Abstract
In this study, novel cellulosome chimeras exhibiting atypical geometries and binding modes, wherein the targeting and proximity functions were directly incorporated as integral parts of the enzyme components, were designed. Two pivotal cellulosomal enzymes (family 48 and 9 cellulases) were thus appended with an efficient cellulose-binding module (CBM) and an optional cohesin and/or dockerin. Compared to the parental enzymes, the chimeric cellulases exhibited improved activity on crystalline cellulose as opposed to their reduced activity on amorphous cellulose. Nevertheless, the various complexes assembled using these engineered enzymes were somewhat less active on crystalline cellulose than the conventional designer cellulosomes containing the parental enzymes. The diminished activity appeared to reflect the number of protein-protein interactions within a given complex, which presumably impeded the mobility of their catalytic modules. The presence of numerous CBMs in a given complex, however, also reduced their performance. Furthermore, a “covalent cellulosome” that combines in a single polypeptide chain a CBM, together with family 48 and family 9 catalytic modules, also exhibited reduced activity. This study also revealed that the cohesin-dockerin interaction may be reversible under specific conditions. Taken together, the data demonstrate that cellulosome components can be used to generate higher-order functional composites and suggest that enzyme mobility is a critical parameter for cellulosome efficiency.
The biological utilization of plant cell wall cellulose requires a number of cellulases and related enzymes to saccharify this biopolymer, which is considered to be the most abundant on Earth. Plant cell wall biomass thus represents an exceptional source of carbon and energy that can potentially be utilized as a low-cost renewable source of mixed sugars for fermentation to biofuels like ethanol. In order to effectively degrade these substrates, aerobic cellulolytic microorganisms commonly secrete copious amounts of complementary cellulases that act synergistically. In contrast, microorganisms that reside in cellulose-rich anaerobic biotopes produce an exceptionally diverse set of cellulases and related enzymes but in much smaller amounts than those produced by aerobic microorganisms. In most cases, these enzymes are gathered in large extracellular complexes called cellulosomes, which in addition to cellulases and hemicellulases, can also contain pectinases, chitinases, proteases, and protease inhibitors.
Since the discovery of these efficient cellulolytic complexes in 1983 (21), the combination of biochemical, biophysical, genomic, and proteomic approaches has been applied to several types of bacterial cellulosomes, which has led to a more generalized view of these intriguing complexes (for reviews, see references 1, 7, and 10). The same building blocks that fabricate cellulosomes are consistently observed, i.e., the cohesin modules of a scaffolding protein bind tightly to dockerin modules borne by the cellulosomal enzymes. Usually, within a given species, the interaction between complementary cohesins and dockerins (of the same type) is nonspecific (24, 39). Thus, a dockerin-bearing enzyme can bind to any of the matching cohesins harbored by the scaffoldin with similar affinity. Nevertheless, proteomic studies have shown that within the complex, some enzymes are abundant, whereas others are very minor components. Thus, the relative amount of the various catalytic subunits vary significantly within the complex (2, 41). The cellulosomes secreted by the mesophilic Clostridium cellulolyticum represents a relatively simple type of multienzyme complex. Its scaffoldin contains eight cohesins in addition to a cellulose-binding module (CBM), and it can thus bind up to eight different enzymes (24). Cohesins and dockerins, however, are not restricted to scaffoldins and enzymes, respectively, since scaffoldins that contain an appended catalytic and/or dockerin module have been reported (8, 9). Furthermore, cellulosomes exhibiting several interacting scaffoldins and more-complex organizations have been described previously (29, 37).
In contrast to the cellulosomal arrangement of enzymes, some hyperthermophilic anaerobic bacteria exhibit a different organization, in which multifunctional cellulases are secreted that comprise several cellulase catalytic modules within the same polypeptide chain. A combination of glycoside hydrolase 48 (GH48) and GH9 catalytic modules has been described in several reports for such bifunctional cellulases (34, 40). The naturally occurring cellulase systems exhibit a diverse variety of molecular architectures, in which the modular elements are mixed and matched in apparent accord with the demands of the given bacterium and its relationships to its particular substrate in its native ecosystem.
Recently, the species-specific characteristics of the cohesin-dockerin interaction was exploited to construct hybrid minicellulosomes that contained up to four different components (11, 12). These artificial “designer cellulosome” complexes comprised a chimeric scaffoldin, possessing an optional CBM and two or three cohesins of divergent specificity (derived from the scaffoldins of C. cellulolyticum, Clostridium thermocellum, and Ruminococcus flavefaciens), and three C. cellulolyticum cellulases, each bearing a dockerin complementary to one of the divergent cohesins. Thus, the two most prominent enzymes (the endoprocessive cellulases Cel48F and Cel9E) as well as less abundant cellulases (Cel5A, Cel8C, Cel9G, and Cel9M) of the cellulosomes produced by C. cellulolyticum were incorporated into the hybrid cellulosomes. These reports demonstrated that the binding of enzyme pairs from C. cellulolyticum onto the hybrid scaffoldins generally induces a substrate-targeting effect due to the scaffoldin-borne CBM which anchors the chimeric cellulosome at the surface of the substrate and a “proximity” effect which triggers additional synergy between the catalytic subunits. In addition, these studies also revealed that the most active complexes on crystalline cellulose contained a GH48 and a GH9 cellulase (C. cellulolyticum Cel48F and Cel9G, respectively) which therefore appear to play a critical role in cellulosome activity, especially on pure crystalline cellulose (12).
To date, the chimeric cellulosomes constructed have been modeled after the overall architecture of native cellulosomes produced by mesophilic clostridia. In the present report, we explored new molecular edifices composed of the same GH48 and GH9 cellulases but displaying divergent geometries. In some cases, the architecture of these novel fabrications was partly inspired by the cellulolytic systems of ruminal bacteria and hyperthermophilic bacteria. In others, the architecture was significantly different from any known bacterial system. The activity of the novel designer cellulosomes on crystalline cellulose was analyzed and compared to a “conventional” designer cellulosome composed of the corresponding parental enzymes. Finally, the relationships between activity, geometry, and conformational flexibility in bacterial cellulosomes are discussed.
MATERIALS AND METHODS
Plasmids and strains.
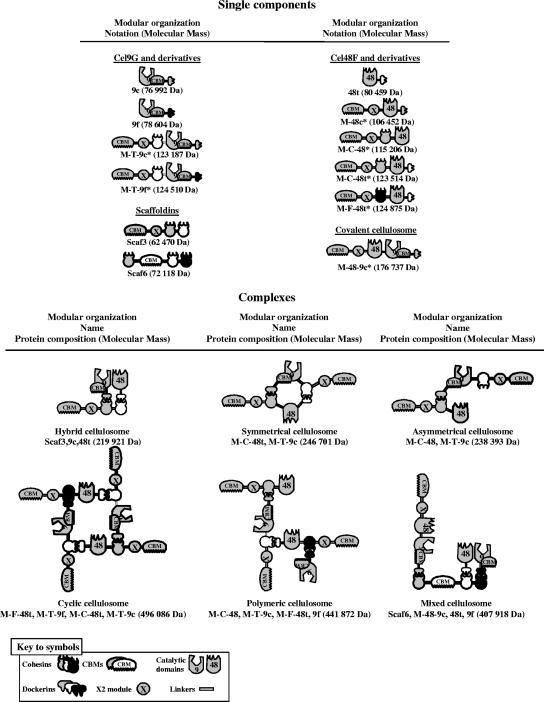
The proteins encoded by the various plasmids constructed in this study are summarized in Fig. 1 (nomenclature of enzymes shown in the legend to Fig. 1). The construction of pETGc (14), pETGf (12), pETFt (11), pETscaf3 (11), and pETscaf6 (12) encoding 9c, 9f, 48t, Scaf3, and Scaf6, respectively (Fig. 1), were described previously.
