Abstract
Fluorescence in situ hybridization (FISH) was used for direct detection of Escherichia coli on pipe surfaces and coupons in drinking water distribution networks. Old cast iron main pipes were removed from water distribution networks in France, England, Portugal, and Latvia, and E. coli was analyzed in the biofilm. In addition, 44 flat coupons made of cast iron, polyvinyl chloride, or stainless steel were placed into and continuously exposed to water on 15 locations of 6 distribution networks in France and Latvia and examined after 1 to 6 months exposure to the drinking water. In order to increase the signal intensity, a peptide nucleic acid (PNA) 15-mer probe was used in the FISH screening for the presence or absence of E. coli on the surface of pipes and coupons, thus reducing occasional problems of autofluorescence and low fluorescence of the labeled bacteria. For comparison, cells were removed from the surfaces and examined with culture-based or enzymatic (detection of β-d-glucuronidase) methods. An additional verification was made by using PCR. Culture method indicated presence of E. coli in one of five pipes, whereas all pipes were positive with the FISH methods. E. coli was detected in 56% of the coupons using PNA FISH, but no E. coli was detected using culture or enzymatic methods. PCR analyses confirmed the presence of E. coli in samples that were negative according to culture-based and enzymatic methods. The viability of E. coli cells in the samples was demonstrated by the cell elongation after resuscitation in low-nutrient medium supplemented with pipemidic acid, suggesting that the cells were present in an active but nonculturable state, unable to grow on agar media. E. coli contributed to ca. 0.001 to 0.1% of the total bacterial number in the samples. The presence and number of E. coli did not correlate with any of physical and/or chemical characteristic of the drinking water (e.g., temperature, chlorine, or biodegradable organic matter concentration). We show here that E. coli is present in the biofilms of drinking water networks in Europe. Some of the cells are metabolically active but are often not detected due to limitations of traditionally used culture-based methods, indicating that biofilm should be considered as a reservoir that must be investigated further in order to evaluate the risk for human health.
The drinking water supplier has to provide the consumer potable water of a quality identical to that leaving the treatment plant. However, it has been well documented that water that reaches the consumer's tap is often of worse microbiological quality than that which left the plant (29). Therefore, the analyses used for monitoring the hygiene of potable water must be rapid to perform and able to detect the major microbial groups of concern. Escherichia coli is still used as the principle indicator for drinking water pollution monitoring (39) and is even considered a superior indicator (11), since enterotoxic and enterohemorrhagic forms are one of the major causes of water-related outbreaks (29). The proportion of waterborne disease outbreaks associated with the distribution system failures has been increasing over the years (26). Moreover, although E. coli is often detected in the drinking water, the source of the contamination is not. Hence, traditional methodology for water sampling and analyses is not always able to ensure public safety regarding both (i) the strategy of sampling and (ii) the choice of the detection method. The sampling strategy is limited to sampling water only, whereas most of the bacteria are attached to the inner surfaces of the pipes forming biofilms. The phenomenon of biofilm formation, or the attachment of microorganisms to the inner surfaces of the drinking water distribution system, has been well documented (see reviews in references 18, 27, and 28). The attachment of organisms to surfaces has been shown to alter their physiology. Attached organisms were found to be generally more active in absorbing nutrients, as well as more resistant to environmental stress such as starvation, heavy metals, and chlorine (2, 21). It has also been shown that bacteria attached to surfaces show greater resistance to disinfection (15, 19, 20, 36). Biofilms in distribution systems may provide a favorable condition for some bacteria, such as opportunistic pathogens (e.g., Legionella spp., Pseudomonas aeruginosa, and Mycobacterium avium), to colonize it and may harbor pathogens, such as Salmonella enterica serovar Typhimurium, which have entered the distribution system (1, 5, 18, 28). It has been shown in lab-scale experiments that E. coli can survive in biofilters (24) and even multiply in the biofilm (17, 34, 40), which again raises the question of its suitability as a drinking water quality indicator. The aim of the present study was to investigate the presence of E. coli in the biofilm of actual water distribution networks in Europe by using a direct detection method. The advantages of an in situ (direct) approach are an undisturbed sample composition and the prevention of cell loss.
Traditional methods for detection of E. coli are based on incubation in nutrient-rich media. Fluorescence in situ hybridization (FISH) is a method wherein a nucleic acid sequence of interest is identified among other sequences by pairing it with a complementary sequence used as a probe. The procedure is quick, and results can be achieved in a couple of hours as opposed to days using culture-based methods. It also should be noted that, depending on environmental conditions (starvation, stress, etc.), bacteria can survive in water in an active but nonculturable state (9) and thus not be detectable by culture-based methods. Methods able to detect bacteria in this nondividing state include the assessment of different cellular functions, namely, efflux pump activity (using SYTO-9 plus ethidium bromide), membrane potential (using [bis-(1,3-dibutylbarbituric acid)trimethine oxonol; DiBAC4(3)]), membrane integrity (using LIVE/DEAD BacLight), cellular respiration (using 5-cyano-2,3-ditolyl tetrazolium chloride reduction), glucose uptake activity (using 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-d-glucose; 2-NBDG), total ATP concentration (determined with BacTiter-Glo) (4), and direct viable count (DVC) in combination with FISH (3). The principle of DVC is an application of a substance inhibiting the cell division but not cell growth (synthesis processes). Thus, the cell count should not increase on behalf of cells capable to divide (3). After staining, the metabolically active cells can be observed as larger and elongated compared to the nonactive cells.
We present here results from the application of FISH techniques for in situ detection of E. coli on inner surfaces of pipes and coupons and biofilm suspensions from pipes and incubators used for collection of biofilm in several large-scale water supply networks around Europe.
MATERIALS AND METHODS
General water analyses.
Unless stated otherwise collection of water samples and water quality analyses were done according to International Organization of Standardization (ISO) standard methods. Assimilable organic carbon (AOC) was measured using methods modified from the van der Kooij method (25). Microbially available phosphorus was determined according to the method of Lehtola et al. (23). Temperature, redox potential, and pH were measured with a WTW Multiline P4 universal meter. Conductivity was measured with EcoScan Con 5 (Eutech Instruments, The Netherlands).
