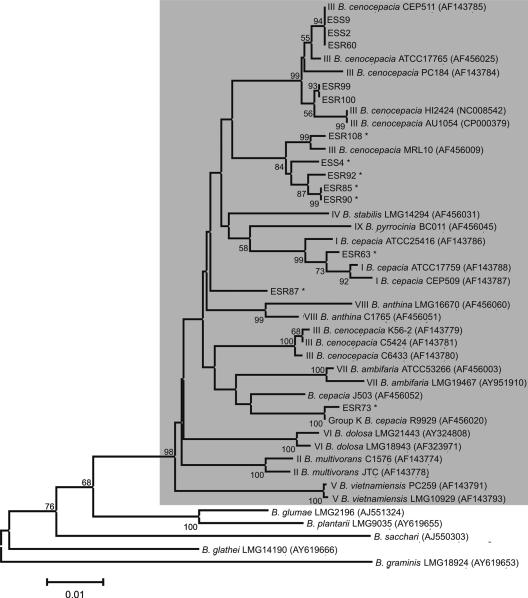
Over the past years, a range of molecular markers have been tested to characterize Burkholderia species from environmental and clinical origins (7, 12, 30, 50, 73). In most of these studies, including a recent multilocus sequence typing (MLST) study (4, 19), the 16S rRNA gene and the recA gene were shown to be good markers by which to characterize and discriminate Burkholderia isolates (2, 13, 64, 69, 81). In the present study, sequencing and phylogenetic analyses were performed on both the 16S rRNA gene (∼1,500 bp) and recA (∼1,000 bp) of all 13 endophytic Burkholderia isolates (Table 2; Fig. 1 and 2). The results showed that the endophytic Burkholderia isolates from sugarcane plants cluster closely with the B. cepacia complex type strains and distantly from other Burkholderia species that do not belong to the B. cepacia complex (Table 2; Fig. 1 and 2). On the basis of a BLAST analysis of both 16S rRNA gene and recA gene sequences, sugarcane isolates ESS2, ESS9, ESR100, ESR99, and ESR60 are closely related (identities ranging from 99 to 100%) to well-characterized B. cenocepacia (gv. III) strains, including strains isolated from CF patients and fully sequenced strains (Table 2). B. cenocepacia (gv. III), as well as B. multivorans (gv. II) and B. dolosa (gv. VI), mostly harbors strains from clinical sources (3). B. cenocepacia strain AU1054 was isolated from the blood of a CF patient and was, on the basis of various genotyping methods, characterized as a representative of the B. cenocepacia PHDC clonal lineage (14). On the basis of 16S rRNA gene analysis, the sugarcane endophytic isolates ESS2 and ESS9 were 100% identical to strain AU1054 (Table 2; Fig. 1). Consistent with the 16S rRNA gene analysis, also the analysis of the recA gene sequences showed that isolates ESS2 and ESS9 presented 100% identity to B. cenocepacia CEP511 (Table 2; Fig. 2), a type strain recovered from a CF patient (Sydney, Australia) and representative of an epidemic strain spread among several patients (55). Similarly, endophytic isolates ESR100, ESR99, and ESR60 were closely (identities, 99 to 100%) related to B. cenocepacia strain HI2424 on the basis of 16S rRNA gene and, in part, recA sequence analyses (Table 2; Fig. 1 and 2). B. cenocepacia strain HI2424 was initially obtained from an onion field but was later also characterized as being another representative of the B. cenocepacia PHDC clonal lineage, which causes extensive infection in CF patients (51). On the basis of analyses of the recA gene sequences, isolates ESR108, ESS4, ESR92, ESR85, and ESR90 clustered and are most closely related to strain MRL-10, an environmental strain classified by MLST (4) as B. cenocepacia (gv. IIIE) (Table 2; Fig. 2). Considering the recA gene phylogeny, isolate ESR87 was less related to a strain of the B. cepacia complex, revealing 97% identity with strain J503 (Table 2) and only 96% identity with B. cenocepacia strain CEP511 (Fig. 2). On the basis of 16S rRNA gene phylogenetic analysis, the endophytic Burkholderia isolates obtained in this study clustered distantly from B. sacchari (AF263278) and B. tropicalis (AJ420332), two isolates obtained previously in Brazil from sugarcane plantation soil and from a sugarcane stem, respectively (Fig. 1). Also on the basis of phylogenetic analysis of the recA sequences, the endophytic Burkholderia isolates obtained in this study clustered distantly from B. sacchari (AJ550303) isolated from sugarcane plantation soil in Brazil (Fig. 2). The presence of B. cenocepacia III-like isolates in endophytic bacterial populations of sugarcane plants supports and further extends the conclusions of previous studies (2, 20, 65) that B. cenocepacia gv. III and lineage III-B occur in natural habitats, which may be a niche for opportunistic human pathogenic bacteria. In vitro assays further showed that all 13 endophytic Burkholderia isolates from sugarcane