Abstract
Insulin gene transcription is limited to the beta cells within the mammalian pancreas and, like insulin secretion, is regulated by glucose. Our previous studies in primary cultured beta cells suggested the presence of a strong glucose-responsive enhancer element between base pairs −341 and −260 of the human insulin promoter, the same region in which a transcriptional repressor had been identified in beta-cell tumor lines. In an attempt to map these promoter activities and resolve these conflicting data, we designed minienhancer constructs spanning this region, and tested them in primary cultured and immortalized cells. One sequence, the Z element (base pairs −292 to −243), functions as both a potent glucose-responsive transcriptional enhancer in primary cultured islet cells and as a transcriptional repressor in immortalized beta and nonbeta cells and in primary fibroblasts. In addition, the Z element binds a novel glucose-responsive protein complex that is found in the nuclei of primary cultured islet cells, but not in the nuclei of tumor cells or primary cultured fibroblasts. These data demonstrate a critical role for the Z element in human insulin gene transcription and its regulation by glucose.
Keywords: transcription/promoter
Insulin, produced by the beta cells in the pancreatic islets of Langerhans, is the key regulator of glucose metabolism in vertebrates. Glucose in turn regulates insulin synthesis and secretion by the beta cell. The increase in insulin synthesis in response to high glucose concentration results, at least in part, from an increase in insulin gene transcription, and therefore depends on the insulin gene promoter (1).
The promoter not only regulates the transcriptional response to glucose but also restricts expression of the insulin gene exclusively to beta cells (2–5). As few as 400 bp of the insulin gene promoter upstream from the transcription start site is sufficient for beta cell-specific expression of a linked reporter gene (2, 3). The cis-acting sequence elements within the rat insulin I (6), rat insulin II (7, 8), and human insulin promoters (9) that are required for beta-cell specificity of insulin gene expression, and the nuclear protein complexes that bind to these elements have been studied extensively in beta-cell tumor lines. Insulin-producing tumor cell lines, however, do not maintain all physiologic features of normal beta cells. Although tumorigenic beta cells produce and secrete insulin, they have lower levels of insulin mRNA when compared with normal beta cells (10, 11), possibly reflecting impaired insulin gene promoter function. Furthermore, the majority of beta-cell tumor lines lack a normal physiologic response to glucose, making them poor models for studying glucose regulation of transcription (12). Given these considerations, it is not surprising that striking differences have been noted in the relative importance of promoter sequence elements between cell lines derived from beta-cell tumors and primary cultured islets. For example, deletion of the sequences between base pairs −341 and −260 of the human insulin promoter produces a marked rise in promoter activity in insulinoma cells (9), but an equally large decrease in primary islet cells (13).
By using primary cultured islets, mapping studies of the rat and human insulin promoters have detected several sequence elements capable of responding to glucose. These sequence elements belong to two classes of sequence motifs, E elements and A elements. E elements contain the core sequence CANNTG, and bind members of the basic helix–loop–helix (bHLH) class of transcription factors (14). A elements are defined by the core sequence TAAT, which constitutes a binding site for homeodomain transcription factors (15). In the human insulin promoter, metabolic response elements have been mapped to the E element, E1 (13), at base pair −100 relative to the transcription start site, and to an A element, A3 (16, 17), at base pair −227 (see Fig. 1A). Although the isolated E1 element in the human insulin promoter can respond by itself to glucose, its function is not independent of other sequence elements. Inclusion of the adjacent A2/C1 site, which by itself shows very weak glucose response, potentiates the response to glucose (13), demonstrating that the metabolic response of the insulin gene promoter requires synergy between different sequence elements.
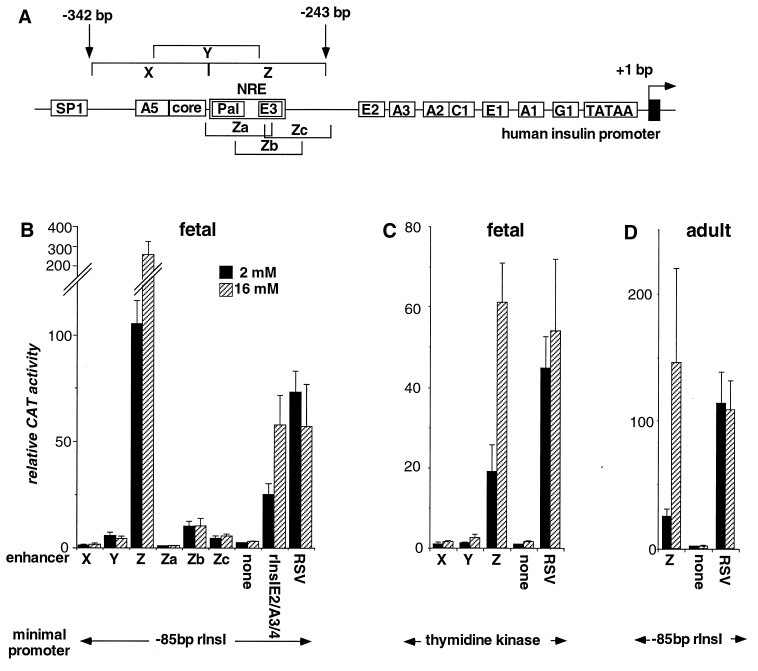
Figure 1.
