Abstract
The worldwide commercial production of the oyster mushroom Pleurotus ostreatus is currently threatened by massive attacks of green mold disease. Using an integrated approach to species recognition comprising analyses of morphological and physiological characters and application of the genealogical concordance of multiple phylogenetic markers (internal transcribed spacer 1 [ITS1] and ITS2 sequences; partial sequences of tef1 and chi18-5), we determined that the causal agents of this disease were two genetically closely related, but phenotypically strongly different, species of Trichoderma, which have been recently described as Trichoderma pleurotum and Trichoderma pleuroticola. They belong to the Harzianum clade of Hypocrea/Trichoderma which also includes Trichoderma aggressivum, the causative agent of green mold disease of Agaricus. Both species have been found on cultivated Pleurotus and its substratum in Europe, Iran, and South Korea, but T. pleuroticola has also been isolated from soil and wood in Canada, the United States, Europe, Iran, and New Zealand. T. pleuroticola displays pachybasium-like morphological characteristics typical of its neighbors in the Harzianum clade, whereas T. pleurotum is characterized by a gliocladium-like conidiophore morphology which is uncharacteristic of the Harzianum clade. Phenotype MicroArrays revealed the generally impaired growth of T. pleurotum on numerous carbon sources readily assimilated by T. pleuroticola and T. aggressivum. In contrast, the Phenotype MicroArray profile of T. pleuroticola is very similar to that of T. aggressivum, which is suggestive of a close genetic relationship. In vitro confrontation reactions with Agaricus bisporus revealed that the antagonistic potential of the two new species against this mushroom is perhaps equal to T. aggressivum. The P. ostreatus confrontation assays showed that T. pleuroticola has the highest affinity to overgrow mushroom mycelium among the green mold species. We conclude that the evolutionary pathway of T. pleuroticola could be in parallel to other saprotrophic and mycoparasitic species from the Harzianum clade and that this species poses the highest infection risk for mushroom farms, whereas T. pleurotum could be specialized for an ecological niche connected to components of Pleurotus substrata in cultivation. A DNA BarCode for identification of these species based on ITS1 and ITS2 sequences has been provided and integrated in the main database for Hypocrea/Trichoderma (www.ISTH.info).
Pleurotus ostreatus is an important edible basidiomycete commonly known as oyster mushroom. This fungus is the third most commercially important edible mushroom worldwide (4). In addition, it is used for the bioconversion of agricultural and industrial lignocellulose debris (2, 32) and as a source of enzymes and other metabolites for industrial and medical applications (13, 26). P. ostreatus can be grown on a wide range of agricultural by-products and industrial wastes (29), although pasteurized straw is most commonly used. Many pests and diseases can cause yield losses in P. ostreatus, and the association of Trichoderma species with the cultivation substratum has long been known to limit production (1). Sharma and Vijay (38) reported green mold of oyster mushroom in North America 10 years ago, and serious cases of green mold diseases of P. ostreatus in commercial operations were detected recently in South Korea (30), Italy (41), Hungary (14), and Romania (20).
Trichoderma green mold infection in edible basidiomycetes has been known for a long time (39). An Agaricus green mold disease started in Northern Ireland in 1985 and rapidly spread over farms across Europe (15, 25). A similar disease appeared in mushroom crops in the United States and Canada (3). The causative agent was originally believed to be Trichoderma harzianum (teleomorph Hypocrea lixii) but was later on clarified to be a new species of Trichoderma, viz., Trichoderma aggressivum, of which two varieties (T. aggressivum var. europaeum and T. aggressivum var. aggressivum) were distinguished from Europe and North America, respectively (35).
The causative agent of the oyster mushroom green mold has been reported to be morphologically and culturally distinct from T. aggressivum (30, 41). Park et al. (30) claimed that two new species (Trichoderma koreana and Trichoderma pleuroti) were responsible for the disease on Pleurotus in South Korea, but they did not provide nomenclaturally valid species descriptions. Hatvani et al. (14) reported that the Hungarian oyster mushroom green mold species has the same internal transcribed spacer 1 (ITS1) and ITS2 sequences as an undescribed species of Trichoderma (Trichoderma sp. strain DAOM 175924) (22) and that its ITS1 and ITS2 sequences were also identical with those deposited for four Trichoderma pathogens of P. ostreatus from South Korea. Recently, Park et al. (31) have formally described two new species causing Pleurotus green mold disease in South Korea, Trichoderma pleurotum and Trichoderma pleuroticola.
The objective of the present study was to use an integrated approach comprising morphological, physiological, and molecular analyses to investigate the evolution of the Trichoderma strains causing Pleurotus green mold disease and to examine reasons for the recent disease outbreaks.
MATERIALS AND METHODS
Fungal strains.
Strains investigated in this study are given in Table 1. They are maintained in the culture collections of the Division of Gene Technology and Applied Biochemistry, Vienna University of Technology, Vienna, Austria, under assigned CPK numbers; at DAOM (Eastern Cereal and Oilseed Research Centre, Ottawa, Canada); and at the Section of Plant Pathology, University of Naples, Portici (NA), Italy. Representative cultures have also been deposited at the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands.
TABLE 1.