plants were able to grow at 37°C (Table 3). It should be emphasized, however, that a complete MLST analysis should be undertaken, as well as infection studies to categorically match the sugarcane isolates with well-characterized clinical isolates of the B. cepacia complex. Nevertheless, 16S rRNA gene and recA gene phylogenetic analyses suggest that the endophytic Burkholderia strains isolated from sugarcane are closely related to well-characterized B. cepacia complex type strains. To date, most studies on the occurrence of B. cepacia complex strains in natural habitats have focused on the maize rhizosphere (19, 20, 63, 65). The results of our study indicate, for the first time, that B. cepacia complex-related isolates are an integral part of the endophytic bacterial community of sugarcane.
FIG. 1.
Neighbor-joining tree of 16S rRNA gene sequences from endophytic bacterial isolates from sugarcane plants. ESS stands for endophyte sugarcane stem, and ESR stands for endophyte sugarcane root. Sequences were obtained from databases, and the accession numbers are in parentheses. The shaded box contains (type) strains belonging to the B. cepacia complex and includes the endophytic isolates from sugarcane plants. The roman numerals in front of the species and strain names are B. cepacia complex genomovar numbers. The isolates from sugarcane plants that produce pyrrolnitrin are indicated by asterisks. The Kimura two-parameter substitution model was used, and the nodes are supported by 1,000 bootstrap replications. Bootstrap values above 50% and the genetic distance scale are shown.
FIG. 2.
Neighbor-joining tree of recA gene sequences from endophytic bacterial isolates from sugarcane plants. ESS stands for endophyte sugarcane stem, and ESR stands for endophyte sugarcane root. Sequences were obtained from databases, and the accession numbers are in parentheses. The shaded box contains (type) strains belonging to the B. cepacia complex and includes the endophytic isolates from sugarcane plants. The roman numerals in front of the species and strain names are B. cepacia complex genomovar numbers. The isolates from sugarcane plants that produce pyrrolnitrin are indicated by asterisks. The Kimura two-parameter substitution model was used, and the nodes are supported by 1,000 bootstrap replications. Bootstrap values above 50% and the genetic distance scale are shown.
Biochemical characterization of endophytic Burkholderia isolates.
Many of the 154 randomly selected bacterial isolates from the rhizosphere, roots, and stems of sugarcane plants produced the plant growth hormone IAA (Table 1). Most of the IAA-producing isolates were found among the stem endophytes, followed by root endophytes and rhizosphere isolates (Table 1). IAA production was detected in 6 of the 13 endophytic Burkholderia isolates, 5 of which were closely related to B. cenocepacia gv. III (Table 2; Fig. 1 and 2). The observation that IAA production is more prevalent among the bacterial endophytes than among rhizosphere bacteria of sugarcane is consistent with the results of a previous study performed with soybean (44). Although the effect and role of IAA production by endophytic bacteria in growth promotion of sugarcane need to be investigated, this trait was considered one of the major mechanisms involved in maize growth promotion by the rhizosphere bacteria Gluconacetobacter azotocaptans DS1, Pseudomonas putida CQ179, and Azospirillum lipoferum N7 under greenhouse conditions (60). Several studies have reported that endophytic microbial communities originate from the soil and rhizosphere (25, 47). The observation that the frequency of IAA-producing bacteria is higher in the stems than in the rhizosphere of sugarcane plants suggests that the plant selects for endophytic bacteria with this trait or that IAA-producing bacteria harbor other traits that allow them to more effectively reach and establish themselves in the inner plant tissue.