Function of the minienhancers in primary cultured islets. Sequence elements in the proximal human insulin promoter and Z element are shown in A. The names used in the text are shown in boxes. The schematic is not to scale. Pal, palindromic sequence. Cultured fetal (B and C) or handpicked adult (D) rat islets were transfected with plasmids containing five tandem copies of the minienhancers shown or a single copy of the Rous sarcoma virus enhancer, linked to the base pair −85 rInsI promoter (B and D), or the base pair −109 thymidine kinase promoter (C) driving the CAT gene in pFOXCAT2 and grown in 2 mM (solid bars) or 16 mM (hatched bars) glucose. CAT activity of cultures transfected with the enhancerless pFOXCAT2 minimal promoter plasmid and grown at 2 mM glucose was set at 1.0. Each data point represents the mean of at least four independent transfections ± SEM.
Considering this interdependence, it is not surprising that multiple cis-acting elements contribute to the basal activity, tissue specificity, and metabolic response of the insulin promoter. In the rat insulin I promoter, no combination of mutations in known metabolic response elements can completely eliminate the promoter’s transcriptional response to glucose (18), suggesting the existence of additional glucose-regulated elements. Likewise, deletion analysis of the human insulin promoter in primary islet cultures suggested that sequences outside of the already mapped E1/A2/C1 and A3 regions contribute to the glucose response of the promoter (13). These experiments also indicated that a strong positive regulatory element maps to the region between base pairs −341 and −260 of the human insulin promoter. Because of the complexity of interactions within the intact promoter, simple deletion analysis does not allow us to determine the functional characteristics of this particular promoter region. Therefore in this study, we used isolated fragments covering the base pair −341-to-−260 region of the human insulin promoter to investigate the ability of this region to respond to glucose and to discriminate between primary cultured islet cells and a variety of tumor cell lines.
MATERIALS AND METHODS
Plasmid Constructions.
All of the minienhancer constructs contain five copies of the minienhancer inserted upstream of the base pair −85 minimal promoter from rat insulin I (rInsI) promoter or upstream of the base pair −109 minimal promoter from the thymidine kinase gene in pFOXCAT2 (13) as described previously (19), except for Rous sarcoma virus, which has only a single copy of the enhancer sequence (18). Sequences of all plasmid consructs were verified by the Sanger dideoxynucleotide sequencing method.
Transfection Assays.
Fetal 21-day gestation Sprague–Dawley rat islets were isolated, transfected, cultured, and harvested as described previously (1).
Adult islets were picked by hand from collagenase-digested adult Sprague-Dawley rat pancreata and cultured overnight in RPMI medium 1640 with 10% fetal bovine serum. Aliquots of 100 adult islets were placed in 12 × 75 mm culture tubes and washed three times with 1 ml of Opti-MEM 1 medium (serum-free, GIBCO/BRL). Plasmid DNA (4 μg) was diluted in 100 μl of Opti-MEM 1. Four micrograms of 25-kDa polyethylenimine (PEI; Aldrich) was diluted in 50 μl of Opti-MEM 1, added to the diluted plasmid DNA, and incubated at room temperature for 15 min. Replication-deficient adenovirus 5 dI-342, produced as previously described (20), was diluted in Opti-MEM 1 to produce a final concentration of 1011 virus particles per ml. The diluted adenovirus (100 μl; approximately 1010 virus particles) were added to the plasmid DNA/PEI mixture and incubated at room temperature for an additional 15 min. The DNA/PEI/adenovirus mixture was then added to the islets and incubated at 37°C for 30 min with gentle agitation, after which the islets were washed three times with 1 ml of RPMI medium 1640 containing 10% fetal bovine serum and incubated for 36 h. Protein extracts of the transfected islets were prepared, and 10 μg of protein was assayed for chloramphenicol acetyltransferase (CAT) activity (6).
βHC9 and βTC3, two transgenic mouse beta-cell lines, and mPAC4.1.2.1 cells, a mouse pancreatic ductal carcinoma cell line, were obtained from D. Hanahan (University of California, San Francisco). Primary fibroblasts were harvested from collagenase-digested pancreata from 21-day gestation fetal Sprague–Dawley rats. All cells were grown in DMEM with 4.5 g/l glucose (GIBCO/BRL) and 10% fetal calf serum. βHC9 cells or primary fibroblasts were transfected at a density of 2 × 106 cells or 1.5 × 104 cells per 60-mm plate with 3.5 μg of plasmid DNA by the Lipofectin technique as described by the manufacturer (GIBCO/BRL). Mouse pancreatic ductal carcinoma cells or a hamster insulinoma cell line, HIT T-15 M.2.2.2 cells were transfected with 10 μg of plasmid DNA at a density of 2.5 × 105 cells or 3 × 106 cells per 10-cm plate by the calcium phosphate technique (6). Cells were harvested, and protein extracts were prepared 48 h after transfection. βHC9 extract (2.5 μg), 25 μg of primary fibroblast extract, 5 μg of mPAC extract, or 1 μg of HIT extract was used to test CAT enzyme activity as previously described (6).
Electrophoretic Mobility Shift Assays (EMSAs).
