Abstract
Engineered microbes are becoming increasingly important as recombinant production platforms. However, the nonfunctionality of membrane-bound cytochrome P450 enzymes precludes the use of industrially relevant prokaryotes such as Escherichia coli for high-level in vivo synthesis of many functional plant-derived compounds. We describe the design of a series of artificial isoflavone synthases that allowed the robust production of plant estrogen pharmaceuticals by E. coli. Through this methodology, a plant P450 construct was assembled to mimic the architecture of a self-sufficient bacterial P450 and contained tailor-made membrane recognition signals. The specific in vivo production catalyzed by one identified chimera was up to 20-fold higher than that achieved by the native enzyme expressed in a eukaryotic host and up to 10-fold higher than production by plants. This novel biological device is a strategy for the utilization of laboratory bacteria to robustly manufacture high-value plant P450 products.
Approximately half of the pharmaceuticals in clinical use today originated from natural compounds (25). The plant kingdom is an invaluable apothecary, for it is the origin of many of the most potent drugs. For the development of human therapeutics, large amounts of plant tissues are required (16). Nevertheless, total chemical synthesis of many plant products is often not feasible given the complexity of enantioselective reactions and low yields. Because of these bottlenecks, the scarcity of natural products has resulted in predicaments in drug development and the depletion of environmental components (25).
Owing to their success in producing large-scale commercial commodities (3, 7), Escherichia coli strains have been engineered to synthesize many important natural products from plants and other hard-to-culture organisms to sustain the availability of drug candidates (10, 13, 17, 21, 26). Low productivity was a major challenge of the recombinant technology which was overcome by using tools based on systems biology (2). One remaining primary disadvantage of the bacterial platform is the nonfunctionality of many key plant enzymes, notably, the membrane-bound cytochrome P450 family (8, 18). This challenge hinders the biosynthesis of many functional molecules by recombinant bacteria. Two major factors prohibit plant P450 enzymes from functioning in E. coli. First is the absence in E. coli of cytochrome P450 reductases (CPRs), which are required by eukaryotic P450 enzymes for electron transfer (27). Second is the translational incompatibility of the membrane signal modules of microsomal P450 enzymes with the bacterium due to the absence of an endoplasmic reticulum (4, 30). A number of studies have attempted to improve the production of functional plant P450 enzymes by E. coli. In most cases, enzyme activity was restored only in vitro, necessitating the use of radioactive substrates to detect the reaction products (6, 11, 23). So far, the reported level of in vivo synthesis of plant hydroxylated products by E. coli has been extremely low (13, 18), and therefore, this strategy has not been suitable for large-scale applications.
Plant estrogen is a target of therapeutic development. Soy isoflavones are currently being tested in chemoprevention clinical trials sponsored by the National Cancer Institute (http://clinicaltrials.gov/ct/show/NCT00513916). Moreover, the isoflavone derivative phenoxodiol (Novogen) is the first new oncology agent among the multiple signal transduction regulators to have entered phase III clinical trials (http://www.phenoxodiol.com/). In plants, isoflavones are synthesized from flavanones, which undergo hydroxylation and novel aryl ring migration mediated by isoflavone synthase (IFS), following spontaneous or enzymatic dehydration (1, 28). To increase the availability of these pharmaceuticals, we engineered a series of artificial P450 enzymes that allowed robust in vivo synthesis of isoflavones by E. coli.
MATERIALS AND METHODS
Plant materials and bacterial strains.
Glycine max seeds were germinated in the dark at 30°C for 3 to 5 days in a growth chamber to obtain etiolated seedlings. Seedlings were wounded by cutting small slits (∼1-mm sections). Incubation in the dark was continued for 8 h before harvesting. Catharanthus roseus plants were purchased from a local nursery. Harvested plant tissues were flash frozen using liquid nitrogen and stored at −70°C until use. E. coli TOP10F′ {Invitrogen; F′ [lacIq Tn10(Tetr)] mcrA (mrr-hsdRMS-mcrBC) 80lacZM15 lacX74 recA1 araD139 (ara-leu)7697 galU galK rpsL (Strr) endA1 nupG } was used for plasmid maintenance. For protein expression and recombinant-isoflavonoid production, E. coli strains TOP10F′, BL21Star (Invitrogen; F− ompT hsdSBB− mB−] gal dcm rne-131 [DE3]), DH5α (Invitrogen; F′ [Φ80dlacZΔM15] Δ[lacZYA-argF]U196 recA1 endA gyrA thi-1 hsdR17[rK− mK−] supE44 relA1), and JM109 (American Type Culture Collection; McrA− recA1 endA1 gyrA96 thi-1 hsdR17[rK− mK−] supE44 relA1 Δ[lac-proAB] [F′ traD36 proAB lacIqZΔM15]) were used.
cDNA isolation and functional expression in Saccharomyces cerevisiae.