Pipe samples.
Only large distribution systems supplying water which meets the European Water Directive EC/93/87 were included in the study. Pipe samples (diameter ≥50 mm; length ca. 500 cm) were collected from Riga (Latvia), Minho region (Portugal), Southampton (United Kingdom), and an anonymous site in France between January 2004 and August 2005. The pipes were cut out from intact water distribution mains during their replacement and sealed at the site under the supervision of a microbiologist. Five pipes were made of cast iron and one was made of concrete, and the pipes were installed more than 25 years ago. Pipes were dissected horizontally into two pieces in order to make the surface accessible for FISH analyses.
Coupon samples.
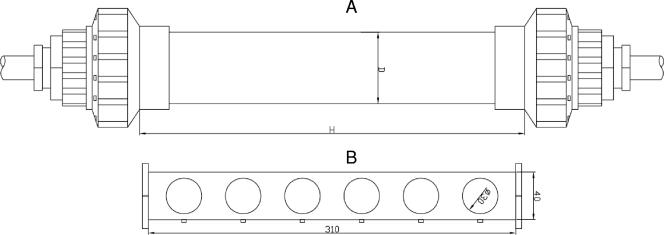
A total of 22 holders (consisting of 25- to 40-mm pipes holding cast iron and polyvinyl chloride [PVC] coupons 3 mm in diameter) were exposed to drinking water (Fig. 1) for several months in the water supply systems. The holders were placed in 15 sites in several locations of water distribution networks (after treatment and at several distances in distribution networks) (Table 1). In Latvia two distribution systems supplying drinking water produced from a surface water source, artificially recharged groundwater, and groundwater sources were used. In France four different distribution systems (described previously by Servais et al. [37]) using surface water for drinking water production were selected. The length of the studied transects varied from 18 to 400 km, and the total number of tanks in all of the distribution systems varied from 2 to 53. The structures of the networks and the water origin (pond, river, and reservoir) were different. The holders were removed from the site and sent to the lab, where they were opened, and the coupons were removed aseptically. The biofilm on coupons and pipe surfaces was fixed by covering the surface with 3 to 4% (vol/vol) formaldehyde for 20 min. After fixation, pipes and coupons were rinsed with sterile water and allowed to dry.
FIG. 1.
Diagram of biofilm sampler, which consists of the coupon holder (B) and the pipe (A) in which the holder with coupons were placed. The pipe was connected to bypass of water distribution systems in Latvia and France.
TABLE 1.
Description of biofilm sampling sites and major water quality parametersa
| Site | Origin | Raw water source | Description of site and water treatment process | Biofilm collector location | Water residence time (h) | Avg temp (°C) | Residual chlorine (mg/liter) | AOC (μg/liter) | BDOC (mg/liter) |
|---|---|---|---|---|---|---|---|---|---|
| A1 | FR | Pound | O3, mineralization, clarification, SF, O3, GAC, ClO2 | Net | 23 | 21.0 | 0 | ND | 1.0 |
| A2 | FR | Pound | O3, mineralization, clarification, SF, O3, GAC, ClO2 | Net | 68 | 18.6 | 0 | ND | 1.3 |
| A3 | FR | Pound | O3, mineralization, clarification, SF, O3, GAC, ClO2 | Net | 77 | ND | 0.01 | ND | 2.2 |
| D1 | FR | Reservoir | NaOCl, MnO4, remineralization, clarification, SF, PO4 injection, O3, NaOCl | Net | 100 | 13.5 | 0.1 | ND | 0.2 |
| D2 | FR | Reservoir | NaOCl, MnO4, remineralization, clarification, SF, PO4 injection, O3, NaOCl | Net | 106 | 13.5 | 0.2 | ND | 0.2 |
| D3 | FR | Reservoir | NaOCl, MnO4, remineralization, clarification, SF, PO4 injection, O3, NaOCl | Net | 130 | 13.0 | 0.1 | ND | 0.1 |
| M1 | FR | Pond | Mineralization, flotation, SF, O3, mineralization, Cl2 | Net | 36 | 15.0 | 0.01 | ND | 0.6 |
| M2 | FR | Pond | Mineralization, flotation, SF, O3, mineralization, Cl2 | Net | 56 | 15.0 | 0.01 | ND | 0.6 |
| T1 | FR | River | Mineralization, flotation, O3, SF, O3, ClO2 | Net | 2 | 17.0 | 0.03 | ND | 0.14 |
| T2 | FR | River | Mineralization, flotation, O3, SF, O3, ClO2 | Net | 6 | 17.0 | <0.1 | ND | 0.23 |
| R1 | LV | River | O3, chemical coagulation, clarification, SF, O3, BAC, Cl2 | Plant before Cl2 | 0 | 2.8 | 0.2 | 205 | ND |
| B1 | LV | Lake | Artificially recharged groundwater, Cl2 | Plant before Cl2 | 0 | 6.7 | <0.1 | 209 | ND |
| B2 | LV | Lake | Artificially recharged groundwater, Cl2 | Net | ND | 5.4 | <0.1 | 236 | ND |
| B3 | LV | Lake | At bottled water production plant using tap water treated with SF, softening, NaOCl, GAC | After addition treatment | ND | 9.2 | <0.1 | 263 | ND |
| C1 | LV | Groundwater | Small local water supply system using iron removal and softening | Net | ND | 11.5 | <0.1 | 117 | ND |
Abbreviations: ND, not determined; FR, France; LV, Latvia; SF, sand filtration; BAC, biologically active carbon; GAC, granular activated carbon.
Biofilm removal protocol from coupons.
Coupons were removed from sampling devices, placed in sterile containers containing 25 to 50 ml of cell-free distilled water, and processed 15 to 25 min later. For some coupons the biofilm was dispersed by a gentle sonication (2 min of ultrasound at 2 W, 20 KHz; Ultrasonic Processor [Cole-Parmer Instruments]). The processor probe was placed 1 cm above the coupon, inside bacterial cell-free distilled water.