Protein extracts were prepared from tumor cells and from primary cells after culturing as described above. After overnight culture in medium containing 5.5 mM glucose, the islets were transferred to medium with the glucose concentrations shown and cultured for 24 h prior to harvest. All nuclear extracts were prepared by the method of Dignam et al. (21).
The probes (for sequences, see Table 1) were labeled with [γ-32P]ATP and T4 polynucleotide kinase. Binding was performed in a 10-μl volume with 2 μg of nuclear extract, 100 pg of labeled probe (approximately 10,000 cpm), 10 mM Hepes (pH 7.8), 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 nM DTT, 3% Ficoll, 100 ng of poly(dI-dC)⋅poly(dI-dC) per μl, and 1% polyvinyl alcohol. After incubation at room temperature for 30 min, the mixtures were electrophoresed on 5% nondenaturing polyacrylamide gels in 0.5× TBE (1× TBE = 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) (22, 23).
Table 1.
Oligonucleotide sequences
| Name | 5′ end base pair | Sequence† |
|---|---|---|
| X | −342 | CCTGCAGCCTCCAGCTCTCCTGGTCTAATGTGGAAAGTGGCCCAGGTGAG |
| Y | −317 | TAATGTGGAAAGTGGCCCAGGTGAGGGCTTTGCTCTCCTGGAGACATTTG |
| Z | −292 | GGCTTTGCTCTCCTGGAGACATTTGCCCCCAGCTGTGAGCAGGGACAGGT |
| Za | −292 | GGCTTTGCTCTCCTGGAGACATTTGCCCCC |
| Zb | −274 | GACATTTGCCCCCAGCTGTGAGCAGGGACA |
| Zc | −269 | TGCCCCCAGCTGTGAGCAGGGACAGGTCTG |
| Za1 | −287 | TGCTCTCCTGGAGACAT |
| Za2 | −281 | CCTGGAGACATTTGCCC |
| Za3 | −276 | AGACATTTGCCCCCA |
| Za4 | −298 | GGTGAGGGCTTTGCTCTCCT |
| M1 | −292 | GGCTGGGCTCTCCTGGAGACATTTGCCCCC |
| M2 | −292 | GGCTTTGCTAGCCTGGAGACATTTGCCCCC |
| M3 | −292 | GGCTTTGCTCTCCTGTCGACATTTGCCCCC |
| M4 | −292 | GGCTTTGCTCTCCTGGAGACAGTTGCCCCC |
| M5 | −292 | GGCTTTGCTCTCCTGGAGACATTTTCCCCC |
| M6 | −292 | GGCTTTGCTAGCCTGTCGACATTTGCCCCC |
| hE1 | −120 | CAGCCCCCAGCCATCTGCCGACCC |
| rE1 | −115 | CTCGCCATCTGCCTA |
| rE2 | −241 | CAGGCCATCTGGCCC |
Position of the base relative to the transcription start site (+1) of the hIns promoter or rInsI promoter for rE1 and rE2.
Substituted bases in the mutated oligonucleotides are underlined.
RESULTS
The Distal Human Insulin Promoter Contains a Strong Positive, Glucose-Responsive Regulatory Element.
A deletion series of the human insulin (hIns) promoter in primary cultured fetal rat islet cells suggested the presence of a positive regulatory element in the region between base pairs −341 and −260 relative to the transcription start site (13). To circumvent the complexity of the intact promoter and to map individual sequence elements, we constructed three minienhancers, each spanning 50 bp of the region (see Fig. 1A and Table 1). Five copies of each minienhancer were linked to a minimal promoter constructed from base pair −85 to base pair +1 of the rInsI promoter driving CAT reporter gene expression. This minimal promoter was chosen because it retains beta-cell specificity but cannot respond to glucose on its own (18). The CAT plasmids were transfected into primary cultured fetal rat islet cells. The cells were harvested 36 h later and assayed for CAT activity (1).
The two most distal minienhancers, X (base pair −342 to −293) and Y (base pair −317 to −268) contain two previously described sequence elements: the A5 (base pair −323 to −314), and the enhancer core element (base pair −317 to −309) (Fig. 1A and Table 1). Both of the minienhancers have only very weak transcriptional activity when transfected into fetal rat islet cells (Fig. 1B). By contrast, a striking activation of transcription is seen with the Z minienhancer (base pair −292 to −243; Table 1). The Z minienhancer also shows a strong response to glucose, with a nearly threefold increase in transcriptional activity from low to high glucose concentrations. The activity of the Z minienhancer even exceeds that of the E2A3/4 (previously called Far-FLAT or FF) minienhancer, a potent, glucose-responsive, cell-specific transcriptional enhancer within the rInsI promoter (1) (Fig. 1B).
To exclude the possibility that activity of the Z minienhancer results from interactions between the Z element and the base pair −85 rInsI promoter, we also tested the multimerized minienhancers linked to a herpes simplex thymidine kinase (TK) minimal promoter. As with the base pair −85 rInsI-linked constructs, the Z minienhancer stimulates transcription and strongly responds to glucose (Fig. 1C), indicating that the Z element acts as an independent transcriptional enhancer also when in the context of a heterologous promoter.
