Abstract
Due to metabolic and morphological changes that can prevent Helicobacter pylori cells in water from growing on conventional media, an H. pylori-specific TaqMan quantitative PCR (qPCR) assay was developed that uses a 6-carboxyfluorescein-labeled probe (A. E. McDaniels, L. Wymer, C. Rankin, and R. Haugland, Water Res. 39:4808-4816, 2005). However, proper internal controls are needed to provide an accurate estimate of low numbers of H. pylori in drinking water. In this study, the 135-bp amplicon described by McDaniels et al. was modified at the probe binding region, using PCR mutagenesis. The fragment was incorporated into a single-copy plasmid to serve as a PCR-positive control and cloned into Escherichia coli to serve as a matrix spike. It was shown to have a detection limit of five copies, using a VIC dye-labeled probe. A DNA extraction kit was optimized that allowed sampling of an entire liter of water. Water samples spiked with the recombinant E. coli cells were shown to behave like H. pylori cells in the qPCR assay. The recombinant E. coli cells were optimized to be used at 10 cells/liter of water, where they were shown not to compete with 5 to 3,000 cells of H. pylori in a duplex qPCR assay. Four treated drinking water samples spiked with H. pylori (100 cells) demonstrated similar cycle threshold values if the chlorine disinfectant was first neutralized by sodium thiosulfate.
The role of Helicobacter pylori, a gram-negative bacterium, in causing chronic gastritis, gastroduodenal ulcers, and gastric cancer in humans has been well established in the last decade (15). Although the pathogenicity of the organism is well recognized, the routes of transmission are not clear (11). Fecal-oral and oral-oral routes have been the routes speculated for person-to-person transmission. Iatrogenic routes as well as vectorial spread by flies have also been proposed (7).
Transmission by drinking water has been suggested as the explanation for a fecal-oral bacterial route, and the presence of H. pylori DNA in drinking water systems has been reported from the United States, England, Germany, Japan, Sweden, Mexico, Gambia, and Peru (4, 8, 23-27, 31, 42). Using a combined fluorescent antibody 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) staining method, Hegarty et al. also demonstrated the presence of respiring H. pylori cells from U.S. surface waters (20). Only one report of live H. pylori isolated and cultured from untreated municipal wastewater has been published (30). However, several laboratory studies have demonstrated that H. pylori can survive in water under a variety of conditions for a period of days to weeks (3, 40, 44). One reason for the inability to culture the bacteria from water could be that the bacterium changes from the normal spiral-shaped bacillary form into the coccoid form when it is exposed to water or to other adverse conditions (2, 6, 10, 12). This coccoid form has been shown to be capable of oxidative metabolism and respiration (13, 19) and has been shown to possess DNA and mRNA, albeit in diminished amounts (34). Whether the coccoid form is viable/infectious is a subject of debate. It has not been possible to resuscitate the coccoid cells and restore their normal morphological state or to grow them in the laboratory, although the spiral form can be grown under suitable culture conditions (6, 40, 44). It has been proposed that the morphological change into the coccoid form is a transitory adaptation to environmental stress for the species to survive and thus may play a role in its survival in and transmission through water. Therefore, it has become necessary to detect H. pylori in this state. In the absence of a culture medium, the current nucleic acid-based methods seem to be the only methods available for detecting H. pylori in water.
Several regular PCR assays are available for H. pylori detection in environmental water (4, 18, 25, 29, 36, 39, 43), with detection sensitivities ranging from 2 to 100 cells per reaction reported by some investigators. Some of these reports have used primer sets that may detect other Helicobacter species. None of these studies presents results in a quantitative manner. The use of quantitative PCR has been reported by Horiuchi et al., who used the 16S rRNA as the target (23), and by McDaniels et al., who used a conserved region from the ureA gene, which is specific for H. pylori (32) and encodes the urease A subunit of the urease protein. A detection sensitivity of 100 cells/ml was reported by the former study, while 10 cells per liter of water was demonstrated by the latter. Because the number of cells in drinking water may be small, with H. pylori DNA being detected by PCR amplification only after concentration of large volumes of drinking water (21, 28, 39), and because the infectious dose of the organism is not known, it becomes necessary to quantify very low numbers. Both false negatives and false positives may arise due to these conditions. To avoid some of these errors, careful controls must be included in any standardized method. The purpose of this study was to develop internal amplification controls (IC) that could be used with the assay developed by McDaniels et al. (32) by multiple laboratories for method validation studies as well as for occurrence studies. In addition, a DNA extraction method that can reliably sample an entire liter of water has been optimized.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Two strains of H. pylori, ATCC 43504 and the clinical isolate H. pylori G12201, were used for most of the experiments. G12201 was obtained from the Baylor College of Medicine, TX, and was named H. pylori Texas. The H. pylori strains were maintained on tryptic soy agar (40 g/liter water)-based 5% blood agar plates. Strains were typically grown for 48 h under microaerophilic conditions (5% CO2) at 37°C before being suspended into phosphate-buffered saline (catalog no. P-3813; Sigma Chemical Co.). Since the majority of the cells change from a rod shape to a coccoid shape at room temperature, the suspension was held on ice to slow the conversion process and was used within the hour.
Enumeration of H. pylori cells.
H. pylori cell suspensions were counted directly by using a hemocytometer (Hauser Scientific, Horsham, PA) under phase contrast (Eclipse 80i; Nikon, Tokyo, Japan) at a total magnification of ×400. At this magnification, both spiral (1 by 2 μm to 1 by 4 μm) and coccoid (1 by 1 μm) forms were visible. Suspensions were typically diluted 1:10 in phosphate-buffered saline before cells were counted. Optical density readings at 640 nm (OD640) were also monitored by a spectrophotometer (MBA 2000; PerkinElmer, Waltham, MA). An OD640 reading of 0.5 corresponded to approximately 9 × 108 cells/ml. The relationship of OD640 to the viable cell count was derived after recording direct microscopic counts and optical density readings at several cell dilutions, with cells obtained from plates incubated for 48 h from different preparations. The correlation between hemocytometer counts and the viable counts was more than 95% (data not shown).
Preparation of DNA from pure cultures and plasmids.
Total genomic DNA from pure cultures of H. pylori was prepared by using a Puregene kit (Gentra, Minneapolis, MN) or a WaterMaster kit (Epicentre Biotechnologies, Madison, WI). The protocols were followed as specified by the manufacturers. DNA was extracted in a final volume of 50 μl of Tris-Cl, pH 8. DNA concentration was measured using a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE) sample adjusted with water to a final concentration of 20 ng/μl water, and 5 μl was used in 25 μl for PCR. Plasmid DNA was prepared using a Qiagen Plasmid Mini kit according to the instructions of the manufacturer (Qiagen Corp., Santa Clarita, CA).