Pleurotus green mold strains used in this study
| Species and strain | Alternative strain no. | Origin | Habitat | NCBI GenBank accession no.d
|
||
|---|---|---|---|---|---|---|
| ITS1 and ITS2 | tef1 | chi18-5 | ||||
| T. pleuroticola strains | ||||||
| DAOM 175924a | CBS 121144 | Canada (Ontario) | On Acer sp. | AY605726 | AY605769 | |
| DAOM 229916 | United States (Wisconsin) | Forest soil, A1 horizon | AY605738 | AY605781 | ||
| CPK 1540 | CBS 121217 | Italy | P. ostreatus substratum | EF392782 | EF392762 | |
| CPK 1541 | Italy | P. ostreatus substratum | EF392783 | |||
| CPK 1542 | Italy | P. ostreatus incubating bales | EF392784 | |||
| CPK 1543 | Italy | P. ostreatus incubating bales | EF392785 | |||
| CPK 1544 | Italy | P. ostreatus incubating bales | EF392786 | EF392763 | ||
| CPK 1545 | Italy | P. ostreatus incubating bales | EF392787 | |||
| CPK 1546 | Italy | P. ostreatus incubating bales | EF392788 | EF392764 | ||
| CPK 1547 | Italy | P. ostreatus incubating bales | EF392789 | |||
| CPK 1548 | Italy | P. ostreatus incubating bales | EF392790 | |||
| CPK 1550 | Italy | Mushroom farm | EF392791 | EF392765 | ||
| CPK 1551 | Italy | Mushroom farm | EF392792 | EF392766 | ||
| CPK 1552 | Italy | Mushroom farm | EF392793 | EF392767 | ||
| CPK 2104 | CBS 121145 | Hungary | P. ostreatus substratum | EF392794 | EF392769 | |
| CPK 2816 | Romania | P. ostreatus substratum | EF601676 | EF601681 | ||
| CPK 2817 | Romania | P. ostreatus substratum | EF601677 | EF601682 | ||
| DZ 56 | CPK 882, CBS 121146 | Iran | Compost for Agaricus bisporus | EF392781 | EF392761 | EF392776 |
| GJS 04-01 | United States (Montana) | Biocontrol of Cercosporab | EF392768 | |||
| GJS 95-81 | The Netherlands | Pleurotus spawn | AF345948 | AF348102 | ||
| CBS 628.77 | The Netherlands | Foodstuff | AF194006 | |||
| JB T2290 | Canada (Quebec) | Elm log | AY605746 | AY605789 | EF392777 | |
| BBA 65638 | Germany | AF194007 | ||||
| GJS 95-14 | New Zealand | Biocontrol of Armillariac | AF055216 | |||
| T. pleurotum strains | ||||||
| CPK 2113a | CBS 121147, DAOM 236051 | Hungary | P. ostreatus substratum | EF392808 | EF392773 | EF392779 |
| CPK 2095 | Hungary | P. ostreatus substratum | EF392796 | |||
| CPK 2096 | Hungary | P. ostreatus substratum | EF392797 | EF392770 | ||
| CPK 2097 | Hungary | P. ostreatus substratum | EF392798 | EF392771 | ||
| CPK 2098 | Hungary | P. ostreatus substratum | EF392799 | EF392778 | ||
| CPK 2099 | Hungary | P. ostreatus substratum | EF392800 | |||
| CPK 2100 | Hungary | P. ostreatus substratum | EF392801 | EF392772 | ||
| CPK 2101 | Hungary | P. ostreatus substratum | EF392802 | |||
| CPK 2102 | Hungary | P. ostreatus substratum | EF392803 | |||
| CPK 2103 | Hungary | P. ostreatus substratum | EF392804 | |||
| CPK 2109 | Hungary | P. ostreatus substratum | EF392805 | |||
| CPK 2110 | Hungary | P. ostreatus substratum | EF392806 | |||
| CPK 2112 | Hungary | P. ostreatus substratum | EF392807 | |||
| CPK 2114 | Hungary | P. ostreatus substratum | EF392809 | |||
| CPK 2116 | CBS 121148 | Hungary | P. ostreatus substratum | EF392810 | EF392774 | EF392780 |
| CPK 2117 | Hungary | P. ostreatus substratum | EF392811 | EF392775 | ||
| CPK 1532 | CBS 121216 | Italy | P. ostreatus substratum | EF392795 | EF601678 | |
| CPK 2814 | Romania | P. ostreatus substratum | EF601674 | EF601679 | ||
| CPK 2815 | Romania | P. ostreatus substratum | EF601675 | EF601680 | ||
Reference strain.
Obtained from G. J. Samuels as T. koningii; used for biocontrol of Cercospora.
Indicated as a biocontrol agent by Dodd et al. (8).
Accession numbers in roman type are newly submitted; those in italics were submitted previously and retrieved for this work.
Morphological analysis.
Cultures were grown on 2% Oxoid malt extract agar (MA) and Oxoid potato dextrose agar (PDA) at 20°C under ambient daylight conditions or in a 12 h:12 h light:dark cycle under fluorescent and near-UV lamps. Colony descriptions are based on observations on MA under ambient daylight conditions, unless otherwise specified. Color codes and terminology are from the Methuen Handbook of Colours (18). Growth rates from 5°C to 40°C at increments of 5° were determined on PDA using the protocol of Samuels et al. (34). Microscopic observations and measurements were made from preparations mounted in lactic acid. Conidiophore structure and morphology were described from macronematous conidiophores taken from the edge of conidiogenous pustules or fascicles when conidia were maturing, usually after 4 to 7 days of incubation. Conidial morphology and measurements were recorded after 14 days.
Metabolic profiles.
Metabolic profiles based on assimilation of carbon sources were performed using Biolog FF MicroPlates (6, 9, 19, 21). Microplates were incubated at 26°C in the dark, and absorbance readings at 490 nm and at 750 nm were analyzed separately. Absorbance data were not corrected for growth in the control well, which was treated as an independent variable in the analyses. Cluster analyses were performed using NTSYS software (33) with a similarity matrix using the product-moment correlation coefficient and employing the unweighted-pair group method using average linkages. SAS was used for analyses of variance (ANOVAs) and canonical variate analyses (36). Univariate ANOVAs were performed on data for each of the 95 different carbon substrata and the control. The substrata were ranked on the ANOVA F values and the degree of significance of the among-species variation in the ANOVAs. The highest ranked variables (probability > F < 0.0001) were selected to perform canonical variate analysis. Wilk's Lambda and Pillai's trace were employed to test the significance of the canonical variate analysis. The total standardized canonical coefficients were used to interpret the three significant eigenvectors obtained from the analysis.
Dual confrontation assays.
To assess the antagonistic potential of T. pleuroticola and T. pleurotum against P. ostreatus and A. bisporus, we isolated pure cultures of the respective mushrooms from the products available on the Austrian market. Three strains of each species were used in dual confrontation tests at 26°C, with T. aggressivum CBS 433.95 and Hypocrea jecorina/Trichoderma reesei QM 6a as positive and negative controls, respectively.
DNA extraction, PCR, and sequencing.
After 5 days of growth on MA at 25 ± 1°C, mycelia were harvested, and genomic DNA was isolated using a QIAGEN DNeasy Plant mini kit by following the manufacturer's protocol.
Amplification of the nuclear rRNA gene cluster, containing the ITS1 and ITS2 and the 5.8S rRNA gene, and of a 0.4-kb fragment of endochitinase chi18-5 (formerly named ech42) was done as described previously (16). An approximately 1-kb portion of the tef1 was amplified and sequenced using primers EF1 [5′-ATGGGTAAGGA(A/G)GACAAGAC-3′] and EF2 [GGA(G/A)GTACCAGT(G/C)ATCATGTT-3′] (28) or as described in Jaklitsch et al. (16).
Purified PCR products for ITS1 and ITS2, tef1, and chi18-5 were subjected to automatic sequencing at MWG (Martinsried, Germany). Sequences were edited manually and deposited in NCBI GenBank and www.ISTH.info.
Phylogenetic analysis.
For the phylogenetic analysis, DNA sequences were aligned using ClustalX and visually edited in Genedoc, version 2.6 (27). The interleaved NEXUS file was formatted using PAUP*, version 4.0b10 (40), and manually formatted for the MrBayes program, version 3.0B4. The Bayesian phylogenetic reconstructions have been performed as described in Jaklitsch et al. (16). According to the protocol of Leache and Reeder (23), posterior probability values lower than 0.95 were not considered significant while values below 0.9 were not shown on the consensus phylogram.
Haplotype networks were constructed manually based on detected shared polymorphic sites and confirmed using statistical parsimony analysis as implemented in TCS, version 2.11 (5), and maximum parsimony analysis using PAUP*, version 4.0b10 (40).
RESULTS
The P. ostreatus-associated strains comprise two phylogenetic Trichoderma species.