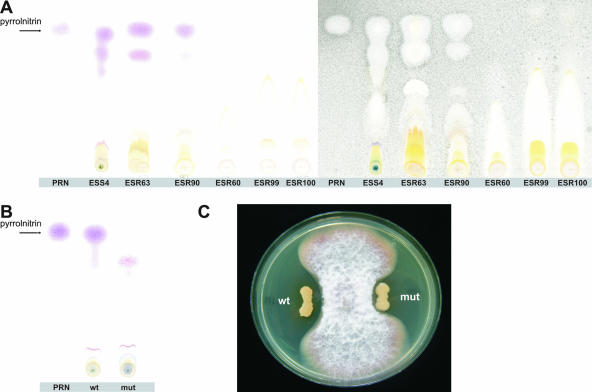
To identify metabolites involved in the growth inhibition of F. moniliforme, the bacterial endophytes were subjected to a range of tests, including enzymatic assays, PCR-based detection of antibiotic genes, and chromatography (TLC and RP-HPLC). All of the bacterial endophytes, except the three Pantoea species, produced extracellular proteases (Table 3). None of the bacterial endophytes produced hydrogen cyanide, a volatile compound found in several antagonistic Pseudomonas species (70, 82), and all of the isolates were negative for chitinase and biosurfactant production (data not shown). PCR-based detection revealed that none of the 18 endophytic isolates harbored specific genes involved in the biosynthesis of the antibiotics 2,4-diacetylphloroglucinol, phenazines, and pyoluteorin; the lack of production of these three antibiotics was confirmed by RP-HPLC analysis of culture extracts of the 18 endophytic isolates (data not shown). PCR-based detection of prnD, one of the four genes involved in pyrrolnitrin biosynthesis (48), and TLC analyses revealed that the endophyte Pseudomonas sp. strain ESR94 and eight of the endophytic Burkholderia isolates produced the antibiotic pyrrolnitrin (Table 3; Fig. 3A). On the basis of 16S rRNA gene and recA gene sequences, the pyrrolnitrin-producing endophytic Burkholderia isolates clustered (Fig. 1 and 2), suggesting that prn genes may be used as an additional molecular marker to further distinguish between strains and species within the B. cepacia complex. Results of the study by Seo and Tsuchiya (74) showed that among a collection of B. cepacia complex strains, 74% were positive in PCR-based detection of the prnC gene; most of the prnC-positive isolates belonged to B. cepacia gv. I, comprising isolates from both clinical and environmental sources. On the basis of their results, Seo and Tsuchiya (74) suggested that restriction fragment length polymorphism analysis of the prnC gene could be used as a marker to distinguish Burkholderia species. However, on the basis of in silico analysis of fully sequenced B. cepacia complex genomes included in the 16S rRNA gene and recA phylogeny in this study (Fig. 1 and 2), the type strains B. cenocepacia HI2424 and AU1054, isolated from onion and from the blood of a CF patient, respectively, do not contain the pyrrolnitrin gene cluster. Furthermore, strains B. ambifaria AMMD, isolated from healthy pea plants, and B. cenocepacia PC184, isolated from CF patients, do contain the genes for pyrrolnitrin biosynthesis. This apparent lack of correlation between the origin of the Burkholderia isolates and the presence of prn genes, suggests that this trait is probably of minor value as an additional marker to distinguish between clinical and environmental Burkholderia isolates.
FIG. 3.