FIG. 1.
Schematic representations of the recombinant proteins and complexes used in this study. The source of the respective module (see the symbol key) is indicated as follows: gray (C. cellulolyticum), white (C. thermocellum), and black (R. flavefaciens). In the shorthand notation for the engineered enzymes, the numbers 9 and 48 refer to the corresponding GH family (GH9 and GH48) of the catalytic module; M designates the tandem CBM3a and X2 modules of the C. cellulolyticum scaffoldin CipC; uppercase letters C, F, and T indicate the source of the cohesin module, C. cellulolyticum, R. flavefaciens, and C. thermocellum, respectively; lowercase letters c, f, and t indicate the source of the dockerin module, C. cellulolyticum, R. flavefaciens, and C. thermocellum, respectively. The names symmetrical, asymmetrical, cyclic, and polymeric are intended both as descriptive terms of the artificial cellulosome-like geometries and to facilitate subsequent narratives in the text. The component compositions are shown schematically and in the shorthand notation are given below the individual elements of the figure. The hybrid and mixed cellulosomes both contain scaffoldins and are thus based on the native complexes. Novel proteins constructed in this study are indicated by an asterisk.
Construction of plasmid pET-M-48c.
To construct plasmid pET-M-48c encoding the hybrid enzyme M-48c (Fig. 1), the DNA encoding the CBM and the X2 module of the miniscaffoldin mini-CipC1 was amplified from pSOS952-cipC1 (26) using forward primer MF1 (5′-TTAGACCCATGGCAGGTACTGGCGTCGTATCAG-3′, with the NcoI site introduced shown in bold type) and reverse primer MF2 (5′-GGTACACCTTGTTTGCTGGACTTGAAACTTTAAGAGAATCGCCAGGGAT-3′, sequence matching with the 3′ extremity of the DNA encoding the X2 module is underlined). The DNA encoding the mature form of 48c was amplified from pSOS952-celF (vector provided by Anne Bélaïch, in which the two internal BamHI sites of the cel48F gene were erased by introducing silent mutations) and the forward and reverse primers MF3 (the 5′-ATCCCTGGCGATTCTCTTAAAGTTTCAAGTCCAGCAAACAAGGTGTACC-3′ sequence matching the 5′ extremity of the DNA encoding the mature form of 48c is underlined) and MF4 (5′-TTGGATCCCTAGTGGTGGTGGTGGTGGTGTTGGATAGAAAGAAGTGCTTT-3′, introducing at the 3′extremity of the gene, a BamHI site in bold type and six His codons underlined), respectively. The two resultant overlapping fragments (overlapping region in italics) were mixed, and a combined fragment was synthesized using primers MF1 and MF4. The fragment was cloned into NcoI-BamHI linearized pET9d (Novagen, Madison, WI), thereby generating pET-M-48c.
Construction of plasmid pET-M-48-9c.
The DNA encoding M-48 (protein M-48c without the C-terminal dockerin module) was amplified using forward primer MF1 and reverse primer FG1 (5′-TTTGGATCCTTTTTTACGCGTCAATAATTTTTCTGGACCTTG-3′), complementary to the DNA encoding the linker that connects the catalytic module of 48c and the dockerin (underlined) and introducing a MluI site (bold italics) and a BamHI site (bold). The amplified fragment was cloned in NcoI-BamHI-linearized pET9d, thereby generating pET-M-48. The DNA encoding enzyme 9c was amplified from pETGc using the primers FG2 (5′-AAAAAACGCGTGCAGGAACATATAACTATGGAG-3′) introducing a MluI site (bold italics) and FG3 (5′-AAAGGATCCCTAGTGGTGGTGGTGGTGGTGGCCTTGAGGTAATTGGGTGAT-3′) introducing a BamHI site (bold) and six His codons (underlined). The DNA synthesized was cloned in MluI-BamHI-linearized pET-M-48, thus generating pET-M-48-9c.
Construction of plasmid pET-M-C-48t.
The DNA encoding 48t was amplified from pET-Ft using primers CF1 (5′-caacctacaaaAGAACTCGAGTCAAGT CCTGCAAACAAGGTG-3′) and CF2 (3′-AAACTCGAGTTAGTGGTGGTGGTGGTGGTG-5′) introducing a XhoI site (bold) at both extremities of the coding sequence. The fragment was cloned in XhoI-linearized pETCip1X (11), resulting in pET-M-C-48t.
Construction of pET-M-C-48.
The DNA encoding the catalytic module of 48t was amplified from pET-Ft using the forward and reverse primers CF1 and CF3 (5′-TTTCTCGAGTGGACCTTGATCTGGGAAGAG-3′) introducing a XhoI site (bold) at both extremities of the fragment. The amplified DNA was cloned in XhoI-linearized pETCip1X, thereby generating pET-M-C-48.
Construction of pET-M-F-48t.
The DNA encoding the cohesin from Ruminococcus flavefaciens was amplified from pETCohrf (12) using the forward and reverse primers R1 (5′-GGCGATTCTCTTAAAGTTACAGTAGCTGGTGGTTTATCCGCTG-3′) and R2 (5′-CAATCCCTCGAGCTTAACAATGATAGCGCCATCAGTAAGAGT-3′, XhoI site in bold). The DNA encoding the X2 module in mini-CipC1 was amplified from pETCip1X using forward and reverse primers R3 (5′-TTAAATAGTGATGCAGGAAGTCTTCC-3′) and R4 (5′-CAGCGGATAAACCACCAGCTACTGTAACTTTAAGAGAATCGCC-3′), respectively. The two resultant overlapping fragments (overlapping region in italics) were mixed, and a combined fragment was synthesized using primers R2 and R3. The resulting fragment was cloned in BamHI-XhoI-linearized pETCip1X, thereby generating pETCipRfX. The pET-M-C-48t plasmid was digested by XhoI, and the fragment corresponding to the coding sequence of 48t was cloned in XhoI-linearized pETCipRfX, thus generating the pET-M-F-48t vector.
Construction of pET-M-T-9c.
The DNA encoding the cohesin 3 from Clostridium thermocellum was amplified from pETscaf3 using forward primer TG1 (5′-CCAAAAGATATCCCTGGCGATCCATCAACACAGCCTGTAACAACACC-3′) introducing an EcoRV site (bold) and reverse primer TG2 (5′-GCTTCTCCATAGTTATAAGTACCTGCGATCCTATCTCCAACATTTACTCC-3′). The DNA encoding the beginning of the coding sequence of the cellulase Cel9G (9c) that contains a unique BglII site was amplified from pETGc using the forward TG3 (5′-GGAGTAAATGTTGGAGATAGGATCGCAGGTACTTATAACTATGGAGAAGC-3′) and reverse TG4 (5′-AAAAAGGCGCCCTAAGACGGTCTTTCCATCTGCAT-3′) primers introducing a NarI site (bold in TG4). The two resultant overlapping fragments (overlapping region in italics) were mixed, and a combined fragment was synthesized using primers TG1 and TG4. The fragment was cloned in EcoRV-NarI-linearized pSOS952-CipC1, thus generating pSOS952-CipT-G. The coding sequence of the modified miniscaffoldin was amplified from pSOS952-CipT-G using forward primer MTG1 (5′-GGAGATATACATATGGCAGGTACTGGCGTCGTATCAGTG-3′) introducing a NdeI site (bold) and the reverse primer TG4. The resulting fragment was digested with NdeI and BglII, whereas pETGc was digested with BglII and XhoI. Both fragments were purified and cloned in a NdeI-XhoI-linearized pET22b(+) (Novagen).