Construction of IC.
A DNA molecule to be used as the IC, corresponding to the amplicon generated by primers HpyF1 and HpyR1 from the H. pylori Texas strain, was made as follows. A PCR amplicon of 135 bp was first generated with primers HpyF1 and HpyR1, using genomic DNA from the H. pylori Texas strain as the target. The sequences of the two oligonucleotides, HpyF1 (5′-GGGTATTGAAGCGATGTTTCCT-3′) and HpyR1 (5′-GCTTTTTTGCCTTCGTTGATAGT-3′), have been described previously (32). This 135-bp fragment was used as the template in two touchdown PCRs (at 65°C to 55°C). In one reaction mixture, the primers HpyF1 and BamH1-F1 were used to generate a 53-bp fragment, and in the other reaction mixture, the primers HpyR1 and BamHI-R1 were used to generate an 89-bp fragment. The sequence of the BamHI-F1 primer was 5′-GGATCCCACGGTTACGAGTT-3′, and the BamHI-R1 primer sequence was 5′-GGATCCCCTATTGAGGCCAA. Both of the primers had a BamHI site incorporated at the 5′ end. The two fragments were gel purified from a 3% agarose gel, using a QIAquick gel extraction kit (Qiagen Corp., Santa Clarita, CA), digested with the restriction enzyme BamHI (Invitrogen, Inc.), cleaned up using a Qiagen PCR purification kit (Qiagen Corp., Santa Clarita, CA), and ligated overnight using T4 DNA ligase (Invitrogen, Inc.). Following ligation, 6 μl of the ligation product was amplified using the HpyF1 and HpyR1 primers. The 135-bp fragment, which should have a new BamHI site incorporated into it, was blunt ended and ligated into a pCC1 vector (8,138 bp), using a copy control cDNA, gene, and PCR cloning kit, and transformed into chemically competent TransforMax EP1300 Escherichia coli, per the instructions of the manufacturer (Epicenter Biotechnologies, Madison, WI). Plasmid DNA from E. coli clones was purified using a QIAprep spin miniprep kit (catalog no. 27104; Qiagen Corp., Santa Clarita, CA.) and sequenced by the core facility of the Children's Hospital Medical Center, Cincinnati, OH. Sequence comparisons were made using BioEdit software (Ibis Therapeutics, Carlsbad, CA).
Optimization of DNA extraction from water.
H. pylori cells were spiked in 1 liter of distilled water. All spiked samples were first filtered through a Nuclepore polycarbonate filter, 47 mm in diameter, with a pore size of 0.45 μm (Whatman, Clifton, NJ). In initial experiments, both 0.45-μm- and 0.2-μm-pore-size filters were examined. No significant differences were observed between the two filters in terms of retention or recovery of either the spiral or the coccoid form of the H. pylori cell, but the flow rate was faster through 0.45-μm filters. It was decided to continue with these filters. Eight methods were examined to determine the most efficient method for the recovery of DNA from the polycarbonate filters. The DNA isolation methods examined included the use of a Puregene DNA isolation kit, a cell and tissue kit (Gentra, Minneapolis, MI), a WaterMaster DNA purification kit with and without inhibitor removal (Epicentre Biotechnologies, Madison, WI), an Ultraclean soil DNA kit (Mo Bio Laboratories, Inc, Carlsbad, CA), a DNA-EZ kit (GeneRight, LLC, New Brunswick, NJ), a DNA-EZ kit with a Qiagen purification column (GeneRight, LLC/Qiagen), and a bead-beating method with 0.3 g of glass beads in 0.1× AE buffer (Qiagen proprietary buffer containing 0.2 μg/ml salmon sperm DNA for 1 min), and a method that combined bead milling and a QIAamp microkit (Qiagen Corp., Santa Clarita, CA).
Protocols according to the manufacturers' instructions were followed with some modifications for some kits. Specifically, all kits that required an isopropanol precipitation of DNA and an ethanol wash were modified to use ice-cold isopropanol and ethanol, and all final resuspension volumes were changed to 30 to 35 μl. An Ultraclean soil DNA kit method was performed according to the manufacturer's alternative protocol in order to obtain maximum yields. A modified DNA-EZ kit was used to capture DNA from the entire lysate, and buffer volumes were scaled up appropriately. A DNA-EZ kit was also tested by substituting a Qiagen purification column for the column provided by GeneRight. The combination bead milling/QIAamp kit method used 0.3 g of glass beads, 300 μl of ATL buffer, 300 μl of AL buffer, and 40 μl of proteinase K for bead milling (Qiagen Corp., Santa Clarita, CA). All lysates were transferred to a QIAamp MinElute column, and procedures were carried out using a QIAamp DNA microkit protocol. A WaterMaster kit was used with and without inhibitor removal. Instead of using a 50-ml conical tube for washing the filter, the filter was placed in a sterile petri dish with the surface that had trapped bacteria facing upward. Washes with 600 μl of tissue and cell lysis buffer were collected in a single 2-ml microcentrifuge tube, and all further procedures were done in 2-ml tubes. This eliminated dividing the sample into two tubes of 1. 5 ml each and later pooling the final DNA solutions, as suggested by the manufacturer.
Contamination occurred consistently while testing with a DNA-EZ kit. Specifically, H. pylori was detected in negative controls (for both columns) with a cycle threshold (CT) value that was sometimes as low as 35.7, which amounts to about 50 cells. The Puregene kit was less sensitive, detecting 400 cells (CT = 40.7). Bead milling in combination with a QIAamp microkit detected 5 cells (CT = 41.7). However, the CT values over multiple cell concentrations varied greatly, suggesting that this method was inconsistent. A Mo Bio kit detected 25 cells (CT = 40.0), and the basic bead milling detected 57 cells (CT = 38.5). Unlike other kits, WaterMaster with or without inhibitor removal consistently detected 10 cells (CT = 38.7) and maintained a strong correlation between CT value and cell concentration. For this reason, a WaterMaster kit without the inhibitor removal step was used for further experiments for drinking water, where there is unlikely to be large interferences from the matrix.