RNAs were isolated from G. max leaflets and flower petals of C. roseus by using the RNeasy plant minikit (QIAGEN). Full-length cDNAs were obtained from RNA materials by reverse transcription and PCR using the SuperScript one-step reverse transcription-PCR kit (Invitrogen). Primers were designed based on the sequences of ifs1 and cpr with GenBank accession numbers AF195798 and X69791, respectively. Gene fragments were cloned into the pYes2.1 TOPO vector (Invitrogen; URA3 Ampr) downstream of the Gal1 promoter sequence and identified by direct sequencing (Biopolymer Resource, Roswell Park Cancer Institute). On the basis of sequence analysis, genes encoding full-length IFS1 (IFS1[1-521]) and CPR (CPR[1-714]) were verified and no mutation was identified. Plasmids harboring IFS1[1-521] were introduced into the yeast S. cerevisiae InvSc1 (Invitrogen; MATa his3D1 leu2 trp1-289 ura3-52[r]). Positive transformants were selected by growth on agar with synthetic complete minimal medium (Invitrogen) and without uracil. The functionality of CPR was determined as described previously (19). To determine the hydroxylation activity of full-length IFS1, which comprises amino acids 1 to 521 (IFS1[1-521]), we conducted in vitro assays using yeast microsomes and whole-cell assays (12). Yeast expression was induced with galactose, and 0.05 mM naringenin and 0.05 mM liquiritigenin were added separately. After 36 h, high-performance liquid chromatography (HPLC) detected the synthesis of genistein and daidzein, which demonstrated the functionality of IFS1[1-521] in yeast.
Generation of P450 constructs.
pTrcMod was used as the expression plasmid for P450 constructs in E. coli. This expression vector was constructed by inserting a DNA fragment containing the multiple cloning sites of Yeplac112 (American Type Culture Collection) into pTrcHis2/LacZ (Invitrogen) between HindIII and SalI sites, downstream of the Ptrc promoter sequence. For the assembly of the P450 constructs, first the CPR gene sequences encoding amino acids 71 to 714 (CPR[71-714]) and 72 to 714 (CPR[72-714]) were separately cloned into pTrcMod between BamHI and KpnI sites, creating pCPR[71-714] and pCPR[72-714], respectively. To create construct C, IFS1[1-521] was inserted into pCPR[71-714] between HindIII and BamHI sites. By using the same restriction sites, IFS1 gene sequences encoding amino acids 1 to 520 (IFS1[1-520]) and 7 to 520 (IFS1[7-520]), IFS1[7-520] fused to a sequence encoding the mammalian peptide ɛ (IFS1[ɛ:7-520]), and the IFS1 gene sequence encoding amino acids 25 to 520 fused to a sequence encoding the mammalian peptide ɛ (IFS1[ɛ:25-520]) were individually inserted into pCPR[72-714] to construct E1, E2, E3, and E4, respectively. To yield E5, IFS1[3-520] was first cloned into pCR2.1-TOPO (Invitrogen). IFS1[ϕ:3-520] was constructed by PCR, which joined IFS1[3-520] with the lacZα leader sequence encoding ϕ. A silent mutation was introduced into the lacZα sequence to remove a HindIII site, and the mutated sequence was fused to IFS1[3-520] to create IFS1[26L:3-520], which was then cloned between HindIII and BamHI sites of pCPR[72-714]. All original or modified cDNA sequences were obtained using PCR with a customized oligonucleotide design (Tables 1 and 2). PCR gene amplifications were performed using an Expand high-fidelity PCR system (Roche). Gene sequences were confirmed by direct sequencing (Biopolymer Resource, Roswell Park Cancer Institute). Restriction endonucleases and T4 DNA ligase from New England Biolabs and Promega were used in subcloning. Subclones were verified by restriction analysis.
TABLE 1.