Design of PNA probe.
After comparison of several sequences published in literature (32, 33) and searching the NCBI BLAST database (http://www.ncbi.nlm.nih.gov/BLAST/), the following sequence: 5′-TCA ATG AGC AAA GGT-3′, published earlier by O'Keefe et al. (31), was selected as the most specific and appropriate for PNA probe synthesis. Based on this sequence a 15-mer PNA probe (ECOLIFILM) was designed, labeled with cyanine dye Cy3 (excitation, 550 nm; emission, 570 nm), and flanked with solubility enhancers. Cy3 was chosen because it provides sufficient signal intensity, even for bacteria living in oligotrophic environments (30), and its emission is not within the autofluorescence wavelength range of that of pipe materials used for drinking water supply (8).
Verification of the probe.
E. coli (ATCC 25922) strains were grown on R2A agar (10), picked and suspended in 1 ml of phosphate-buffered saline (PBS; 7 mM Na2HPO4, 3 mM NaH2PO4, 130 mM NaCl [pH 7.2]) at a concentration of 107 −108 cells/ml, and vortex mixed. Cell suspensions were pelleted by centrifugation at 6,000 rpm for 3 min, the supernatant was removed, and the cells were then resuspended in PBS. This washing procedure was repeated two more times. After the last wash, about 200 μl of cell suspension was spread onto a clean microscope slide and allowed to dry. To the dried cells 3 to 4% (vol/vol) formaldehyde was applied, and the cells were fixed for 20 min. After fixation the microscope slide was rinsed with water and allowed to dry.
FISH using PNA probes.
A portion (50 to 500 μl) of PNA hybridization mix consisting of hybridization buffer (50 mM Tris-HCl, 10% [wt/vol] 50% dextran sulfate, 0.1 mM NaCl, 30% [vol/vol] formamide, 30% [vol/vol] tetra-sodium pyrophosphate, 0.2% [wt/vol] polyvinylpyrrolidone, 0.2% [wt/vol] Ficoll 400, 5 mM disodium EDTA, 0.1% [vol/vol] Triton X-100) containing 200 nM fluorescently labeled PNA probe was applied to the dry coupon, pipe, or filter surfaces and covered with cover glass. The sample was incubated at 57°C for 60 or 90 min in a tight vessel containing water vapor to avoid concentration effects due to evaporation. The samples were immersed in a vessel containing prewarmed (57°C) washing buffer (5 mM Tris, 15 mM NaCl, 0,1% Triton X-100 [pH 10]) and incubated for 30 min. After that the samples were removed from the vessel, rinsed with water, and allowed to dry.
Total bacterial number.
The total bacterial count was determined immediately after FISH analyses. For this, DAPI (4′,6′-diamidino-2-phenylindole) was applied as counterstain. On the surface, a 1:1 mixture of DAPI (10 μg/ml) and Triton X-100 (0.1%) was applied so that the final concentration of DAPI was 5 μg/ml, followed by incubation for 15 to 20 min, and then the excess liquid was removed. The surface was then rinsed and air dried.
Epifluorescence microscopy.
Microscopy examination was conducted by using an epifluorescence microscope (Leica DMLB) equipped with a 50-W power supply, mercury lamp, several filter sets, and a camera (CoolSNAP Pro; Media Cybernetics, Inc.). For detection of E. coli with an ECOLIFILM probe, a narrow-range Y3 filter (excitation, 545 ± 30 nm; emission, 610 ± 75 nm; dichromatic mirror, 565 nm) was used. For DAPI-stained cells, a filter (excitation, 340 to 380; emission, >425 nm; dichromatic mirror, 400 nm) was used. Samples were examined by using a 1,000× oil immersion or 400× dry objective lens. Images were analyzed by using Image-Pro Plus version 4.5 (Media Cybernetics, Inc.) for Windows.
For samples to be identified as E. coli positive, the following criteria were considered: (i) bright signal in the CY filter channel is emitted by the cell, (ii) the cell stains positive for DAPI, (iii) the cell does not appear in other channels where Cy3 is not emitting light (to distinguish the false positives from autofluorescence), and (iv) the cell morphology resembles that of E. coli. The number of counted viewing fields was 20 to 100. For the total bacterial count, all blue-stained cells were counted in randomly chosen microscopic viewing fields delineated by the eyepiece micrometer. Either a minimum of 300 bacteria was counted or as many viewing fields so that a coefficient of variation of <30% was obtained.
DVC.
In a sterile container containing 9 ml of 0.5× R2A medium and pipemidic acid mixture, the final concentration of 10 μl/ml was prepared from stock solution (10 g/liter in 0.05 M NaOH), to which 1 ml of sonicated biofilm suspension was added, followed by vortex mixing for 30 s or, alternatively, a coupon was immersed and incubated for 8 h at room temperature. The cells were then fixed in the mixture or on the coupon surface to reach the final formaldehyde concentration of 3 to 4%. An aliquot of a sonicated sample was filtered on an Anodisc 25 filter (25-mm diameter; pore size, 0.2 μm; Whatman International, Ltd., Great Britain). Bacteria collected on filters or coupons surfaces were hybridized with the probe as described above, followed by counterstaining with DAPI, if necessary.
Two samples were performed for every DVC experiment. FISH was performed for the first replicate, and FISH-DVC was performed for the second replicate. The lengths of FISH-positive cells were measured in both samples. A sample was considered DVC positive if the size difference between samples was at least 1.5-fold.
Culture methods and enzymatic methods for water and biofilm analyses.
Heterotrophic plate counts in water and sonicated biofilm samples were determined after incubation for 7 days at 20°C on R2A agar (32a).
Culture methods.