TLC analysis of pyrrolnitrin production by endophytic bacteria from sugarcane plants and growth inhibition of F. moniliforme by Burkholderia sp. strain ESS4 and its prnD mutant. (A) Pyrrolnitrin is detected in cell extracts of cultures of the endophytic bacterial strains ESS4, ESR63, and ESR90 from sugarcane plants by spraying TLC plates with Ehrlich's reagent. Pure pyrrolnitrin (PRN; Rf is 0.48) was included as a control and is indicated by an arrow. TLC plates were subsequently sprayed with a spore suspension of the fungus Cladosporium cucumerinum, and the white zones indicate areas where fungal growth was inhibited. (B) TLC analysis of cell extracts from cultures of wild-type (wt) strain ESS4 and its prnD mutant (mut); pyrrolnitrin was detected in the wild-type strain but not in the mutant. (C) Dual-culture assay with Burkholderia strain ESS4 (wt), its prnD mutant (mut), and the fungus F. moniliforme (inoculated in the center).
Role of pyrrolnitrin in growth inhibition of F. moniliforme by endophytic Burkholderia.
Pyrrolnitrin [3-chloro-4-(2′-nitro-3′-chlorophenyl)pyrrole] is an antifungal metabolite with activities against a range of plant and human pathogenic fungi (37, 49). Pyrrolnitrin was originally described for Burkholderia pyrrocinia (1) and later for Pseudomonas, Enterobacter, Myxococcus, and Serratia (36). Pyrrolnitrin plays a key role in the antifungal activities of specific Pseudomonas strains (49, 66) and has been postulated to play a role in the natural suppressiveness of some soils against root rot caused by Rhizoctonia solani (32). To further clarify the relative importance of pyrrolnitrin production by the endophytic Burkholderia isolates in growth inhibition of F. moniliforme, site-directed mutagenesis of the prnD gene was performed with Burkholderia isolate ESS4. The partial prnD gene sequence of strain ESS4 obtained in this study (accession number EF602550) was 94% identical to that of type strain B. cepacia AMMD (CP000441). Site-directed mutagenesis resulted in 6 (out of 15) putative prnD mutants. Disruption of the prnD gene in these mutants was confirmed by PCR analysis, and loss of pyrrolnitrin production was confirmed in TLC analysis (Fig. 3B). Assays with F. moniliforme showed that the prnD mutant was slightly less effective at inhibition of hyphal growth than its parental strain (Fig. 3C), suggesting that most of the activity against F. moniliforme is related to other, as yet unknown, metabolites. Burkholderia species are well known for their production of a range of antifungal metabolites, including phenylacetic acid, hydrocinnamic acid, 4-hydroxyphenylacetic acid, and 4-hydroxyphenylacetate methyl ester (39, 57). The role of these compounds, if present in the isolated strains, in the control of fungal diseases and endophytic colonization of sugarcane remains to be investigated.
Conclusion.
The endophytic bacterial community associated with sugarcane harbors multiple genera with potential for plant growth promotion and disease control. On the basis of 16S rRNA gene and recA sequence and phylogenetic analyses, most of the selected endophytic isolates obtained from sugarcane plants belonged to the genus Burkholderia and are closely related to clinical isolates of the B. cepacia complex. These results exemplify and support the conclusions of Parke and Gurian-Sherman (62) that accurate identification and classification of antagonistic and environmental Burkholderia isolates are essential before application to seeds and planting material for disease control and plant growth promotion. The relatively high frequency of different B. cepacia complex species associated with sugarcane plants reinforces the hypothesis of Berg et al. (5) that plant-associated environments may act as a reservoir for opportunistic human pathogenic bacteria. To categorically match the endophytic Burkholderia isolates from sugarcane with well-characterized clinical isolates of the B. cepacia complex, a complete MLST analysis should be conducted, followed by infection and epidemiological studies.
Acknowledgments
We thank the Brazilian institutions CAPES (National Council of Research, Brazil, proc. no. 3771/05-9) and FAPESP (Foundation for Research Assistance, São Paulo State, Brazil, proc. no. 03/01436-5) for financial support. We thank CTC (Sugarcane Technology Center) for conducting the field experiments in Brazil.