Once the kit was optimized, water samples were spiked with either E. coli KS10 cells or cells from the H. pylori Texas strain or with both strains simultaneously. Typically the E. coli strain was grown overnight in Luria-Bertani (LB) broth containing 12.5 μg chloramphenicol/ml. The next morning, this culture was used to inoculate 3 ml of fresh broth containing 12.5 μg/ml of chloramphenicol prepared from an ethanolic stock solution of 25 mg/ml. Cells were allowed to grow for 2 h, at the end of which a reading at OD640 was taken. Using the relationship OD640 of 0.5 equaled 2.5 × 108 cells, the cells were diluted in distilled water so that 105 cells/ml, 104 cells/ml, and 103 cells/ml were obtained, giving concentrations of 1,000, 100, and 10 cells, respectively, in 10 μl of each of these dilutions. Typically, 1-liter water samples were spiked with 10 cells of E. coli as the matrix spike. A 10-μl aliquot was also plated on chloramphenicol agar plates, and the plates were incubated overnight at 37°C to obtain the actual number of cells added. After being spiked with the desired number of H. pylori cells, each of the samples was filtered, and the DNA was extracted by using a WaterMaster kit. DNA was extracted in a final volume of 33 μl of water, and 11 μl was used for each 25-μl PCR. A duplicate PCR was run for each sample, and if the CT values were similar (standard deviation of ≤ 1), then the third aliquot was not run. With 14 μl being used up for reagents, a maximum volume of 11 μl could be used as the template, and this volume was used to get the best sensitivity in the 25-μl PCR. The number of cells used for generating the data points was therefore taken as one-third of the total cells added (e.g., 33.33 for 100 cells; 3.3 for 10).
Fast PCR.
Fast PCR was used to generate fragments for the preparation of IC. The maximum reaction mixture volume for the instrument (30 μl) was used to maximize products for cloning purposes. It contained 15 μl of 2× Gene Amp Fast PCR mixture (Applied Biosystems, Foster City, CA), 5 μl of template DNA, and primers at a final concentration of 1 μM. Reactions were performed in an ABI 9800 Fast Thermal Cycler (Applied Biosystems, Foster City, CA), and the reaction time was 45 min for touchdown PCR, used to generate the individual fragments, and 20 min for regular PCR, used to generate the initial amplicon and the final amplicon, using the HpyF1 and HpyRI primers. The cycling conditions for touchdown PCR were as follows: an initial single cycle at 95°C for 10 s, followed by five cycles of three-temperature cycling at 95°C for 0 s, 65°C for 0 s, and 72°C for 20 s, followed by 21 cycles of three-temperature cycling at 95°C for 0 s, 65°C for 0 s, and 72°C for 20 s, with the annealing temperature of 65°C incrementally decreased by 0.5°C in subsequent cycles from the initial 65°C to 55°C, followed by 15 cycles of three-temperature cycling at 95°C for 0 s, 55°C for 0 s, and 72°C for 20 s and a final single cycle at 72°C for 5 min. Regular PCR was performed using the default parameters of the instrument and consisted of the following steps: an initial single cycle at 95°C for 10 s, followed by 30 cycles of two-temperature cycling at 94°C for 0 s and 64°C for 15 s, followed by a final cycle at 72°C for 10 s.
TaqMan assay.
The duplex PCR was performed in 25-μl volumes in 0.2-ml optical-grade PCR tubes (Applied Biosystems, Foster City, CA). Each 25 μl of reaction mixture contained 1 μl of probe and primer mixture, 0.5 μl of 5% bovine serum albumin, and 12.5 μl of TaqMan Universal Master mixture, which is a 2× proprietary mixture containing AmpliTaq Gold DNA polymerase, AmpErase uracil N-glycosylase (UNG), amounts of each deoxynucleoside triphosphate (dNTPs) with dUTP, the passive reference dye ROX, and optimized buffer components (Applied Biosystems, Foster City, CA). The master mixture was used without any modification. The final primer concentration in the mixture was 1 μM (for each primer) and 40 nM for each of the probes. The probe that recognized the H. pylori ureA gene, TaqMan probe 1, was labeled with 6-FAM (6-carboxyfluorescein) at the 5′end and with TAMRA (6 carboxytetramethylrhodamine) at the 3′ end (5′-FAM-CTCGTAACCGTGCATACCCCTATTGAG-TAMRA3′). TaqMan probe 2, which recognized the modified fragment, was labeled with VIC dye (Applied Biosystems proprietary dye) at the 5′ end (5′-VIC-CTCGTAACCGTGGGATCCCCTATTGAG-TAMRA-3′). Both of the probes were made by Applied Biosystems (Foster City, CA). The volume of template used was 11 μl. Depending on the experiment, the template consisted of purified genomic DNA, plasmid DNA, or whole-cell suspensions in water. The copy number calculations were determined as previously described, with the size of the H. pylori genome taken at 1,700 kb and the plasmid pKS10 (vector, 8.139 kb plus an insert of 0.135 kb) at 8.274 kb (38). Thus, 1 fg of genomic DNA amounted to 0.54 copies, while 1 fg of plasmid amounted to 110 copies. The standard cycling condition consisted of an initial single cycle at 95°C for 10 min to activate the AmpliTaq Gold, followed by 50 cycles of two-temperature cycling consisting of 15 s at 95°C and 1 min at 60°C. PCR was performed with an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster city, CA) per instructions provided in the instrument's manual. The CT baseline value was set at 0.06, and the number of cycles run was 45. The numbers were arrived at empirically after performing multiple repeats of the assay over a period of days.
Chlorine measurements.
One-liter water samples were collected from four different distribution systems located in Ohio and in Kentucky. Total chlorine was measured using a Hach Free-and-Total chlorine pocket colorimeter (Hach, Co., Loveland, CO) by the N,N-diethyl-p-phenylenediamine procedure for chlorine measurements (14). The water sample was divided into two aliquots of 0.5 liters each, and Na-thiosulfate was added to a final concentration of 0.1% to 1%. Each aliquot was spiked with 100 cells of H. pylori and 10 cells of E. coli.
Statistical analyses.
The precision of CT measurements for cell, plasmid, or genomic DNA standards was expressed as a percent coefficient of variation (%CV; the standard deviation was expressed as a percentage of the mean). Analysis of covariance tests were performed using Statistical Analysis Software (SAS/STAT User's Guide version 6.4, SAS Institute, Inc., Cary, NC) with procedures PROC MIXED and PROC GLM.
RESULTS
Construction of IC.
