Abstract
An analysis of the molecular diversity of N2 fixers and denitrifiers associated with mangrove roots was performed using terminal restriction length polymorphism (T-RFLP) of nifH (N2 fixation) and nirS and nirK (denitrification), and the compositions and structures of these communities among three sites were compared. The number of operational taxonomic units (OTU) for nifH was higher than that for nirK or nirS at all three sites. Site 3, which had the highest organic matter and sand content in the rhizosphere sediment, as well as the lowest pore water oxygen concentration, had the highest nifH diversity. Principal component analysis of biogeochemical parameters identified soil texture, organic matter content, pore water oxygen concentration, and salinity as the main variables that differentiated the sites. Nonmetric multidimensional scaling (MDS) analyses of the T-RFLP data using the Bray-Curtis coefficient, group analyses, and pairwise comparisons between the sites clearly separated the OTU of site 3 from those of sites 1 and 2. For nirS, there were statistically significant differences in the composition of OTU among the sites, but the variability was less than for nifH. OTU defined on the basis of nirK were highly similar, and the three sites were not clearly separated on the basis of these sequences. The phylogenetic trees of nifH, nirK, and nirS showed that most of the cloned sequences were more similar to sequences from the rhizosphere isolates than to those from known strains or from other environments.
Mangrove ecosystems play an important role as refuge, feeding, and breeding areas for many organisms and sustain an extensive food web based on detritus. Additionally, mangroves export nutrients to adjacent marine ecosystems such as sea grass communities and coral reefs (28). In the last 20 years, more than 50% of the world's mangroves have been cleared, mainly for aquaculture, timber production, and urban development (48, 49). Mexico lost about 70,000 ha of mangroves between 1993 and 2000 (56).
Mangroves of semiarid areas are generally nitrogen deficient but are nonetheless highly productive. This apparent paradox can be explained by the high rate of biological nitrogen-fixing activity in sediments, the rhizosphere of the mangrove trees, decomposing leaves, and aerial roots and bark. These contribute from 40 to 60% of the total nitrogen required by the ecosystem (for a review, see reference 28). While nitrogen fixation in sediments is likely to be limited by insufficient energy sources, the mangrove rhizosphere sustains high rates of nitrogen-fixing activity (57, 70), which may contribute significantly to the health and sustenance of the ecosystem by supplying most of its nitrogen requirements (23, 27, 28, 70). The high diazotrophic activity associated with mangrove roots is probably due to root exudates, which are a source of carbon and energy for the bacteria, low nitrogen concentrations, and microaerophilic conditions, all necessary for the expression of regulatory and structural nitrogenase genes (21). It is not known if these conditions in the rhizosphere can be conducive to diazotroph diversity. Nitrogen-fixing bacteria identified as members of the genera Azospirillum, Azotobacter, Rhizobium, Clostridium, Klebsiella, Vibrio, and Phyllobacterium have been isolated from the rhizosphere of various mangrove species (10, 27, 57). However, the true extent of the diversity of diazotrophs associated with mangrove roots has not been determined and it is possible that the organisms that have been isolated are poorly represented in the natural environment.
Denitrification is a dissimilatory process in which oxidized nitrogen is used as an alternative electron acceptor for energy production when oxygen is limiting and consists of four reaction steps in which nitrate is reduced to dinitrogen gas (71). The occurrence of denitrifying bacteria in the mangrove rhizosphere is of interest because it could imply loss of fixed nitrogen via denitrification, and nitrogen is frequently a limiting nutrient in such systems (27, 28). The fluctuations in oxygen tension that occur in the rhizosphere due to tidal cycles (one mangrove tree may be exposed to two flooding and two dry periods in 24 h) probably favor the growth and establishment of bacteria capable of using alternate electron acceptors and thus obtaining energy without the need of oxygen. Such conditions and a supply of carbon and energy sources through root exudates might be conducive to denitrification, as demonstrated in the case of sea grasses such as Halodule uninervis and Thalassia hemprichii that are in subtidal to very low interstitial habitats (60), and allow the sustenance of a denitrifying community associated with mangrove roots. Denitrifying bacteria have been isolated from mangrove sediments (34), but none have been isolated from mangrove roots. This work represents the first attempt to study the community of denitrifiers in that system.
Several authors have used the nitrite reductase genes nirK and nirS, the key enzymes in the denitrification process that code for copper and cytochrome cd1-containing nitrite reductases, respectively, to study the genetic diversity of denitrifiers in a variety of natural habitats and laboratory settings. These include groundwater (67), biofilm reactors (13), soils (47), river sediments (63), estuarine sediments (42), marine sediments (5, 6, 35), and seawater (9, 29).
The nifH gene, which encodes the Fe protein component of nitrogenase, has been used as a functional gene to characterize diazotrophic communities in many different habitats and settings such as marine plankton, termite hindguts, microbial mats, terrestrial soils, the open ocean, lakes, rivers, estuaries, the rhizosphere of plants, and bioreactors (for a review, see reference 69).
This work is an attempt to assess the composition and structure of denitrifying and N2-fixing bacteria associated with the roots of the black mangrove Avicennia germinans in an undisturbed semiarid mangrove ecosystem and to test whether the biogeochemical properties of the rhizosphere influenced the composition and structure of these bacterial communities. To accomplish this, the molecular diversity of nirS, nirK, and nifH was investigated using terminal restriction fragment length polymorphism (T-RFLP).
MATERIALS AND METHODS
Study area and sampling.
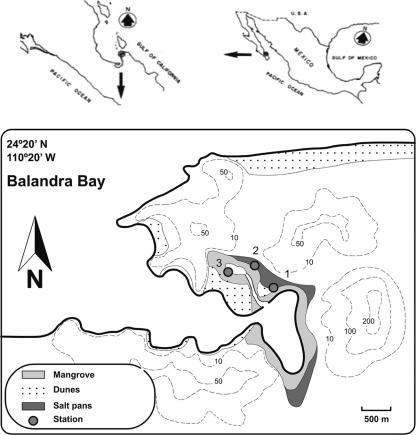
Thirty- to 40-cm-high seedlings of the black mangrove A. germinans, with the root system and surrounding soil intact (three seedlings per site), were collected from the semiarid mangrove ecosystem at Balandra, in Baja California Sur, México, in the summer of 2004 during low tide (Fig. 1). The three sampling sites were chosen because they were located in the intertidal zone, and all of them had seedlings of A. germinans. Sites 1 and 2 were in the high intertidal zone, and site 3 was in the mid-intertidal zone.
FIG. 1.
Location of the mangrove in Balandra Bay, B.C.S., México. (Modified from reference 27 with permission of the publisher.)
The rhizosphere samples (soil that surrounds the roots of plants), having two replicates per plant, were processed for nutrient analysis (NH4+, NO3−, and organic matter content). The whole root system of every tree was put aside and washed twice with sterile seawater. Some of the roots were used for isolation of denitrifying bacteria and others for immediate DNA extraction. Oxygen, pH, and salinity in the rhizosphere pore water were measured in situ using portable field instruments: a plantlet was dug out using a soil core sampler made out of polyvinyl chloride pipe, and the water that filtered into the hole was collected. Dissolved oxygen was measured with an oxygen meter (830A, equipped with Orion DO 083010 probe; Thermo Electron Corp.), pH with a field pH-meter (B-213; Horiba, Germany), and salinity with a refractometer (Aquafauna Biomarine).
The Balandra mangrove site, located at 24°20′N, 110°20′W approximately 20 km north of the metropolitan area of the city of La Paz in a semiarid area of the southern Baja California Peninsula (25) (Fig. 1), receives no freshwater, except for the small annual rainfall of 157 mm and has an average annual temperature of 29°C. It is populated by three mangrove species: Rhizophora mangle (L.), red mangrove; A. germinans (L.), Stern, black mangrove; and Laguncularia racemosa Gaertn., white mangrove.
Biogeochemical parameters of pore water and sediment samples.
Concentrations of NH4+ and NO3− and organic matter content in the soil that surrounded the roots were determined as described by Hernandez-López and Vargas-Albores (26), based on the method of Strickland and Parsons (61), but adapted to reading on microplates. Organic matter content was determined as described by NOM 021 SEMARNAT (55) with a chemical titration method modified from Walkley and Black (65). Soil texture was determined as described by NOM 021 SEMARNAT (55) with a method based on the use of a hydrometer (16). Dissolved iron and copper in soil were analyzed according to Van Loon (65).
Isolation of denitrifying bacteria from the roots of mangrove trees.