We also thank Marco Kruijt for valuable assistance and suggestions during the course of this work.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Arima, K., H. Imanaka, M. Kousaka, A. Fukuda, and G. Tamura. 1964. Pyrrolnitrin, a new antibiotic substance, produced by Pseudomonas. Agric. Biol. Chem. 28:575-576. [Google Scholar]
- 2.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, A., E. Mahenthiralingam, P. Drevinek, P. Vandamme, J. R. Govan, D. J. Waine, J. J. LiPuma, L. Chiarini, C. Dalmastri, D. A. Henry, D. P. Speert, D. Honeybourne, M. C. J. Maiden, and C. G. Dowson. 2007. Environmental Burkholderia cepacia complex isolates in human infections. Emerg. Infect. Dis. 13:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, A., E. Mahenthiralingam, K. M. Thickett, D. Honeybourne, M. C. J. Maiden, J. R. Govan, D. P. Speert, J. J. LiPuma, P. Vandamme, and C. G. Dowson. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, G., L. Eberl, and A. Hartmann. 2005. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7:1673-1685. [DOI] [PubMed] [Google Scholar]
- 6.Bevivino, A., C. Dalmastri, S. Tabacchioni, and L. Chiarini. 2000. Efficacy of Burkholderia cepacia MCI 7 in disease suppression and growth promotion of maize. Biol. Fertil. Soils 31:225-231. [Google Scholar]
- 7.Bevivino, A., V. Peggion, L. Chiarini, S. Tabacchioni, C. Cantale, and C. Dalmastri. 2005. Effect of Fusarium verticillioides on maize-root-associated Burkholderia cenocepacia populations. Res. Microbiol. 156:974-983. [DOI] [PubMed] [Google Scholar]
- 8.Blanco, Y., D. Pinon, M. E. Legaz, and C. Vicente. 2005. Antagonism of Gluconacetobacter diazotrophicus (a sugarcane endosymbiont) against Xanthomonas albilineans (pathogen) studied in alginate-immobilized sugarcane stalk tissues. J. Biosci. Bioeng. 99:366-371. [DOI] [PubMed] [Google Scholar]
- 9.Boddey, R. M., S. Urquiaga, B. J. R. Alves, and V. Reis. 2003. Endophytic nitrogen fixation in sugarcane: present knowledge and future applications. Plant Soil 252:139-149. [Google Scholar]
- 10.Bonsall, R. F., D. M. Weller, and L. S. Thomashow. 1997. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 63:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bric, J. M., R. M. Bostock, and S. E. Silverstone. 1991. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 57:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campana, S., G. Taccetti, N. Ravenni, F. Favari, L. Cariani, A. Sciacca, D. Savoia, A. Collura, E. Fiscarelli, G. De Intinis, M. Busetti, A. Cipolloni, A. d'Aprile, E. Provenzano, I. Collebrusco, P. Frontini, G. Stassi, M. Trancassini, D. Tovagliari, A. Lavitola, C. J. Doherty, T. Coenye, J. R. W. Govan, and P. Vandamme. 2005. Transmission of Burkholderia cepacia complex: evidence for new epidemic clones infecting cystic fibrosis patients in Italy. J. Clin. Microbiol. 43:5136-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, C. H., D. E. Stead, and R. H. A. Coutts. 2003. Development of a species-specific recA-based PCR test for Burkholderia fungorum. FEMS Microbiol. Lett. 224:133-138. [DOI] [PubMed] [Google Scholar]
- 14.Chen, J. S., K. A. Witzmann, T. Spilker, R. J. Fink, and J. J. LiPuma. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643-649. [DOI] [PubMed] [Google Scholar]
- 15.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 16.Coenye, T., P. Vandamme, J. J. LiPuma, J. R. W. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 41:2797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Compant, S., B. Duffy, J. Nowak, C. Clement, and E. A. Barka. 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71:4951-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compant, S., B. Reiter, A. Sessitsch, J. Nowak, C. Clement, and E. Ait Barka. 2004. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71:1685-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalmastri, C., A. Baldwin, S. Tabacchioni, A. Bevivino, E. Mahenthiralingam, L. Chiarini, and C. Dowson. 2007. Investigating Burkholderia cepacia complex populations recovered from Italian maize rhizosphere by multilocus sequence typing. Environ. Microbiol. 9:1632-1639. [DOI] [PubMed] [Google Scholar]