To further map this region, we synthesized three overlapping oligonucleotides of 30-bp length covering the 50-bp Z-element region (Table 1), and tested them in CAT reporter plasmids analogous to those with the X, Y, and Z minienhancers. None of the shorter minienhancers produces significant transcriptional activation or responds to glucose (Fig. 1B). These results suggest that the activity and glucose response of the Z element depend on at least two separate sequence elements within the Z-element region.
To exclude the possibility that activity of the Z element is limited to fetal islet cells, the Z minienhancer was tested in adult rat islets. As in fetal islets, the Z minienhancer strongly transactivated the reporter gene in isolated adult islets, indicating that the Z element is an important activator of insulin gene transcription in both fetal and mature islet cells (Fig. 1D).
The Z Element Functions as an Enhancer Only in Primary Cultured Islets.
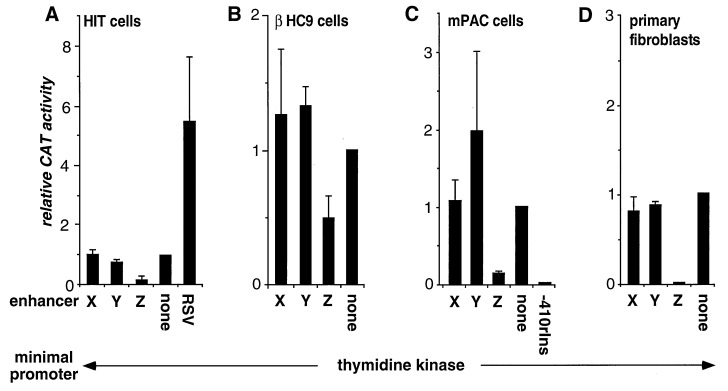
In the hamster insulinoma cell line H1T-T15 M2.2.2, Boam et al. (9) found that deletion of the Z-element region between base pairs −281 and −260 caused a 25-fold activation of the hIns promoter relative to the base pair −281 promoter, and therefore concluded that a negative response element lies between base pairs −281 and −260. We, like Boam et al., observed a sixfold decrease in promoter activity with the Z minienhancer when compared with an enhancerless control vector (Fig. 2A). As in primary cultured islet cells, the X and the Y minienhancers had very weak activity in HIT cells (Fig. 2A).
Figure 2.
Function of the minienhancers in tumor cell lines and primary cultured fibroblasts. Cells were transfected with plasmids containing five tandem copies of the minienhancers shown or a single copy of the Rous sarcoma virus enhancer, linked to the base pair −109 thymidine kinase (TK) promoter driving the CAT gene in pFOXCAT2. In C, the base pair −410 rInsI promoter was linked directly to the CAT gene. CAT activity of cells transfected with the enhancerless pFOXCAT2TK plasmid was arbitrarily set at 1.0. Each data point represents the mean of at least four independent transfections ± SEM.
To exclude the possibility that the observed repressor effect of Z element could be a unique feature of HIT cells, we also transfected two other immortalized beta-cell lines with the same constructs. βTC3 and βHC9 cells are both derived from transgenic mice that express the simian virus 40 large tumor antigen (T antigen) under control of the insulin promoter (24, 25). Beta tumor cells (βTCs) are generated from fully developed beta-cell tumors, and beta hyperplasia cells (βHCs) from pretumorigenic, hyperplastic islets. In contrast to tumorigenic beta cells, βHC are thought to maintain normal islet physiology and response to glucose (25). However, as in HIT cells, the Z minienhancer represses transcription in both βHC9 and βTC3 cells (Fig. 2B, and data not shown), suggesting fundamental differences of insulin gene regulation between immortalized beta cells and native islet cells. We also observed transcriptional repression by the Z element in αTC1.6 cells, a cell line derived from alpha cell tumors produced by expression of a simian virus 40 large T antigen transgene, results similar to those obtained in immortalized beta cells (data not shown). We also assayed for activity of the Z minienhancer in non-islet cell lines and found that the Z element acts as a transcriptional repressor in mPAC4.1.2.1 cells, a pancreatic ductal carcinoma cell line (Fig. 2C) and in BHK21 cells, a cell line from hamster kidney (data not shown).
These data raised the possibility that repression of transcription by the Z element is a unique feature of tumor cells or cells expressing the simian virus 40 large T antigen. To explore this hypothesis, we tested the Z element in primary cultured rat fibroblasts. In contrast to primary islet cells, the Z minienhancer causes a marked reduction in transcriptional activity (Fig. 2D), similar to the effect observed in tumor cell lines.
The Z Element Binds a Glucose-Responsive Protein Complex.