To isolate denitrifying bacteria, A. germinans roots prepared as described by Holguin et al. (27) were inoculated into serum bottles containing enrichment medium consisting of four components prepared as follows. Solution A was prepared with 1.5 g of KNO3, 8.6 g of peptone, 20 g of NaCl, 3.0 g of MgSO4·7H2O, and 0.02 g of CaCl2 in distilled water (970 ml) to prepare 1 liter of medium at pH 7.0. After autoclaving, 10 ml each of solutions B (sodium acetate, 0.45 g ml−1), C (ethanol, 0.25 g ml−1), and D (microelements from HGB medium [27]) was added. After incubation for 7 days at 30°C, 100 μl of enrichment culture was transferred to screw-cap test tubes filled with modified Patureau medium (44), which consisted of five solutions prepared as follows. Solution A, containing 0.409 g of KNO3, 3.0 g of MgSO4, 20.0 g of NaCl, 0.212 g of (NH4)2SO4, 0.161 mg of yeast extract, 0.02 of g CaCl2, and distilled water (860 ml) to prepare 1 liter of medium, and solution B (100 ml of 0.1 M phosphate buffer at pH 7.0) were mixed with 10 ml each of enrichment medium solutions B and D and 20 ml of solution C. After 7 days of incubation, the bacteria were spread on solid Patureau modified medium and incubated until colonies appeared. Each colony morphotype was tested for denitrification activity as described by Hernandez-López and Vargas-Albores (26). Denitrification was presumed positive if neither NO2− nor NO3− could be detected or showed only traces in the culture supernatant after isolate incubation for 5 days under static conditions in modified Patureau medium. Nitrite in the culture supernatant was measured as described by Bendschneider and Robinson (3), and nitrate was measured as described by Morris and Riley (39), using a flow injection analysis system (LACHAT; QuikChem 8000, Milwaukee, WI). The final criteria for considering an isolate a denitrifier were detection of nirS or nirK by PCR followed by BLAST sequence analysis. Bacteria were identified by ACCULAB (Newark, DE) through sequencing 500 bp of the 16S RNA gene by automated dideoxy terminator sequencing chemistry.
The nitrogen-fixing isolates described in this paper were isolated from A. germinans seedling roots (15): Paracoccus sp. strain AG4BC, Aeromonas sp. strain LR7YC, Pseudomonas sp. strain LR6A, and Pseudomonas sp. strain LR6B. To verify the presence of the nifH gene in these isolates, it was amplified by PCR and the sequence was analyzed with BLAST. The diazotrophic activity of the strains was verified with the acetylene reduction assay. The results showed the strains fixed as much N2 as Azospirillum brasilense, a widely used diazotroph in agriculture (2).
DNA extraction, primer design, and PCR amplification.
To extract DNA, the mangrove seedling roots were washed with sterile seawater, cut into small fragments, and ground in a mortar with a 0.39 M phosphate buffer solution at pH 7.6. The supernatant was collected, and the cell pellet was recovered after centrifugation for 2 min at 500 × g and ground again in the same buffer solution. This procedure was repeated twice. The three supernatant fractions were collected and mixed and centrifuged at 10,000 × g for 30 min, and the pellet was recovered and stored at −70°C.
DNA was extracted from the pellet by the method described by Schwieger et al. (54), except that the freeze-thaw cycles included freezing the samples at −70°C instead of in liquid nitrogen. Phenolic compounds and humic acids were removed from the extracted DNA using the PowerSoils DNA isolation kit (MO Bio Laboratories, Carlsbad, CA).
Primers were designed using Primer Select software (DNAstar, Madison, WI). It was decided to design new nirK and nirS primers because the number of published sequences of these genes in the database has increased since the latest designs (6, 35). The nirS primers were designed to amplify an approximately 660-bp region by comparing the available sequences of Paracoccus denitrificans U75413, Alcaligenes eutrophus X91394, Pseudomonas stutzeri X56813, Azoarcus tolulyticus AY078272, Pseudomonas aeruginosa AE004488, Pseudomonas fluorescens AF114792, Acidovorax sp. strain AY078273, Thauera selenatis AY078264, Thauera chlorobenzoica AY078263, and the γ-Proteobacteria isolate GPR248400. The nirK primers were designed to amplify an approximately 404-bp region, based on the previously published sequences of Bradyrhizobium japonicum BA000040, Rhizobium hedysari RHU65658, Rhodobacter sphaeroides RSU62291, Pseudomonas aureofaciens PANIRKA, Alcaligenes sp. strain AB046603, Hypomicrobium denitrificans AB076606, and Alcaligenes xylosoxidans AF051831.
In order to maximize the number of nifH sequences amplified from diazotrophs associated with mangrove roots, it was decided to construct different primers from those already published (45) that are specific for free-living, heterotrophic diazotrophs. The available sequences of Bradyrhizobium sp. strain NC_004463, Rhizobium sp. strain K00487, Methanosarcina mazei AY029234, Azospirillum brasilense M64344, Synechococcus sp. strain U22146, and Anabaena sp. strain J05111 were compared, and primers were designed to amplify a fragment of approximately 606 bp.
The nifH, nirK, and nirS primers were created based on sequences from unrelated microorganisms. Since these genes are usually highly conserved, despite not having degenerated positions, we expected to obtain an ample spectrum of products from groups within eubacterial nifH, nirK, and nirS genes. The following primers were used for amplification: for nirK, 5′-ACAACGTCGACTTCCACGCC-3′ (primer F; positions 792 to 810 correspond to Hyphomicrobium denitrificans AB076606) and 5′-GCCGACCGTGCCGTTGAAGA-3′ (primer R; positions 1345 to 1364 correspond to Hyphomicrobium denitrificans AB076606); for nirS, 5′-TGAACGTCAAGGAAACCGGCCA-3′ (primer F; positions 1058 to 1080 correspond to Paracoccus denitrificans U75413) and 5′-AGCTTCAGGGTCTTGTCGTCG-3′ (primer R; positions 1345 to 1364 correspond to Paracoccus denitrificans U75413); and for nifH, 5′-TCTACGGAAAGGGCGGTATCGG-3′ (primer F; positions 181 to 203 correspond to Bradyrhizobium sp. strain NC_004463) and 5′-GGCACGAAGTGGATCAGCTG-3′ (primer R; positions 777 to 796 correspond to Bradyrhizobium sp. strain NC_004463). PCR was performed with a Mastercycle gradient (Eppendorf AG, Hamburg, Germany). The PCR parameters for nirK and nirS were 94°C for 2 min, 94°C for 45 s, 65°C for 45 s, and 72°C for 1 min for 35 cycles, followed by 72°C for 20 min. For nifH, the PCR parameters were 94°C for 2 min, 94°C for 45 s, 57.8°C for 45 s, and 72°C for 1 min for 35 cycles, followed by 72°C for 20 min. For T-RFLP analysis, the forward primer of each gene was 5′-end labeled with high-performance liquid chromatography-grade fluorescent label 6-carboxyfluorescein (6-FAM; IDT, Coralville, IA). PCR amplification was performed under the same conditions as specified for nonlabeled primers. The PCR mixtures (20 μl) contained 2 μl of 10× buffer, 0.4 μl of 10 mM deoxynucleoside triphosphates (dNTPs), 1.2 μl of MgCl2 (25 mM), 20 pmol of each primer, 0.2 μl of Taq polymerase (Promega), and 100 ng of DNA. Colony PCR (64) was used to amplify genes from the bacterial strains isolated from mangrove roots, increasing the initial temperature time of the PCR to 5 min.
Each PCR product was visualized after electrophoresis in 1.5% Tris-borate-EDTA (TBE) agarose gels. The PCR products were purified using Qiaquick spin columns (QIAGEN, Valencia, CA) according to the manufacturer's instructions and quantified using a spectrophotometer at A260 (UV-2800; UNICO).
T-RFLP analysis.
To select restriction endonucleases capable of resolving as many target genes as possible, an in silico digestion of the nirK, nirS, and nifH regions to be amplified was performed with the RestrictionMapper 3.0 software. For nirK and nirS, MspI, RsaI, Mn1I, MboII, and HhaI were used, and for nifH, MspI, BsaHI, BstUI, BarI, Mn1I, MboII, RsaI, and HhaI were used.
Based on the resulting arrays of terminal restriction fragment (T-RF) size distributions, the enzymes MspI, HhaI (New England Biolabs, Beverly, MA), and RsaI (Invitrogen, Carlsbad, CA) were selected for nirK and nirS; for nifH, MspI, MnII, and HhaI (New England Biolabs) were selected. The PCR product (∼375 ng) in a volume of 30 μl was digested separately with the different enzymes following the manufacturer's instructions.
The digested products were purified using Centri-sep columns (Princeton Separations, NJ), dried with a centrifugal evaporator (RC1010; Jouan, Winchester, VA), and resuspended in 9.8 μl of deionized formamide with 0.2 μl of GeneScan-500 LIZ internal size standard (Applied Biosystems, Foster City, CA). Aliquots of 10 μl were placed in 96-well plates (ThermoFast 96; ABGene, Epsom, United Kingdom) and sealed with adhesive tape (Applied Biosystems). The plates were centrifuged so the sample would settle on the bottom of the well, denatured by heating at 94°C for 2 min, and immediately transferred onto ice.
Fragments were analyzed with an automated sequencer (ABI 3730; Applied Biosystems) based on detection of the 5′-end fluorescent label 6-FAM. The size and intensity of each T-RF (peak height and area) were calculated automatically using GeneMapper version 3.0 software (Applied Biosystems). The obtained T-RFs are hereafter called operational taxonomic units (OTU).
Cloning and sequencing.
The amplified PCR products were ligated to the pCR2.1-TOPO vector using the Original TA cloning kit (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. A number of clones from each experiment were selected randomly for further analysis.