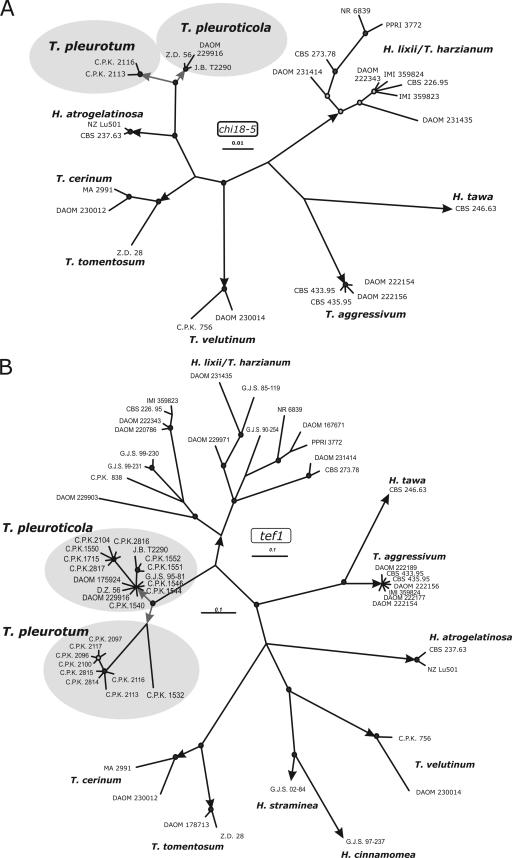
Strains from oyster mushroom-producing farms having severe green mold infections from Hungary, Romania, Italy, and South Korea exhibited the same or highly similar ITS1 and ITS2 sequences as the previously recognized putative new species “Trichoderma cf. aureoviride DAOM 175924” (NCBI GenBank accession no. AY605726) (22) and the recently described Pleurotus green mold agent from Korea T. pleuroticola CNUMH 601 (NCBI GenBank DQ164409) (31). We have previously documented that strain DAOM 175924 forms a separate phylogenetic branch in the vicinity of T. harzianum and T. aggressivum in the Harzianum clade (22, 24). We applied two criteria of Dettman et al. (7) employing multilocus genealogies to determine if the various isolates associated with green mold on Pleurotus represented a single species or more than one distinct species. The criterion of genealogical concordance requires that the clade must be present in the majority of single-locus trees. The genealogical nondiscordance criterion recognizes a clade as an independent evolutionary lineage when it is reliably supported by at least one single-locus genealogy and if it is not contradicted by any other single gene tree determined by the same methods. To do this, we amplified and sequenced fragments from three different phylogenetic markers, i.e., a fragment spanning the ITS1-5.8S rRNA-ITS2 region of the rRNA gene cluster, a fragment covering the fourth and fifth introns and the last long exon of the translation elongation factor 1-alpha (tef1) gene, and a fragment including a portion of the fifth exon of the chi18-5 gene encoding a family 18 chitinase. In order to compose the sample set for phylogenetic analysis, the resulting sequences were subjected to the sequence similarity search tool implemented in TrichoBLAST (17; also www.ISTH.info). No identical hits except “Trichoderma sp. strain DAOM 175924” were detected, but the highest sequence homology was shown for species from the Harzianum clade (10). As shown in Fig. 1, the phylogenies obtained from independent analyses of tef1 and chi18-5 markers placed the Pleurotus green mold isolates and conspecific strains in one supported hypthetical taxonomic unit close to H. lixii/T. harzianum. On the chi18-5 tree the hypothetical taxonomic unit node of Pleurotus-associated isolates and allied strains further bifurcated into two significantly supported clades. A similar divergence also takes place on the tef1 tree: one clade comprises strain DAOM 175924, most of the Italian, one of the Hungarian, and two Romanian Pleurotus green mold strains, as well as environmental isolates from Iran, North America, and New Zealand; the second clade includes most of the Hungarian and two Romanian green mold strains, together with one strain isolated from soft rot of wood in Germany. A single Italian isolate, CPK 1532, from a Pleurotus farm occupies a basal position to this clade.
FIG. 1.
Bayesian analyses of the phylogenetic position of Pleurotus green mold species based on their chi18-5 and tef1 sequences. Posterior probability coefficients are given at respective nodes and shown only if the branch was highly supported (>0.95). Arrows indicate branches leading to currently recognized species. GenBank accession numbers for T. pleurotum and T. pleuroticola are given in Table 1. Accession numbers for other sequences may be retrieved from GenBank using the searches “species+strain+endochitinases 42” and “species+strain+translation elongation” for chi18-5 and tef1, respectively.
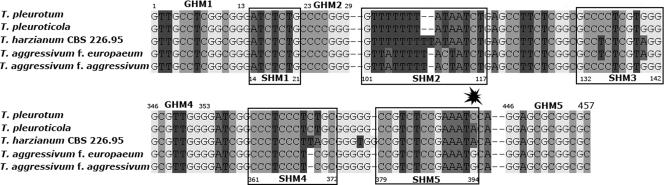
Visual analysis of ITS1 and ITS2 sequences (Fig. 2) support the relationship of the studied strains to the Harzianum clade. All strains from Pleurotus farms were segregated into two ITS2 alleles which differed from each other by one single nucleotide polymorphism (position 394 from the first nucleotide of the first genus-specific hallmark) (11). This divergence strictly corresponds to the two significant clades in the analyses of tef1 and chi18-5. Both of these alleles were 5 to 6 nucleotides different from the type allele of T. harzianum (CBS 226.95) and from the two known alleles of T. aggressivum.
FIG. 2.
ITS-based oligonucleotide BarCode for identification of mushroom green mold species. GHM1 to GMH5 and SHM1 to SHM5 indicate positions of genus- and species-specific hallmarks as indicated in Druzhinina et al. (11). The star shows the position of the diagnostic substitution inside SHM5 for T. pleurotum, T. pleuroticola, T. aggressivum, and H. lixii/T. harzianum. Type sequences were retrieved using accession numbers given in the study of Druzhinina et al. (11).
The concordant divergence of three loci showing two clades of Pleurotus green mold strains indicates the presence of two phylogenetic species. Because of the identity of the ITS1 and ITS2 sequences of strains DAOM 175924 and CNUMH 601 and of strains CPK 1532 and CNUMH 501 (NCBI GenBank DQ164405) and based on the similar ecological characterizations, we assume that the detected new species correspond to T. pleuroticola type culture CNUMH 601 and T. pleurotum type culture CNUMH 501, which have been formally described by Park et al. (31). Since ex-type cultures of these species were not deposited in publicly accessible culture collections and therefore were not available for this study, we refer to strains DAOM 175924 and CPK 1532 as reference strains for T. pleuroticola and T. pleurotum, respectively. For rapid molecular identification, oligonucleotide BarCodes based on differences in ITS1 and ITS2 sequences among T. pleurotum, T. pleuroticola, and related species have been implemented in the database for the Hypocrea/Trichoderma DNA oligonucleotide BarCode program TrichOKEY (11, 12; also www.ISTH.info).
Biogeography of T. pleuroticola and T. pleurotum.