Construction of pET-M-T-9f.
The pETGf vector was digested with NcoI and XhoI, and the fragment corresponding to the 3′extremity of the gene was cloned in a NcoI-XhoI-linearized pET-M-T-9c, thus generating pET-M-T-9f.
Positive clones were verified by DNA sequencing. Escherichia coli BL21(DE3) strain (Novagen) was used as the production host.
Production and purification of recombinant proteins.
The production and purification of 9c, 9f, 48t, Scaf3, and Scaf6 were performed as previously described (12). E. coli BL21(DE3) strains overproducing M-T-9c, M-T-9f, M-48c, M-C-48, M-C-48t, M-F-48t, and M-48-9c were grown in toxin flasks at 37°C in Luria-Bertani medium supplemented with glycerol (12 g/liter) and the appropriate antibiotic until an A600 of 1.5 was reached. The culture was then cooled to 18°C (M-48c, M-C-48, M-C-48t, and M-F-48t) or 16°C (M-T-9c, M-T-9f, and M-48-9c), and isopropyl thio-β-d-galactoside was added to a final concentration of 200 μM (M-48c, M-C-48, M-C-48t, M-F-48t, and M-48-9c) or 100 μM (M-T-9c and M-T-9f). After 16 h, the cells were harvested by centrifugation (3,000 × g, 15 min), resuspended in 30 mM Tris-HCl, pH 8.0, 1 mM CaCl2, 0.1 mg/ml DNase I (Roche, Mannheim, Germany) and broken in a French press. M-T-9c, M-T-9f, M-48c, M-C-48, M-C-48t, M-F-48t, and M-48-9c, which all contain a C-terminal His tag, were purified on nickel-nitrilotriacetic acid resin (Qiagen, Vanloo, The Netherlands). The purification of the recombinant proteins was achieved on Q-Sepharose fast flow (GE Healthcare, Uppsala, Sweden) equilibrated in 30 mM Tris-HCl, pH 8.0, 1 mM CaCl2. The proteins of interest were eluted by a linear gradient of 0 to 500 mM NaCl in 30 mM Tris-HCl, pH 8.0, and 1 mM CaCl2.
The purified proteins were dialyzed by ultrafiltration against 10 mM Tris-HCl, pH 8.0, and 1 mM CaCl2 and stored at −80°C. The concentration of the proteins was estimated by absorbance at 280 nm in 6 M guanidine hydrochloride and 25 mM sodium phosphate, pH 6.5, using the program ProtParam tool (www.expasy.org/tools/protparam.html).
Nondenaturing PAGE.
Samples (10 μM final concentration) were mixed at room temperature in 20 mM Tris-maleate, pH 6.0, 1 mM CaCl2, and 4-μl volumes of the samples were subjected to polyacrylamide gel electrophoresis (PAGE) (4 to 15% gradient) using a Phastsystem apparatus (GE Healthcare).
Gel filtration analyses.
Gel filtration experiments were carried out using an Ákta FPLC system (GE Healthcare). Samples (100 μl, 10 μM) were injected into a Superdex 200 (10 × 300) GL analysis grade column (GE Healthcare) equilibrated in 50 mM potassium phosphate, pH 7.0, and 150 mM NaCl. A flow rate of 0.5 ml/min was used during chromatography.
Stability of the complexes.
Proteins were mixed at a final concentration of 10 μM in 20 mM Tris-maleate, pH 6.0, 1 mM CaCl2, and 0.01% NaN3 and incubated at 37°C. Samples were extracted at 0, 1, 6, and 24 h and subjected to nondenaturing PAGE as described above and to sodium dodecyl sulfate-PAGE (SDS-PAGE) (10%) after the addition of loading buffer (25%, vol/vol) and 5 min of boiling using a vertical electrophoresis system (Hoefer, San Francisco, CA).
Enzyme activity.
Phosphoric acid swollen (PAS)-cellulose was prepared from Avicel PH101 (Fluka, Buchs, Switzerland) as previously described (35). Kinetic experiments were performed by incubating at 37°C aliquots (40 μl) of the protein samples (10 μM or 5 μM, in 20 mM Tris-maleate, pH 6.0, and 1 mM CaCl2) with 4 ml of PAS-cellulose or microcrystalline cellulose Avicel PH101 at 3.5 g/liter in 20 mM Tris-maleate, pH 6.0, 1 mM CaCl2, and 0.01% (wt/vol) NaN3. In these experiments, the final concentration was 0.1 μM for the covalent cellulosome and the dimeric complexes that contain one GH48 and one GH9 catalytic module (i.e., hybrid, symmetrical, and asymmetrical cellulosomes), and it was 0.05 μM for tetrameric complexes that contain two GH48 and two GH9 catalytic modules (i.e., cyclic, polymeric and mixed cellulosomes). Thus, the cumulative concentration of GH48 and GH9 cellulases was kept constant in all kinetic experiments (0.1 μM). Aliquots (900 μl) were extracted at 0, 20, 40, 60, and 80 min (PAS-cellulose) or 0, 1, 6, and 24 h (Avicel), centrifuged, and examined for soluble reducing sugars by the Park and Johnson method (25) using glucose as the standard.
RESULTS
Design, preparation, and characterization of new enzyme components.
Seven new engineered enzymes were designed and produced in E. coli. Their modular organization is presented in Fig. 1. These novel cellulases are derived from either Cel48F or Cel9G from C. cellulolyticum, both of which were previously shown to play a crucial role in cellulose degradation (12). Each engineered enzyme was appended with the potent cellulose-binding family 3a CBM (CBM3a) and the first X2 module of the scaffoldin CipC from the same bacterium. The presence of a CBM3a in the enzyme chimeras provides the major substrate-targeting function for the assembled complex. The CBM3c of Cel9G was maintained in all GH9 cellulases. CBM3c was previously shown to be a “catalytic” or “helper” CBM (14, 18, 30) and occurs structurally linked to the GH9 catalytic domain in this particular type of enzyme (22). The inclusion of a CBM3c serves to convert the GH9 enzyme from a simple endoglucanase to a processive endoglucanase. CBM3c displays weak or no affinity for cellulose, in contrast to the other family 3 CBMs (CBM3a and CBM3b), which are known to strongly interact with crystalline cellulose (32). As indicated in Fig. 1, a bifunctional enzyme containing both GH48 and GH9 catalytic modules combined with the CBM3a and X2 modules of CipC was also produced (termed covalent cellulosome). In most cases, cohesins and/or dockerins from either C. cellulolyticum, C. thermocellum, or R. flavefaciens were also introduced, in order to enable the engineered cellulases to interact selectively and in different ways with additional complementary components and to generate cellulolytic complexes with various geometries.