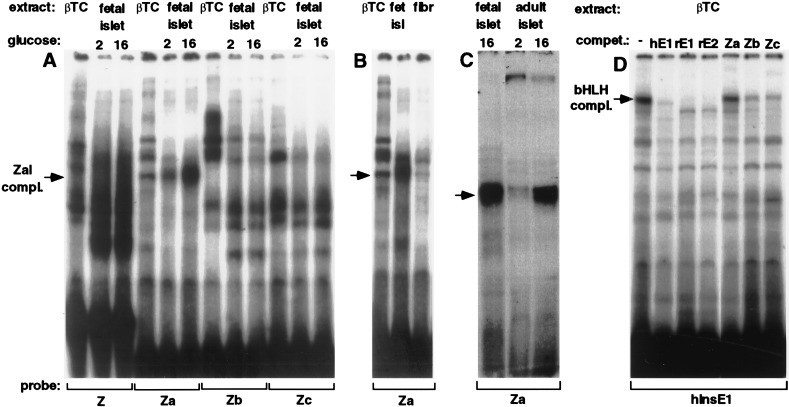
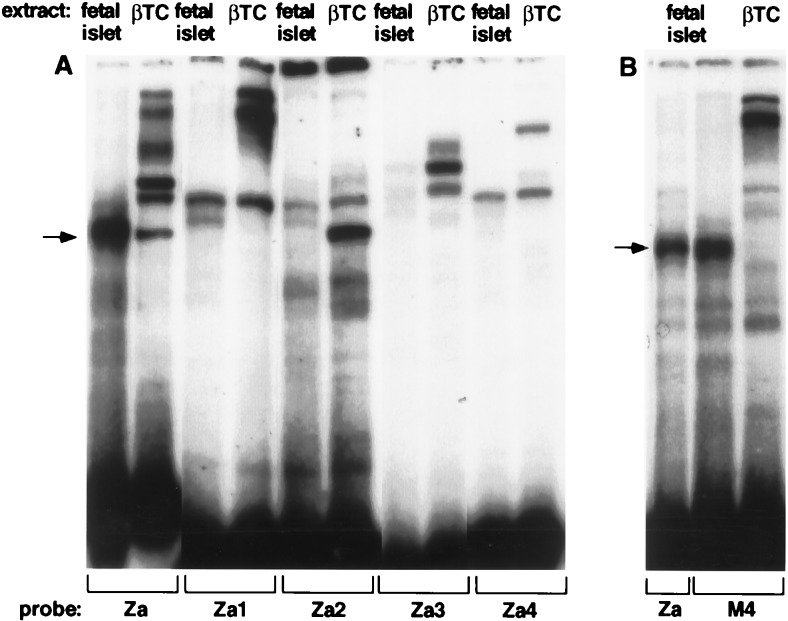
To test for glucose effects on nuclear DNA-binding proteins, we made nuclear extracts from both fetal and adult rat islets after growth in various glucose concentrations. The fetal islets were grown and harvested in the same way as the transfected islet cultures. Employing EMSA, we tested nuclear extracts for the ability to bind labeled double-stranded oligonucleotides spanning the Z-element region (Table 1). As seen in Fig. 3A, a large number of protein complexes bind to the Z element. Interestingly, the most distal part of the Z element (Za) binds a fast-migrating complex that markedly increases in abundance in response to high glucose concentration (ZaI complex). A narrower band, which in some gels had a slightly faster mobility than the ZaI complex, is present in nuclear extracts from βTC cells (Fig. 3A) and from rat primary fibroblasts (Fig. 3B). This finding suggests that the glucose-responsive complex may be distinct from the complex present in extracts from βTC cells and primary fibroblasts. By EMSA, there was no obvious difference in the complexes that bound to the Zb and Zc element between βTC and primary fibroblast nuclear extracts (data not shown).
Figure 3.
Binding of primary islet and βTC nuclear extracts, and of the islet E box binding complex to the Z element region. The binding specificities of nuclear proteins were tested by EMSA. See Table 1 for the sequences of the probes and competitors. Nuclear extract from primary cultured fetal rat islet cells grown in 2 mM or 16 mM glucose (A and C), βTC cells (A–D), primary cultures rat fibroblasts (B), or cultured adult rat islet cells grown in 2 mM or 16 mM glucose (C) was added to the 32P-labeled probes shown. (D) The binding specificities of the βTC nuclear hInsE1 binding complexes were tested by adding a 20-fold molar excess of the unlabeled competitors shown: human insulin E1 (hE1) rat insulin I E1 (rE1), and rat insulin I E2 (rE2). The resulting protein-DNA complexes were separated by polyacrylamide gel electrophoresis. The glucose-responsive complex (ZaI) is indicated by the arrows (A–C).
To test whether extracts from adult primary islets contain a complex of identical mobility and glucose response as fetal islet extracts, we directly compared both nuclear extracts in an EMSA, using the Za oligonucleotide as a probe. As shown in Fig. 3C, adult islets contain the same complex as fetal islets, and they show an even stronger glucose response.
The Glucose-Responsive Complex Does Not Bind to E-Boxes.
To identify potential transcription-factor binding sites, we searched the Z element for known recognition sites of DNA-binding proteins, and identified a palindromic sequence motif between base pairs −284 and −275 and two E-boxes, one directly adjacent to the palindrome (base pair −274 to −268) and a second one further 3′ between base pairs −263 and −258 (E3) (see Fig. 1A). In beta-cell nuclei, among other proteins, E-boxes bind a protein complex containing the ubiquitously expressed bHLH proteins E12/E47 (26–29) and BETA2 (30), a protein of limited tissue distribution.