Coliforms and E. coli were analyzed by using a multiple-tube most-probable-number (MPN) method (LVS EN ISO 9308:2:1990) or membrane filtration (MF) method (LVS EN ISO 9308:1, 2001) using selective culture medium for the rapid quantitative detection of E. coli and coliform bacteria by optical differentiation of colonies on membrane filters (Rapid E. coli 2 Agar; Bio-Rad, France) and TTC-Tergitol 7 agar (Bio-Rad). The results were expressed as the MPN of bacteria per square centimeter of biofilm.
Enzymatic methods.
For the detection of coliforms and E. coli, 1 to 10 ml of sonicated biofilm suspension was tested with qualitative Colilert or Colisure assay (IDEXX, Laboratories, Inc.).
PCR.
E. coli ATCC 25922 was obtained from the American Type Culture Collection. Pseudomonas fluorescens P17 MSCL 559 was obtained from the culture collection of the University of Latvia. Staphylococcus aureus MSSA476 was obtained from the Biomedical Research and Study Center of Latvia. The samples were prepared as follows. Cells were grown in LB medium overnight. The cell suspension was serially diluted with LB medium, and the concentration of viable cells was validated for the prepared samples by plating them on LB plates. Then, 1 μl of intact bacterial suspension was used as a template for PCR. The biofilm samples were processed as for cultivation-based methods (see above), and then 1 μl of the resulting suspension was used as a template for PCR. An upstream primer (5′-AGAGGATGACCAGCAACAC-3′) and a downstream primer (5′-TACGCATTTCACCGCTAC-3′) were designed to amplify a 404-bp portion of E. coli gene for 16S rRNA, which included the ECOLIFILM binding site, and used for PCR analysis. The concentration of each primer was 0.2 μM, the concentration of each nucleotide was 0.2 mM, and Mg2+ ions were used at 1.6 mM; 2.5 U of Taq DNA polymerase (Fermentas, Lithuania) was taken per reaction, and the amplification conditions were as follows: 10 min of denaturation at 94°C, followed by 35 cycles of 94°C for 20 s, 50°C for 20 s, and 72°C for 30 s, with a final extension step at 72°C for 3 min. The cyclic reaction was performed by using MasterCycler personal PCR machine (Eppendorf, Germany). The PCR products were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining.
RESULTS
The specificity of the probe to E. coli was proved earlier (31); therefore, it was only verified that ECOLIFILM does not bind to Aquaspirillum spp. and Pseudomonas fluorescens. Experiments using glass slides spiked with E. coli and these typical drinking water bacteria showed that the probe was specific and produced a bright fluorescent signal (data not shown).
Pipes.
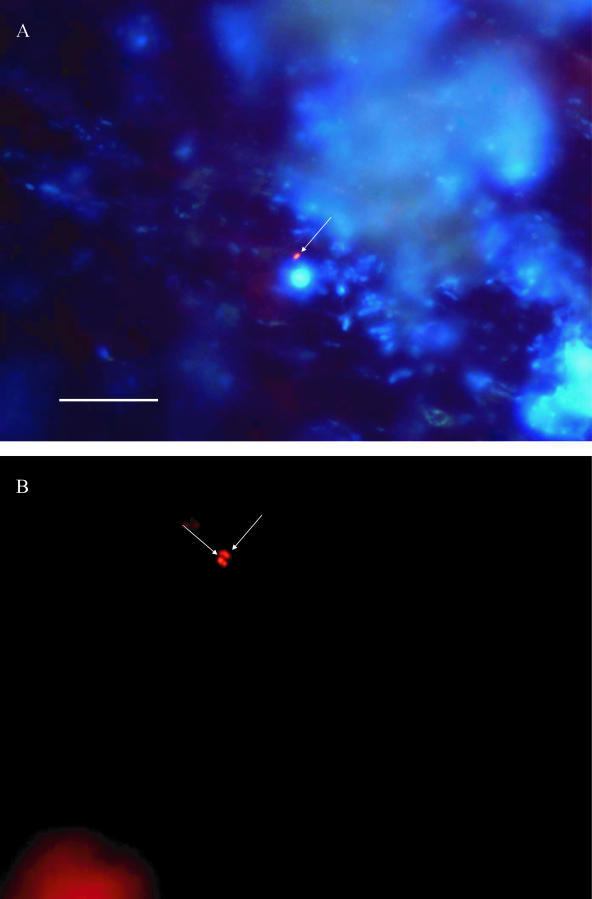
In total, four cast iron pipes from England, Latvia, and France, and one concrete pipe from Portugal were examined. On four pipes FISH was performed directly on the surface of the pipe after they were cut open; for one cast iron pipe, examination was not possible due to the high background fluorescence of the pipe material. Part of the biofilm suspension was removed and examined with a culture method (MPN or MF) and an enzymatic method (Colisure). The Chromagar method indicated the presence of E. coli in one of five pipes, whereas all pipes were E. coli positive when analyzed by FISH (data not shown). Direct examination of the pipe surfaces showed that E. coli was attached to the biofilm as single cells (Fig. 2). Microcolonies or cell clusters were not observed.
FIG. 2.
In situ epifluorescent image of biofilms on the surface of pipes from one of the sites in Latvia (A) and England (B). The arrows point to E. coli cells. FISH was performed directly on the surfaces of the pipes that were exposed to drinking water for more than 25 years. Combined photographs of E. coli and all bacteria detected with DAPI in blue filter (excitation, 360/20 nm; emission, >425 nm) are shown in panel A. In panel B, photographs of E. coli detected by FISH in yellow filter (Y3: excitation, 535/50 nm; emission, 610/75 nm) are shown. Bar, 10 μm.
Water parameters in collector installation sites.
The different parameters of water from biofilm collector installation sites are shown in Table 1. In the samples from Latvia the average temperature was generally low at all sites and did not exceed 12°C. The residual chlorine level in the water supplies was ≤0.2 mg/liter and contained enough nutrients for bacterial growth (AOC > 100 μg/liter) (Table 1). The temperature was generally higher in France, where it reached up to 21°C. The residual chlorine was very low (≤0.1 mg/liter) in all of the networks with exception of D2 (reservoir). The total bacterial number varied from 105 to 108 cell/cm2, and the heterotrophic plate count (HPC) varied between 102 and 107 CFU/cm2 (Table 2) in both networks. None of the factors generally considered important for bacterial regrowth (temperature, biofilm retention time, AOC or biodegradable dissolved organic carbon (BDOC), and chlorine) had a significant effect on the HPC and total bacterial number.