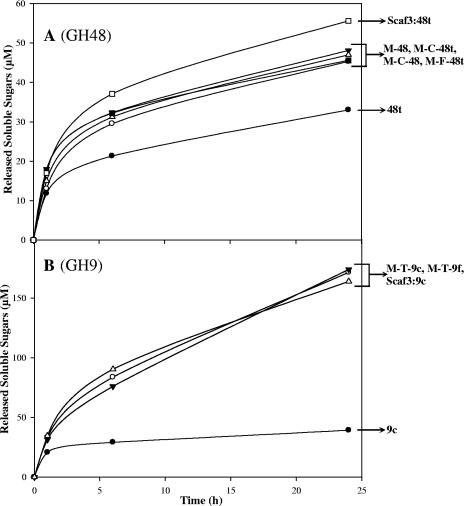
The activity of the recombinant enzymes was assayed on microcrystalline cellulose (Avicel) and “amorphous” PAS-cellulose using standard conditions (11, 12). As shown in Fig. 2A, the fusion of the GH48 catalytic module of Cel48F to the CBM3a and the X2 module of the scaffoldin CipC induced a 1.4-fold increase in Avicelase activity. Neither the introduction of a cohesin between the X2 module and the catalytic module nor the deletion/replacement of the C-terminal dockerin module altered the activity of the engineered cellulases which remained approximately 1.4-fold more active than the parental enzyme (48t). Nevertheless, the binding of 48t to Scaf3 generated a complex that was slightly more active (approximately 20%) than the engineered Cel48F.
FIG. 2.
Avicel hydrolysis by the recombinant enzymes alone. The amounts of reducing sugars were determined after 0, 1, 6 and 24 h of incubation at 37°C. (A) GH48 cellulases. The curves are labeled as follows: •, 48t; ○, M-48c; ▾, M-C-48; ▵, M-C-48t; ▪, M-F-48t; and □, Scaf3:48t complex. (B) GH9 cellulases. The curves are labeled as follows: •, 9c; ○, M-T-9c; ▾, M-T-9f; and ▵, the Scaf3:9c complex. The enzyme concentration was 0.1 μM in all kinetic experiments. The data are the means of three independent experiments (variation within 5% of the mean).
For Cel9G-based enzymes, the enhancement of the activity was 4.4-fold compared to the parental enzyme 9c (Fig. 2B). Furthermore, the activity of the engineered Cel9G was similar to that observed for the Scaf3/Gc complex, thus reflecting a more prominent CBM effect than for the GH48 cellulase.
The specific activity of the various engineered and parental enzymes was also determined on PAS-cellulose (not shown). In contrast to the enhanced activity on crystalline cellulose, the activity of the modified cellulases on amorphous cellulose was drastically reduced compared to the activities of the parental enzymes. In the case of the GH48 cellulases, the four engineered enzymes appended with the CBM3a of the scaffoldin exhibited threefold-reduced specific activities (2.8 to 3.2 IU/μmol), compared to parental enzyme 48t (8.9 IU/μmol). Similarly, the specific activities of engineered enzymes that contained both a CBM3a and a cohesin, M-T-9f (4.5 IU/μmol) and M-T-9c (5.1 IU/μmol), were reduced twofold compared to the specific activity of parental enzyme 9c (9.9 IU/μmol).
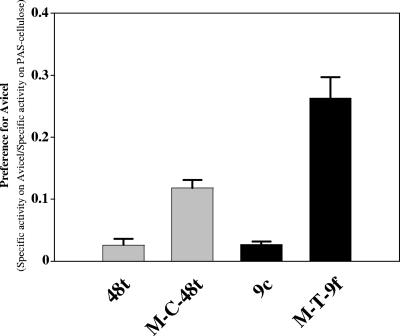
The introduction of the CBM3a into either Cel48F or Cel9G thus significantly modified the substrate specificity of both cellulases. As shown in Fig. 3, the “preference” for Avicel versus PAS-cellulose, defined as the ratio between the specific activity on Avicel (calculated after 24 h of hydrolysis) and the specific activity on PAS-cellulose, was increased 4.2- and 9.7-fold for M-C-48t and M-T-9f, respectively, compared to the parental enzymes. In both types of enzymes, the activity on PAS-cellulose was reduced, whereas the Avicelase activity was improved. A similar observation has previously been reported for hybrid cellulosomes composed of the same GH48 and GH9 enzymes (11). In the latter study, binding of the Cel48F/Cel9G enzyme pair to a hybrid scaffoldin appended with a CBM3a induced reduction (40%) of the activity on PAS-cellulose compared to the corresponding free enzyme system, whereas hydrolysis of Avicel by the complex was improved fourfold. It thus appears that the identical properties were induced upon covalent linkage of the CBM3a to the enzymes. A plausible explanation for this observation is that at high concentrations of reactive sites (PAS-cellulose), the strong CBM3-mediated binding to the substrate reduces the motion of the associated enzyme(s) and hampers activity on such a substrate.
FIG. 3.
Preference of parental versus engineered enzymes for Avicel over PAS-cellulose. The enzymes are indicated at the bottom of the graph. The data represent the ratio of the specific activity on Avicel/specific activity on PAS-cellulose. The specific activity on Avicel was estimated after 24 h of hydrolysis (data from Fig. 2). The standard deviations are indicated by the error bars.
Complex formation and reversibility of the cohesin-dockerin interaction.
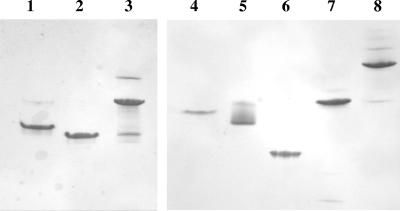
The different complexes listed in Fig. 1 were generally assembled by mixing stoichiometric amounts of the desired components. CaCl2 was added to the buffers, since the cohesin-dockerin interaction is known to be calcium dependent (13, 39). In all cases, the formation of complexes was verified by nondenaturing electrophoresis and was found to be total or nearly total for dimeric complexes as well as for the “mixed cellulosome” (see scheme and designations in Fig. 1). Two examples of nondenaturing PAGE are shown in Fig. 4. The complexes were also routinely analyzed by gel filtration (data not shown), and the apparent molecular masses obtained with this technique were in good agreement with the expected sizes of the various complexes.
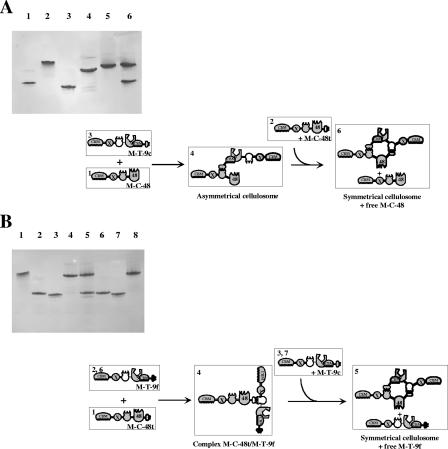
FIG. 4.
Electrophoretic mobility of components and assembled complexes on nondenaturing gels. Lane 1, M-C-48 alone; lane 2, M-T-9c alone; lane 3, asymmetrical cellulosome containing M-C-48 and M-T-9c; lane 4, Scaf6 alone; lane 5, 48t alone; lane 6, 9f alone; lane 7, M-48-9c alone; lane 8, mixed cellulosome containing Scaf6, 48t, 9f, and M-48-9c. In each lane, the concentrations of the indicated proteins were 10 μM, except in lane 8 where 5 μM of each component was used. Similar quality gels were obtained for all complexes used in this study.