The presence of the desired gene insert in the plasmid was verified by colony PCR. Plasmids were isolated with the QIAprep Miniprep kit (QIAGEN, Valencia, CA) and sequenced with vector-specific primers by BIOtech (Cornell University, Ithaca, NY). The sequences thus obtained were compared with nirK, nirS, and nifH sequences from GenBank (February 2006) using BLAST.
Sequences were aligned with CLUSTALW and translated into amino acids using MEGA 3.1 (The Pennsylvania State University, University Park). Alignments were compared with reference sequences from the database and hand corrected when necessary. Phylograms were edited with the Treeview software program. Phylogenetic trees were based on available sequences in GenBank, using all clone sequences from all three sites as well as sequences from the isolates obtained in this study. Neighbor-joining phylogenies (52) were constructed with Protdist and Neighbor (Phylip version 3.66 beta) (18) by using percent dissimilarity distances and pairwise deletion of gaps. They were compared with neighbor-joining trees constructed by using the amino acid sequence distance measurement Poisson and gamma distribution correction for multiple substitutions, in order to identify frameshifts (MEGA 3.1 software) (33). Bootstrapping was used to estimate reliability of phylogenetic trees with 1,000 replicate trees.
Data analysis.
Principal component analysis (PCA) performed with Primer 5.0 (Primer-E Ltd., Plymouth, United Kingdom) and MVSP 3.1 (KCS, Wales, United Kingdom) provided a means to separate and group sediment samples based on their biogeochemical properties. T-RFLP data were used to calculate diversity with the Shannon-Weaver diversity index and dominance with the Simpson index (31).
The T-RFLP raw data sets comprised peaks that reflected the sizes of terminal fragments present (measured in base pairs) and the area and height of each peak (measured in fluorescence units).
The relative abundances of T-RFs were standardized in percent by calculating the ratio of a given peak height to the normalized total peak height of each sample and analyzed with Primer 5.0. Peaks with an area less than 1% of the total were reassigned as zero, and the proportion of each remaining peak was recalculated. Peaks with an area less than 5% were removed. The data set was analyzed with a 5% threshold in order to remove any bias caused by the amount of PCR product (50).
To examine community patterns for the root-associated diazotrophic and denitrifying bacteria based on the T-RFLP data, nonmetric multidimensional scaling (MDS) was applied using the Bray-Curtis coefficient, ideal for the construction of similarity matrices and group analysis (12, 50). For comparative purposes, the data were converted to presence/absence of peaks (binary data) and to relative abundance (area of peaks), transforming them with the fourth root.
An MDS plot is used to examine statistically significant differences among samples as a horizontal distance. An important component of the plot is a measure of the goodness of fit of the final plot, termed the “stress.” A stress greater than 0.2 indicates that the plot is close to random, a stress less than 0.2 indicates a useful two-dimensional picture, and a stress less than 0.1 corresponds to an ideal ordination with no real prospect of misinterpretation (12). Stress was calculated as described by Kruskal (32) with the Primer 5.0 software.
Analysis of similarity (ANOSIM) (50) was used to examine statistical significance among samples and to test the null hypothesis that there was no difference in the composition of OTU among our three study sites. A level of significance (P value) was also produced for the analysis. This statistical test compared the variabilities of the OTU among the different sites and within sites. Similarity percentage (SIMPER) was used to analyze the average contribution of individual OTU to the average dissimilarity among samples. The similarity of the 1% threshold data was examined, comparing both presence/absence and relative abundance of fragments.
Nucleotide sequence accession numbers.
Gene sequences from mangrove isolates and environmental samples have been deposited in the EMBL nucleotide sequence database under the following accession numbers: EU035272 to EU035284 for mangrove isolates and DQ176978 to DQ176996 and DQ176999 to DQ177041 (nifH), DQ177043 to DQ177090 and DQ177094 to DQ177095 (nirK), and DQ177096 to DQ177146 (nirS) for environmental samples.
RESULTS
Biogeochemical properties of rhizosphere and pore water.
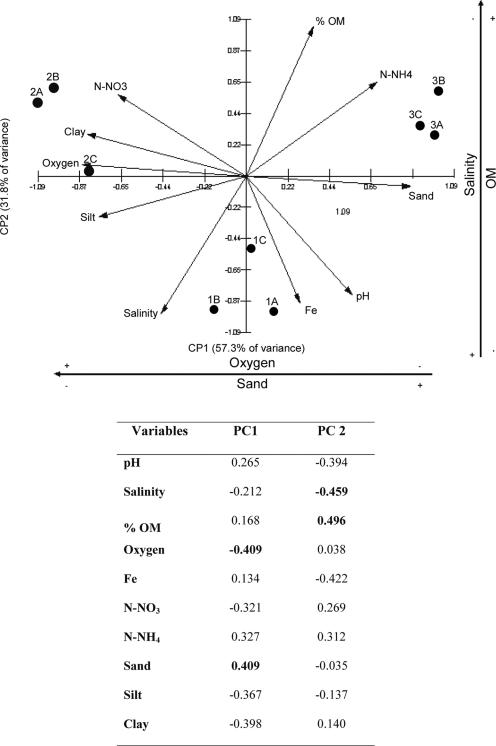
PCA of rhizosphere and pore water biogeochemical parameters (Table 1) separated the three sites into three very-well-defined groups and revealed clear differences among the three sites (Fig. 2). The data were reduced to two principal components, PC1 and PC2, which explained 89% of the variation in the parameters. Soil texture and pore water oxygen concentration were separated from other biogeochemical parameters with regard to PC1, which explained 57% of the variation. PC2 separated organic matter and pore water salinity and explained 32% of the variation. Soil texture, organic matter content in sediment, pore water oxygen concentration, and pore water salinity were thus the main variables that explained differences among the three sites (Fig. 2).
TABLE 1.
Biogeochemical properties of the rhizosphere and pore water in three mangrove sites at Balandraa
| Site | pH | % Organic matter | Oxygen concn (mg/liter)b | Salinity (%)b | Concn (mg/kg) of:
|
% Soil type
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fe | N-NO3 | N-NH4 | Sand | Silt | Clay | |||||
| 1 | 8.26 ± 0.07 | 0.7 ± 0.2 | 0.9 ± 0.1 | 76 ± 1 | 6.7 ± 4.7 | 0.24 ± 0.07 | 1.87 ± 0.2 | 50.2 ± 12.7 | 26 ± 10 | 23.8 ± 2.3 |
| 2 | 7.52 ± 0.38 | 3.06 ± 0.75 | 1.93 ± 0.06 | 68.6 ± 1.1 | 0.62 ± 0.06 | 0.68 ± 0.30 | 4.65 ± 0.6 | 9.5 ± 2 | 34.6 ± 6.1 | 55.8 ± 5.8 |
| 3 | 8.05 ± 0.07 | 4.88 ± 0.57 | 0.26 ± 0.01 | 60.6 ± 2.1 | 3.42 ± 0.60 | 0.22 ± 0.02 | 26.47 ± 5.7 | 77.5 ± 0 | 10.0 ± 0 | 12.5 ± 0 |
Values are means ± standard errors.
Values correspond to pore water.
FIG. 2.
Ordinate PCA plot based on biogeochemical parameters from three different sites. % OM, percentage of organic matter. PC1 was represented by sediment texture and oxygen, while PC2 was represented by % OM and salinity. The directions of arrows indicate the relative loadings on the first and second principal components. The table shows the variables that determine the differences between the sites.
The biogeochemical properties of the three sites varied considerably. Site 1 had relatively high sand (55%) and clay (24%) content, with the lowest content of organic matter of all sites (Table 1). Site 2 had a high percentage of clay (76%) and a relatively high pore water concentration of oxygen (1.9 mg/ml), compared to concentrations of 0.9 and 0.3 mg/ml from sites 1 and 3. Site 3 was composed mainly of fine sand (77%) and had the lowest pore water salinity and oxygen concentration of the three, but had the highest percentage of organic matter. All three sites had very high pore water salinity (61 to 76 ppm).
Isolation of denitrifying bacteria from the roots of A. germinans.
Five isolates of denitrifying bacteria were identified by the 16S RNA gene as Vibrio sp. strain 9B, Arthrobacter sp. strain 61K, Corynebacterium sp. strain 63K, Corynebacterium sp. strain 12A, and Oceanomonas sp. strain 5A. The isolate 64K could not be identified because it produces excessive amounts of polysaccharides, which affects DNA extraction. The reason for the small number of isolates obtained was probably because the isolation medium utilized included only acetate and ethanol as carbon sources instead of the wide range of compounds exuded by mangrove roots (A. germinans exudes propionic, succinic, malic, fumaric, benzoic, and palmitic acids, among others) (G. Holguin and M. Bacilio, unpublished results).
All isolates were identified only to the genus level, since in all cases the percentage of genetic distance to the closest match, defined as the number of nucleotide differences between two sequences, was too great to make a species-level identification. Further characterization of the diazotrophic and denitrifying isolates, consisting of denitrification and acetylene reduction assays as well as PCR amplification of nifH, nirK, and nirS, revealed that the diazotrophs Paracoccus sp. strain AG4BC and Aeromonas sp. strain LR7YC were also denitrifiers and that all of the denitrifying isolates were also nitrogen fixers.