TABLE 2.
Results of coupon analyses at the sites of Latvia and France
| Site | Biofilm collector installation date | Biofilm collector retention times in water supply (days) | Avg HPC (cells/cm2) | CFU/cm2 as determined by culture methods
|
Total no. of bacteria (cells/cm2) | |
|---|---|---|---|---|---|---|
| E. coli | Coliforms | |||||
| A1 | January 2005 | 68 | 6.9 × 103 | 0 | 0 | 9.84 × 106 |
| A2 | January 2005 | 68 | 4.8 × 103 | 0 | 0 | 6.82 × 106 |
| A3 | January 2005 | 68 | 9.4 × 104 | 0 | 0 | 1.30 × 107 |
| D1 | January 2005 | 119 | 1.0 × 105 | 0 | 0 | 4.4 × 106 |
| May 2005 | 175 | 3.7 × 104 | 0 | 0 | ≥108 | |
| D2 | January 2005 | 119 | 1.2 × 104 | 0 | 0 | 6.4 × 106 |
| May 2005 | 175 | 1.5 × 104 | 0 | 0 | 8.3 × 107 | |
| D3 | January 2005 | 119 | 1.2 × 104 | 0 | 0 | 8.0 × 107 |
| M1 | January 2005 | 119 | 1.7 × 104 | 0 | 0 | 1.6 × 105 |
| M2 | January 2005 | 119 | 6.1 × 104 | 0 | 0 | 5.5 × 105 |
| T1 | January, 2005 | 119 | 2.9 × 102 | 0 | 0 | 4.3 × 107 |
| May 2005 | 175 | 2.2 × 103 | 0 | 0 | 2.1 × 107 | |
| T2 | January 2005 | 119 | 4.6 × 103 | 0 | 0 | 1.3 × 107 |
| May 2005 | 175 | 4.7 × 104 | 0 | 0 | 1.2 × 107 | |
| R1 | December 2005 | 173 | 8.5 × 104 | 0 | 0 | 1.7 × 107 |
| December 2005 | 166 | 1.3 × 106 | 0 | 1.1 | 1.6 × 107 | |
| B1 | May 2006 | 35 | 6.9 × 107 | 0 | 0 | ≥108 |
| B2 | December 2005 | 145 | 2.7 × 104 | 0 | 0 | 8.9 × 106 |
| B3 | December 2005 | 101 | 8.7 × 106 | 0 | 0 | 1.3 × 107 |
| C1 | December 2005 | 41 | 4.8 × 104 | 0 | 0 | 2 × 105 |
Biofilm collectors/coupons.
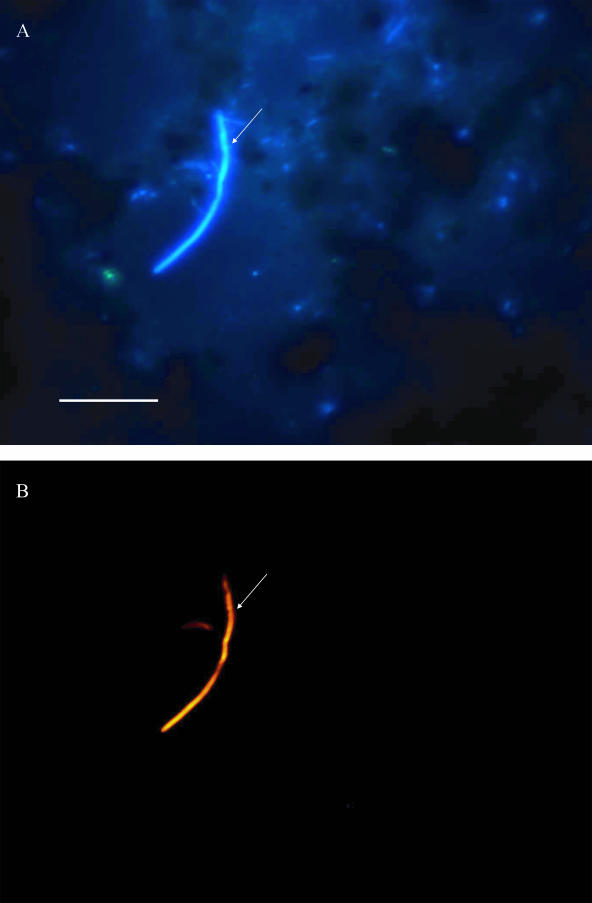
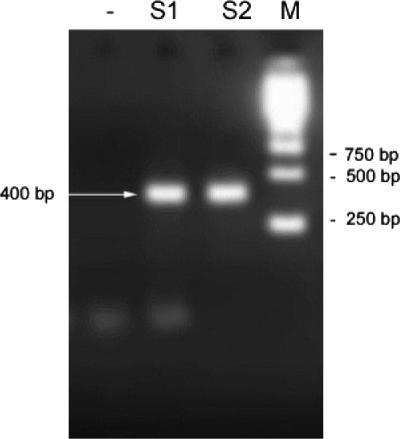
A total of 44 coupons (cast iron, PVC, or stainless steel) from Latvia (two water distribution systems) and France (four water distribution systems) were examined for the presence of E. coli after they were placed in a bypass of the water supplies for 1 to 6 months. Coupons were examined by FISH and by culture and enzymatic methods. The results showed that 58% of samples were positive as determined by FISH, while none were positive with the MPN and MF methods (Table 3). To examine whether the cells are viable or dead, some E. coli organisms were incubated in the presence of pipemidic acid and a low concentration of nutrients for gentle resuscitation. In most of the sonicated coupons the cells were viable and metabolically active. Figure 3 shows an example of an elongated E. coli cell, indicating metabolic activity. These cells were not culturable in the rich media used in the Colilert and MPN tests. The presence of E. coli was confirmed also by PCR (Fig. 4). The amplification of negative controls (S. aureus and P. fluorescens) with both primers gave no signal (data not shown). Figure 4 shows two samples, one marked as not spiked with E. coli cells (S1) and another spiked with 2,000 to 6,000 E. coli cells/ml (S2), used as templates for PCR, resulted in an ∼400-bp PCR product showing that the E. coli genome DNA was present in the initial sample. This fragment was also sequenced and corresponded to a 402-bp fragment of E. coli 16S rRNA containing the probe-binding 15-bp sequence. Different dilutions were also amplified and confirmed the presence of the bacterium (data not shown).