In the case of the “cyclic cellulosomes” and “polymeric cellulosomes,” however, the desired complexes could not be obtained without specific precautions. For instance, if the engineered cellulases M-F-48t, M-T-9c, M-T-9f, and M-C-48t were mixed sequentially, no tetrameric complex was obtained, and only the dimeric complexes M-F-48t/M-T-9f and M-C-48t/M-T-9c were observed, in which each cellulase is bound twice to its partner (data not shown). This result suggested that significant reversibility of the cohesin-dockerin systems used prevented the formation of the desired tetrameric complex. The affinity constant of the cohesin-dockerin interaction for both mesophilic bacteria is in the range of 109 M−1 (12, 13), whereas for C. thermocellum, the affinity constant (KA) value is estimated to be ≥1010 M−1 (12). Despite its high affinity, the rapid reversibility of the cohesin-dockerin interaction was, however, demonstrated in the case of C. cellulolyticum, when M-C-48, M-T-9c, and M-C-48t were mixed sequentially. As shown in Fig. 5A, M-T-9c interacts initially with M-C-48, but when M-C-48t is finally added to the mixture, the initial complex totally dissociates, and the dimeric complex M-T-9c/M-C-48t and free M-C-48 are observed. Interestingly, the higher-affinity C. thermocellum cohesin-dockerin interaction was also found to be reversible under these conditions (Fig. 5B). Mixing the assembled dimeric complex M-T-9f/M-C-48t with M-T-9c generated the complete release of M-T-9f and the formation of the symmetrical cellulosome M-C-48t/M-T-9c. To our knowledge, such a rapid dissociation of the cohesin/dockerin complex in the absence of calcium-chelating reagent (13, 39) has not been observed previously for either C. cellulolyticum or C. thermocellum. It is hypothesized that a trimeric complex is transiently formed when the third component is added to the initial dimeric complex (Fig. 5), leading to an elevated local concentration of the competing cohesin and the ultimate release of the initially bound enzyme. The reversibility of the cohesin-dockerin interaction was investigated further, and the data are available in the supplemental material.
FIG. 5.
Analysis of the reversibility of the cohesin-dockerin interaction. (A) Electrophoretic mobility of components and assembled complexes in C. cellulolyticum. Lane 1, M-C-48 alone; lane 2, M-C-48t alone; lane 3, M-T-9c alone; lane 4, complex M-C-48/M-T-9c (asymmetrical cellulosome); lane 5, complex M-C-48t/M-T-9c (symmetrical cellulosome); lane 6, asymmetrical cellulosome mixed with M-C-48t resulting in symmetrical cellulosome plus free M-C-48. In each lane, equimolar concentrations (10 μM) of the indicated proteins were used. (B) Electrophoretic mobility of components and assembled complexes in C. thermocellum. Lane 1, M-C-48t alone; lane 2, M-T-9f alone; lane 3, M-T-9c alone; lane 4, complex M-C-48t/M-T-9f; lane 5, complex M-C-48t/M-T-9f mixed with M-T-9c resulting in complex M-C-48t/M-T-9c (symmetrical cellulosome) plus free M-T-9f; lane 6, M-T-9f alone; lane 7, M-T-9c alone; lane 8, complex M-C-48t/M-T-9c (symmetrical cellulosome). In each lane, equimolar concentrations (10 μM) of the indicated proteins were used. Schematic representations of the assembly and dissociation of the indicated complexes are shown below each gel. See the legend to Fig. 1 for details of subunits, modular components. and shorthand notation. The numbers in the top left-hand corner indicate the corresponding lanes in the nondenaturing gel.
Taking into account these data, the assembly of the cyclic cellulosome therefore required a two-step procedure, comprising the initial formation of the dimeric complexes M-F-48t/M-T-9c and M-C-48t/M-T-9f, which were then mixed to form the final tetrameric complex (Fig. 6). The architecture of the cyclic cellulosome was also verified by incubating this tetrameric complex with either 9c, 9f, or 48t. Analysis of the above mixtures by nondenaturing PAGE (data not shown) confirmed that all the cohesins contained in the cyclic cellulosome were occupied, since the added single enzymes remained in the free state, and the band with altered mobility corresponding to the cyclic cellulosome remained unchanged. Thus, with respect to the molecular mass observed by gel filtration (Fig. 6) and the lack of available cohesins, the schematic representation of the cyclic cellulosome reported in Fig. 1 is assumed to reflect the overall organization of this tetrameric complex. Similarly, the “polymeric cellulosome” was also fabricated using a defined sequential procedure, in which the dimeric complex M-F-48t/M-T-9c was initially assembled prior to the addition of the cellulases M-C-48 and 9f. The results (not shown) were essentially identical to those obtained for the cyclic cellulosome.
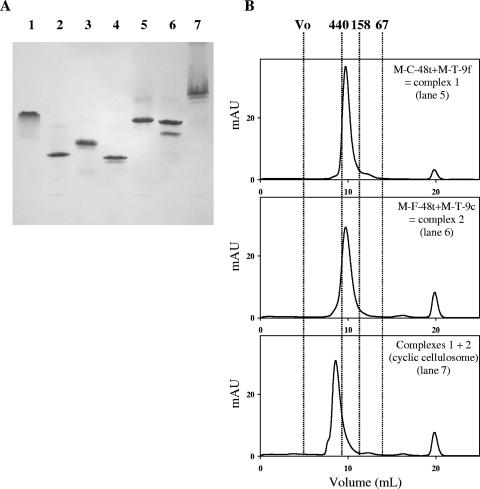
FIG. 6.
Assembly of the cyclic cellulosome. (A) Electrophoretic mobility of components and assembled complexes. Lane 1, M-C-48t alone; lane 2, M-T-9f alone; lane 3, M-F-48t alone; lane 4, M-T-9c alone; lane 5, complex 1 (M-C-48t plus M-T-9f); lane 6, complex 2 (M-F-48t plus M-T-9C); lane 7, complex 1 plus complex 2 (cyclic cellulosome). In lanes 1 to 6, equimolar concentrations (10 μM) of the indicated proteins were used. In lane 7, complexes 1 and 2 were mixed at a final concentration of 5 μM. (B) Gel filtration analysis of the assembled complexes. Injected proteins (100 μl) and the corresponding lanes in panel A are indicated on each chromatogram. mAU refers to milli absorbance units at 280 nm. Vertical lines indicate the positions of molecular mass markers: blue dextran or void volume (>2 MDa), ferritin (440 kDa), aldolase (158 kDa), and bovine serum albumin (67 kDa). For each chromatogram, the indicated proteins were at a concentration of 10 μM, except for cyclic cellulosome, where 5 μM concentrations of complexes 1 and 2 were used. The peak observed at 20 ml in all chromatograms corresponds to the maleic acid contained in the sample buffer. Note that the formation of complex 2 generates a major and minor band on nondenaturing PAGE (panel A, lane 6), but a single peak at the expected mass on gel filtration (panel B). Since the mobilities of the bands differ from those observed for free components (panel A, lanes 3 and 4), it is assumed that the two bands correspond to two different conformations of complex 2.
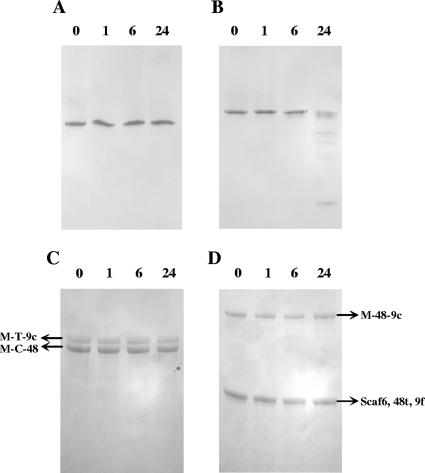
The stability of the various complexes at 37°C was explored by subjecting the samples to nondenaturing PAGE (Fig. 7A and B) and SDS-PAGE (Fig. 7C and D). Most complexes were found to be very stable over a 24-h period of incubation at 37°C as exemplified by the “asymmetrical cellulosome” (Fig. 7A and C). However, the “mixed cellulosome” preparation was found to be notably less stable than the other engineered complexes. After 24 h of incubation, the intensity of the band of altered mobility corresponding to the tetrameric complex was reduced by approximately 60% (Fig. 7B), although analysis by SDS-PAGE did not reveal any significant proteolysis of its components (Fig. 7D). It thus seems likely that prolonged incubation at 37°C induced a partial dissociation of this particular complex.