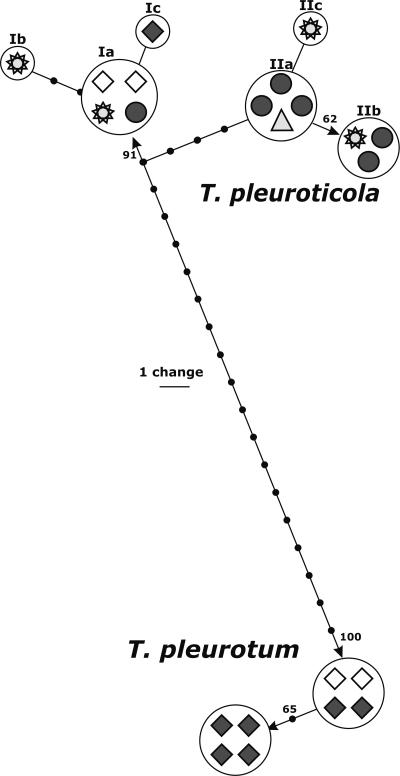
We performed a detailed analysis of the tef1 alleles of T. pleuroticola and T. pleurotum to investigate possible biogeographic traits in the distribution of the isolates associated with Pleurotus. Figure 3 shows the distribution of individual tef1 alleles among isolates from different locations. The scheme was constructed from one of 100 saved most parsimonious trees obtained using a heuristic search implemented in PAUP*, version 4b10. Six Hungarian and two Romanian strains of T. pleurotum showed almost no intraspecific variability since two groups of tef1 sequences (four isolates each) were separated by only one A↔G transition and one indel in one of several 5′-AnTn-3′ spans of the intron. In contrast, two major alleles of T. pleuroticola (Fig. 3, I and II) were distinguished based on five diagnostic transitions. Six tested Italian strains of T. pleuroticola isolated from cultivated Pleurotus substratum were found to be polymorphic; one strain has the tef1 allele Ia identical to strain GJS 04-01 known to be a biocontrol agent from Montana used against Cercospora in sugar beet; three strains share the same allele (IIa) with strain DZ56 isolated from Agaricus compost in Iran, and the two remaining strains have the tef1 allele (IIb) identical to that of reference strain DAOM 175924 isolated from Acer sp. in Canada. GJS 95-81 isolated from Pleurotus spawn in The Netherlands has one position that differs from the type allele. The only Hungarian isolate of T. pleuroticola (CPK 2104) belongs to the first major allele. Thus, the absence of any biogeographical pattern for the distribution of T. pleuroticola and, moreover, the mixed composition of the Italian sample suggest the presence of a distribution vector for the species.
FIG. 3.
Distribution of individual tef1 alleles among isolates from different locations. The scheme was manually constructed based on one of the 100 saved most parsimonious trees obtained using a heuristic search implemented in PAUP*, version 4b10 (39). ⧫, isolates from Hungary; ⋄, isolates from Romania; •, isolates from Italy; star, isolates from North America; ▴, single isolate from Iran. Arabic numbers correspond to bootstrap coefficients; roman numbers show main tef1 alleles.
Evolution of T. pleurotum was accompanied by a loss of certain carbon utilization traits.
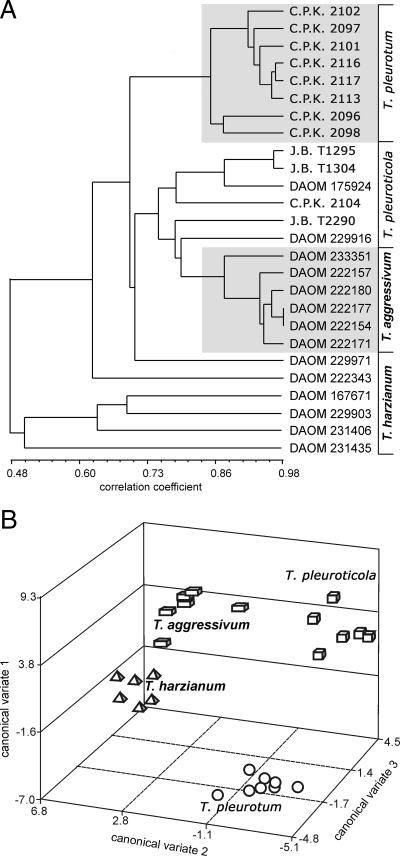
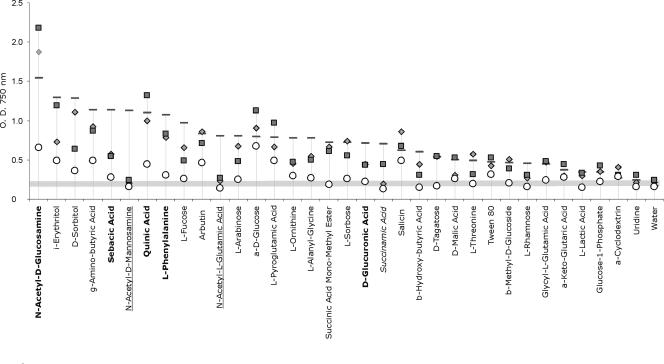
The results of a cluster analysis based on optical density at 750 nm (assimilation and growth) for 95 carbon sources after 96 h of incubation are presented in Fig. 4A. The strains of T. pleurotum formed a “monophenetic” grouping in the cluster analysis, whereas T. pleuroticola and T. aggressivum were not clearly separated. In univariate ANOVAs, however, highly significant differences among all the species (P of <0.001) were seen for 34 substrata. A canonical variate analysis was performed for the 12 most significant substrata (P of <0.0001). All three canonical variates were highly significant (P of <0.001) (Table 2), and a plot of T. pleuroticola, T. pleurotum, T. aggressivum, and T. harzianum on the three canonical variates (Fig. 4B) clearly distinguishes T. pleuroticola and T. pleurotum from each other, as well as from the closely related species in the Harzianum clade.
FIG. 4.
(A) Cluster analysis of Trichoderma strains based on 750-nm optical density readings (mycelial growth) after a 96-h incubation; “monophenetic” taxa are shaded. (B) Canonical variate analysis of 750-nm optical density readings after a 96-h incubation.
TABLE 2.