- 20.Dalmastri, C., L. Pirone, S. Tabacchioni, A. Bevivino, and L. Chiarini. 2005. Efficacy of species-specific recA PCR tests in the identification of Burkholderia cepacia complex environmental isolates. FEMS Microbiol. Lett. 246:39-45. [DOI] [PubMed] [Google Scholar]
- 21.De Souza, J. T., and J. M. Raaijmakers. 2003. Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol. Ecol. 43:21-34. [DOI] [PubMed] [Google Scholar]
- 22.De Souza, J. T., D. M. Weller, and J. M. Raaijmakers. 2003. Frequency, diversity, and activity of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in Dutch take-all decline soils. Phytopathology 93:54-63. [DOI] [PubMed] [Google Scholar]
- 23.Divan Baldani, V. L., J. I. Baldani, and J. Döbereiner. 2000. Inoculation of rice plants with the endophytic diazotrophs Herbaspirillum seropedicae and Burkholderia spp. Biol. Fertil. Soils 30:485-491. [Google Scholar]
- 24.Dong, Z., M. Heydrich, K. Bernard, and M. E. McCully. 1995. Further evidence that the N2-fixing endophytic bacterium from the intercellular spaces of sugarcane stems is Acetobacter diazotrophicus. Appl. Environ. Microbiol. 61:1843-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elvira-Recuenco, M., and J. W. L. van Vuurde. 2000. Natural incidence of endophytic bacteria in pea cultivars under field conditions. Can. J. Microbiol. 46:1036-1041. [DOI] [PubMed] [Google Scholar]
- 26.Estrada-De Los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Estrada-De Los Santos, P., P. Mavingui, B. Cournoyer, F. Fontaine, J. Balandreau, and J. Caballero-Mellado. 2002. A N2-fixing endophytic Burkholderia sp. associated with maize plants cultivated in Mexico. Can. J. Microbiol. 48:285-294. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 29.Feng, Y., D. Shen, and W. Song. 2006. Rice endophyte Pantoea agglomerans YS 19 promotes host plant growth and affects allocations of host photosynthates. J. Appl. Microbiol. 100:938-945. [DOI] [PubMed] [Google Scholar]
- 30.Fiore, A., S. Laevens, A. Bevivino, C. Dalmastri, S. Tabacchioni, P. Vandamme, and L. Chiarini. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 31.Fisher, P. J., O. Petrini, and H. M. L. Scott. 1992. The distribution of some fungal and bacterial endophytes in maize (Zea mays L.). New Phytol. 122:299-305. [DOI] [PubMed] [Google Scholar]
- 32.Garbeva, P., K. Voesenek, and J. D. Elsas. 2004. Quantitative detection and diversity of the pyrrolnitrin biosynthetic locus in soil under different treatments. Soil Biol. Biochem. 36:1453-1463. [Google Scholar]
- 33.Goldemberg, J. 2006. The ethanol program in Brazil. Environ. Res. Lett. (Online.) doi: 10.1088/1748-9326/1/1/014008. [DOI]
- 34.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Mol. Biol. Res. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 36.Hammer, P. E., D. S. Hill, S. T. Lam, K. H. Van Pee, and J. M. Ligon. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63:2147-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill, D. S., J. I. Stein, N. R. Torkewitz, A. M. Morse, C. R. Howell, J. P. Pachlatko, J. O. Becker, and J. M. Ligon. 1994. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl. Environ. Microbiol. 60:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iniguez, A. L., Y. Dong, and E. W. Triplett. 2004. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol. Plant-Microbe Interact. 17:1078-1085. [DOI] [PubMed] [Google Scholar]