FIG. 7.
Stability of the asymmetrical and mixed cellulosomes. Samples containing 10 μM asymmetrical complex (A and C) or 5 μM mixed cellulosomes (B and D) were incubated at 37°C. At 0, 1, 6, and 24 h (time of incubation indicated above the lanes), the aliquots were subjected to nondenaturing PAGE (A and B) or to SDS-PAGE (C and D).
Activity of the complexes toward Avicel.
To facilitate the comparison between the various complexes, the final concentration of all GH48 and GH9 enzymes was maintained at 0.1 μM in the test tubes. Thus, the concentrations of dimeric complexes that contain only one GH48 and one GH9 enzyme, such as the “symmetrical cellulosome,” were 0.1 μM, whereas the concentrations of the tetrameric cellulosomes composed of two GH48 and two GH9 engineered cellulases per complex (for instance, the “cyclic cellulosome”) were adjusted to 0.05 μM during the kinetic experiments.
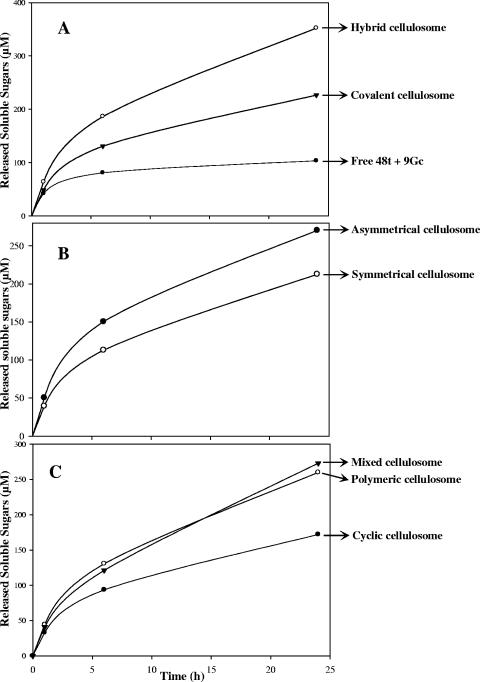
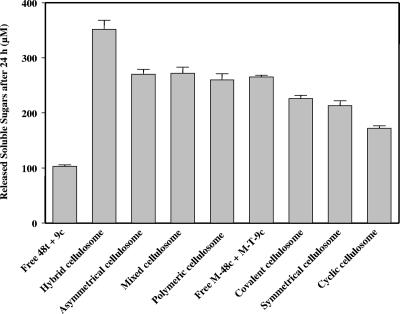
On crystalline cellulose, the covalent cellulosome (Fig. 8A) was 2.2-fold more active than the mixture of free 9c and 48t but significantly less efficient (36%) than the hybrid cellulosome. In fact, at the examined times, the amount of soluble sugars released by the covalent cellulosome corresponded to approximately the mean value between the free enzyme system and the noncovalent complex. These data indicate that despite a rather unusual organization for the selected cellulosomal GH48 and GH9 enzymes, both catalytic modules are functional in the covalent cellulosome. A similar bifunctional enzyme has previously been created (27), using GH48 and GH9 catalytic modules of noncellulosomal enzymes. The hybrid enzyme that also contains an efficient CBM3b was found to be twofold more active than the mixture of free enzymes was. Nevertheless, our data show that the hybrid cellulosome is noticeably more adapted for the degradation of this crystalline substrate than the combination of its major functional modules (i.e., the two catalytic modules together with the potent cellulose-binding CBM3a) in a single polypeptide chain.
FIG. 8.
Kinetic studies of Avicel hydrolysis by the various engineered cellulosomes. The amounts of reducing sugars were determined after 0, 1, 6, and 24 h of incubation at 37°C. The data show the means of four independent experiments (variation within 5% of the mean).
Figure 8B shows the kinetics obtained with the asymmetrical and symmetrical cellulosomes. The first complex was formed via a single docking system, whereas in the symmetrical cellulosome, the engineered cellulases are bound to each other by two divergent cohesin/dockerin devices, located at both sides of the catalytic entities (Fig. 1). The asymmetrical complex was found to be 1.27-fold more active than the symmetrical cellulosome. This indicates that bridging the two enzymes at two different positions in the peptide chain alters the activity of the dimeric complex, resulting in a reduction of the total hydrolysis of the crystalline substrate. At the end of the reaction kinetics, the range of the amount of soluble sugars released by the symmetrical complex (213 ± 9 μM) was similar to that observed for the single protein, i.e., the covalent cellulosome (226 ± 6 μM).
The Avicelase activity of the tetrameric complexes was also investigated, and the results are shown in Fig. 8C. The kinetics obtained for the polymeric cellulosomes and the mixed cellulosomes were relatively similar, though their architecture is rather different (Fig. 1). The range of the amount of released soluble sugars from the polymeric cellulosomes and the mixed cellulosomes after 24 h of incubation (260 ± 11 μM and 273 ± 11 μM, respectively) was the same as that observed for the asymmetrical cellulosome (269 ± 9 μM). Compared to the hybrid cellulosome (Scaf3:48t/9c), their respective activities are reduced by approximately 25% on crystalline cellulose. The complex termed cyclic cellulosome displayed the lowest activity on Avicel (approximately 50% of the activity of the hybrid cellulosome). This result clearly indicates that this type of artificial organization, which diverges from the existing cellulase systems described so far, is less adapted to the degradation of the crystalline substrate.
DISCUSSION
In light of recent reports on the intricate architecture of the R. flavefaciens cellulosome (29), the clostridial complexes appear to represent relatively simplistic forms of bacterial cellulosomes. Indeed, the concept of bacterial cellulosome has noticeably evolved over the past decade. The combination of different approaches, including genomic and proteomic approaches, has revealed that the geometry, size, composition, and location of these multienzyme complexes vary considerably among cellulosome-producing bacteria (8, 20, 29). Furthermore, in some hyperthermophilic bacteria, such as Caldocellulosiruptor saccharolyticus or Anaerocellum thermophilum (34, 40), the most essential cellulosomal modules can be produced as a single polypeptide chain.
To date, the cellulosome chimeras constructed essentially mimicked the overall organization of the native cellulosomes produced by C. cellulolyticum and other related mesophilic clostridia. Nevertheless, the possibility of preparing chimeric cellulosome components that possess divergent cohesin-dockerin pairs enables us to control in vitro the exact composition of artificial cellulosome systems that display novel geometries. It is thus imperative to examine the cellulose-degrading activities of such artificial complexes within the context of the native systems in order to determine whether improved activities can be achieved. To fabricate these new designer cellulosomes, Cel48F and Cel9G from C. cellulolyticum were appended with the tandem CBM3a and X2 modules from the scaffoldin CipC (as a substrate-targeting dyad) and with an optional cohesin and/or dockerin from either C. cellulolyticum, R. flavefaciens, or C. thermocellum. The introduction of CBM3a into either Cel48F or Cel9G improved their activity on Avicel at the apparent expense of their activity on more tractable substrates, such as PAS-cellulose. Indeed, GH48 and GH9-CBM3c enzymes appended with an efficient CBM3 (generally CBM3b) have been discovered in various bacteria (4, 15, 28, 31). Acetivibrio cellulolyticus was also found to produce an enzyme-containing scaffolding protein that exhibits one GH9 module (not associated with a CBM3c), one CBM3b, and seven cohesin modules (8). This is the only known example of an enzyme module as an integral part of a scaffoldin protein. Such naturally occurring variations in the cellulosome organizations formed the initial basis for the types of unconventional designer cellulosomes devised in this study.