Forward primer sequences for gene constructs
| Construct | Forward primer sequencea |
|---|---|
| pCPR[71-714] | AAAGGATCCACAATGTCTTCCGGATCGGGTAAAAAAGTCG |
| pCPR[72-714] | AAAGGATCCACATCTTCCGGATCGGGTAAAAAAGTCG |
| IFS1[1-521] | GGGAAGCTTATGTTGCTGGAACTTGCACTTGG |
| IFS1[1-520] | GGGAAGCTTATGTTGCTGGAACTTGCACTTGG |
| IFS1[7-520] | GGGAAGCTTATGCTTGGTTTGTTTGTGTTAGCTTTGTT |
| IFS1[ɛ:7-520] | GGGAAGCTTATGGCTCTGTTATTAGCAGTTTTTCTTGGTTTGTTTGTGTTAGCTTTGTT |
| IFS1[ɛ:25-520] | GGGAAGCTTATGGCTCTGTTATTAGCAGTTTTTAAATCAAAAGCACTTCGCCACCTCCCA |
| IFS1[ϕ:3-520] | GGGAAGCTTATGACCATGATTACGCCAAGTTTGG |
Restriction sites are underlined.
TABLE 2.
Reverse primer sequences for gene constructs
| Construct | Reverse primer sequencea |
|---|---|
| pCPR[71-714] | AAAGGTACCTCACCAGACATCTCGGAGATACCTT |
| pCPR[72-714] | AAAGGTACCTCACCAGACATCTCGGAGATACCTT |
| IFS1[1-521] | AAAGGATCCTTAAGAAAGGAGTTTAGATGCAACGCC |
| IFS1[1-520] | AAAGGATCCAGAAAGGAGTTTAGATGCAACGCC |
| IFS1[7-520] | AAAGGATCCAGAAAGGAGTTTAGATGCAACGCC |
| IFS1[ɛ:7-520] | AAAGGATCCAGAAAGGAGTTTAGATGCAACGCC |
| IFS1[ɛ:25-520] | AAAGGATCCAGAAAGGAGTTTAGATGCAACGCC |
| IFS1[ϕ:3-520] | AAAGGATCCAGAAAGGAGTTTAGATGCAACGCC |
Restriction sites are underlined.
In vivo P450 activity measurement.
E. coli strains (TOP10F′, BL21Star, DH5α, and JM109) were transformed with plasmids harboring the P450 constructs. Positive transformants were selected on Luria-Bertani (LB; Sigma) agar plates containing 100 μg of ampicillin/ml and 1% glucose. For each construct, six colonies were chosen and plated onto fresh LB agar plates with ampicillin and glucose to confirm colony-forming ability. The presence of gene constructs was confirmed by restriction analysis. All liquid cultures were supplemented with 1% glucose and 100 μg of ampicillin/ml, and the bacteria were grown in 250-ml flasks with 300-rpm orbital shaking. Initially, strains at a starting optical density at 600 nm (OD600) of 0.1 were seeded into LB medium from 3-ml overnight cultures and grown until the cultures reached an OD600 of 0.8. At this point, protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the strains were grown for 3 h at 30°C. Cells were collected by centrifugation and resuspended in either Terrific broth (TB; Difco) or M9 minimal medium (1× M9 salts [Difco], 6 nM thiamine, 1 μM MgSO4), supplemented with 1 mM IPTG and the flavanone substrate naringenin or liquiritigenin (Indofine) at a concentration of 0.05 mM. Because phenylpropanoic products are excreted from E. coli into the culture medium (18), the in vivo reaction products were analyzed by extracting the culture medium with ethyl acetate. The organic phase was evaporated in vacuo, the residues were dissolved in methanol-HCl (9:1, vol/vol) solution, and the solution was continuously mixed for 1 h and incubated at 55°C for 15 min. The solution was then evaporated, and the residue was redissolved in methanol for HPLC analysis. HPLC was performed using an Agilent 1100 series instrument and a reverse-phase ZORBAX SB-C18 column (4.6 by 150 mm). Compounds were separated by isocratic elution with methanol-0.1% formic acid in water (1:1, vol/vol) at a flow rate of 1.0 ml/min. Products were identified by matching the retention times, cochromatography patterns, and UV spectra of standard genistein and daidzein (Indofine).
RESULTS
Engineering of one-component artificial isoflavone synthase.