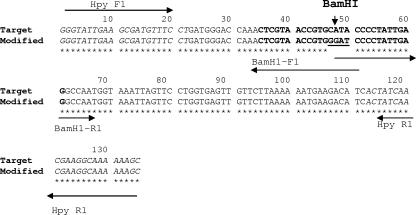
An IC that could be used to measure false-negative PCR results was constructed by PCR mutagenesis (Fig. 1). This amplicon corresponds to the amplicon generated from the target template of H. pylori but includes a newly generated BamHI site in the probe region so that it can be recognized by a different probe. Plasmids from four of the clones produced in the transformation reaction were sequenced to verify that the desired site was mutated. In the correctly mutated clone, the plasmid had four bases changed, from position 47 to 50 of the wild-type amplicon generated from H. pylori genomic DNA, and CATA was replaced by GGAT (Fig. 1). This clone was designated KS10, and the plasmid was designated pKS10. Both the plasmid and the target DNA were amplified using the same primer set, HpyF1 and HpyR1, in a duplex TaqMan PCR assay.
FIG. 1.
Sequence of the modified fragment of 135 bp of the IC and the target sequence. The italicized sequences are the primer binding sites for HPYF1 and HPYR1, as labeled. The underlined sequence is the region where the target amplicon has been modified. The sequences in bold are the probe binding sites for the target (FAM probe) and the modified fragment (VIC probe).
The PCR amplicon generated from plasmid pKS10 was recognized by TaqMan probe 2, labeled with VIC dye, but not with TaqMan probe 1. Similarly, TaqMan probe 1 did not recognize pKS10. Plasmid DNA from E. coli KS10 was prepared in 10-fold dilutions so that the detection limit of the probe could be determined. Probe 2 was able to detect five copies (0.05 fg DNA) of the plasmid in three different experiments, while probe 1 was able to detect 10 copies (20 fg) of the H. pylori genome (Fig. 2). Plasmid DNA was linearized using the restriction enzyme HindIII. The standard curves obtained with the linearized and nonlinearized DNA were similar, and their regression lines were y = 42.87 − 3.49x (R2 = 0.99; average %CV = 0.68) and y = 43.03 − 3.71x (R2 = 0.99; average %CV = 0.59).
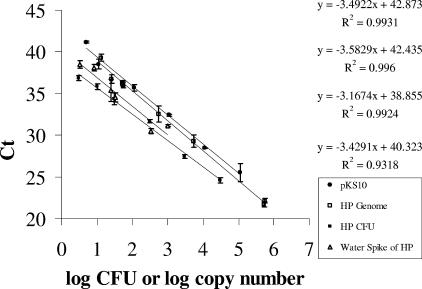
FIG. 2.
Comparison of different molecules for the standard curve. Standard curves were generated by 10 serial dilutions of genomic DNA, internal control plasmid pKS10 DNA, or H. pylori (HP) cells, each containing one copy of the ureA gene or its modified form, analyzed with an ABI 7000 system. Each 25 μl of PCR mixture, containing 11 μl of DNA or whole cells, was tested in a single-plex reaction using either the FAM probe (genome and whole cells) or the VIC probe (plasmid). The regression lines calculated for each set of data points are shown, in order from top to bottom, pKS10, H. pylori genome, H. pylori cells, and water spike of H. pylori. The standard deviations of the data points are denoted in the graph and range from ±0.08 to ±0.97.
Comparison of molecules for the generation of standard curves.
In order to determine the appropriate molecule for the generation of standard curves, known quantities of pKS10 plasmid DNA, H. pylori genomic DNA, and H. pylori whole cells were used in TaqMan PCRs. The concentrations of the DNA from the plasmid and the H. pylori genome were obtained by measuring the absorbance in a NanoDrop spectrophotometer. The number of H. pylori cells was obtained by taking the average of direct microscopic counts and the cell count obtained from OD640 that was optimized, as described above (see Materials and Methods). The standard curves generated from each of the molecules were plotted in parallel and compared to a standard curve generated with a known number of H. pylori cells spiked in water from which the DNA was recovered by a WaterMaster kit. An example of a representative set of curves obtained from the TaqMan PCR, using either the VIC probe or the FAM probe, is presented in Fig. 2. Each standard curve was obtained from data points taken from three independent experiments with replicate samples in each. The standard curves generated were linear over a range of 5 orders of magnitude with genomic DNA and plasmid pKS10 (Fig. 2). The curves generated with genomic DNA and plasmid pKS10 had similar slopes (P = 0.59), and their estimated regression lines were y = 42.87 − 3.49x (R2 = 0.93; average %CV = 1.55) and y = 42.43 − 3.58x (R2 = 0.93; average %CV = 0.68), while that generated by spiking whole cells directly into the reaction mixture was y = 39.22 − 3.28x (R2 = 0.99; average %CV = 1.12). Compared to the regression line generated by known quantities of H. pylori spiked in water, y = 40.308 − 3.43x (R2 = 0.93; average %CV = 1.68), it appeared that either the genomic DNA, the plasmid DNA, or the whole cells would be a suitable standard because there were no significant differences between the slopes (P > 0.05) based on analysis of covariance tests. Interestingly, the intercept obtained with the regression line of H. pylori spiked with water was lower than that generated by genomic DNA or plasmid DNA, which meant that more copies of the ureA gene were obtained than predicted from one copy number per cell of H. pylori.
PCR efficiency and recovery efficiency of H. pylori DNA from water.
In order to get an accurate estimate of the target molecules in the water sample, other control molecules are needed: controls to test the efficiency of the PCR itself (PCR positive control) and controls for the effect of the water matrix, such as for natural organic matter, minerals, and other inhibitory compounds, on the recovery of DNA from the H. pylori cells, as well as on the PCR. With each new PCR batch, plasmid pKS10 DNA at 1,000 copies was added directly to a tube containing PCR master mixture. A CT value of 31.9 was routinely obtained. This served as the PCR positive control.