- 39.Kang, Y., R. Carlson, W. Tharpe, and M. A. Schell. 1998. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 64:3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karthikeyan, M., R. Bhaskaran, K. Radhika, S. Mathiyazhagan, V. Jayakumar, R. Sandosskumar, and R. Velazhahan. 2005. Endophytic Pseudomonas fluorescens Endo2 and Endo35 induce resistance in black gram (Vigna mungo L. Hepper) to the pathogen Macrophomina phaseolina. J. Plant Interact. 1:135-143. [Google Scholar]
- 41.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 42.Koeuth, T., J. Versalovic, and J. R. Lupski. 1995. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res. 5:408. [DOI] [PubMed] [Google Scholar]
- 43.Kuiper, I., G. V. Bloemberg, S. Noreen, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 2001. Increased uptake of putrescine in the rhizosphere inhibits competitive root colonization by Pseudomonas fluorescens strain WCS 365. Mol. Plant-Microbe Interact. 14:1096-1104. [DOI] [PubMed] [Google Scholar]
- 44.Kuklinsky-Sobral, J., W. L. Araujo, R. Mendes, I. O. Geraldi, A. A. Pizzirani-Kleiner, and J. L. Azevedo. 2004. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 6:1244-1251. [DOI] [PubMed] [Google Scholar]
- 45.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 46.Lacava, P. T., W. L. Araujo, J. Marcon, W. Maccheroni, and J. L. Azevedo. 2004. Interaction between endophytic bacteria from citrus plants and the phytopathogenic bacteria Xylella fastidiosa, causal agent of citrus-variegated chlorosis. Lett. Appl. Microbiol. 39:55-59. [DOI] [PubMed] [Google Scholar]
- 47.Lamb, T. G., D. W. Tonkyn, and D. A. Kluepfel. 1996. Movement of Pseudomonas aureofaciens from the rhizosphere to aerial plant tissue. Can. J. Microbiol. 42:1112-1120. [Google Scholar]
- 48.Lee, J. K., E. L. Ang, and H. Zhao. 2006. Probing the substrate specificity of aminopyrrolnitrin oxygenase (PrnD) by mutational analysis. J. Bacteriol. 188:6179-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ligon, J. M., D. S. Hill, P. E. Hammer, N. R. Torkewitz, D. H. Hans-Joachim, and K. H. van Pée. 2000. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag. Sci. 56:688-695. [Google Scholar]
- 50.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 52.Lodewyckx, C., J. Vangronsveld, F. Porteous, E. R. B. Moore, S. Taghavi, M. Mezgeay, and D. der Lelie. 2002. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 21:583-606. [Google Scholar]
- 53.Loiret, F. G., E. Ortega, D. Kleiner, P. Ortega-Rodes, R. Rodes, and Z. Dong. 2004. A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J. Appl. Microbiol. 97:504-511. [DOI] [PubMed] [Google Scholar]
- 54.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao, S., S. J. Lee, H. Hwangbo, Y. W. Kim, K. H. Park, G. S. Cha, R. D. Park, and K. Y. Kim. 2006. Isolation and characterization of antifungal substances from Burkholderia sp. culture broth. Curr. Microbiol. 53:358-364. [DOI] [PubMed] [Google Scholar]
- 58.Mavrodi, O. V., B. B. M. Gardener, D. V. Mavrodi, R. F. Bonsall, D. M. Weller, and L. S. Thomashow. 2001. Genetic diversity of phlD from 2, 4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology 91:35-43. [DOI] [PubMed] [Google Scholar]
- 59.McSpadden Gardener, B. B., K. L. Schroeder, S. E. Kalloger, J. M. Raaijmakers, L. S. Thomashow, and D. M. Weller. 2000. Genotypic and phenotypic diversity of phlD-containing Pseudomonas strains isolated from the rhizosphere of wheat. Appl. Environ. Microbiol. 66:1939-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehnaz, S., and G. Lazarovits. 2006. Inoculation effects of Pseudomonas putida, Gluconacetobacter azotocaptans, and Azospirillum lipoferum on corn plant growth under greenhouse conditions. Microb. Ecol. 51:326-335. [DOI] [PubMed] [Google Scholar]