Total standardized canonical coefficients
| Rank | Substrate | Canonical variate 1 | Canonical variate 2 | Canonical variate 3 |
|---|---|---|---|---|
| 1 | l-Sorbose | 1.11 | 0.19 | 1.00 |
| 2 | N-Acetyl-d-glucosamine | 0.05 | −2.26 | 1.52 |
| 3 | l-Threonine | 2.99 | 1.89 | −4.36 |
| 4 | N-Acetyl-l-glutamic acid | −2.87 | −1.53 | −0.28 |
| 5 | Sebacic acid | −1.85 | 2.36 | 0.73 |
| 6 | Quinic acid | 0.94 | 0.74 | 0.75 |
| 7 | d-Tagatose | 2.38 | 0.83 | 1.02 |
| 8 | l-Fucose | 1.39 | 1.40 | −1.31 |
| 9 | Succinic acid mono-methyl ester | 0.40 | −0.50 | −2.11 |
| 10 | Glycyl-l-glutamic acid | 1.39 | −1.93 | −0.22 |
| 11 | β-Hydroxybutyric acid | 1.84 | 1.14 | −0.11 |
| 12 | d-Glucuronic acid | −4.43 | 0.21 | 5.25 |
Metabolic profiles of T. pleuroticola, T. pleurotum, and related species in the Harzianum clade on the 34 highly significant substrata (P of <0.001) are compared in Fig. 5 and in the table in the supplemental material. In general, T. harzianum had the highest growth rate on a majority of the carbon sources (18/34). However, this did not reflect an overall higher growth rate or preferred temperature optima, since T. pleuroticola had the highest growth rates on the most readily assimilated compounds such as N-acetyl-d-glucosamine and α-d-glucose. Notably, all three species causing mushroom green mold diseases did not grow on N-acetyl-d-mannosamine or on N-acetyl-l-glutamic acid (Fig. 5) whereas T. harzianum readily assimilated these carbon sources. T. pleurotum exhibited generally impaired or slow growth on the majority of the carbon sources, and the species was unable to grow on d-tagatose, succinic acid mono-methyl ester, d-glucuronic acid, α-d-glucose-1-phosphate, and β-methyl-d-galactoside, which were all assimilated by the other species. In addition, growth of T. pleurotum on N-acetyl-d-glucosamine, sebacic acid, quinic acid, l-phenylalanine, and arbutin was significantly slower than growth of the other three species. The highest assimilation rates for T. pleuroticola occurred on N-acetyl-d-glucosamine and quinic acid, which could be useful to differentiate between the two causative agents of Pleurotus green mold disease. The carbon assimilation profile for T. pleuroticola was very similar to that of T. aggressivum. The two species are distinguished by the inability of T. aggressivum to assimilate α-ketoglutaric acid, l-malic acid, and succinamic acid.
FIG. 5.
Mean growth of mushroom green mold species on carbon sources for which statistically significant differences among the species were detected. ⧫, T. aggressivum; ▪, T. pleuroticola; ○, T. pleurotum; —, H. lixii/T. harzianum. The order of the carbon sources is the rank of the growth on 95 carbon sources and water, based on optical density at 750 nm at 96 h for the mean of six strains of H. lixii/T. harzianum. Carbon sources utilized differently by T. pleurotum and other species are in shown in boldface, underlining indicates cases in which all green mold species were different from H. lixii/T. harzianum, and use of italics indicates the case in which T. aggressivum and T. pleurotum were different from two other species.
Antagonism of T. pleurotum and T. pleuroticola against P. ostreatus in vitro.
Results of dual confrontation tests with P. ostreatus and A. bisporus are shown in Fig. 6. A. bisporus was vulnerable to all Trichoderma strains tested. The culture of this mushroom developed slowly and after 10 days was partly overgrown by Trichoderma reesei QM 6a (teleomorph H. jecorina) used as a negative control since it is a tropical species without notable mycoparasitic ability. All strains of the three green mold species covered the whole plate including the colony of A. bisporus after 5 to 7 days. The isolates of T. pleuroticola and T. pleurotum produced intense conidiation locally over the colony of A. bisporus. The P. ostreatus culture was fully resistant against T. reesei QM 6a, with the contact area between the two opposing colonies characterized by the well-expressed inhibition area. T. aggressivum was able to overgrow about one-third of the radius of the P. ostreatus colony, but the overgrowth stopped after 12 days of incubation. A clearly melanized barrage reaction in the confrontation zone was seen on the reverse of the plate. T. pleurotum also caused a strong antagonistic response from P. ostreatus although it was able to overgrow the majority of the Pleurotus colony by the end of the experiment. The confrontation zone between two opposing colonies was marked by intensive melanization of P. ostreatus hyphae and abundant conidiation of T. pleurotum (Fig. 6). In contrast to the T. pleurotum, T. pleuroticola did not cause any visible antagonistic reaction from the P. ostreatus culture and was able to overgrow the Pleurotus colony within 4 to 6 days.
FIG. 6.
Dual confrontation assays between cultures of A. bisporus and P. ostreatus and mushroom green mold species observed after 10 days of incubation on PDA. H. jecorina/T. reesei QM 6a was used as a negative control.
Morphology of T. pleuroticola and T. pleurotum.
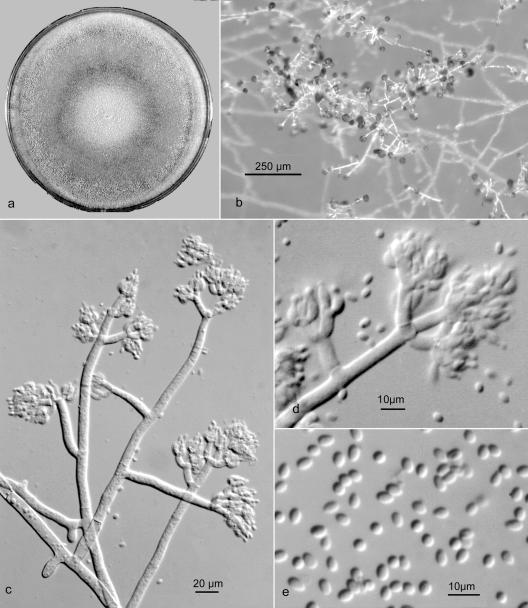
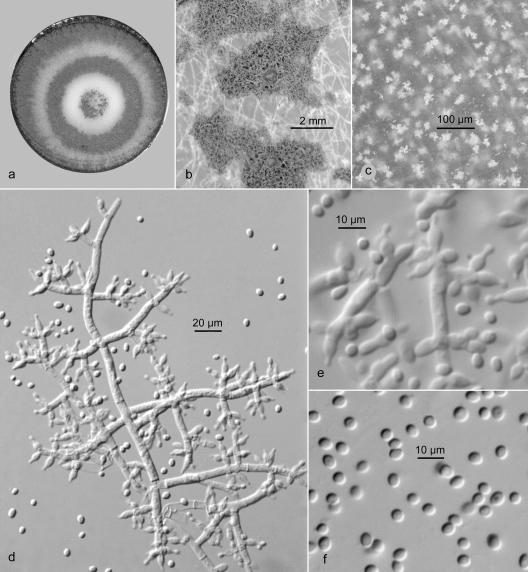
The close genetic relationship of T. pleuroticola and T. pleurotum, as revealed by gene sequence analysis, led us to expect that these two species would have very similar morphologies. However, and also in accordance with Park et al. (31), this was found not to be the case. The most striking difference was that T. pleurotum isolates exhibited a gliocladium-like conidiophore branching (Fig. 7), whereas T. pleuroticola showed a typical pachybasium-like conidiophore (Fig. 8) developing in fascicles or pustules more characteristic of species in the Harzianum clade. Subtle differences were also apparent in conidial size and growth rates.
FIG. 7.
T. pleurotum CPK 2113 (reference culture). (a) Colony on PDA after 10 days at 25°C. (b) Prostrate conidiophore with gliocladium-like branches. (c) Conidiophore with crowded apical branches and phialides. (d) Terminal branches with appressed phialides. (e) Ellipsoid conidia.
FIG. 8.
T. pleuroticola strain DAOM 175924 (reference culture). (a) Colony on PDA after 10 days at 25°C. (b) Flat conidiogenous pustules. (c) Crystals in reverse on PDA after 14 days. (d) Conidiophore with paired or verticillate branches and phialides. (e) Phialides. (f) Mostly subglobose conidia.