Optimization of PCR.
Pseudomonas sp. strain LR6A was used as positive control for amplification of nifH, and Aeromonas sp. strain LR7YC and A. brasilense Cd1 were used for nirK and nirS, respectively. All positive controls and root DNA samples yielded the desired PCR-amplified products: an ∼660-bp fragment for nirS, an ∼404-bp fragment for nirK, and an ∼606-bp fragment for nifH. To optimize PCR conditions, the optimum temperature of annealing (using gradient PCR) and the optimum concentrations of magnesium and template DNA for all three genes were determined. Cloning and sequencing of the amplified products showed that nondesired targets were not being amplified. In the case of the few mispriming products, it was verified that the band corresponded to the gene by cloning and sequencing the expected size fragment (data not shown).
Selection of restriction endonucleases for T-RFLP.
In spite of the fact that similar restriction patterns were found for all of the enzymes, the number of fragments varied. For nifH, the highest OTU resolution was given by MnlI digestion and provided 78 OTU, while for nirS the highest level of resolution was obtained with HhaI, generating 19 OTU. Subsequently, the OTU generated by those enzymes were analyzed further. For nirK, the highest OTU resolution was given by RsaI; however, the OTU generated by this enzyme could not be validated with the obtained clones, so the OTU obtained with MspI were used for further analysis.
Assignment of clones and isolates to T-RFs and analysis of nifH, nirK, and nirS gene T-RFLP fragments.
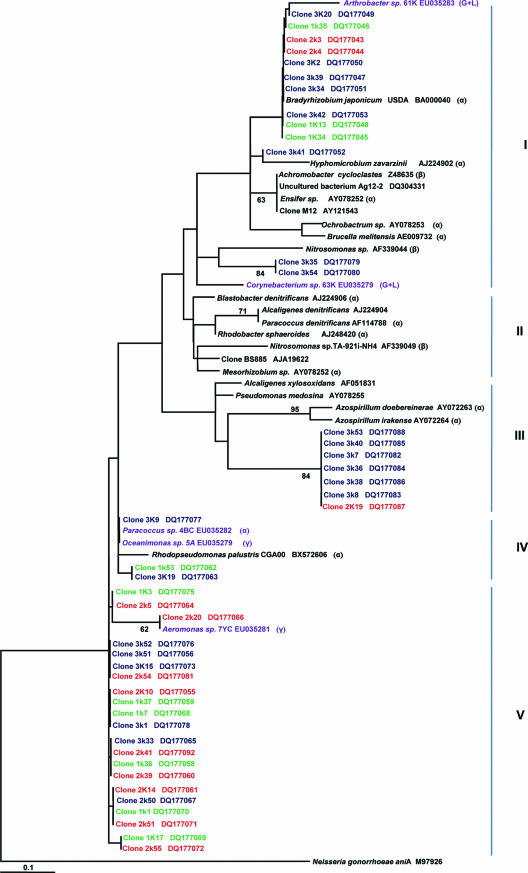
To validate the T-RF fragments, in a computer simulation, nifH, nirK, and nirS sequences from all clones obtained and from the cultivated isolates were cleaved with three restriction endonucleases for each gene. The lengths of these theoretical T-RFs were calculated, and clones and isolates were assigned to peaks found in the electrograms (data not shown). Clones corresponded with few exceptions to fragments that represented the dominant T-RFs within the community. In some cases, one clone was assigned to a certain T-RF, but generally, several clones were assigned to the same T-RF. The phylogenetic analysis of the nifH, nirK, and nirS sequences generally showed that clones assigned to the same T-RF were grouped in the same cluster (see Fig. 4) (data not shown). For nifH, the mangrove rhizosphere isolates Oceanomonas sp. strain 5A, Vibrio sp. strain 9B, strain 64K, and Paracoccus sp. strain AG4BC were represented in all three sites and assigned the same T-RF (data not shown). These results concur with the phylogenetic tree, where the isolates are all grouped in cluster I. Isolates Aeromonas sp. strain LR7YC, Arthrobacter sp. strain 61K, and Corynebacterium sp. 12A were assigned separate T-RFs.
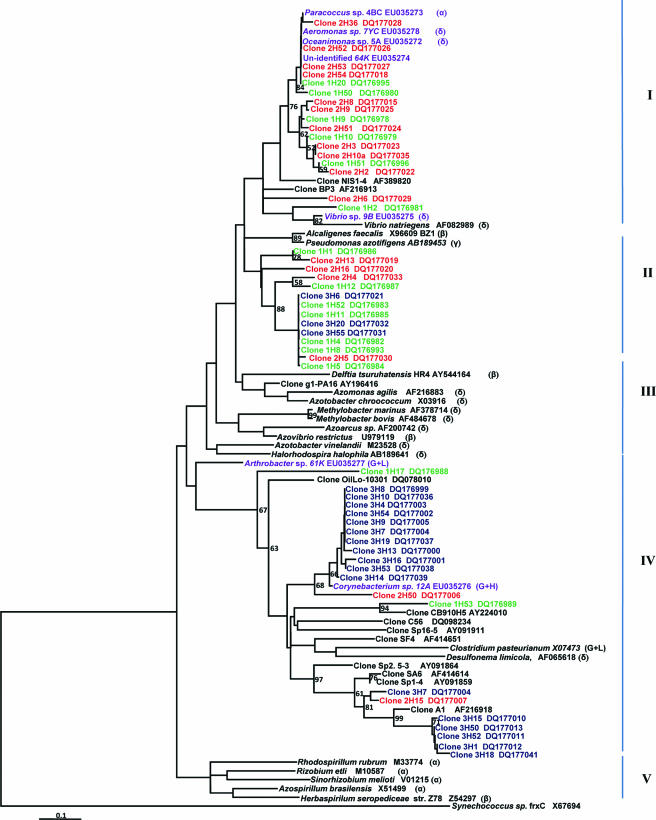
FIG. 4.
Neighbor-joining phylogenetic tree of nifH constructed using 52 cloned and partial amino acid sequences of 204 amino acids obtained from the three sites. They were compared with 31 sequences from GenBank and the sequences of our cultivated diazotrophic and denitrifying mangrove root isolates (purple). Sequences in bold are those obtained in this study, and they were designated according to the site they were taken from: site 1, 1H (green); site 2, 2H (red); site 3, 3H (blue). We used the frxC sequence from Synechococcus sp. (X67694) as an outgroup. Bootstrap values greater than 500 from 1,000 replicate trees are reported at the nodes. The scale represents 10 mutations per 100 amino acid positions. G+H and G+L indicate gram-positive bacteria with high- and low-GC contents, respectively.
For nirK, the dominant T-RFs were shared by all sites and clones were assigned to most peaks. Clones from cluster V were assigned the same T-RF (no. 24, cut with MspI) in all three sites. In site 3, clones from different clusters (I and III) were assigned the same T-RF. The mangrove isolates could not be assigned to T-RFs (data not shown). For nirS, one dominant peak (no. 116, cut with HhaI) was shared by all three sites. Clones from cluster VI and the only nirS isolate, Corynebacterium sp. strain 12A, also from cluster VI, were all assigned to T-RF 84 (cut with HhaI) at the three sites (data not shown). For nifH, the highest number of OTU and the highest diversity index values were found at site 3. The dominance indices were similar at all sites. For nirK, the diversity index values were the same for all three sites. The highest nirS diversity value was found at site 2 (Table 2).
TABLE 2.
Genetic diversity of nifH, nirK, and nirS OTU in bacteria associated with mangrove roots
| Gene | Station | No. of OTU | Ha | 1-λ′b |
|---|---|---|---|---|
| nifH | 1 | 21 | 1.92 | 0.78 |
| 2 | 18 | 1.59 | 0.70 | |
| 3 | 39 | 2.05 | 0.77 | |
| nirK | 1 | 7 | 1.07 | 0.53 |
| 2 | 7 | 1.07 | 0.53 | |
| 3 | 10 | 1.07 | 0.52 | |
| nirS | 1 | 4 | 1.03 | 0.57 |
| 2 | 8 | 1.90 | 0.83 | |
| 3 | 7 | 1.61 | 0.75 |
The Shannon-Weaver diversity index was calculated with the formula H = −Σ(pi)(log2 pi), where p is the ratio of different OTU to the total number of OTU.
The dominance Simpson index was calculated with the formula 1 − λ′ = 1 − [ΣiNi(Ni − 1)]/[N(N − 1)], where Ni is the individual number of OTU (i), and N is their total number.