TABLE 3.
Results from detection of E. coli on coupons by FISH (with or without resuscitation) and culture-based (MPN or membrane filtration) or enzyme-based (Colilert or Colisure; IDEXX, Ltd.) analysis after exposure to drinking water in France and Latvia for 1 to 6 monthsa
| Site | Coupon material | FISH | Culture or enzymatic method |
|---|---|---|---|
| A1 | CI | - | ND |
| PVC | - | - | |
| A2 | CI | - | ND |
| PVC | - | ND | |
| PVC | - | - | |
| A3 | CI | - | ND |
| PVC | - | ND | |
| PVC | - | - | |
| D1 | CI | + | - |
| D2 | CI | + | - |
| PVC | + | ND | |
| PVC | + | - | |
| D3 | CI | + | - |
| PVC | + | - | |
| CI | + | ND | |
| PVC | + | ND | |
| M1 | CI | + | ND |
| CI | - | ND | |
| PVC | - | - | |
| PVC | - | - | |
| PVC | - | - | |
| M2 | CI | + | - |
| CI | - | - | |
| CI | - | - | |
| PVC | + | ND | |
| T1 | CI | - | ND |
| PVC | + | - | |
| CI | + | - | |
| PVC | + | ND | |
| CI | - | ND | |
| T2 | CI | + | ND |
| CI | + | - | |
| PVC | + | - | |
| R1 | SS | - | - |
| SS | + | - | |
| SS | + | ND | |
| SS | - | ND | |
| B1 | SS | + | - |
| B2 | SS | + | - |
| B3 | SS | + | - |
| SS | + | - | |
| SS | + | - | |
| C1 | SS | + | ND |
+, Positive; −, negative; ND, not determined; CI, cast iron; SS, stainless steel.
FIG. 3.
Epifluorescence image of metabolically active E. coli in biofilm sample from water distribution networks in France. Biofilm was removed from a cast iron coupon (T2; retention time, 119 days) by sonification and subjected to resuscitation procedure for 8 h in 50% R2A media with pipemidic acid. The total bacteria number was detected with DAPI (A) in blue filter (excitation, 360/20 nm; emission, >425 nm) and E.coli detected with FISH methods in yellow filter (Y3: excitation, 535/50 nm; emission, 610/75 nm) (B). The arrows point to an elongated E. coli cell. Bar, 10 μm.
FIG. 4.
PCR product (400 bp) resulting from amplification of biofilm material (from site B2 in Latvia) not spiked with E. coli cells (S1) and spiked with 2,000 to 6,000 E. coli cells/ml (S2). A negative control is also shown (−).
E. coli was found in all networks except one network in France (site A). This network received water from a protected forest pond, and the water was treated with two-stage ozonation and granular activated carbon filtration and contained a high BDOC level. The approximate bacterial number was determined for some of the samples from several sites and was in range of 200 to 500 cells/cm2; thus, E. coli contributed from 0.001% to 0.1% of the total bacterial number. No correlation was found between the presence of E. coli and the absence or presence of various water quality parameters (AOC, BDOC, temperature, redox potential, chlorine, or inorganic nutrient concentration) or other parameters (pipe material type, hydraulic residence type, or water source).
DISCUSSION
Routinely, only water samples are examined, although most of the bacteria in drinking water distribution networks are attached to the surfaces of pipes forming biofilms (see, for example, reference 5). Several studies at the lab scale have demonstrated that even in the presence of detectable levels of chlorine, E. coli can be incorporated in biofilms (34), survive in the biofilm for up to 40 days (1, 17), and persist there in a metabolically active form (13, 40). Some authors suggest that E. coli can even grow in drinking water distribution systems (7) because these bacteria can adapt to the oligotrophic environment. If appropriate nutrients are not available, E. coli can live solely on the metabolic products of other bacteria (38).
Most of the studies that have examined the presence of E. coli in biofilms have used culture-based methods (41). These methods have limitations, including duration of incubation, antagonistic organism interference, lack of specificity, and poor detection of slow-growing or nondividing microorganisms (35). Plate count methods also result in some inaccuracy since the cells can be clumped together and intertwined with other biofilm components (24). It is worthwhile to note here that methods using microbial growth will be unable to detect nondividing cells at all. Therefore, the number of E. coli in the drinking water distribution network could be underestimated. The presence of E. coli was not indicated by the traditional culture-based methods in the present study, a finding in agreement with previous findings showing that cultivation-independent detection methods detect 10 to 20 times more cells (6).