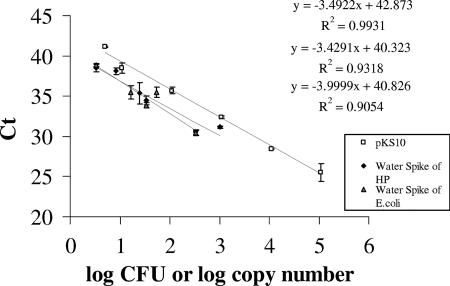
Plasmid pKS10 has been cloned in E. coli EP1300. These cells can be added directly to the water sample, and the DNA recovered can be used to determine the matrix inhibitory effects. Before E. coli cells can be used as a surrogate for H. pylori, however, it must be determined whether E. coli can be recovered from water as well as H. pylori. The recovery of both of these organisms from water was efficient, using a WaterMaster kit (Fig. 3), and the regression lines of H. pylori, y = 40.31 − 3.43x (R2 = 0.93; average %CV = 1. 68) and E. coli, y = 40.83 − 3.99x (R2 = 0.91; average %CV = 1.32) had slopes that were not significantly different (P = 0.5275). The results demonstrate that the recovery of H. pylori was not different from that of E. coli cells. The recovery was ≥100% compared to a standard curve generated by plasmid pKS10, where one copy of plasmid is equivalent to one copy of the ureA gene.
FIG. 3.
Comparison of recovery of H. pylori (HP) and E. coli KS10 cells from water. Standard curves were generated with DNA recovered from water that was spiked with H. pylori cells or E. coli KS10 cells at the concentrations shown in the figure. H. pylori DNA was evaluated with the FAM dye, while E. coli KS10 was evaluated with the VIC dye, in single-plex TaqMan PCR. A standard curve obtained from different concentrations of plasmid pKS10 (VIC probe) was also plotted in parallel (R value). Data points are averages of samples from three different experiments.
The number of E. coli cells containing IC compatible with target H. pylori.
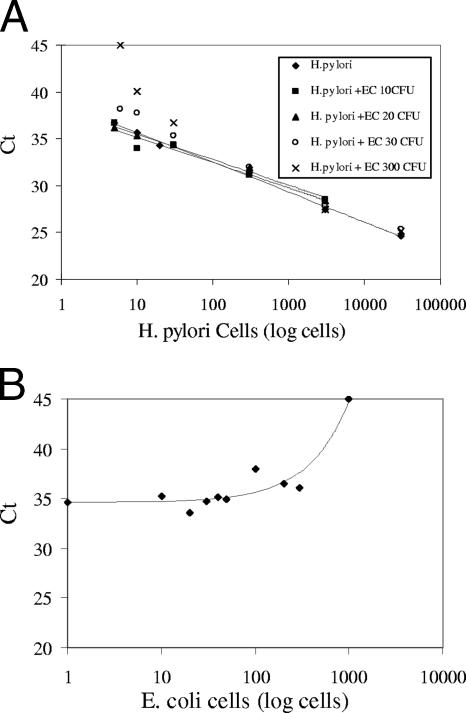
Before E. coli KS10 can be used as a matrix spike, the number of cells to be added needs to be determined. Since the target and the IC are coamplified by the same set of primers, it becomes necessary to titrate the amounts of the IC to be added such that it would not compete with the target H. pylori DNA molecules beyond the limits of detection. H. pylori cells diluted in water at different concentrations, ranging from 3,000 to 5 cells, were added directly to the PCR mixture along with 10, 20, 30, or 300 cells of E. coli KS10 (Fig. 4A). There was no major change in the CT values for as little as five H. pylori cells in the presence of ≤20 cells of E. coli KS10 (36.7, 36.7, and 36.2 [Table 1]). When the ratio of H. pylori to E. coli cells was 5:30, the CT value of H. pylori shifted from 36.7 to 38.12; at 10:30, the CT value shifted from 35.7 to 37.7 (a relative shift of 1.4 and 2, respectively [Table 1], with a fluorescence signal obtained most of the time); and at a ratio of 30:300, the CT value shifted from 34.3 to 36.7 (a 2.4 relative shift, with a distorted signal). Ten and five cells of H. pylori could not be detected in the presence of 300 cells of E. coli. In order to get a finer titration at the lower limits of detection, different concentrations of E. coli KS10, ranging from 10 to 1,000 cells, were added to 20 cells of H. pylori (Table 2 and Fig. 4B). In the presence of up to 50 cells of E. coli, 20 cells of H. pylori cells could be detected, after which the CT value began to shift toward 40 and the fluorescence signal became distorted.
FIG. 4.
Detection of H. pylori in the presence of IC. (A) To different amounts of H. pylori cells (5 to 3,000 cells), either 10 (▪), 20 (▴), 30 (○), or 300 (×) E. coli KS10 cells were added. The curve denoted by a ♦ was generated with no E. coli cells added. No trend line was added to data points generated with 30 and 300 cells. (B) To 20 cells of H. pylori, 0, 10, 20, 30, 40, 50, 100, 300, or 1,000 E. coli KS10 cells were added. The CT value obtained for each data point is shown in Table 1 for panel A and in Table 2 for panel B.
TABLE 1.
Coamplification of E. coli cells containing IC and H. pylori ureAa
| No. of H. pylori cells |
CT value ± SD for the no. of E. coli cells added
|
||||
|---|---|---|---|---|---|
| 0 | 10 | 20 | 30 | 300 | |
| 30,000 | 24.6 ± 0.72 | 25.9 ± 0.21 | 25.4 ± 0.22 | 25.2 ± 0.33 | |
| 3,000 | 27.4 ± 0.11 | 28.5 ± 0.17 | 28.5 ± 0.89 | 27.9 ± 0.36 | 27.5 ± 0.08 |
| 300 | 31.7 ± 0.19 | 31.2 ± 0.13 | 31.9 ± 0.08 | 31.9 ± 0.42 | 31.7 ± 0.52 |
| 30 | 34.3 ± 0.78 | 34.4 ± 0.32 | 34.4 ± 0.35 | 35.3 ± 1.1 | 36.7 ± 0.54* |
| 10 | 35.7 ± 0.29 | 35.2 ± 0.25 | 35.4 ± 0.22 | 37.7 ± 1.9 | UD |
| 5 | 36.7 ± 1.0 | 36.7 ± 1.02 | 36.2 ± 0.22 | 38.1 ± 1.6 | UD |
*, the amplification curve was distorted. UD, undetermined, and no CT values were obtained. The CT values ± standard deviations (SD) generated from the H. pylori cells (using the FAM probe) are averages of duplicate experiments with two replicates performed for each. The average %CV was less than 2%.
TABLE 2.