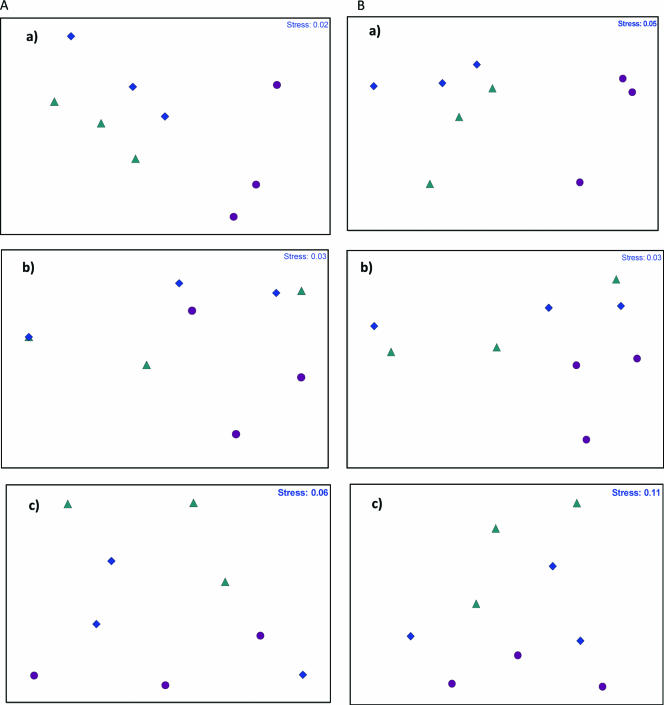
MDS analysis of the OTU for nifH, nirK, and nirS using the Bray-Curtis coefficient revealed that the stress values for analysis based both on the presence/absence of OTU and on their relative abundance were very small (≤0.1) (Fig. 3). Analysis based both on the presence/absence of OTU and on their relative abundance gave the same pattern of distribution and grouping. However, MDS plots calculated using the latter method discriminated better the differences among the three sites (Fig. 3).
FIG. 3.
MDS plot based on Bray-Curtis similarities of the T-RFLP data consisting of the presence/absence of OTU (A) and their relative abundance (B) over a 1% threshold. Triangles, site 1; rhombuses, site 2; circles, site 3. Panels a, nifH; panels b, nirK; panels c, nirS.
MDS analysis for nifH revealed clear separation between the communities of site 3 and those of sites 1 and 2 (Fig. 3). The nirK and nirS OTU from the three sites did not separate clearly into groups (Fig. 3). These results were corroborated for the three genes by group analysis (results not shown).
The ANOSIM analysis for the nifH OTU, which allowed significance testing of the data groups, gave a test statistic (global R) of 0.80 for P < 0.01 based on relative abundance, revealing statistically significant differences among the OTU of the three sites (R can range from −1 to 1; objects that are more dissimilar among groups than within groups will be indicated by an R greater than 0; an R of 0 indicates the null hypothesis is true) (Table 3). When we compared the data in pairs, we found statistically significant differences between the OTU from sites 1 and 2, 1 and 3, and 2 and 3. However, the OTU from sites 1 and 2 were more similar to one another than the other sites (Table 3).
TABLE 3.
ANOSIM of the nifH, nirK, and nirS OTU from the three sites following application of a 1% thresholda
| Gene and station comparison |
R statistic of 1% threshold data based on:
|
|
|---|---|---|
| Presence/absence | Relative abundance | |
| nifH | ||
| Station 1 vs 2 | 0.15 | 0.41 |
| Station 1 vs 3 | 0.91 | 1.0 |
| Station 2 vs 3 | 0.85 | 1.0 |
| Global R | 0.62 | 0.80 |
| nirK | ||
| Station 1 vs 2 | −0.333 | −0.259 |
| Station 1 vs 3 | 0.204 | 0.333 |
| Station 2 vs 3 | 0.056 | 0.037 |
| Global R | −0.012 | 0.012 |
| nirS | ||
| Station 1 vs 2 | 0.07 | 0.33 |
| Station 1 vs 3 | 0.44 | 0.52 |
| Station 2 vs 3 | 0.02 | 0.33 |
| Global R | 0.18 | 0.42 |
Similarity was calculated based on presence/absence and relative abundance. P < 0.01 in all cases for the pairwise comparisons.
For nirK, the global R based on presence/absence showed the OTU among the sites were not significantly different (Table 3). The analysis based on relative abundance revealed significant differences among the sites, but the R value was very low. For nirS, the OTU among the sites were significantly different but the global R was lower than for nifH. Pairwise comparison of the nirS OTU from the sites demonstrated those from sites 1 and 3 were more different from each other than those from sites 1 and 2 or 2 and 3. SIMPER analysis confirmed the results obtained by ANOSIM analysis for all three genes (Table 4).
TABLE 4.
SIMPER analysis of nifH, nirK, and nirS OTU
| OTU | % Dissimilarity of OTU between sites:
|
|||||
|---|---|---|---|---|---|---|
| Presence/absence
|
Relative abundance
|
|||||
| 1 and 2 | 1 and 3 | 2 and 3 | 1 and 2 | 1 and 3 | 2 and 3 | |
| nifH | 39 | 63 | 62 | 35 | 65 | 65 |
| nirK | 33 | 42 | 38 | 25 | 33 | 29 |
| nirS | 49 | 53 | 50 | 49 | 53 | 50 |
Phylogenetic analysis of nifH, nirK, and nirS sequences.
Comparison of 52 nifH cloned sequences showed that they were 79% to 96% similar, while the 44 nirK and 32 nirS clone sequences were 81% to 100% and 79% to 100% similar, respectively.
The phylogenetic tree for nifH constructed by the neighbor-joining method showed five clusters (I to V) (Fig. 4) congregated in two major divisions. The first division grouped clusters I to IV, while the second group comprised cluster V. Clones from clusters I displayed 88 to 99% similarity to the mangrove root isolates, Paracoccus sp. strain LR4BC (α-Proteobacteria), strain 64K, Oceanimonas sp. strain 5A (γ-Proteobacteria), Vibrio sp. strain 9B, and Aeromonas sp. strain LR7YC (both γ-Proteobacteria), and grouped with two clones from the S. alterniflora rhizosphere (36, 37). Cluster II was formed by clones from the three sites. None of the mangrove clones grouped in clusters III and V, represented by known species of α-, β-, and γ-Proteobacteria. In cluster IV, some clones branched together with the mangrove isolate Corynebacterium sp. strain 12A (Actinobacteria, gram positive, high GC content) while others branched with clones from sea grass beds (1), the S. alterniflora rhizosphere (36, 37), oligotrophic oceans (68), assemblages of diazotrophs (clones AY091859, AY091864, and AY091911; http://www.ncbi.nlm.nih.gov/), Chesapeake bay microbial communities (clone DQ09234), and oil-contaminated marine sediments (clone DQ078010) (40), as well as Delsufonema limicola (γ-Proteobacteria) and Clostridium pasteurianum (gram positive, high GC content). All sequences from cluster IV branched with the mangrove isolate Arthrobacter sp. strain 61K (Actinobacteria, gram positive, high GC content).
In general, the tree showed that clones from sites 1 and 2 were inclined to group together and that most of the clones from site 3 clustered separately from those of sites 1 and 2. The mangrove root cloned sequences were more similar to sequences from the isolates than to sequences from previously characterized strains. Sequences from clusters I and II might represent novel sequences of diazotrophs.
The phylogenetic tree of nirK generated five clusters (I to V) (Fig. 5), with all of the clones grouping in clusters I, III, IV, and V. Clones from cluster I were highly similar to the mangrove isolate Arthrobacter sp. strain 61K (Actinobacteria, gram positive, low GC content) and Corynebacterium sp. strain 63K (Actinobacteria, gram positive, high GC content), while those clones from clusters IV and V were similar to the isolates Paracoccus sp. strain AG4BC (α-Proteobacteria), Oceanimonas sp. strain 5A (γ-Proteobacteria), and Aeromonas sp. strain LR7YC (γ-Proteobacteria).
FIG. 5.
Neighbor-joining phylogenetic tree of nirK constructed using 44 cloned and partial amino acid sequences of 140 amino acids obtained from the three sites. They were compared with 21 sequences from GenBank and the sequences of our cultivated diazotrophic and denitrifying mangrove root isolates (purple). The clones were designated according to the site they were taken from: site 1, 1K (green); site 2, 2K (red); site 3, 3K (blue). We used the aniA sequence from Neisseria gonorrhoeae (M97926) as an outgroup. Bootstrap values greater than 500 from 1,000 replicate trees are reported at the nodes. The scale represents 10 mutations per 100 amino acid positions.