FISH methods have already been used for the detection of E. coli in biofilms. To date, several probes have been developed for the detection of E. coli in drinking water samples (32-33, 38, 40). In the present study, the published sequence 5′-TCA ATG AGC AAA GGT-3′ (31) was used to design the probe ECOLIFILM applied for direct detection of bacteria on surfaces and in sonicated biofilm samples. Due to the secondary structure, this 16S rRNA region of E. coli is not readily accessible by DNA probes (14). The target has to be denatured prior to in situ hybridization, which can be achieved by treatment with, for example, extremes of pH or heat. Such treatments generally lead to a loss of morphology. Therefore, a compromise has to be found between the intensity of the hybridization signal and the preservation of morphology. The DNA probe has several other limitations, including electrostatic repulsion between negatively charged backbones, resulting in slower and weaker binding (reviewed by Good and Nielsen [16]) and low fluorescence when not easily accessible parts of the ribosome are used as targets (14). A significant improvement in the sensitivity of the FISH method was achieved by using a PNA probe. PNA molecules are DNA mimics in which the negatively charged sugar-phosphate backbone of DNA is replaced with a noncharged polyamide backbone. PNA probes contain the same nucleotide bases and follow standard Watson-Crick base-pairing rules while hybridizing to complementary nucleic acid sequences (12). PNA probes have a hydrophobic surface; thus, the hybridization can occur at a low salt concentration, which, in turn, supports recoiling of the rRNA secondary structure (31). PNA probes do not encounter the electrostatic repulsion because of noncharged backbone. Hence, they hybridize to the targets rapidly and tightly and with high specificity, as shown here.
Examination of field samples from real distribution networks can be very challenging, because such samples can contain a lot of impurities of different origin that can complicate the discrimination of the target cells. The use of the DVC method prior to FISH dramatically improved the detection of E. coli cells. In sonicated samples, metabolically active cells were elongated, allowing much better discrimination of the cells, in particular in biofilm samples from cast iron material, which contained many impurities. However, when DVC was applied to the undisturbed biofilms (whole coupons), only a slight increase in cell length and volume was observed. This could be due to poor penetration of the biofilm by nutrient and antibiotic mixture. Nevertheless, it improved the discrimination of the target cells from the fluorescent particles of different origins and significantly facilitated the enumeration of FISH-positive bacteria.
Since E. coli in all cases was observed as a single cell, it is likely that the bacteria are sparsely distributed and present in low concentration, which does not indicate extensive pollution. Although some studies have shown that E. coli may grow in controlled laboratory-scale experiments (17, 34, 40), in general it is assumed that E. coli is not multiplying in the drinking water distribution network. The scarce distribution of individual cells observed in the present study might support the general assumption that the bacteria are most likely not multiplying since no microcolonies were detected.
In the present study E. coli was found on coupons from five of six water supply systems. The exact expression of results as the number of E. coli per surface area is problematic because only a small percentage (<0.5%) of sample surface area can be examined, but, according to our estimations, it could reach up to 0.1% of the total bacterial number. All water systems were supplied with surface water that received extensive treatment (clarification, filtration, and three-step disinfection) or used artificially recharged groundwater. It was not possible to identify factors (nutrient level, temperature, length of networks, type of pipe material, bacterial number in biofilm, etc.) that would explain the presence or absence of E. coli in the biofilm. The only difference in the water supply system (site A) in which biofilm samples did not contain E. coli was high temperature (>15°C) and high nutrient level (BDOC > 1.0 mg/liter) in the drinking water. According to earlier findings, coliform bacteria tend to grow in water with high temperature and BDOC or AOC levels (22). However, in our study this was the only network in which E. coli was not found. Further research is required to elucidate this finding.
In summary, we have shown that E. coli is brought into the distribution network through a malfunction in the water treatment process or as intrusion through pipes and becomes trapped in the biofilm, where the cells remain in a viable, metabolically active form. However, these cells are not detected because, routinely, only water samples are analyzed, and this is done using culture or enzymatic methods which do not detect active but nonculturable bacteria.
Acknowledgments
This study has been undertaken as part of a research project that is supported by the European Union within the Fifth Framework Energy, Environment, and Sustainable Development Programme (EVK1-2002-00108).
The authors are solely responsible for this work; it does not represent the opinion of the European Community, and the Community is not responsible for any use that might be made of data appearing herein.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Armon, R., J. Starosvetzky, T. Abel, and M. Green. 1997. Survival of Legionella pneumophila and Salmonella typhimurium in biofilm systems. Water Sci. Technol. 35:293-300. [Google Scholar]
- 2.Backer, K. 1984. Protective effect of turbidity on Escherichia coli during chlorine disinfection. Worcester Consortium for Higher Education, Worchester, MA.
- 3.Baudart, J., J. Coallier, P. Laurent, and M. Prevost. 2002. Rapid and sensitive enumeration of viable diluted cells of members of the family Enterobacteriaceae in freshwater and drinking water. Appl. Environ. Microbiol. 68:5057-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berney, M., H. Weilenmann, and T. Egli. 2006. Flow-cytometric study of vital cellular functions in Escherichia coli during solar disinfection (SODIS). Microbiology 152:1719-1729. [DOI] [PubMed] [Google Scholar]
- 5.Berry, D., C. Xi, and L. Raskin. 2006. Microbial ecology of drinking water distribution systems. Curr. Opin. Biotechnol. 17:297-302. [DOI] [PubMed] [Google Scholar]
- 6.Bjergbaek, L. A., and P. Roslev. 2005. Formation of nonculturable Escherichia coli in drinking water. J. Appl. Microbiol. 99:1090-1098. [DOI] [PubMed] [Google Scholar]
- 7.Camper, A., G. McFeters, W. Characklis, and W. Jones. 1991. Growth kinetics of coliform bacteria under conditions relevant to drinking water distribution systems. Appl. Environ. Microbiol. 57:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y., M. Le Puil, J. Biggerstaff, A. Randall, A. Schulte, and J. Taylor. 2003. Direct estimation of biofilm density on different pipe material coupons using a specific DNA-probe. Mol. Cell Probes 17:237-243. [DOI] [PubMed] [Google Scholar]
- 9.Colwell, R., and D. Grimes. 2000. Nonculturable microorganisms in the environment. ASM Press, Washington, DC.
- 10.Eaton, A. D., L. S. Clesceri, and A. E. Greenberg. 1995. Standard methods for the examination of water and wastewater. APHA, Washington, DC.