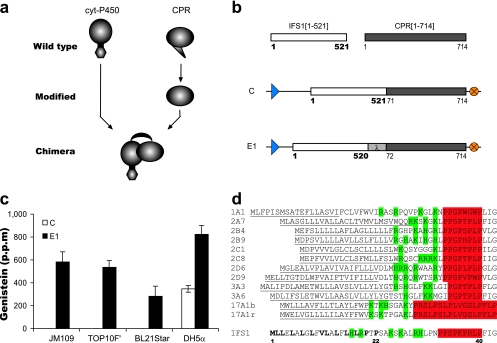
A soluble P450 enzyme from Bacillus megaterium (P450BM-3) was previously shown to contain both heme and reductase domains on a single polypeptide (self-sufficient). Because of this feature, P450BM-3 exhibits the highest turnover rate among known P450 enzymes (27). We imitated the unique architecture of the bacterial enzyme to design an artificial plant P450 (Fig. 1a). cDNAs encoding IFS1[1-521] (15) and full-length CPR, comprising amino acids 1 to 714 (CPR[1-714]), were derived from G. max and C. roseus, respectively. Catharanthus CPR was chosen because it enhances the catalysis by different plant P450 enzymes in vivo in engineered yeast strains (19). A total of 71 N-terminal residues of CPR[1-714] were deleted to create CPR[72-714] in order to reduce membrane association without compromising catalytic activities (18, 22). To engineer the one-component enzyme, first the stop codon of IFS1[1-521] was removed to create IFS1[1-520]. Next, the 3′ terminus of the modified IFS1 cDNA was adjoined at the 5′ terminus to CPR[72-714] cDNA through an artificial linker (λ) to create a translational fusion (Fig. 1b). The λ fragment, encoding a glycine-serine-threonine (GST) sequence, was chosen for use in engineering a GSTSSGSG junction that would prevent the formation of secondary structures. For comparison, we also attempted to functionalize IFS[1-521] in E. coli by CPR coexpression. To allow transcription initiation, a start codon was attached in front of the truncated CPR[72-714] cDNA to create CPR[71-714], which was fused to IFS1[1-521] in construct C (Fig. 1b). However, even though CPR was coexpressed from construct C, we anticipated that the low turnover rate of construct C would result in low-level in vivo catalysis because in order to facilitate electron shuttling, microsomal P450 enzymes rely on both electrostatic and hydrophobic forces to interact with the P450 redox partners (27). To test the functionality and in vivo catalysis, E. coli strains JM109, TOP10F′, BL21Star, and DH5α were used as whole-cell biocatalysts. The characterization of the in vivo conversion of naringenin to genistein demonstrated that construct C did not yield P450 activity in any strain except DH5α. However, the expression in DH5α yielded genistein at only 300 ppm (Fig. 1c). The one-component enzyme construct, E1, enabled genistein synthesis by all the E. coli strains (Fig. 1c). The effect of the increased turnover rate could be observed in the 2.4-fold increase in production by DH5α expressing E1 over that by the strain expressing the C construct. However, the overall in vivo catalysis by the E1 enzyme still suffered from low efficiency and resulted in isoflavonoid synthesis at levels up to only 800 ppm, suggesting nonoptimal functionality of the artificial system.
FIG. 1.
Design and functional activity of cytochrome P450 constructs. (a) General illustration of the construction of a one-component enzyme chimera from the parental cytochrome P450 (cyt-P450) and CPR proteins. (b) Construction of gene ensembles encoding a cytochrome P450, IFS1[1-521], and a CPR, CPR[1-714]. Numbers correspond to the amino acids encoded by the first and last codons of the gene sequences. All gene ensembles were flanked by a trc promoter (blue arrowhead) and an rrnB transcriptional terminator sequence (orange circle). C indicates the construct coexpressing the native full-length IFS1[1-521] and the truncated CPR[71-714] containing a start codon. The E1 one-component enzyme chimera was constructed through the formation of a translational fusion between the native IFS1[1-520] and the truncated CPR[70-714] through a linker (λ). (c) In vivo redox activity of E. coli strains expressing C and E1 upon the addition of naringenin substrate to accumulate genistein in TB medium. (d) N-terminal sequence analysis of mammalian cytochrome P450 enzymes together with IFS1 through proline-rich-region alignment. Underlined sequences represent the membrane recognition signal domain of mammalian P450 enzymes. Positively charged residues are indicated in green, and the proline-rich motif is indicated in red. For IFS1, hydrophobic amino acids ahead of the proline-rich motif are in boldface letters.
Fine-tuning the membrane recognition signal of artificial P450 enzymes.