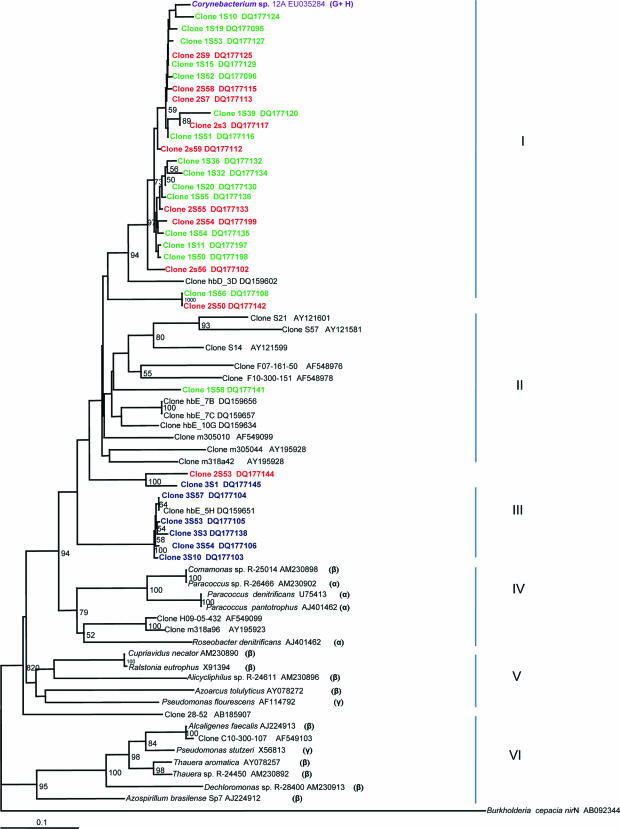
In general, the nirK tree showed that there were two main divisions: in the first division (clusters I, II, and III), 20 of the mangrove clones were grouped with α- and β-Proteobacteria, while the second division (clusters IV and V) grouped 24 of the mangrove clones and displayed very high similarity to three of the mangrove root isolates. The phylogenetic tree of nirS generated six clusters (I to VI) congregated in two major divisions (Fig. 6). The first division consisted of clusters I to V, and the second one consisted of cluster VI. All of the mangrove clones grouped in clusters I, II, and III. Clones from sites 1 and 2 formed cluster I and showed 90 to 98% similarity to the only nirS mangrove isolate, Corynebacterium sp. strain 12A. They also displayed 83 to 87% similarity to a clone obtained from a coastal aquifer (53). Clones from clusters II and III displayed similarity to clones from other environments (35, 47, 53, 67). Clusters IV, V, and VI were formed by known strains belonging to α-, β-, and γ-Proteobacteria. We observed the same tendency in the nirS tree as in the nifH tree: i.e., clones from sites 1 and 2 clustered together, while those from site 3 formed separate groups. Sequences from groups 1 and 2 might represent novel sequences of denitrifiers.
FIG. 6.
Neighbor-joining phylogenetic tree of nirS constructed using 32 cloned and partial amino acid sequences of 240 amino acids obtained from the three sites. They were compared with 33 sequences from GenBank and the sequence from our cultivated nirS denitrifying mangrove root isolate (purple). The clones were designated according to the site they were taken from: site 1, 1S (green); site 2, 2S (red); site 3, 3S (blue). We used the nirA sequence from Burkholderia cepacia (AB092344) as an outgroup. Bootstrap values greater than 500 from 1,000 replicate trees are reported at the nodes. The scale represents 10 mutations per 100 amino acid positions.
DISCUSSION
The conditions that prevail in the mangrove rhizosphere might be conducive to denitrification and allow the sustenance of a denitrifying community in that habitat. This work represents the first attempt to explore this hypothesis.
The high levels of diazotrophic activity associated with the mangrove rhizosphere (27, 28, 57, 70) imply that this habitat fosters conditions that may favor the maintenance of substantial diazotroph diversity. Previous to this work, there was no information to support this hypothesis. To explore these questions, T-RFLP analysis of nifH, nirK, and nirS was done for three sites having distinct biogeochemical parameters. There are few studies that involve the parallel analyses of both diazotrophic and denitrifying communities in any type of environment (38, 51).
Analysis of nifH, nirK, and nirS gene T-RFLP fragments.
nifH had the largest number of OTU of all three genes and displayed the highest diversity. No statistically significant difference between nirK and nirS diversity values was found. A study based on DNA-DNA hybridization with probes targeting nifH, nirK, and nirS found that the abundance of diazotrophs in rhizosphere of forest plants was close to that of denitrifiers having nirS (the signal intensity for nirS and nifH was the same). Denitrifiers having nirK were not as abundant (the signal intensity for nirK was statistically lower than those of nirK and nifH), suggesting that the conditions in the rhizosphere of forest plants were more favorable to nirS than to nirK (38).
Most reports on characterization of nirK and nirS denitrifying communities have found higher diversity for nirS than for nirK in marine sediments (5, 42), forest soils (38, 47), nitrate- and uranium-contaminated groundwater (67), and culture collections of denitrifiers (14). One exception is an analysis of the diversity of transcripts of the nirK and nirS genes in rhizosphere samples of three legumes, in which nirK transcripts were detected but nirS transcripts were not (59). Based on the former results, it is not known if nirK has an inherently low diversity in natural habitats or if the restricted diversity of nirK sequences is due to characteristics of the studied soils and sediments (47).
Biogeochemical properties and gene diversity.
PCA separated site 3 from the other sites by its high percentage of sand and organic matter, as well as a low pore water oxygen concentration and salinity (Table 1). Site 2 was separated from the other two sites by its high content of clay and low concentration of oxygen, while site 1 was separated by its low organic matter content and high salinity. These observations are supported by T-RFLP data analysis, which clearly separated the OTU of site 3 from those of sites 1 and 2 (Fig. 3 and Tables 3 and 4), and by the phylogenetic tree for nifH (Fig. 5) in which the majority of the clones from site 3 clustered with each other and clones from sites 1 and 2 grouped together.
The biogeochemical diversity between the three sites might explain the differences in the structure of the nifH OTU for the three sites: some studies have suggested a link between functional gene diversity and ecosystem biogeochemistry (9, 35). Soil texture, the main variable that segregated the sites in our work, can greatly influence the composition of its bacterial communities (8, 20, 22, 24, 30, 58). As revealed by T-RFLP analyses of bacterial community structures in different particle size fractions of topsoil samples, Sessitsch et al. (58) found fine soil particles (clay fraction) supported higher microbial diversity than larger particles. It has been suggested that fine particles provide a protective habitat for microorganisms by excluding predators (protozoa) (17, 46). This is contrary to our results, in which the highest nifH diversity was associated with sand-rich soil (site 3).
Despite the contrast in results between Sessitsch and collaborators (58) and this work, other factors besides soil texture known to influence the composition of bacterial communities in soil (41, 62) could explain the higher diversity and uniqueness of the nifH OTU found at site 3. This site probably offered conditions more favorable to microbial growth than the other sites, and its high organic matter content and low oxygen concentration could have favored nitrogen fixers in particular. High organic matter content can increase sediment water retention (43) as well as provide carbon and energy sources to sustain microbial growth. A shortage of carbon can limit nitrogen fixation. Zuberer and Silver (70) demonstrated that nitrogen-fixing activity in plant-free sediments of a Florida mangrove increased greatly with the addition of various carbon sources, indicating an energy limitation for nitrogenase. The low oxygen concentration, probably caused by the high rate of aerobic microbial respiration supported by the availability of energy and carbon, would also favor nitrogen fixation: most prokaryotes fix nitrogen under anaerobic or microaerophylic conditions (21). In spite of its high sand content that could have otherwise limited water retention (25), site 3 probably had higher water retention than sites 1 and 2 due to its location in the mid-intertidal zone, as opposed to sites 1 and 2, which were located in the high intertidal zone. Its location would limit the time of exposure of site 3 to solar radiation during low tide and thus water loss through evaporation. Additionally, the proximity of site 3 to the inner lagoon (Fig. 1) would moisten the rhizosphere through capillary action.
Site 2, with the highest pore water oxygen concentration, had the lowest number of nifH OTU and the lowest diversity, but the highest dominance. This could mean that this site selected for diazotrophs capable of fixing nitrogen under aerobic conditions. This is supported by the nifH phylogenetic tree, which shows that clones from clusters I and II, mainly from site 2 (Fig. 4), are related to Azotobacter chroococcum (88 to 93% similarity) and Azomonas agilis (93 to 94% similarity), both known to fix nitrogen under aerobic conditions (66).
The diversities of nirK OTU were the same in all three sites; this result was supported by the MDS and ANOSIM analyses. For nirS, the highest diversity was found in site 2 followed by site 3 (Table 2). This might be because site 2 had the highest content of nitrate of all sites and a relatively high content of organic matter (3.1%). According to Zumft (71), the dominant exogenous signals that induce the synthesis of denitrification systems are low oxygen tension and the presence of a respirable N oxide. In addition to these factors, other studies have found that nitrate (9, 35) and the availability of carbon (6) can control denitrifying community structure.
Phylogenetic analysis of nifH, nirK, and nirS sequences.
Most of the mangrove root bacterial sequences branched with γ- or δ-Proteobacteria, and a few branched with α-Proteobacteria. As in these results, most of the nifH genes of diazotrophs associated with the rhizosphere of the salt marsh grasses S. alterniflora and Juncus roemerianus were from γ-Proteobacteria and the α- and β-Proteobacteria were poorly represented (7, 37). Besides the high abundance of Proteobacteria in the nifH clones, 12 clones displayed very high similarity to the mangrove isolate Corynebacterium sp. strain 12A, a gram-positive strain with high GC content, which belongs to the Actinobacteria.
Some of the nifH clones from cluster IV (Fig. 4) displayed 74 to 98% similarity to Desulfonema limicola. This is in agreement with studies that showed that diazotrophic sulfate-reducing bacteria are abundant and diverse in the Spartina rhizosphere and are important contributors to N2 fixation in Spartina vegetated sediments (7, 37). The contribution of nitrogen to the mangroves of this bacterial group has not been elucidated, although it is presumed to be important (28).