Coamplification of 20 H. pylori cells in the presence of E. coli cells with ICa
| No. of E. coli cells |
CT values ± SD for
|
|
|---|---|---|
| FAM probe | VIC probe | |
| 0 | 34.6 ± 0.52 | ND |
| 10 | 35.3 ± 0.17 | 37.2 ± 0.37 |
| 20 | 33.6 ± 0.30 | 37.1 ± 0.86 |
| 30 | 34.7 ± 0.93 | 35.5 ± 0.12 |
| 40 | 35.1 ± 2.17 | 35.3 ± 0.10 |
| 50 | 34.9 ± 0.52 | 34.9 ± 0.18 |
| 100 | 38.0* | 33.3 ± 0.75 |
| 200 | 36.5* | 32.8 ± 0.16 |
| 300 | 36.1* | 32.2 ± 0.16 |
| 1,000 | UD | 30.8 ± 0.07 |
To each PCR tube containing 20 cells of H. pylori, 0 to 1,000 cells of E. coli KS10 were added. The CT values ± standard deviations (SD) are averages of duplicate experiments, with two replicates performed for each. The average %CV was less than 2%. *, amplification curves were distorted. UD, undetermined, and no CT values were obtained.
Evaluation of different water samples.
Chlorine is known to affect the DNA of bacterial cells (16). When the DNA was extracted from water samples spiked with H. pylori by a WaterMaster kit and evaluated with a duplex PCR assay, it was found that the presence of 0.1% sodium thiosulfate used to neutralize the chlorine resulted in an equivalent CT value for all the water samples. Furthermore, the CT value was similar to the CT value observed for double-distilled water spiked with the same number of cells (Table 3).
TABLE 3.
Comparison between DNA detection in the presence and absence of chlorine residue
| Water sample (500 ml) source | Total chlorine residue (mg/liter) |
CT value (FAM probe)a
|
|
|---|---|---|---|
| 100 H. pylori cells | 100 H. pylori cells in the presence of Na-thiosulfate | ||
| Kentucky | 1.50 | 40.63 | 34.11 |
| Ohio-1 | 1.20 | 39.29 | 34.98 |
| Ohio-2 | 1.58 | 39.13 | 34.70 |
| Ohio-3 | 0.86 | 37.17 | 34.09 |
| Distilled water | 0 | 34.30 | 34.36 |
CT values are averages of duplicate samples.
DISCUSSION
There is a need for a sensitive method that would reliably detect a few cells of H. pylori in water samples as a means to further determine whether water is one of the sources of infection for this bacterium. qPCR provides diagnostic microbiology laboratories with a powerful tool with which to quantify and achieve a high degree of sensitive and specific detection of targets relative to that of regular PCR. However, specific controls must be used in order to avoid false-negative results due to matrix inhibitory effects of the field samples, the failure of the method to perform optimally, or false positives due to cross-contamination of samples.
Positive controls are necessary to verify if the method is adequately amplifying the target to its limits of detection and if false negatives are absent. Such controls need to be used at several stages in the method: (i) as a matrix spike to test the degree of inhibition from the matrix, if any, and thus give an estimate of the efficiency of nucleic acid extraction from the sample; (ii) as a PCR inhibitor control to test whether the PCR part of the reaction is performing optimally, thus indicating that all the reagents have been added correctly; and (iii) as a method control to determine if the reagents as well as the instruments are performing optimally. One or more kinds of internal controls can be used in the method (37). Such controls can be exogenously added. When naked DNA is being used as an exogenous positive control, it can be used only to test the efficiency of the downstream part of the method, e.g., the PCR. However, if the DNA is incorporated into a surrogate organism, then it can be used as a matrix spike, and the efficiency of the whole method for the particular matrix can be determined. In the method proposed, it was decided to use a modified DNA fragment as the PCR-positive control and a whole organism, an E. coli strain, into which the modified form of the amplicon that is detected in the H. pylori target was cloned, as a matrix spike. Since the modified DNA in the plasmid is quantified similarly to the H. pylori genomic target, the DNA structure surrounding the target does not appear to affect the PCR amplification efficiency. This molecule was used as the PCR-positive control in a single-plex assay with each PCR run. The use of a VIC dye-labeled-modified fragment as a positive control can also eliminate contamination issues, which can result in false positives that may arise due to the detection of the positive control in a negative field sample if H. pylori DNA was used.
H. pylori is difficult to grow in the laboratory and may not easily lend itself to interlaboratory validation. Although plasmids have been found in H. pylori and used in H. pylori mutagenesis, it is not clear whether recombinant plasmids can be routinely maintained in the organism and used as necessary (5, 21). A well-defined bacterial system was needed that would routinely produce a single-copy plasmid. Because E. coli can be detected using the DNA extraction system of a WaterMaster kit at the same level of sensitivity as for H. pylori, E. coli may be an adequate surrogate for H. pylori. The use of a modified form of the target amplicon that had a change in the probe region allowed for the use of the same set of primers but with a differentially labeled probe. Thus, unnecessary competition arising from the use of two sets of primers was avoided. A second advantage is the elimination of the need to collect and extract DNA from duplicate samples because this approach allows one sample to be used for both the control and the target detections simultaneously, with the use of two fluorescent dyes in a multiplex PCR. This approach also reduces the chances of variability that may arise from comparing two different aliquots, albeit from the same water source. Initially, the modified form of the target was cloned into a multicopy plasmid. However, the addition of a few cells of E. coli containing this plasmid shifted the CT value of low numbers of H. pylori cells beyond the limits of detection. This unexpected result can be explained by the fact that in an actively dividing standard E. coli cell, a multicopy plasmid can reach up to 100 copies per cell (41). Thus, the addition of 10 cells of this strain amounted to having 1,000 copies of the modified target. Since it is likely that if H. pylori was present in water it would be at low levels, the modified ureA fragment was inserted into a single-copy plasmid that would allow the detection of 10 copies from 10 cells. The copy control vector pCC1, which contains both the E. coli F factor-based single-copy origin of replication and the high-copy oriV origin of replication, proved to be convenient for this since it could be produced in a single copy in EP1300 cells (Epicentre Biotechnologies) or, when induced by an induction solution, produced in multiple copies. The latter was used for generating large amounts of plasmid to be used as standard molecules. Cloning the fragment in a single-copy plasmid provided an additional amount of quality control by keeping cross-contamination due to IC to a minimum.
After carefully titrating the number of copies of the internal control to be used in the PCR so that the target could still be detected within the limits of detection, it was decided to keep the internal control at 10 to 20 cells. At this level, there would be little competition with even as low as five cells of H. pylori. Although there is the risk of suppressing the signal with the VIC probe from using the IC at 10 to 20 cells, if greater than 300 cells of H. pylori cells are present in the field sample, it would not lead to a false-negative result. In such an event, the positive result generated from the H. pylori DNA by the FAM signal would be taken as valid because the IC amplification becomes unnecessary (22). The use of IC at a high concentration may also lead to nondetection of low-grade inhibition from the matrix, which in turn may lead to nondetection of a low number of target samples (35).