The incompatibility of the membrane recognition signal of eukaryotic P450 leads to nonfunctionality in E. coli (4). However, unlike mammalian P450 enzymes, plant P450 enzymes have not been extensively characterized. When the N terminus of IFS1 was aligned with corresponding regions of various characterized mammalian P450 enzymes (31), several distinct regions were recognized (Fig. 1d). IFS1 starts with a domain spanning around 20 residues rich in hydrophobic amino acids. Next to this region, a domain enriched with positively charged residues, followed by a proline-rich cluster, comprises around 20 residues. Studies of mammalian P450 enzymes have demonstrated that the N-terminal hydrophobic segment serves as a membrane anchor and that the positively charged domain halts translocation into the endoplasmic reticulum membranes. Moreover, it was speculated that the proline-rich domain serves as a hinge bridging the anchor module and the cytoplasmic heme-containing domain (29). To search for one-component enzyme chimeras that displayed optimum activity in E. coli, we created several tailor-made membrane recognition signals at the N terminus.
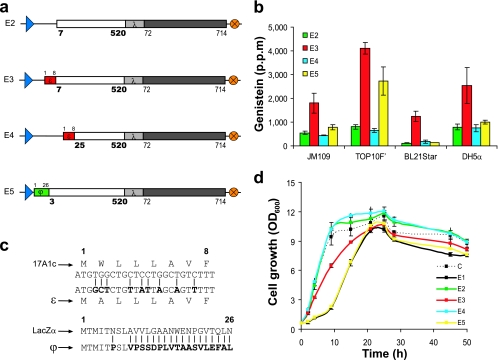
The first reconstruction of the membrane signal was performed by removing six N-terminal residues of the E1 enzyme to generate the E2 enzyme (Fig. 2a). This approach was tested because N-terminal truncations of mammalian P450 enzymes have been used previously to enhance protein solubility in E. coli for crystallization purposes (29). Our results demonstrated that all bacterial strains expressing E2 had only marginal increases in genistein production compared with those expressing E1 (Fig. 2b). Therefore, the removal of the six residues of the native membrane signal did not significantly improve catalysis by the one-component enzyme chimera.
FIG. 2.
Tuning the membrane recognition signal of the artificial one-component P450. (a) Construction of the reengineered one-component P450 enzymes (E2 to E5 enzymes) based on the E1 enzyme. (b) In vivo redox activity of E. coli expressing the fine-tuned artificial E2, E3, E4, and E5 P450 enzymes upon the addition of naringenin substrate to accumulate genistein in TB medium. (c) Synthetic sequences, ɛ and ϕ, used to replace the deleted N-terminal domain of the native IFS1. Nucleotide or amino acid substitutions in parental sequences (17A1c and LacZα) are indicated by boldface letters. (d) Growth profiles of the E. coli TOP10F′ strain expressing C, E1, E2, E3, E4, and E5 during the bioconversion of naringenin substrate in TB medium.
The expression of the bovine 17α-hydroxylase in E. coli was enabled by codon modifications in the sequence encoding the N-terminal peptide (4). To explore the feasibility of improving catalysis by the one-component plant P450 enzyme chimera by using a mammalian leader sequence (Fig. 1c), we created E3 by combining E2 with a DNA fragment encoding the eight-residue synthetic mammalian peptide ɛ (Fig. 2a). The expression of E3 improved genistein production by all strains. The highest level of production obtained was that by TOP10F′. Compared with the productivity catalyzed by E2 expression, catalysis by E3 resulted in a fivefold improvement in productivity, with genistein synthesized at 4,000 ppm (Fig. 2b). On the basis of these results, we postulated that a chimera with the further removal of the native P450 anchor, followed by replacement with ɛ, would display similar or improved in vivo activity. We created the E4 enzyme by removing 24 N-terminal residues of the E1 enzyme and replacing them with ɛ (Fig. 2a). Surprisingly, bacteria expressing E4 generated genistein at a low level similar to the levels generated by bacteria expressing E1 or E2 (Fig. 2b). This result suggests that substantial removal up to the stop signal sequence of IFS1 severely reduced the in vivo activity of the one-component enzyme chimera. We also examined the effect of the minimum removal of the native membrane module and the utility of an E. coli native gene product as a leader sequence. The E5 enzyme was generated by replacing two N-terminal residues of the E1 enzyme with 26 residues of ϕ (Fig. 2c), a modified E. coli LacZα fragment for α-complementation (Fig. 2a). The in vivo assays demonstrated that the expression of E5 resulted in the synthesis of relatively high levels of genistein (2,700 ppm) by E. coli TOP10F′ (Fig. 2b). However, the levels of improvement in genistein synthesis by all other strains were not pronounced. These data showed that by using ϕ as a leader sequence, the functionality of the one-component enzyme in E. coli could be improved, even though replacement with the bacterial sequence resulted in a range of activities in different E. coli strains. Overall, by fine-tuning modifications of the membrane signal, we identified an artificial construct (the E3 enzyme) that robustly performed in vivo catalysis in E. coli. Variation of the membrane recognition signal also corresponded to variability in growth kinetics of the E. coli biocatalysts (Fig. 2d). E. coli bacteria expressing one-component enzyme chimeras with short membrane signals (E2 and E4) or expressing the nonfunctional construct C initially exhibited a high exponential-growth rate (∼1.1 h−1). However, the expression of the one-component enzyme constructs that still contained the native plant membrane signal (E2 and E5) resulted in a low initial growth rate (∼0.2 h−1). Furthermore, the initial growth rate of TOP10F′ expressing the most robust chimera, E3, was intermediate and was calculated to be ∼0.7 h−1.