We therefore asked whether oligonucleotides spanning the Z element could bind this heterodimeric bHLH protein complex. As shown in Fig. 3D, a slow-migrating complex is formed when an end-labeled oligonucleotide containing the hIns gene E1 E-box is added to nuclear extract from βTC cells. This complex has previously been shown to contain the E12/E47/BETA2 heterodimer (27, 30). To test the binding specificity of the complex, several 5′ sequence elements were added as unlabeled competitors. As expected, the rInsI gene E-boxes, E1 and E2, compete for binding of the complex to the hIns E1 site (Fig. 3D). The Zb and Zc elements, both of which contain the second potential E-box (E3), can compete weakly for the hIns E1 binding complex, but the Za element cannot (Fig. 3D). This competition experiment demonstrates that whereas E12/47/BETA2 heterodimers may bind to the glucose-responsive Z element in beta-cell nuclei, they cannot bind the Za sequence and are not responsible for the glucose-responsive ZaI complex.
By using antibodies against known islet cell transcription factors, we attempted to identify the proteins contained in the glucose-responsive complex. Antibodies against the bHLH proteins E12/E47, Meso1, BETA2 and the homeodomain proteins NKX6.1, PDX1, CDX3, and PAX6 all fail to supershift the glucose-responsive complex in an EMSA using nuclear extract from fetal rat islets (data not shown). These results suggest that unidentified transcription factors may bind to the Za site.
Fine Mapping the Protein Binding Site.
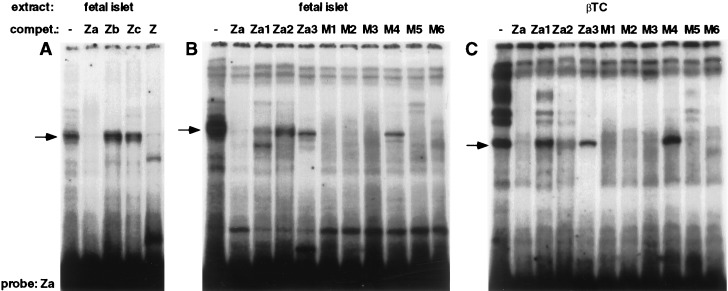
To further delineate the binding site for the glucose-responsive ZaI complex, four 15- to 17-bp oligonucleotides (Za1 to Za4) were constructed to cover the Za region (Table 1). Using the Za element as a radioactively labeled probe, we added these oligonucleotides as unlabeled competitors in an EMSA with extracts from fetal rat islets. As expected, the Z and Za oligonucleotides compete for binding of the ZaI complex (Fig. 4A). In contrast, all shorter oligonucleotides fail to compete efficiently for binding of the ZaI complex (Fig. 4B, and data not shown), although the Za1 probe competes weakly. In addition, when the same oligonucleotides were used as labeled probes, they did not bind a complex of similar mobility (Fig. 5A). This experiment suggests that the glucose-responsive ZaI complex either requires a larger binding site or depends on more than one binding site within the region.
Figure 4.
Binding of primary islet and βTC nuclear extract to the Za element region. The binding specificities of nuclear proteins were tested by EMSA. See Table 1 for the sequences of the probe and competitors. Nuclear extract from primary cultured fetal rat islet cells grown in 16 mM glucose (A and B) and from βTC (C) was added to the 32P-labeled Za probe. The binding specificities of the nuclear Za binding complexes were tested by adding a 150-fold molar excess of the unlabeled competitors shown. The glucose-responsive complex (ZaI) is indicated by the arrows.
Figure 5.
Distinguishing primary islet and βTC nuclear complexes binding to the Za region. The binding specificities of nuclear proteins were tested by EMSA. See Table 1 for the sequences of the probe and competitors. Nuclear extract from βTC or from primary cultured fetal rat islet cells grown in 16 mM glucose was added to the 32P-labeled probes shown. The glucose-responsive complex (ZaI) is indicated by the arrows.
To identify the bases responsible for protein binding, six different mutations with a C-to-A or G-to-T conversion were introduced along the Za-region (Table 1). None of the mutations introduced into the Za oligonucleotide completely abolishes binding of the competitor to the glucose-responsive ZaI complex; only mutant 4 fails to compete for part of the complex (Fig. 4B).
The Glucose-Responsive Complex Is Found Only in Primary Cultured Islet Cells.
Based on the EMSA using nuclear extracts from primary islets compared with tumorigenic beta cells, it appeared that the ZaI complex may be distinct from the complex contained in βTC cells (Fig. 3A). To distinguish the complex seen in primary cells from that seen in tumor cells, we performed an EMSA by using βTC nuclear extract with the same competitors as described in Fig. 4B. In contrast to the competition experiment using extracts from fetal rat islets, the 17-bp Za2 oligonucleotide competes more effectively for the complex in βTC cells, and mutation 4 renders the Za element unable to compete for the Za-binding complex (Fig. 4C). This experiment provides additional evidence that the ZaI complex contained in primary islets may be distinct from the complex of similar mobility in tumor cells, and identifies the thymidine at base pair −271 as a crucial base for binding of the tumor cell complex.
To substantiate that the two complexes bind to different sites, we used the four oligonucleotides spanning the Za region and mutant 4 as probes in an EMSA with extracts from fetal rat islets or βTC cells. As suggested by the competition experiments, βTC cells but not fetal islets contain the complex binding to the Za2 oligonucleotide (Fig. 5A). Likewise, mutant 4 binds a complex of similar mobility to the glucose-responsive complex in primary islets, but not in βTC nuclear extracts (Fig. 5B). These results clearly show that the glucose-responsive ZaI complex is only present in primary islets and not in cells derived from beta-cell tumors.