Many of the current methods described in the literature do not sample the entire liter of sample collected (23, 32, 42). In this method, we examined several commercial DNA extraction kits. A WaterMaster kit allowed us to test the entire liter of sample with greater than 95% recovery of cells. An unexpected observation was that more copies of the ureA gene from H. pylori cells were obtained than that predicted from the number of cells spiked. Since the cells were derived from 48-h plates of H. pylori (where ≥99% of the cells are in the spiral form), the cells may be in the process of DNA replication and cell division, resulting in an increase in the copy number of some of the genes of the cells without impacting the turbidity of the media or the microscopic counts. The cells present in the water distribution system are likely to be in the coccoid form. This form has been shown to be a viable but unculturable form, with diminished DNA (1, 28). The effectiveness of a WaterMaster kit in extracting the DNA from these cells and the subsequent detection by qPCR does not seem to be different from that for the spiral cells (unpublished results). Thus, the standard curve generated with the purified plasmid can be used to estimate the copy number in the field sample.
When water samples were collected from different distribution systems and spiked with a fixed number of H. pylori cells, the presence of residual chlorine seemed to have affected the observed CT value. It is known that chlorine and chlorine-releasing agents bind directly to the DNA, forming chlorinated derivatives of nucleotide bases, which has a deleterious effect on the growth of bacteria (16, 17). In one study, at a concentration of 2.6 mg/liter (50 μM), hypochlorous acid completely inhibited the growth of E. coli and DNA synthesis by 96% within 5 min (33). The binding of chlorine to the DNA may have resulted in increased CT values. Another explanation could be that the chlorine residue interfered with the fluorescence signal of the qPCR. A change in fluorescence intensities of E. coli cells containing enhanced green fluorescent protein on exposure to active chlorine has been reported by Burnett and Beuchat (9). Additional work is needed to clarify the effect of chlorine in qPCR assays. Other components in the water matrix might also have an affect on the CT values. In any case, water samples from distribution systems should be collected in the presence of a chlorine-neutralizing agent such as 0.1% sodium thiosulfate.
In conclusion, the method described in this study, which uses a specific internal standard as a positive control and a new method of sample preparation, could form the basis for an accurate and robust screening tool for the detection of H. pylori DNA in water at levels as low as 5 to 10 cells/liter of water.
Acknowledgments
We thank Orin Shanks, John Olszewski, Richard Haughland, Kevin Oshima, Gene Rice, and Jim Sinclair of the EPA for valuable discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Adams, B. L., T. C. Bates, and J. D. Oliver. 2003. Survival of Helicobacter pylori in a natural freshwater environment. Appl. Environ. Microbiol. 69:7462-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benaissa, M., P. Babin, N. Quellard, L. Pezennec, Y. Cenatiempo, and J. L. Fauchere. 1996. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect. Immun. 64:2331-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beneduce, L., G. Spano, M. Libergoli, M. Labonia, and S. Massa. 2003. Survival of Helicobacter pylori in well water. World J. Microbiol. Biotechnol. 19:505-508. [Google Scholar]
- 4.Benson, J. A., K. A. Fode-Vaughan, and M. L. Collins. 2004. Detection of Helicobacter pylori in water by direct PCR. Lett. Appl. Microbiol. 39:221-225. [DOI] [PubMed] [Google Scholar]
- 5.Bereswill, S., R. Schonenberger, A. H. van Vliet, J. G. Kusters, and M. Kist. 2005. Novel plasmids for gene expression analysis and for genetic manipulation in the gastric pathogen Helicobacter pylori. FEMS Immunol. Med. Microbiol. 44:157-162. [DOI] [PubMed] [Google Scholar]
- 6.Bode, G., F. Mauch, and P. Malfertheiner. 1993. The coccoid forms of Helicobacter pylori. Criteria for their viability. Epidemiol. Infect. 111:483-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, L. M. 2000. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol. Rev. 22:283-297. [DOI] [PubMed] [Google Scholar]
- 8.Bunn, J. E., W. G. MacKay, J. E. Thomas, D. C. Reid, and L. T. Weaver. 2002. Detection of Helicobacter pylori DNA in drinking water biofilms: implications for transmission in early life. Lett. Appl. Microbiol. 34:450-454. [DOI] [PubMed] [Google Scholar]
- 9.Burnett, S. L., and L. R. Beuchat. 2002. Comparison of methods for fluorescent detection of viable, dead, and total Escherichia coli O157:H7 cells in suspensions and on apples using confocal scanning laser microscopy following treatment with sanitizers. Int. J. Food Microbiol. 74:37-45. [DOI] [PubMed] [Google Scholar]
- 10.Catrenich, C. E., and K. M. Makin. 1991. Characterization of the morphologic conversion of Helicobacter pylori from bacillary to coccoid forms. Scand. J. Gastroenterol. Suppl. 181:58-64. [PubMed] [Google Scholar]
- 11.Cave, D. R. 1997. How is Helicobacter pylori transmitted? Gastroenterology 113:S9-S14. [DOI] [PubMed] [Google Scholar]
- 12.Cellini, L., N. Allocati, E. Di Campli, and B. Dainelli. 1994. Helicobacter pylori: a fickle germ. Microbiol. Immunol. 38:25-30. [DOI] [PubMed] [Google Scholar]
- 13.Cellini, L., I. Robuffo, E. Di Campli, S. Di Bartolomeo, T. Taraborelli, and B. Dainelli. 1998. Recovery of Helicobacter pylori ATCC 43504 from a viable but not culturable state: regrowth or resuscitation? APMIS 106:571-579. [PubMed] [Google Scholar]
- 14.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton. 1998. Standard methods for the examination of water and wastewater, 20th ed., chapter 4500Cl. American Public Health Association, Washington, DC.