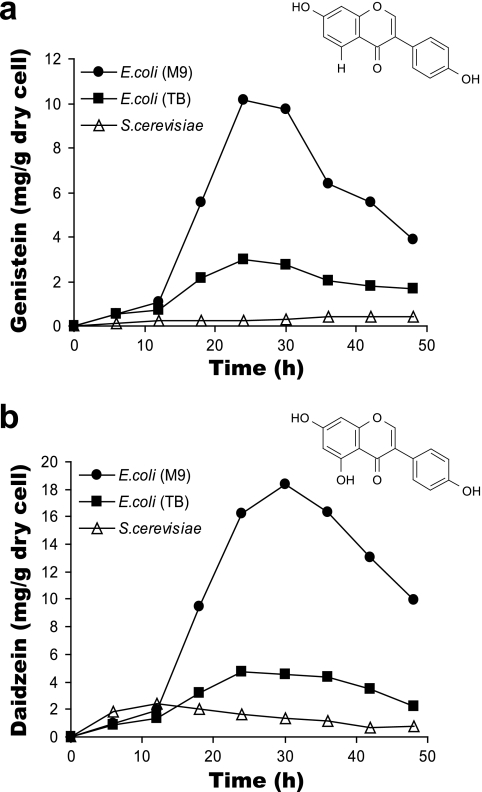
The productivity of E. coli TOP10F′ expressing the highly active chimera (E3) was compared with that of the yeast expressing the native IFS1 together with CPR. Monitoring production levels from the E. coli biocatalysts cultured in minimal and rich media showed that higher levels of isoflavone synthesis were achieved in minimal-medium cultures, even though biomass generation was reduced (Fig. 3). The recombinant E. coli bacteria also generated significantly larger amounts of genistein and daidzein than the recombinant yeast (Fig. 3). This result demonstrates that the engineered P450 machinery resulted in a higher level of in vivo isoflavone synthesis by E. coli than by a eukaryotic host, such as yeast. This difference is more pronounced when one takes into account the fact that the yeast biomass was ∼5-fold greater (OD600 of ∼15) than that of E. coli grown in minimal medium. The maximum amounts of genistein and daidzein produced by the recombinant E. coli strain were ∼10 and ∼18 mg/g (dry weight) of cells (∼5,000 and ∼10,000 ppm), respectively, in 24 h. The highest levels of genistein and daidzein synthesized by yeast were ∼0.5 and ∼2 mg/g (dry weight) of cells (∼1,000 and ∼2,000 ppm). Therefore, catalysis by the artificial P450 resulted in an increase of up to ∼5-fold in the volume of production over that catalyzed by the native P450 expressed in yeast. On the basis of specific biomass production, this value correlates to a ∼20-fold increase over the production catalyzed by the wild-type enzyme.
FIG. 3.
Comparison of isoflavone production profiles of recombinant E. coli TOP10F′ expressing the artificial E3 P450 enzyme and S. cerevisiae expressing native IFS1 and CPR. (a) Bioconversion of naringenin for genistein accumulation. (b) Bioconversion of liquiritigenin for daidzein accumulation. Recombinant E. coli was cultured in M9 minimal salt medium or TB medium. Recombinant S. cerevisiae was cultured in synthetic complete minimal yeast medium.