- 61.Oliveira, A. L. M., S. Urquiaga, J. Döbereiner, and J. I. Baldani. 2002. The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil 242:205-215. [Google Scholar]
- 62.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 63.Payne, G. W., A. Ramette, H. L. Rose, A. J. Weightman, T. H. Jones, J. M. Tiedje, and E. Mahenthiralingam. 2006. Application of a recA gene-based identification approach to the maize rhizosphere reveals novel diversity in Burkholderia species. FEMS Microbiol. Lett. 259:126-132. [DOI] [PubMed] [Google Scholar]
- 64.Payne, G. W., P. Vandamme, S. H. Morgan, J. J. LiPuma, T. Coenye, A. J. Weightman, T. H. Jones, and E. Mahenthiralingam. 2005. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 71:3917-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pirone, L., L. Chiarini, C. Dalmastri, A. Bevivino, and S. Tabacchioni. 2005. Detection of cultured and uncultured Burkholderia cepacia complex bacteria naturally occurring in the maize rhizosphere. Environ. Microbiol. 7:1734-1742. [DOI] [PubMed] [Google Scholar]
- 66.Raaijmakers, J. M., M. Vlami, and J. T. de Souza. 2002. Antibiotic production by bacterial biocontrol agents. Antonie Leeuwenhoek 81:537-547. [DOI] [PubMed] [Google Scholar]
- 67.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1998. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.) Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 68.Raid, R. N. 2006. Pokkah boeng disease of sugarcane. Agronomy Department, University of Florida, Gainesville. http://edis.ifas.ufl.edu/pdffiles/SC/SC00400.pdf.
- 69.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramette, A., Y. Moenne-Loccoz, and G. Defago. 2003. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol. Ecol. 44:35-43. [DOI] [PubMed] [Google Scholar]
- 71.Rosenblueth, M., and E. Martínez-Romero. 2006. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 19:827-837. [DOI] [PubMed] [Google Scholar]
- 72.Saiman, L., and J. Siegel. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salles, J. F., F. A. De Souza, and J. D. van Elsas. 2002. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl. Environ. Microbiol. 68:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seo, S. T., and K. Tsuchiya. 2004. PCR-based identification and characterization of Burkholderia cepacia complex bacteria from clinical and environmental sources. Lett. Appl. Microbiol. 39:413-419. [DOI] [PubMed] [Google Scholar]
- 75.Sessitsch, A., T. Coenye, A. V. Sturz, P. Vandamme, E. A. Barka, J. F. Salles, J. D. Van Elsas, D. Faure, B. Reiter, and B. R. Glick. 2005. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 55:1187-1192. [DOI] [PubMed] [Google Scholar]
- 76.Sessitsch, A., J. G. Howieson, X. Perret, H. Antoun, and E. Martínez-Romero. 2002. Advances in Rhizobium research. Crit. Rev. Plant Sci. 21:323-378. [Google Scholar]
- 77.Strobel, G. A. 2003. Endophytes as sources of bioactive products. Microbes Infect. 5:535-544. [DOI] [PubMed] [Google Scholar]
- 78.Sturz, A. V., B. R. Christie, and J. Nowak. 2000. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19:1-30. [Google Scholar]
- 79.Tabacchioni, S., A. Bevivino, C. Dalmastri, and L. Chiarini. 2002. Burkholderia cepacia complex in the rhizosphere: a minireview. Ann. Microbiol. 52:103-117. [Google Scholar]
- 80.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanlaere, E., T. Coenye, E. Samyn, C. Van den Plas, J. Govan, F. De Baets, K. De Boeck, C. Knoop, and P. Vandamme. 2005. A novel strategy for the isolation and identification of environmental Burkholderia cepacia complex bacteria. FEMS Microbiol. Lett. 249:303-307. [DOI] [PubMed] [Google Scholar]
- 82.Voisard, C., C. Keel, D. Haas, and G. Defago. 1989. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]