- 15.Cover, T. L., and M. J. Blaser. 1992. Helicobacter pylori and gastroduodenal disease. Annu. Rev. Med. 43:135-145. [DOI] [PubMed] [Google Scholar]
- 16.Dennis, W. H. J., V. P. Olivieri, and C. W. Kruse. 1978. Reaction of uracil with hypochlorous acid. Biochem. Biophys. Res. Commun. 83:168-171. [DOI] [PubMed] [Google Scholar]
- 17.Dukan, S., and D. Touati. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 178:6145-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engstrand, L., A. M. Nguyen, D. Y. Graham, and F. A. el-Zaatari. 1992. Reverse transcription and polymerase chain reaction amplification of rRNA for detection of Helicobacter species. J. Clin. Microbiol. 30:2295-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gribbon, L. T., and M. R. Barer. 1995. Oxidative metabolism in nonculturable Helicobacter pylori and Vibrio vulnificus cells studied by substrate-enhanced tetrazolium reduction and digital image processing. Appl. Environ. Microbiol. 61:3379-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegarty, J. P., M. T. Dowd, and K. H. Baker. 1999. Occurrence of Helicobacter pylori in surface water in the United States. J. Appl. Microbiol. 87:697-701. [DOI] [PubMed] [Google Scholar]
- 21.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 22.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horiuchi, T., T. Ohkusa, M. Watanabe, D. Kobayashi, H. Miwa, and Y. Eishi. 2001. Helicobacter pylori DNA in drinking water in Japan. Microbiol. Immunol. 45:515-519. [DOI] [PubMed] [Google Scholar]
- 24.Hulten, K., H. Enroth, T. Nystrom, and L. Engstrand. 1998. Presence of Helicobacter species DNA in Swedish water. J. Appl. Microbiol. 85:282-286. [DOI] [PubMed] [Google Scholar]
- 25.Hulten, K., S. W. Han, H. Enroth, P. D. Klein, A. R. Opekun, R. H. Gilman, D. G. Evans, L. Engstrand, D. Y. Graham, and F. A. el-Zaatari. 1996. Helicobacter pylori in the drinking water in Peru. Gastroenterology 110:1031-1035. [DOI] [PubMed] [Google Scholar]
- 26.Klein, P. D., D. Y. Graham, A. Gaillour, A. R. Opekun, and E. O. Smith for the Gastrointestinal Physiology Working Group. 1991. Water source as risk factor for Helicobacter pylori infection in Peruvian children. Lancet 337:1503-1506. [DOI] [PubMed] [Google Scholar]
- 27.Krumbiegel, P., I. Lehmann, A. Alfreider, G. J. Fritz, D. Boeckler, U. Rolle-Kampczyk, M. Richter, S. Jorks, L. Muller, M. W. Richter, and O. Herbarth. 2004. Helicobacter pylori determination in non-municipal drinking water and epidemiological findings. Isotopes Environ. Health Stud. 40:75-80. [DOI] [PubMed] [Google Scholar]
- 28.Kusters, J. G., M. M. Gerrits, J. A. Van Strijp, and C. M. Vandenbroucke-Grauls. 1997. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 65:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, C., P. R. Musich, T. Ha, D. A. J. Ferguson, N. R. Patel, D. S. Chi, and E. Thomas. 1995. High prevalence of Helicobacter pylori in saliva demonstrated by a novel PCR assay. J. Clin. Pathol. 48:662-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, Y., T. E. Redlinger, R. Avitia, A. Galindo, and K. Goodman. 2002. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl. Environ. Microbiol. 68:1436-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazari-Hiriart, M., Y. Lopez-Vidal, S. Ponce-de-Leon, J. J. Calva, F. Rojo-Callejas, and G. Castillo-Rojas. 2005. Longitudinal study of microbial diversity and seasonality in the Mexico City metropolitan area water supply system. Appl. Environ. Microbiol. 71:5129-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniels, A. E., L. Wymer, C. Rankin, and R. Haugland. 2005. Evaluation of quantitative real time PCR for the measurement of Helicobacter pylori at low concentrations in drinking water. Water Res. 39:4808-4816. [DOI] [PubMed] [Google Scholar]
- 33.McKenna, S. M., and K. J. Davies. 1988. The inhibition of bacterial growth by hypochlorous acid. Possible role in the bactericidal activity of phagocytes. Biochem. J. 254:685-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narikawa, S., S. Kawai, H. Aoshima, O. Kawamata, R. Kawaguchi, K. Hikiji, M. Kato, S. Iino, and Y. Mizushima. 1997. Comparison of the nucleic acids of helical and coccoid forms of Helicobacter pylori. Clin. Diagn. Lab. Immunol. 4:285-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenstraus, M., Z. Wang, S. Y. Chang, D. DeBonville, and J. P. Spadoro. 1998. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J. Clin. Microbiol. 36:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki, K., Y. Tajiri, M. Sata, Y. Fujii, F. Matsubara, M. Zhao, S. Shimizu, A. Toyonaga, and K. Tanikawa. 1999. Helicobacter pylori in the natural environment. Scand. J. Infect. Dis. 31:275-279. [DOI] [PubMed] [Google Scholar]
- 37.Sen, K., G. S. Fout, R. Haugland, C. Moulton, A. Grimm, G. Di giovanni, M. A. Feige, J. Best, G. Lott, J. Scheller, E. Reiley, K. Conell, and M. Marshall. 2004. Quality assurance/quality control guidance for laboratories performing PCR analyses on environmental samples, publication no. 815-B-04-001. Office of Water (4607), U.S. Environmental Protection Agency, Cincinnati, OH.
- 38.Sen, K. 2000. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J. Clin. Microbiol. 38:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shahamat, M., M. Alavi, J. E. Watts, J. M. Gonzalez, K. R. Sowers, D. W. Maeder, and F. T. Robb. 2004. Development of two PCR-based techniques for detecting helical and coccoid forms of Helicobacter pylori. J. Clin. Microbiol. 42:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shahamat, M., U. Mai, C. Paszko-Kolva, M. Kessel, and R. R. Colwell. 1993. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl. Environ. Microbiol. 59:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summers, D. 1998. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 29:1137-1145. [DOI] [PubMed] [Google Scholar]
- 42.Watson, C. L., R. J. Owen, B. Said, S. Lai, J. V. Lee, S. Surman-Lee, and G. Nichols. 2004. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J. Appl. Microbiol. 97:690-698. [DOI] [PubMed] [Google Scholar]
- 43.Weiss, J., J. Mecca, E. da Silva, and D. Gassner. 1994. Comparison of PCR and other diagnostic techniques for detection of Helicobacter pylori infection in dyspeptic patients. J. Clin. Microbiol. 32:1663-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West, A. P., M. R. Millar, and D. S. Tompkins. 1992. Effect of physical environment on survival of Helicobacter pylori. J. Clin. Pathol. 45:228-231. [DOI] [PMC free article] [PubMed] [Google Scholar]