- 11.Edberg, S., E. Rice, R. Karlin, and M. Allen. 2000. Escherichia coli: the best biological drinking water indicator for public health protection. Symp. Ser. Soc. Appl. Microbiol. 29:106S-106S. [DOI] [PubMed] [Google Scholar]
- 12.Egholm, M., O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden, and P. E. Nielsen. 1993. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 365:566-568. [DOI] [PubMed] [Google Scholar]
- 13.Fass, S., M. L. Dincher, D. J. Rasoner, D. Gatel, and J.-C. Block. 1996. Fate of Escherichia coli experimentally injected in a drinking water distribution pilot system. Water Res. 30:2215-2221. [Google Scholar]
- 14.Fuchs, B., G. Wallner, W. Beisker, l. I. Schwipp, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, P., and M. R. W. Brown. 1995. Phenotypic plasticity and mechanisms of protection of bacterial biofilms from antimicrobial agents, p. 118-132. In H. E. Lappin-Scott and J. W. Costerton (ed.), Microbial biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 16.Good, L., and P. Nielsen. 1997. Progress in developing PNA as a gene-targeted drug. Antisense Nucleic Acid Drug Dev. 7:431-437. [DOI] [PubMed] [Google Scholar]
- 17.Keevil, C. 2001. Continuous culture models to study pathogens in biofilms. Methods Enzymol. 334:104-122. [DOI] [PubMed] [Google Scholar]
- 18.Keevil, C. W. 2002. Pathogens in environmental biofilms, p. 2339-2356. In G. Bitton (ed.), Encyclopedia of environmental microbiology. Wiley, New York, NY.
- 19.Keevil, C., C. Mackerness, and J. Colbourne. 1990. Biocide treatment of biofilms. Int. Biodeterioration 26:169-179. [Google Scholar]
- 20.LeChevallier, M., C. Cawthon, and R. Lee. 1988. Inactivation of biofilm bacteria. Appl. Environ. Microbiol. 54:2492-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeChevallier, M., T. Hassenauer, A. Camper, and G. McFeters. 1984. Disinfection of bacteria attached to granular activated carbon. Appl. Environ. Microbiol. 48:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeChevallier, M. W., N. J. Welch, and D. B. Smith. 1996. Full-scale studies of factors related to coliform regrowth in drinking water. Appl. Environ. Microbiol. 62:201-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehtola, M. J., I. T. Miettinen, T. Vartianen, and P. J. Martikainen. 1999. A new sensitive bioassay for determination of microbially available phosphorus in water. Appl. Environ. Microbiol. 65:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J., S. McLellan, and S. Ogawa. 2006. Accumulation and fate of green fluorescent labeled Escherichia coli in laboratory-scale drinking water biofilters. Water Res. 40:3023-3028. [DOI] [PubMed] [Google Scholar]
- 25.Miettinen, I. T., T. Vartainen, and P. J. Martikainen. 1999. Determination of assimilable organic carbon in humus-rich drinking waters. Water Res. 33:2277-2282. [Google Scholar]
- 26.Moe, C., and R. Rheingans. 2006. Global challenges in water, sanitation and health. J. Water Health 4:41-57. [PubMed] [Google Scholar]
- 27.O'Toole, G., H. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 28.Parsek, M., and P. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 29.Percival, S., J. Walker, and P. R. Hunter. 2000. Microbiological aspects of biofilms and drinking water. CRC Press, Inc., Boca Raton, FL.
- 30.Pernthaler, A., C. Preston, J. Pernthaler, E. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry-O'Keefe, H., S. Rigby, K. Oliveira, D. Sorensen, H. Stender, J. Coull, and J. Hyldig-Nielsen. 2001. Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods 47:281-292. [DOI] [PubMed] [Google Scholar]
- 32.Prescott, A., and C. Fricker. 1999. Use of PNA oligonucleotides for the in situ detection of Escherichia coli in water. Mol. Cell Probes 13:261-268. [DOI] [PubMed] [Google Scholar]
- 32a.Reasoner, D. J., and E. E. Geldreicht. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regnault, B., S. Martin-Delautre, M. Lejay-Collin, M. Lefevre, and P. Grimont. 2000. Oligonucleotide probe for the visualization of Escherichia coli/Escherichia fergusonii cells by in situ hybridization: specificity and potential applications. Res. Microbiol. 151:521-533. [DOI] [PubMed] [Google Scholar]
- 34.Robinson, P., J. Walker, C. Keevil, and J. Cole. 1995. Reporter genes and fluorescent probes for studying the colonization of biofilms in a drinking water supply line by enteric bacteria. FEMS Microbiol. Lett. 129:183-188. [DOI] [PubMed] [Google Scholar]
- 35.Rompre, A., P. Servais, J. Baudart, M. de-Roubin, and P. Laurent. 2002. Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J. Microbiol. Methods 49:31-54. [DOI] [PubMed] [Google Scholar]
- 36.Saby, S., A. Vidal, and H. Suty. 2005. Resistance of Legionella to disinfection in hot water distribution systems. Water Sci. Technol. 52:15-28. [PubMed] [Google Scholar]
- 37.Servais, P., A. Anzil, D. Gatel, and J. Cavard. 2004. Biofilm in the Parisian suburbs drinking water distribution system. J. Water Supply Res. Technol.-AQUA 53:313-323. [Google Scholar]
- 38.Szewzyk, U., W. Manz, R. Amann, K. H. Schleifer, and T. A. Stenstrom. 1994. Growth and in-situ detection of a pathogenic Escherichia coli in biofilms of a heterotrophic water-bacterium by use of 16S-ribosomal-RNA-directed and 23S-ribosomal-RNA-directed fluorescent oligonucleotide probes. FEMS Microbiol. Ecol. 13:169-175. [Google Scholar]
- 39.WHO. 1996. Guidelines for drinking-water quality. World Health Organization, Geneva, Switzerland.
- 40.Williams, M. M., and E. B. Braun-Howland. 2003. Growth of Escherichia coli in model distribution system biofilms exposed to hypochlorous acid or monochloramine. Appl. Environ Microbiol. 69:5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wingender, J., and H. Flemming. 2004. Contamination potential of drinking water distribution network biofilms. Water Sci. Technol. 49:277-286. [PubMed] [Google Scholar]