DISCUSSION
The Z Element as a Positive Regulator of Insulin Gene Transcription.
We have shown that the Z element functions as a potent transcriptional enhancer in primary cultured islet cells, but represses transcription in immortalized beta and nonbeta cells and primary fibroblasts. These results are not inconsistent with the work of K. Docherty and colleagues (9, 31), who delineated a negative regulatory element between base pairs −281 and −260 of the hIns promoter, because those studies were performed exclusively in tumor-cell lines.
Based on our data, two simple models could account for the opposite behavior of the Z element in the primary cultured islet cells and in the other cells tested. First, a repressor that is present in the other cells may be absent from primary cultured beta cells, allowing a ubiquitous activator to function. Alternatively, the activator may be expressed only in primary islet cells, where it overrides the effect of a ubiquitous repressor. Our data are most consistent with the second model, although we cannot rule out the former. The ZaI complex, which binds to the proximal portion of the Z element, is present in nuclear extracts from fetal and adult primary islets, but is absent from βTC or primary fibroblast nuclear extracts. The intensity of this band correlates with the activity of the Z element, suggesting that it could function as an activator. The Z element also binds several nuclear complexes that appear to be ubiquitous; one or more of these could function as repressors.
It should be noted that the isolated Za region that binds the ZaI complex has virtually no activity in primary islet cells when it is separated from the remainder of the larger Z element. If the ZaI complex functions as a transcriptional activator, as we propose, it must do so by interacting with proteins that bind to other sequences within the Z element. In this regard, the ZaI complex may function similarly to the bHLH proteins that bind to the insulin-promoter E elements but only act as transcriptional activators in cooperation with proteins binding to the adjacent A3 (13, 19, 32) or C1 (29, 33, 34) elements.
What explains the specific expression of the ZaI complex in primary cultured islet cells? We first considered the possibility that expression of ZaI could be extinguished by the profound changes that occur when cells are immortalized or by the actions of the simian virus 40 large T oncogene that is active in HIT and βTC cells. Our data, however, argue against this possibility, because both transcriptional repression by the Z element and the absence of the ZaI complex also were observed in primary fibroblasts that were not immortalized. Alternatively, the ZaI complex may be expressed only in completely differentiated beta cells or islet cells, but not in less well-differentiated insulinoma cell lines, possibly explaining their impaired insulin production (11, 12). Finally, the expression of the ZaI complex could be regulated by the proliferation state of the cells. In contrast to both tumor cell lines and primary fibroblasts, primary islet cells do not divide in culture. This model suggests the interesting possibility that the transcription of the human insulin gene could be directly linked to the proliferation state of the beta cell through the activity of the ZaI complex binding to the Z element.
The Z Element Confers Glucose Responsiveness to the Insulin Gene Promoter.
In an earlier study we identified the E1 and A2/C1 elements as glucose-responsive elements in the hIns promoter in primary islet cultures (13). Deletion analysis, however, indicated that these elements do not account for the full glucose responsiveness of the promoter. In this context we have mapped an additional glucose-responsive element to the Z-element region and shown that the Z element functions as a strong glucose-responsive element when isolated from the remainder of the promoter.
The remarkable increase in binding of the ZaI complex in response to glucose strongly suggests that it contributes to the transcriptional activation of the Z element by glucose, although the Za binding site does not respond to glucose by itself and requires the presence of adjacent elements to confer full activity and metabolic response to the Z minienhancer. The ability of adjacent elements to synergize and enhance glucose response has been well-demonstrated in the rInsI promoter, where the E2-A3/4 minienhancer functions as an independent glucose-responsive transcriptional unit (1, 16). In this example, glucose regulates binding of the bHLH protein dimers that bind to the E2 element (18). The Z element contains several potential E elements, one of which (E3) can compete for binding of the bHLH complex found in beta-cell nuclei. The competition analysis, however, demonstrates that the ZaI complex is distinct from the E element binding complex.
Other investigators have mapped glucose responsiveness to the A elements in the insulin promoter, including the A3 element in the hIns promoter (16, 17) (see Fig. 1A). The A3 element binds the homeodomain transcription factor PDX1, which is phosphorylated in cells grown in high glucose (35), as well as several other homeodomain proteins. Addition of antibodies against these beta-cell homeodomain proteins including PDX1 does not alter the mobility of the ZaI complex in EMSA. Therefore, the protein(s) in the ZaI complex appear to represent a novel glucose-regulated protein(s).
In conclusion, this study supports the concept that activity of the insulin promoter and its response to metabolic signals are controlled by the interactions of a number of nuclear proteins that bind to different sequence elements along the promoter. This model allows the beta cell to differentially respond to metabolic signals by modulating the transcriptional response through more than one pathway.
Acknowledgments
We thank Juehu Wang, Janet Lau, and Yi Zhang for excellent technical assistance, and our laboratory colleagues for critical readings of the manuscript. This work was supported by grants from the National Institutes of Health (DK48281) and the Greenwall Foundation to M.S.G.; M.S. is a recipient of postdoctoral fellowships from the Deutsche Forschungsgemeinschaft and the Juvenile Diabetes Foundation International, and S.C.G. is a recipient of a National Institutes of Health Research Service Award (DK0937702).