The various engineered enzymes designed in this study served as building blocks to generate the cellulosome-like chimeras with the desired geometries. None of the newly designed cellulosomes (that lacked a define scaffoldin component) was found to be as active as the analogous “conventional” hybrid cellulosome (that contained a scaffoldin). The activities of the dimeric and tetrameric complexes tested are summarized in Fig. 9. The combined data indicate that two classes of complexes were generated in this study. The most active group includes the dimeric complex termed “asymmetrical cellulosome” and the tetrameric complexes termed “mixed polymeric cellulosomes” and “polymeric cellulosomes.” Their activity is reduced by approximately 20 to 25% compared to the hybrid cellulosome, but it was similar to the Avicelase activity of the analogous free CBM-containing enzyme pair M-48c plus M-T-9c, which cannot interact with each other owing to the lack of a matching cohesin-dockerin pair (Fig. 9). These data suggest that the previously reported “proximity” effect (11) was strongly impaired by the geometries exhibited by these complexes. The second series of complexes includes the covalent, symmetrical, and cyclic cellulosomes, in which the observed Avicelase activities are even more diminished (35 to 52%) compared to the activity of the hybrid cellulosome. The mobility of enzymes in the latter three complexes appears to be limited, owing either to multiple cohesin-dockerin interactions in the symmetrical and cyclic cellulosome configurations or to their covalent coupling in the covalent cellulosome. Taken together, these data suggest that increasing restriction of the enzyme mobility within the complexes proportionally affects the activity. This is consistent with recent small angle X-ray scattering analyses performed on hybrid cellulosomes which revealed an important conformational flexibility in designer cellulosomes, largely due to the mobility of the incorporated enzymes (17). In the present study, limitation in enzyme mobility would primarily reflect supplementary binding to the other cellulosome partners. Such a phenomenon is clearly revealed by the comparative activities of the asymmetrical versus symmetrical cellulosomes or by those of the polymeric versus cyclic cellulosomes. In both cases, the two complexes have almost the same protein composition and essentially differ by the number of protein-protein interactions within the complex.
FIG. 9.
Comparative solubilization of microcrystalline cellulose Avicel by the various complexes and free enzyme systems. The composition of the complexes and free enzyme systems is indicated at the bottom of the graph. The data represent the amount of released soluble sugars after 24 h of reaction by the given enzyme system and are summarized from Fig. 8. Error bars indicate the standard deviations.
Superfluous binding of the complexes to the substrate may also reduce the mobility of the catalytic subunits. It was previously shown that introduction of a second CBM3a at the C terminus of Scaf3 causes a decrease in the Avicelase activity of the resulting designer cellulosome (11). The complexes generated in the present study contain one to four CBM3a, whereas the hybrid cellulosome contains only one CBM3a, borne by the scaffoldin. Indeed, the least efficient complex, the cyclic cellulosome, has four distinct CBM3a. Nevertheless, the Avicelase activity of the various complexes cannot be correlated directly to the number of CBM3a they contain. For instance, the polymeric complex, which contains three CBM3a, displays a higher Avicelase activity than either the symmetrical cellulosome (two CBM3a) or the covalent cellulosome (one CBM3a).
The results of this study indicate that the intrinsic mobility of the catalytic subunits and targeting of the complex to the crystalline substrate by means of a single CBM are essential characteristics of this system. These properties indeed match native cellulosome organization selected by mesophilic clostridia but do not apply to the elaborate cellulosomes produced by A. cellulolyticus or ruminal bacteria that form a vast network of interacting scaffoldins, catalytic subunits, and cell-surface-anchoring proteins (29, 36, 37). Nevertheless, the GH48 and GH9 enzymes, selected as the model components for our studies on designer cellulosomes, are common components of the relatively simple, natural cellulosomes produced by the mesophilic clostridia. The homologous native enzymes that participate in the more-complex cellulosomes may be better adapted for this particular configuration. It is, however, worth noting that long linkers (up to 550 residues) are frequently reported for scaffoldins participating in these complex cellulosomes (38), whereas in cellulosomes produced by mesophilic clostridia, the linkers connecting the various modules in the single scaffolding protein are usually about 10 residues long (19, 24, 33). Thus, in more intricate cellulosome systems, the particularly long linkers of their interacting scaffoldins may help maintain conformational flexibility and compensate for the stringency induced by multiple cohesin-dockerin interactions and perhaps also multiple cellulosome-substrate interactions.
The notably reduced activity of the covalent cellulosome is also likely to reflect the restricted mobility of its catalytic modules. The association of the cellulosome components by means of cohesin-dockerin interactions probably permits more conformational flexibility than a direct connection to the other partners through a covalent linker. Indeed, binding to cohesin was shown previously to trigger a pleating of the enzyme linker that connects the catalytic and dockerin modules, but some limited flexibility was still found to persist (16). Furthermore, two recent studies have shown that type I dockerins from C. thermocellum may bind in two ways to the cognate cohesin, generating two different orientations for the associated catalytic module with respect to the cohesin/dockerin interface (5, 6). This observation indicates a direct contribution of the cohesin-dockerin interaction in cellulosome plasticity.
In view of the above, our results indicate that enzyme mobility within the complex is a critical parameter for cellulosome activity. The plasticity of these large multienzymes machines is probably dictated by the recalcitrant nature of plant cell wall polysaccharides which are heterogeneous, insoluble, and partly crystalline and that changes constantly during the degradation process. Our data also indicate that the designer cellulosome technology allows the fabrication of a versatile and essentially unlimited number of complexes with desired geometries, which may serve various biotechnological purposes. Furthermore, such molecular edifices could include enzymes that do not form complexes in vivo, since it has been recently shown that incorporation of a noncellulosomal enzyme into a hybrid cellulosome can improve its activity (23).
Supplementary Material
Acknowledgments
We thank Odile Valette for expert technical assistance. We are grateful to Sandrine Pagès, Céline Boileau, and Raphael Lamed for helpful discussions.
This work was supported by the CNRS, the Conseil Général des Bouches du Rhône, and the Provence-Alpes-Côte d'Azur region. We are grateful to TOTAL SA for financial support. Additional support was provided by the Agence Nationale de la Recherche (grant ANR-05-BLAN-0259-01). E.A.B. acknowledges financial support from the Israel Science Foundation (442/05). F.M. holds a fellowship from the French government agency Ministère de l'Enseignement Supérieur et de la Recherche.