Detailed notes on diagnostic morphology of the two species follow.
Trichoderma pleurotum (Fig. 7a to f).
Colonies producing limited aerial mycelium and sparse conidiation from effuse conidiophores on MA, initially greenish white (27:A:2), becoming grayish green (26-27:B-C:3-4), later dull green (26:E-F:4-5), and finally in age on MA dull gray green (25-26:E-F:3) with conidiation evenly distributed in small irregular fascicles; on PDA with more abundant effuse conidiation becoming dark green in age (25:F:3-4). Reverse on MA more or less colorless; on PDA often developing light yellow sectors (4:A:3-5), and in age more or less conspicuously colored dull yellowish brown to reddish brown (5:D:4 to 8:D:5). Conidiophores arising from the substratum, usually unbranched near the base, over most of the length bearing gliocladium-like primary branches at nearly right angles, primary branches approximately equal in length, usually unbranched near the base, with branches arising singly and irregularly or in whorls of up to four near the apex, the apex of the conidiophore and primary branches usually terminated by a whorl of three or four branches rebranching once or twice and bearing crowded whorls of phialides; conidiophore and branches relatively broad, up to 8 μm diameter toward the base, terminal branches mostly 4 to 13 μm long and 3 to 4 μm wide. Phialides arising almost exclusively in verticils of four to seven on terminal branches, nearly ampulliform, often curved, sharply constricted at the conidium-bearing apex, mostly 4.5 to 7.5 by 2.5 to 4.0 μm, solitary phialides rare, intercalary phialides not observed, phialides persistent in age. Conidia predominately ellipsoid, occasionally obovoid with one end pointed, less often subglobose, 2.5 to 4.8 by 1.7 to 2.6 μm (average, 3.6 by 2.1 μm), pale green viewed microscopically, dark green in mass, smooth walled. Chlamydospores few, subglobose.
Morphologically, T. pleurotum has a conidiophore branching pattern that is unique within the Harzianum clade, in that conidiophores are mostly solitary and more or less prostrate, branching in an irregular fashion, with branches scattered, arising separately and bearing crowded whorls of appressed phialides at the apex resembling the conidiophore in Gliocladium. Conidia are mostly ellipsoid and longer than in most other species in the clade.
Trichoderma pleuroticola (Fig. 8a to f).
Colonies producing limited aerial mycelium on MA, on PDA producing fasciculate, white aerial mycelium, conidiophores forming small pustules that coalesce in broad concentric zones, initially greenish gray (25:B:2-3), soon grayish green (25:C:3-4 to 25:E:4-5), in age on MA dull to dark green (26:E-F:4-5), in age on PDA darker green, or with brighter green, flat pustules fringed by white mycelium and renewed conidiation. Reverse more or less uncolored at first, in age dull yellowish on MA, on PDA typically turning dark brown (8:F:5), abundant, small, yellow crystals often developing in PDA after 7 days of incubation. Conidiophores branching in a more or less pyramidal fashion with branches increasing in length toward the base, branches arising singly or paired toward the base of the conidiophore main axis, near the apex often three to four verticillate, primary branches branching in a pattern similar to the main axis, conidiophore and branches comparatively narrow and flexuous, main axis up to 5.5 μm wide at the base, gradually narrowing to 2.5 to 3.0 μm at the apex, terminal branches cylindrical, 6 to 14 by 2.5 to 3.3 μm. Phialides paired or three to four verticillate at the apex of the terminal branches, or arising separately and scattered along the sides of the conidiophore and branches, ampulliform to lageniform, abruptly narrowing to a conidium bearing collulum less than 1 μm wide, mostly 4.2 to 9.5 by 3.0 to 4.2 μm, or terminal phialides acerose and up to 20 μm long. Short, cylindric intercalary phialides occasionally produced beneath septa on terminal branches or from sides of phialides. Phialides seceding in older cultures. Conidia subglobose to broadly ellipsoid, less often obovoid and pointed at the base, 2.6 to 5.0 by 2.4 to 3.7 μm (average, 3.7 by 2.8 μm), bright green viewed microscopically, dark gray green in mass, smooth walled. Chlamydospores usually in chains or clusters, subglobose, 4 to 10 μm diameter, pale greenish.
Morphologically, the conidiophore in T. pleuroticola is organized in essentially the same fashion as in T. harzianum and related species in the Harzianum clade. The conidiophore is branched at regular intervals with branches increasing in length to the base, and branches and phialides arise mostly in uncrowded verticils. Conidia are significantly larger in T. pleuroticola than in T. harzianum, but the most distinctive morphological feature of T. pleuroticola is the production by most strains of a dark brown pigment and yellow crystals in the agar on PDA.
DISCUSSION
In this paper, we provide evidence that the causal agent for the oyster mushroom green mold disease, which recently started to spread in Europe and Asia, is actually two different although genetically closely related species of Trichoderma which correspond to recently described taxa T. pleurotum and T. pleuroticola (31). Both species are also closely related to the H. lixii/T. harzianum species aggregate and to T. aggressivum, which is the causal agent of green mold disease of Agaricus. These findings are in accordance with those reported by Park et al. (30), who used parsimony analysis of ITS1 and ITS2, rpb2, and tef1 sequences to separate the Pleurotus pathogenic strains into two clusters which they called “Trichoderma sp. strain K1” and “strain K2.” Unfortunately, sequences from their study, except for ITS1 and ITS2 from six isolates, have not been deposited in public databases and thus could not be included in our analysis. The available ITS1 and ITS2 sequences indicate that their six strains of Trichoderma sp. strain K1 isolated from Pleurotus substrata (rice straw, cotton waste, and sawdust) from four locations in South Korea correspond to T. pleuroticola (reference GenBank accession numbers DQ164409 and DQ164410 for strains CNU601 and CNU646, respectively) subsequently described by the same authors (31). Similarly, strain K2 represents T. pleurotum, 14 strains of which were isolated from the same substrata in six locations (reference GenBank accession numbers DQ164405, DQ164406, DQ164407, and DQ164408 for strains CNU501, CNU523, CNU538 and CNU571, respectively). According to Park et al. (30, 31) both species coexist in South Korean Pleurotus farms with no clear dominance of one or the other species.