ABBREVIATIONS
- βTC
beta tumor cell
- βHC
beta hyperplasia cell
- bHLH
basic helix–loop–helix
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility shift assay
- hIns
human insulin
- rInsI
rat insulin I
- PEI
polyethylenimine
- large T antigen
large tumor antigen
References
- 1. German M S, Moss L G, Rutter W J. J Biol Chem. 1990;265:22063–22066. [PubMed] [Google Scholar]
- 2.Walker M D, Edlund T, Boulet A M, Rutter W J. Nature (London) 1983;306:557–581. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- 3.Edlund T, Walker M D, Barr P J, Rutter W J. Science. 1985;230:912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D. Nature (London) 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 5.Cordle S R, Whelan J, Henderson E, Masuoka H, Weil P A, Stein R. Mol Cell Biol. 1991;11:2881–2886. doi: 10.1128/mcb.11.5.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlsson O, Edlund T, Moss J B, Rutter W J, Walker M D. Proc Natl Acad Sci USA. 1987;84:8819–8823. doi: 10.1073/pnas.84.24.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whelan J, Poon D, Weil P A, Stein R. Mol Cell Biol. 1989;9:3253–3259. doi: 10.1128/mcb.9.8.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowe D T, Tsai M-J. Mol Cell Biol. 1989;9:1784–1789. doi: 10.1128/mcb.9.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boam D S, Clark A R, Docherty K. J Biol Chem. 1990;265:8285–8296. [PubMed] [Google Scholar]
- 10.Hammonds P, Schofield P N, Ashcroft S J H. FEBS Lett. 1987;213:149–154. doi: 10.1016/0014-5793(87)81481-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Shen L, Najafi H, Kolberg J, Matschinsky F, Urdea M, German M. Proc Natl Acad Sci USA. 1997;94:4360–4365. doi: 10.1073/pnas.94.9.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen D A, Welsh M, Casadaban M J, Steiner D F. J Biol Chem. 1985;260:13585–13589. [PubMed] [Google Scholar]
- 13.Odagiri H, Wang J, German M S. J Biol Chem. 1996;271:1909–1915. doi: 10.1074/jbc.271.4.1909. [DOI] [PubMed] [Google Scholar]
- 14.Ephrussi A, Church G, Tonegawa S, Gilbert W. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 15.Gehring W J, Affolter M, Burglin T. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 16.Melloul D, Ben-Neriah Y, Cerasi E. Proc Natl Acad Sci USA. 1993;90:3865–3869. doi: 10.1073/pnas.90.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen H V, Serup P, Leonard J, Michelsen B K, Madsen O D. Proc Natl Acad Sci USA. 1994;91:10465–10469. doi: 10.1073/pnas.91.22.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.German M, Wang J. Mol Cell Biol. 1994;14:4067–4075. doi: 10.1128/mcb.14.6.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.German M S, Moss L G, Wang J, Rutter W J. Mol Cell Biol. 1992;12:1777–1788. doi: 10.1128/mcb.12.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curiel D T, Agarwal S, Wagner E, Cotten M. Proc Natl Acad Sci USA. 1991;88:8850–8854. doi: 10.1073/pnas.88.19.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignam J D, Lebowitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fried M, Crothers D M. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garner M M, Revzin A. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efrat S, Linde S, Kofod H, Spector D, Delannoy M, Grant S, Hanahan D, Baekkeskov S. Proc Natl Acad Sci USA. 1988;85:9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radvanyi F, Christgau S, Baekkeskov S, Jolicoeur C, Hanahan D. Mol Cell Biol. 1993;13:4223–4232. doi: 10.1128/mcb.13.7.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aronheim A, Ohlsson H, Park C W, Edlund T, Walker M D. Nucleic Acids Res. 1991;19:3893–3899. doi: 10.1093/nar/19.14.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.German M S, Blanar M A, Nelson C, Moss L G, Rutter W J. Mol Endocrinol. 1991;5:292–299. doi: 10.1210/mend-5-2-292. [DOI] [PubMed] [Google Scholar]
- 28.Cordle S R, Henderson E, Masuoka H, Weil P A, Stein R. Mol Cell Biol. 1991;11:1734–1738. doi: 10.1128/mcb.11.3.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheih S, Tsai M. J Biol Chem. 1991;266:16708–16714. [PubMed] [Google Scholar]
- 30.Naya F J, Stellrecht C M, Tsai M J. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 31.Clark A R, Wilson M E, Leibiger I, Scott V, Docherty K. Eur J Biochem. 1995;232:627–632. [PubMed] [Google Scholar]
- 32.German M S, Wang J, Chadwick R B, Rutter W J. Genes Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- 33.Sharma A, Stein R. Mol Cell Biol. 1994;14:871–879. doi: 10.1128/mcb.14.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwung Y P, Gu Y Z, Tsai M J. Mol Cell Biol. 1990;10:1784–1788. doi: 10.1128/mcb.10.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macfarlane W M, Smith S B, James R F, Clifton A D, Doza Y N, Cohen P, Docherty K. J Biol Chem. 1997;272:20936–44. doi: 10.1074/jbc.272.33.20936. [DOI] [PubMed] [Google Scholar]