In their extensive survey of nifH phylotypes across different habitats, Zehr et al. (69) concluded that the nifH phylotypes obtained were usually not closely related to sequences from previously characterized strains and environments. Accordingly, 26% of the clones branched separately, forming a separate division and displaying no similarity to known strains or to clones from other environments, suggesting that they are unique, representative novel sequences of diazotrophs (cluster II; Fig. 4). However, the other eight clones were very similar (80 to 95% similarity) to clones recovered from the salt marsh grass S. alterniflora (36) (cluster IV; Fig. 4).
Chèneby et al. (11) screened a collection of denitrifying isolates by amplified 16S rRNA gene restriction analysis and found that Agrobacterium was the most represented genus in the maize rhizosphere, followed by Streptomyces (Actinobacteria, gram positive, high GC content). Other cultivation-dependent and -independent approaches have shown that α-Proteobacteria predominate in the maize rhizosphere. Within that group, the rhizobium-related phylotypes accounted for 30% of the clone library and 83% of the culture collection. In this study, none of the clones displayed similarity to Rhizobium spp. The Actinobacteria are probably an abundant group in the mangrove rhizosphere: 24 of the nirS clones displayed 90 to 98% identity to the isolate Corynebacterium sp. strain 12A, and 13 nirK clones were 96 to 99% similar to the isolate Arthrobacter sp. strain 61K (Fig. 5 and 6).
Most of the nirK clones (probably representatives of α- and γ-Proteobacteria) grouped together with the mangrove root isolates and displayed much higher similarity to the latter than to known strains or clones obtained from other environments (Fig. 5), suggesting they may be from novel denitrifiers. Generally, nirK clones obtained from the environment were not closely related to sequences from previously described strains or isolates (5, 35, 47). Unlike nirK, some researchers have found that nirS clones were related to known cultivated denitrifiers (35, 47). However, Braker et al. (5) found that nirS sequences had little relationship to any strain with known nirS sequences or to isolates that were mostly close relatives of P. stutzeri.
We cannot discard the possibility that some of our nirK, nirS, or nifH sequences are multiple copies of the genes. Also, our analysis did not test the potential artifacts from PCR directly. However, T-RFLP profiles have been shown to be relatively stable to variability in PCR conditions (4). In silico restriction cleavage of our clones and isolates and comparison of the calculated T-RF lengths to T-RFLPs derived from mangrove roots samples revealed simulated T-RFs and T-RFs from natural samples were highly analogous.
Seven of 10 isolates had nifH together with either nirK or nirS: 5 had nirK, and 1 had nirS. Rösch et al. (51) found that several species of Azospirillum and Herbaspirillum, Gluconacetobacter diazotrophicus, Rhodospirillum rubrum, Ralstonia eutropha, and P. stutzeri had either the nirS or nirK gene together with the nifH gene (assessed by PCR amplification). Several species of Hyphomicrobium had both nirK and nifH, assessed by Southern blot hybridization (19).
In sum, our findings reveal that denitrifiers and nitrogen fixers coexist in the mangrove rhizosphere, although mangrove roots sustain a higher diversity of nifH than of nirK or nirS. It seems contradictory that a nitrogen-starved system would also support denitrification, which implies loss of fixed nitrogen. However, in a system subjected to fluctuations in oxygen tension, denitrification offers the enormous advantage of getting energy without the need for oxygen, a benefit that probably surpasses the drawback of losing some of the fixed nitrogen. According to our results, the biogeochemical characteristics of the rhizosphere can determine the structure of the diazotrophic community associated with mangrove roots. The fact that in the case of the denitrifying community there was no link between site and diversity of genes determined by T-RFLP is probably caused by the low number of diagnostic OTU for these genes as compared to nifH. This large difference in resolution may very well explain why no diversity patterns were found in response to biogeochemical characteristics of the environment.
Acknowledgments
This work was supported by CONACyT, project 41367-Z and fellowship no. 181882.
We thank Esther Angert and Julie Frey for supplying the equipment and for their valuable contributions to the T-RFLP technique. We thank Anatol Eberhard for helpful suggestions and revising the manuscript and Taylor Morey for editing.
A.L.F.-M. wants to dedicate this work to the memory of Gina Holguin, an advisor and friend without whom this study could not have succeeded. She participated in this work in memory of the late Juan Holguin Franco.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Bagwell, C. E., J. R. La Rocque, G. W. Smith, S. W. Polson, M. J. Friez, J. W. Longshore, and C. R. Lovell. 2002. Molecular diversity of diazotrophs in oligotrophic tropical seagrass bed communities. FEMS Microbiol. Ecol. 39:113-119. [DOI] [PubMed] [Google Scholar]
- 2.Bashan, Y., and G. Holguin. 1997. Azospirillum-plant relationships: environmental and physiological advances (1990-1996). Can. J. Microbiol. 43:103-121. [DOI] [PubMed] [Google Scholar]
- 3.Bendschneider, K., and R. J. Robinson. 1952. A new spectrophotometric determination of nitrite in sea water. J. Mar. Res. 11:87-96. [Google Scholar]
- 4.Blackwood, C. B., T. Marsh, S.-H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braker, G., H. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. M., M. J. Friez, and C. R. Lovell. 2003. Expression of nifH genes by diazotrophic bacteria in the rhizosphere of short form Spartina alterniflora. FEMS Microbiol. Ecol. 43:411-417. [DOI] [PubMed] [Google Scholar]
- 8.Buyer, J. S., D. P. Roberts, and E. Russek-Cohen. 1999. Microbial community structure and function in the spermosphere as affected by soil and seed type. Can. J. Microbiol. 45:138-144. [Google Scholar]
- 9.Castro-Gonzalez, M., G. Braker, L. Farias, and O. Ulloa. 2005. Communities of nirS-type denitrifiers in the water column of the oxygen minimum zone in the eastern South Pacific. Environ. Microbiol. 7:1298-1306. [DOI] [PubMed] [Google Scholar]
- 10.Cevallos, M. A., S. Encarnación, A. Leija, Y. Mora, and J. Mora. 1996. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-β-hydroxybutyrate. J. Bacteriol. 178:1646-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chèneby, D., S. Perrez, C. Devroe, S. Hallet, Y. Couton, F. Bizouard, G. Luretig, J. C. Germon, and L. Philippot. 2004. Denitrifying bacteria in bulk and maize-rhizospheric soil: diversity and N2O-reducing abilities. Can. J. Microbiol. 50:469-474. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, K. R. 1993. Non-parametric multivariate analysis of changes in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 13.Cole, A. C., M. J. Semmens, and T. M. LaPara. 2004. Stratification of activity and bacterial community structure in biofilms grown on membranes transferring oxygen. Appl. Environ. Microbiol. 70:1982-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne, M. S., A. Arunakumari, B. A. Averill, and J. M. Tiedje. 1989. Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl. Environ. Microbiol. 55:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davila-Lule, A., A. Flores-Mireles, C. Villicaña, M. A. Ruiz, A. Eberhard, M. Gronquist, A. Carrillo, P. Vazquez, and G. Holguin. 2004. Plant growth-promoting bacteria isolated from mangrove roots produce acyl homoserine lactones, p. 96. 22nd Latin-Am. Conf. Rhizobiol., Miguel Pereira, Río de Janeiro, Brazil, 13 to 15 September 2004.
- 16.Day, P. R. 1965. Particle fractionation and particle-size analysis, p. 545-567. In C. A. Black (ed.), Methods of soil analysis. American Society of Agronomy, Madison, WI.
- 17.Elliot, E. T., R. V. Anderson, D. C. Coleman, and C. V. Cole. 1980. Habitable pore space and microbial trophic interactions. Oikos 35:327-335. [Google Scholar]
- 18.Felsenstein, J. 1993. PHYLIP inference package, version 3.5. Department of Genetics, University of Washington, Seattle.
- 19.Fesefeldt, A., K. Kloos, H. Bothe, H. Lemmer, and C. G. Gliesche. 1997. Distribution of denitrification and nitrogen fixation genes in Hyphomicrobium spp. and other budding bacteria. Can. J. Microbiol. 44:181-186. [Google Scholar]
- 20.Girvan, M. S., J. Bullimore, J. N. Pretty, A. M. Osborn, and A. S. Ball. 2003. Soil type is the primary determinant of the composition of the total and active bacterial communities of arable soils. Appl. Environ. Microbiol. 69:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glick, B. R., C-L- Patten, G. Holguin, and D. M. Penrose. 1999. Biochemical and genetic mechanisms used by plant growth promoting bacteria. Imperial College Press, London, United Kingdom.