There are several indications that the infection is introduced to farms via the substratum for mushroom cultivation, and differences in species distribution may be due to the use of certain substrata which, depending on the manufacturer, may consist of cereal straw, sawdust, bagasse, or waste cotton. In our sample T. pleuroticola dominates in samples from Italian Pleurotus farms while T. pleurotum is abundant among Hungarian isolates. Although wheat straw is used as a major component for the Pleurotus substratum in both countries, the difference in species composition may be due to the addition of pulverized “tufo” in Italian farms, which is a natural calcareous rock of volcanic origin that raises the substratum pH to around 8. To the best of our knowledge, there is no such technological stage in Hungarian farms, where wheat straw is moisturized in the open air before use as Pleurotus substratum. The hypothesis of a possible reduction of T. pleurotum infection by the alkalization of the substratum may be further supported by the fact that Pleurotus green mold is not reported to be a severe problem in the United States, where the addition of lime to increase pH to 7.5 is widely practiced (http://mushroomspawn.cas.psu.edu/). However, this treatment seems to be ineffective against T. pleuroticola. There may be another explanation for the occurrence of the two Pleurotus-associated green mold species in mushroom farms. T. pleuroticola is also frequently isolated from soil and plant debris, and we report environmental strains from Canada, the United States, Europe, and New Zealand. It seems to have a global occurrence, although possibly favoring a temperate climate. T. pleuroticola infections may therefore have multiple origins and even be due to introductions from the surrounding environment. In contrast, T. pleurotum, just like T. aggressivum, has so far never been isolated from areas outside of mushroom farms. Seaby (37) reported evidence that T. aggressivum could be carried by red pepper mites into Agaricus mushroom farms. Another possibility would be that T. pleurotum could be an endophyte of plants used for preparation of the mushroom substratum (possibly wheat, rice, and cotton). The vectors for T. pleurotum and T. pleuroticola into mushroom farms are currently under investigation in our laboratories. The consistent cooccurrence of these two species in mushroom farms in Romania, Italy, Hungary, and South Korea is interesting. Yet recent metagenomic studies on the occurrence of Trichoderma in Austrian soils frequently reveals the presence of T. pleuroticola but never of T. pleurotum (M. A. Friedl and I. S. Druzhinina, unpublished data), thus suggesting that these two species occupy different ecological and trophic niches in nature.
The large phenetic divergence of T. pleuroticola and T. pleurotum, morphologically and metabolically, in spite of the very close phylogenetic relationship, is a unique finding, as fungi are believed to develop phenotypic differences only after accumulation of some genetic distance, which gives rise to “cryptic” species which can hardly be phenotypically distinguished. The fact that T. pleurotum occupies a more terminal position than T. pleuroticola in all gene trees and that the latter exhibits similar morphological and metabolic characteristics as its phylogenetically close members in the Harzianum clade of Hypocrea/Trichoderma (T. harzianum and T. aggressivum) suggest that this change in morphology is due to a loss rather than a gain of gene function. Kullnig-Gradinger et al. (22), comparing the morphotypes and phylogeny of Trichoderma species have speculated that the switch from fungicolous to saprophytic habitats was accompanied by the expression of the pachybasium-like conidiophore morphology. In this sense, the return to gliocladium-like morphology may be advantageous under the conditions of the natural niche of T. pleurotum. The gliocladium-like morphology is rare in the Harzianum clade although it is known for the anamorph of Hypocrea tawa.
Even though there is a consistent association between T. aggressivum and Agaricus, on one hand, and T. pleuroticola/T. pleurotum and Pleurotus, on the other hand, our confrontation assays show that the two newly described species pose a potential threat to mushroom-producing farms: both T. pleuroticola and T. pleurotum were able to inhibit and then overgrow Agaricus culture, while Pleurotus showed some resistance toward T. aggressivum. Since T. pleuroticola is frequently isolated from various soils and plant debris, we consider this species as the most dangerous agent of mushroom green mold disease in general.
Finally, this study also places some caveats on the use of some Trichoderma isolates as biocontrol agents. Two of the T. pleuroticola isolates of this study were obtained from other researchers as biofungicides against soil-borne diseases (Table 1). In view of the present identification of T. pleuroticola as a causative agent of oyster mushroom green mold, this application could be problematic. However, the fact that infections by T. pleuroticola and T. pleurotum—although probably common for decades (see reference 39)—only recently increased dramatically suggests there is a special trigger for the infections, which may involve the source of the substrate used for cultivation, its preparation, or other conditions of mushroom cultivation. This, in turn, implies that if this trigger can be determined, the risk of infection can be managed. With the molecular tools presented in this paper, the processes involved in the preparation of substrata for the cultivation of P. ostreatus can be investigated, and infections can be traced back to their sources. Oligonucleotide probes based on diagnostic polymorphisms in tef1 sequences offer the development of real-time PCR techniques for quantitative detection as well and are in preparation in our laboratories.
Supplementary Material
Acknowledgments
This work was supported partly by Austrian Science Fund grants FWF P-12748-MOB and FWF P-16601 to C.P.K. and by grant OTKA F68381 from the Hungarian Scientific Research Fund to L.K., as well as by the Austrian Exchange Service and the Hungarian National Office for Research and Technology under the bilateral project WTZ HU03/2007-TéT A02/2006. L.K. is a grantee of the János Bolyai Research Scholarship (Hungarian Academy of Sciences).
We also thank Parivash Shoukouhi for contributions to the physiological and molecular studies undertaken at the Eastern Cereal and Oilseed Research Center.
Footnotes
Published ahead of print on 7 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anonymous. 2005. Consensus document on the biology of Pleurotus spp. (oyster mushroom). Series on harmonisation of regulatory oversight in biotechnology, no. 34 Organisation for Economic Co-operation and Development, Environment Directorate, Paris, France.
- 2.Ballero, M., E. Mascia, A. Rescigno, and E. S. D. Teulada. 1990. Use of Pleurotus for transformation of polyphenols in waste waters from olive presses into proteins. Micol. Ital. 19:39-41. [Google Scholar]
- 3.Castle, A., D. Speranzini, N. Rghei, G. Alm, D. Rinker, and J. Bissett. 1998. Morphological and molecular identification of Trichoderma isolates on North American mushroom farms. Appl. Environ. Microbiol. 64:133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, R. 1996. Functional properties of edible mushrooms. Nutr. Rev. 54:91-93. [DOI] [PubMed] [Google Scholar]
- 5.Clement, M., D. Posada, and K. A. Crandall. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657-1659. [DOI] [PubMed] [Google Scholar]
- 6.dela Cruz, T. E. E., B. E. Schulz, C. P. Kubicek, and I. S. Druzhinina. 2006. Carbon source utilization by the marine Dendryphiella species D. arenaria and D. salina. FEMS Microbiol. Ecol. 58:343-353. [DOI] [PubMed] [Google Scholar]
- 7.Dettman, J. R., D. J. Jacobson, and J. W. Taylor. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703-2720. [DOI] [PubMed] [Google Scholar]
- 8.Dodd, S., R. N. Crowhurst, A. G. Rodrigo, G. J. Samuels, R. A. Hill, and A. Stewart. 2000. Examination of Trichoderma phylogenies derived from ribosomal DNA sequence data. Mycol. Res. 104:23-34. [Google Scholar]
- 9.Druzhinina, I. S., M. Schmoll, B. Seiboth, and C. P. Kubicek. 2006. Global carbon utilization profiles of wild-type strains, mutants and transformants of Hypocrea jecorina. Appl. Environ. Microbiol. 72:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Druzhinina, I. S., A. G. Kopchinskiy, and C. P. Kubicek. 2006. The first one hundred of Trichoderma species is characterized by molecular data. Mycoscience 47:55-64. [Google Scholar]
- 11.Druzhinina, I. S., A. G. Kopchinskiy, M. Komon, J. Bissett, G. Szakacs, and C. P. Kubicek. 2005. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 42:813-828. [DOI] [PubMed] [Google Scholar]
- 12.Druzhinina, I. S., and A. G. Kopchinskiy. 2006. TrichOKEY v. 2—a DNA oligonucleotide BarCode program for the identification of multiple sequences of Hypocrea and Trichoderma, p. 53-59. In W. Meyer and C. Pearce (ed.), Proceedings of the 8th International Mycological Congress, Cairns, Australia. Medimond-Monduzzi, Bologna, Italy.