Footnotes
Published ahead of print on 28 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bayer, E. A., J. P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 2.Blouzard, J. C., C. Bourgeois, P. de Philip, O. Valette, A. Bélaïch, C. Tardif, J. P. Bélaïch, and S. Pagès. 2007. Enzyme diversity of the cellulolytic system produced by Clostridium cellulolyticum explored by two-dimensional analysis: identification of seven genes encoding new dockerin-containing proteins. J. Bacteriol. 189:2300-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Bronnenmeier, K., K. Kundt, K. Riedel, W. H. Schwarz, and W. L. Staudenbauer. 1997. Structure of the Clostridium stercorarium gene celY encoding the exo-1,4-β-glucanase Avicelase II. Microbiology 143:891-898. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho, A. L., F. M. Dias, T. Nagy, J. A. Prates, M. R. Proctor, N. Smith, E. A. Bayer, G. J. Davies, L. M. Ferreira, M. J. Romao, C. M. Fontes, and H. J. Gilbert. 2007. Evidence for a dual binding mode of dockerin modules to cohesins. Proc. Natl. Acad. Sci. USA 104:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho, A. L., F. M. Dias, J. A. Prates, T. Nagy, H. J. Gilbert, G. J. Davies, L. M. Ferreira, M. J. Romao, and C. M. Fontes. 2003. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc. Natl. Acad. Sci. USA 100:13809-13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demain, A. L., M. Newcomb, and J. H. Wu. 2005. Cellulase, clostridia, ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 1999. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J. Bacteriol. 181:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 2000. A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J. Bacteriol. 182:4915-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 11.Fierobe, H. P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Belaich, R. Lamed, Y. Shoham, and J. P. Belaich. 2002. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 12.Fierobe, H. P., F. Mingardon, A. Mechaly, A. Belaich, M. T. Rincon, S. Pages, R. Lamed, C. Tardif, J. P. Belaich, and E. A. Bayer. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280:16325-16334. [DOI] [PubMed] [Google Scholar]
- 13.Fierobe, H. P., S. Pages, A. Belaich, S. Champ, D. Lexa, and J. P. Belaich. 1999. Cellulosome from Clostridium cellulolyticum: molecular study of the Dockerin/Cohesin interaction. Biochemistry 38:12822-12832. [DOI] [PubMed] [Google Scholar]
- 14.Gal, L., C. Gaudin, A. Belaich, S. Pages, C. Tardif, and J. P. Belaich. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilad, R., L. Rabinovich, S. Yaron, E. A. Bayer, R. Lamed, H. J. Gilbert, and Y. Shoham. 2003. CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammel, M., H. P. Fierobe, M. Czjzek, S. Finet, and V. Receveur-Brechot. 2004. Structural insights into the mechanism of formation of cellulosomes probed by small angle X-ray scattering. J. Biol. Chem. 279:55985-55994. [DOI] [PubMed] [Google Scholar]
- 17.Hammel, M., H. P. Fierobe, M. Czjzek, V. Kurkal, J. C. Smith, E. A. Bayer, S. Finet, and V. Receveur-Brechot. 2005. Structural basis of cellulosome efficiency explored by small angle X-ray scattering. J. Biol. Chem. 280:38562-38568. [DOI] [PubMed] [Google Scholar]
- 18.Irwin, D., D. H. Shin, S. Zhang, B. K. Barr, J. Sakon, P. A. Karplus, and D. B. Wilson. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, S., Y. Barak, R. Lamed, E. A. Bayer, and M. Morrison. 2006. The functional repertoire of prokaryote cellulosomes includes the serpin superfamily of serine proteinase inhibitors. Mol. Microbiol. 60:1344-1354. [DOI] [PubMed] [Google Scholar]
- 21.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandelman, D., A. Belaich, J. P. Belaich, N. Aghajari, H. Driguez, and R. Haser. 2003. X-ray crystal structure of the multidomain endoglucanase Cel9G from Clostridium cellulolyticum complexed with natural and synthetic cello-oligosaccharides. J. Bacteriol. 185:4127-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mingardon, F., A. Chanal, A. M. López-Contreras, C. Dray, C. Tardif, E. A. Bayer, and H. P. Fierobe. 2007. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl. Environ. Microbiol. 73:3822-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagès, S., A. Bélaïch, H. P. Fierobe, C. Tardif, C. Gaudin, and J. P. Bélaïch. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, J. T., and M. J. Johnson. 1949. A submicrodetermination of glucose. J. Biol. Chem. 181:149-151. [PubMed] [Google Scholar]
- 26.Perret, S., L. Casalot, H.-P. Fierobe, C. Tardif, F. Sabathe, J.-P. Belaich, and A. Belaich. 2004. Production of heterologous and chimeric scaffoldins by Clostridium acetobutylicum ATCC 824. J. Bacteriol. 186:253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riedel, K., and K. Bronnenmeier. 1998. Intramolecular synergism in an engineered exo-endo-1,4-beta-glucanase fusion protein. Mol. Microbiol. 28:767-775. [DOI] [PubMed] [Google Scholar]
- 28.Riedel, K., J. Ritter, S. Bauer, and K. Bronnenmeier. 1998. The modular cellulase CelZ of the thermophilic bacterium Clostridium stercorarium contains a thermostabilizing domain. FEMS Microbiol. Lett. 164:261-267. [DOI] [PubMed] [Google Scholar]
- 29.Rincon, M. T., T. Cepeljnik, J. C. Martin, R. Lamed, Y. Barak, E. A. Bayer, and H. J. Flint. 2005. Unconventional mode of attachment of the Ruminococcus flavefaciens cellulosome to the cell surface. J. Bacteriol. 187:7569-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakon, J., D. Irwin, D. B. Wilson, and P. A. Karplus. 1997. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4:810-818. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez, M. M., F. I. Pastor, and P. Diaz. 2003. Exo-mode of action of cellobiohydrolase Cel48C from Paenibacillus sp. BP-23. A unique type of cellulase among Bacillales. Eur. J. Biochem. 270:2913-2919. [DOI] [PubMed] [Google Scholar]
- 32.Shimon, L. J., S. Pages, A. Belaich, J. P. Belaich, E. A. Bayer, R. Lamed, Y. Shoham, and F. Frolow. 2000. Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 A resolution. Acta Crystallogr. Sect. D 56:1560-1568. [DOI] [PubMed] [Google Scholar]
- 33.Shoseyov, O., M. Takagi, M. A. Goldstein, and R. H. Doi. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc. Natl. Acad. Sci. USA 89:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Te'o, V. S., D. J. Saul, and P. L. Bergquist. 1995. celA, another gene coding for a multidomain cellulase from the extreme thermophile Caldocellum saccharolyticum. Appl. Microbiol. Biotechnol. 43:291-296. [DOI] [PubMed] [Google Scholar]
- 35.Walseth, C. S. 1952. Occurrence of cellulase in enzyme preparations from microorganisms. Tech. Assoc. Pulp Pap. Ind. 35:228-233. [Google Scholar]
- 36.Xu, Q., Y. Barak, R. Kenig, Y. Shoham, E. A. Bayer, and R. Lamed. 2004. A novel Acetivibrio cellulolyticus anchoring scaffoldin that bears divergent cohesins. J. Bacteriol. 186:5782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, Q., E. A. Bayer, M. Goldman, R. Kenig, Y. Shoham, and R. Lamed. 2004. Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell surface-anchoring scaffoldin and a family 48 cellulase. J. Bacteriol. 186:968-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Q., W. Gao, S.-Y. Ding, R. Kenig, Y. Shoham, E. A. Bayer, and R. Lamed. 2003. The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell surface anchoring protein. J. Bacteriol. 185:4548-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yaron, S., E. Morag, E. A. Bayer, R. Lamed, and Y. Shoham. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121-124. [DOI] [PubMed] [Google Scholar]
- 40.Zverlov, V., S. Mahr, K. Riedel, and K. Bronnenmeier. 1998. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile Anaerocellum thermophilum with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 144:457-465. [DOI] [PubMed] [Google Scholar]
- 41.Zverlov, V. V., J. Kellermann, and W. H. Schwarz. 2005. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5:3646-3653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.