- 22.Gonzalez-Acosta, B., Y. Bashan, N. Y. Hernandez-Saavedra, F. Ascencio, and G. De la Cruz-Agüero. 2007. Seasonal seawater temperature as the major determinant for populations of culturable bacteria in the sediments of an intact mangrove in an arid region. FEMS Microbiol. Ecol. 55:311-321. [DOI] [PubMed] [Google Scholar]
- 23.Gotto, J. W., and B. F. Taylor. 1976. N2 fixation associated with decaying leaves of the red mangrove (Rhizophora mangle). Appl. Environ. Microbiol. 31:781-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths, B. S., K. Ritz, and L. A. Glover. 1996. Broad-scale approaches to the determination of soil microbial community structure: application of the community DNA hybridization technique. Microb. Ecol. 31:269-280. [DOI] [PubMed] [Google Scholar]
- 25.Hausenbuiller, R. L. 1972. Soil science: principle and practices. W. C. Brown Company, Dubuque, IA.
- 26.Hernández-Lopez, J., and F. Vargas-Albores. 2003. A microplate technique to quantify nutrients (NO2−, NO3−, NH4+, PO43−) in seawater. Aquac. Res. 34:1201-1204. [Google Scholar]
- 27.Holguin, G., M. A. Guzman, and Y. Bashan. 1992. Two new nitrogen-fixing bacteria from the rhizosphere of mangrove trees: their isolation, identification and in vitro interaction with rhizosphere Staphylococcus sp. FEMS Microbiol. Ecol. 101:207-216. [Google Scholar]
- 28.Holguin, G., P. Vazquez, and Y. Bashan. 2001. The role of sediments microorganisms in the productivity, conservation and rehabilitation of the mangrove ecosystems: an overview. Biol. Fertil. Soils 33:265-278. [Google Scholar]
- 29.Jayakumar, D. A., C. A. Francis, S. W. A. Naqvi, and B. B. Ward. 2004. Diversity of nitrite reductase genes (nirS) in the denitrifying water column of the coastal Arabian Sea. Aquat. Microbiol. Ecol. 34:69-78. [Google Scholar]
- 30.Kennedy, N., E. Brodie, J. Connolly, and N. Clipson. 2004. Impact of lime, nitrogen and plant species on bacterial community structure in grassland microcosms. Environ. Microbiol. 6:1070-1080. [DOI] [PubMed] [Google Scholar]
- 31.Krebs, C. J. 1994. Ecological methodology, 2nd ed., Benjamin Cummings, Menlo Park, CA.
- 32.Kruskal, J. B. 1964. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 298:1-27. [Google Scholar]
- 33.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 34.Lin, Y. T., and W. Y. Shieh. 2006. Zobellella denitrificans gen. nov., sp. nov., and Zobellella taiwanensis sp. nov., denitrifying bacteria capable of fermentative metabolism. Int. J. Sys. Evol. Microbiol. 56:1209-1215. [DOI] [PubMed] [Google Scholar]
- 35.Liu, X., S. M. Tiquia, G. Holguin, L. Wu, S. C. Nold, A. H. Devol, K. Luo, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone of the Pacific Coast of Mexico. Appl. Environ. Microbiol. 69:3549-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovell, C. R., Y. M. Piceno, J. M. Quattro, and C. E. Bagwell. 2000. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66:3814-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lovell, C. R., M. J. Friez, J. W. Longshore, and C. E. Bagwell. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mergel, A., O. Schmitz, T. Mallmann, and H. Bothe. 2001. Relative abundance of denitrifying and dinitrogen-fixing bacteria in layers of forest soil. FEMS Microbiol. Ecol. 36:33-42. [DOI] [PubMed] [Google Scholar]
- 39.Morris, A. W., and J. P. Riley. 1963. The automatic determination of nitrate in sea water. Deep-Sea Res. 12:765-772. [Google Scholar]
- 40.Musat, F., J. Harder, and F. Widdel. 2006. Study of nitrogen fixation in microbial communities of oil-contaminated marine sediment microcosm. Environ. Microbiol. 8:1834-1843. [DOI] [PubMed] [Google Scholar]
- 41.Nannipieri, P., J. Asche, M. T. Ceccherini, L. Landi, G. Pietramellara, and G. Renella. 2003. Microbial diversity and soil functions. Eur. J. Soil Sci. 54:655-670. [Google Scholar]
- 42.Nogales, B., K. N. Timmis, D. B. Nedwell, and A. M. Osborn. 2002. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortiz-Villanueva, B., and C. A. Ortiz-Solorio. 1984. Edafología. Universidad Autonoma de Chapingo, Chapingo, México.
- 44.Patureau, D., N. Bernet, and R. Moletta. 1996. Study of the denitrifying enzymatic system of Comamonas sp. strain SGLY2 under various aeration conditions with a particular view on nitrate and nitrite reductases. Curr. Microbiol. 32:25-32. [Google Scholar]
- 45.Piceno, Y. M., P. A. Noble, and C. R. Lovell. 1999. Spatial and temporal assessment of diazotroph assemblage composition in vegetated salt marsh sediments using denaturing gradient gel electrophoresis analysis. Microb. Ecol. 38:157-167. [DOI] [PubMed] [Google Scholar]
- 46.Postma, J., and J. A. van Veen. 1990. Habitable pore space and survival of Rhizobium leguminosarum, biovar trifolii introduced into soil. Microb. Ecol. 19:146-161. [DOI] [PubMed] [Google Scholar]
- 47.Priemé, A., G. Braker, and J. M. Tiedje. 2002. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl. Environ. Microbiol. 68:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Primavera, J. H. 2000. Development and conservation of Philippine mangroves: institutional issues. Ecol. Econ. 35:91-106. [Google Scholar]
- 49.Primavera, J. H., R. S. Sadaba, M. J. H. L. Lebata, and J. P. Altamirano. 2004. Handbook of mangroves in the Philippines—Panay. Southeast Asian Fisheries Development Center Aquaculture Department, Iloilo, Philippines.
- 50.Rees, G. N., D. S. Baldwin, G. O. Watson, S. Perryman, and D. L. Nielsen. 2004. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Leeuwenhoek 86:339-347. [DOI] [PubMed] [Google Scholar]
- 51.Rösch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 53.Santoro, A. E., A. B. Boehm, and C. A. Francis. 2006. Denitrifier community composition along a nitrate and salinity gradient in a coastal aquifer. Appl. Environ. Microbiol. 72:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Secretaría de Medio Ambiente y Recursos Naturales (SEMARNAT). 2002. Norma oficial 021-SEMARNAT-2000, que establece las especificaciones de fertilidad, salinidad y clasificación de suelos. Estudios, muestreo y análisis. Metodo AS-07. Diario Oficial de la Federación, 31 December 2002. SEMARNAT, Tlalpan, Mexico.
- 56.Secretaría de Medio Ambiente y Recursos Naturales (SEMARNAT). 2003. Norma oficial de emergencia NOM-022-SEMARNAT-2003, que establece las especificaciones para la preservación, conservación y restauración del manglar. Diario Oficial de La Federación, 10 April 2003. SEMARNAT, Tlalpan, Mexico.
- 57.Sengupta, A., and S. Chaudhuri. 1991. Ecology of heterotrophic dinitrogen fixation in the rhizosphere of mangrove plant community at Ganges river estuary in India. Oecologia 87:560-564. [DOI] [PubMed] [Google Scholar]
- 58.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma, S., M. K. Aneja, J. Mayer, J. C. Munch, and M. Schloter. 2005. Diversity of transcripts of nitrite reductase genes (nirK and nirS) in rhizospheres of grain legumes. Appl. Environ. Microbiol. 71:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shieh, W. Y., and J. T. Yang. 1997. Denitrification in the rhizosphere of the two seagrasses Thalassia hemprichii (Ehrenb.) Aschers and Halodule uninervis (Forsk.) Aschers. J. Exp. Mar. Biol. Ecol. 218:229-241. [Google Scholar]
- 61.Strickland, J. D., and T. R. Parsons. 1972. A practical handbook of seawater analysis. Bull. Fish. Res. Board Can. 167:1-310. [Google Scholar]
- 62.Suzuki, C., T. Kunito, T. Aono, C. T. Liu, and H. Oyaizu. 2005. Microbial indices of soil fertility. J. Appl. Microbiol. 98:1062-1074. [DOI] [PubMed] [Google Scholar]
- 63.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trower, M. K. 1996. A rapid PCR-based colony screening protocol for cloned inserts. Methods Mol. Biol. 58:329-333. [DOI] [PubMed] [Google Scholar]
- 65.Van Loon, J. C. 1985. Selected methods of trace metal analysis, p. 94-96. In P. J. Elving, J. D. Winefordner, and I. M. Kolthoff (ed.), Chemical analysis, vol. 80. Wiley Interscience Publication, Mississauga, Canada. [Google Scholar]
- 66.White, D. 1999. The physiology and biochemistry of prokaryotes. Oxford University Press, New York, NY.
- 67.Yan, T., M. W. Fields, L. Wu, Y. Zu, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity and characterization of nitrite reductase gene fragments (nirK and nirS) from nitrate- and uranium-contaminated groundwater. Environ. Microbiol. 5:13-24. [DOI] [PubMed] [Google Scholar]
- 68.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]
- 70.Zuberer, D., and W. S. Silver. 1978. Biological dinitrogen fixation (acetylene reduction) associated with Florida mangroves. Appl. Environ. Microbiol. 35:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]