- 13.Gunde-Cimerman, N. 1999. Medicinal value of the genus Pleurotus (Fr.) P. Karst. (Agaricales s.l. Basidiomycetes). Int. J. Med. Mushrooms 1:69-80. [Google Scholar]
- 14.Hatvani, L., Z. Antal, L. Manczinger, A. Szekeres, I. S. Druzhinina, C. P. Kubicek, A. Nagy, E. Nagy, C. Vágvölgyi, and L. Kredics. 2007. Green mould diseases of Agaricus and Pleurotus spp. are caused by related but phylogenetically different Trichoderma species. Phytopathology 97:532-537. [DOI] [PubMed] [Google Scholar]
- 15.Hermosa, M. R., I. Grondona, and E. Monte. 1999. Isolation of Trichoderma harzianum Th2 from commercial mushroom compost in Spain. Plant Dis. 83:591. [DOI] [PubMed] [Google Scholar]
- 16.Jaklitsch, W. M., M. Komon, C. P. Kubicek, and I. S. Druzhinina. 2006. Hypocrea crystalligena sp. nov., a common European species with a white-spored Trichoderma anamorph. Mycologia 98:500-514. [DOI] [PubMed] [Google Scholar]
- 17.Kopchinskiy, A., M. Komon, C. P. Kubicek, and I. S. Druzhinina. 2005. TrichoBLAST: a multilocus database for Trichoderma and Hypocrea identifications. Mycol. Res. 109:657-660. [DOI] [PubMed] [Google Scholar]
- 18.Kornerup, A., and J. H. Wanscher. 1978. Methuen handbook of colour, 3rd ed. Eyre Methuen, London, United Kingdom.
- 19.Kraus, G., I. Druzhinina, J. Bissett, H. J. Prillinger, G. Szakacs, W. Gams, and C. P. Kubicek. 2004. Trichoderma brevicompactum sp. nov. Mycologia 96:1057-1071. [PubMed] [Google Scholar]
- 20.Kredics, L., L. Hatvani, Z. Antal, L. Manczinger, I. S. Druzhinina, C. P. Kubicek, A. Szekeres, A. Nagy, C. Vágvölgyi, and E. Nagy. 2006. Green mould disease of oyster mushroom in Hungary and Transylvania. Acta Microbiol. Immunol. Hung. 53:306-307. [Google Scholar]
- 21.Kubicek, C. P., J. Bissett, C. M. Kullnig-Gradinger, I. S. Druzhinina, and G. Szakacs. 2003. Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genet. Biol. 38:310-317. [DOI] [PubMed] [Google Scholar]
- 22.Kullnig-Gradinger, C. M., G. Szakacs, and C. P. Kubicek. 2002. Phylogeny and evolution of the fungal genus Trichoderma—a multigene approach. Mycol. Res. 106:757-767. [Google Scholar]
- 23.Leache, A. D., and T. W. Reeder. 2002. Molecular systematics of the eastern fence lizard (Sceloporus undulatus): a comparison of parsimony, likelihood and Bayesian approaches. Syst. Biol. 51:44-68. [DOI] [PubMed] [Google Scholar]
- 24.Lieckfeldt, E., C. M. Kullnig, C. P. Kubicek, G. J. Samuels, and T. Börner. 2001. Trichoderma aureoviride: phylogenetic position and characterization. Mycol. Res. 105:313-322. [Google Scholar]
- 25.Mamoun, M. L., J.-M. Savoie, and J.-M. Olivier. 2000. Interactions between the pathogen Trichoderma harzianum Th2 and Agaricus bisporus in mushroom compost. Mycologia 92:233-240. [Google Scholar]
- 26.Marzullo, L., R. Cannio, P. Giardina, M. T. Santini, and G. Sannia. 1995. Veratryl alcohol oxidase from Pleurotus ostreatus participates in lignin biodegradation and prevents polymerization of laccase-oxidized substrates. J. Biol. Chem. 270:3823-3827. [DOI] [PubMed] [Google Scholar]
- 27.Nicholas, K. B., and H. B. Nicholas, Jr. 1997. Genedoc: a tool for editing and annotating multiple sequence alignments. http://www.psc.edu/biomed/genedoc.
- 28.O'Donnell, K., E. Cigelnik, and H. L. Nirenberg. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465-493. [Google Scholar]
- 29.Pani, B. K., S. N. Panda, and S. R. Das. 1997. Utilization of some agricultural bioproducts and other wastes for sporophore production of oyster mushroom. Orissa J. Hortic. 25:36-39. [Google Scholar]
- 30.Park, M. S., K. S. Bae, and S. H. Yu. 2004. Molecular and morphological analysis of Trichoderma isolates associated with green mold epidemic of oyster mushroom in Korea. J. Huazhong Agric. Univ. 23:157-164. [Google Scholar]
- 31.Park, M. S., K. S. Bae, and S. H. Yu. 2006. Two new species of Trichoderma associated with green mold of oyster mushroom cultivation in Korea. Mycobiology 34:11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puniya, A. K., K. G. Shah, S. A. Hire, R. N. Ahire, M. P. Rathod, and R. S. Mali. 1996. Bioreactor for solid-state fermentation of agro-industrial wastes. Indian J. Microbiol. 36:177-178. [Google Scholar]
- 33.Rolf, F. J. 1997. NTSYSpc numerical taxonomy and multivariate analysis system, version 2.00. Exeter Software, Setauket, New York.
- 34.Samuels, G. J., O. Petrini, K. Kuhls, E. Lieckfeldt, and C. P. Kubicek. 1998. The Hypocrea schweinitzii complex and Trichoderma sect. Longibrachiatum. Stud. Mycol. 41:1-54. [Google Scholar]
- 35.Samuels, G. J., S. L. Dodd, W. Gams, L. A. Castlebury, and O. Petrini. 2002. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94:146-170. [PubMed] [Google Scholar]
- 36.SAS Institute, Inc. 1989. SAS/STAT user's guide, version 6, 4th ed., vol. 1. SAS Institute, Inc., Cary, NC.
- 37.Seaby, D. A. 1989. Further observations on Trichoderma. Mushroom J. 197:147-151. [Google Scholar]
- 38.Sharma, S. R., and B. Vijay. 1996. Yield loss in Pleurotus ostreatus spp. caused by Trichoderma viride. Mushroom Res. 5:19-22. [Google Scholar]
- 39.Sinden, J., and E. Hauser. 1953. Nature and control of three mildew diseases of mushrooms in America. Mushroom Sci. 2:177-180. [Google Scholar]
- 40.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer, Sunderland, MA.
- 41.Woo, S. L., P. Di Benedetto, M. Senatore, K. Abadi, S. Gigante, I. Soriente, S. Ferraioli, F. Scala, and M. Lorito. 2004. Identification and characterization of Trichoderma species aggressive to Pleurotus in Italy. J. Zhejiang Univ. Agric. Life Sci. 30:469-470. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.