DISCUSSION
Since P450 enzymes are powerful catalysts, attempts have recently been made to engineer bacterial P450 enzymes in order to attain improved functions for potential industrial applications (14, 24). Plant-derived P450 enzymes are indispensable and integral in the synthesis of drugs and functional chemicals. However, the utilization of plant P450 enzymes to synthesize plant pharmaceuticals by means of biotechnology has been prevented because currently, methods to isolate P450 enzymes from plants result in low yields and the loss of enzyme activities. Heterologous expression of plant P450 enzymes by using rapidly growing industrial bacteria such as E. coli may present a viable solution to this dilemma. However, the nonfunctionality of plant P450 enzymes in E. coli presents a rate-limiting step in the recombinant approach.
We initially attempted to functionalize the plant P450 isoflavone synthase in E. coli by coexpression with a plant P450 reductase in order to compensate for the lack of P450 redox partner proteins in E. coli. However, this expression system generated only low levels of isoflavonoid in DH5α and no isoflavonoid in the other common E. coli strains tested. The low turnover rate of the wild-type P450 was improved by the construction of a one-component artificial enzyme that mimics the architecture of the bacterial P450BM-3, an enzyme that exhibits the highest turnover rate of any known P450 enzyme. The increased turnover rate of the artificial enzyme was indicated by the elevated synthesis of isoflavone, which demonstrated the importance of domain interaction between the cytochrome P450 and its redox partner protein.
The other primary challenge of plant P450 expression in E. coli is the incompatibility of the membrane recognition signal. In order to explore the feasibility of further improving the in vivo redox activity of the artificial enzyme, the sequence of the N-terminal domain of the P450 was fine-tuned by splicing at different locations and inserting modified sequences. The fine-tuning strategy resulted in the identification of a chimeric P450 that was better able than the wild-type enzyme expressed in yeast to synthesize plant-derived isoflavones. The expression of the most prominent chimera in bacteria catalyzed the production of higher levels of P450 products than the expression of the wild-type enzyme in yeast, which is a widely used eukaryotic expression host of membrane-bound P450 enzymes. Moreover, the bacterial synthesis also compared well with that by soy plants engineered to improve isoflavone contents (∼1 and ∼3 mg/g of seed weight for genistein and daidzein, respectively [20, 32]). Therefore, not only could the artificial enzyme design be used to increase the availability of plant estrogen pharmaceuticals, but it also could potentially be applied across a diverse range of plant P450 enzymes to enable high-level synthesis of other scarce plant chemicals by industrial microbes such as E. coli.
From the perspective of biosynthetic engineering, the robust expression of the chimeric P450 allows the reconstitution of complete plant biosynthetic pathways (5) in E. coli strains that contain membrane-bound P450 enzymes. This platform not only presents a new strategy to synthesize natural pharmaceuticals but can also be applied to mutasynthesis strategies for novel chemical structures (9).
Acknowledgments
We acknowledge financial support of this work by the U.S. National Science Foundation (BES-0331404). E. Leonard acknowledges the New York State Professional Development Award and Mark Diamond Research Funds (SUM-05-14).
We thank Birger Lindberg Møller for useful discussions.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Akashi, T., T. Aoki, and S. Ayabe. 1999. Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 121:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper, H., K. Miyaoku, and G. Stephanopoulos. 2005. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23:612-616. [DOI] [PubMed] [Google Scholar]
- 3.Altaras, N. E., and D. C. Cameron. 1999. Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli. Appl. Environ. Microbiol. 65:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, H. J., M. P. Arlotto, and M. R. Waterman. 1991. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. USA 88:5597-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbulis, I. E., and B. Winkel-Shirley. 1999. Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA 96:12929-12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, O. A., R. J. Peters, and R. Croteau. 2003. Monoterpene biosynthesis pathway construction in Escherichia coli. Phytochemistry 64:425-433. [DOI] [PubMed] [Google Scholar]
- 7.Causey, T. B., S. Zhou, K. T. Shanmugam, and L. O. Ingram. 2003. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: homoacetate production. Proc. Natl. Acad. Sci. USA 100:825-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, M. C., and J. D. Keasling. 2006. Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2:674-681. [DOI] [PubMed] [Google Scholar]
- 9.Chemler, J. A., Y. Yan, E. Leonard, and M. A. Koffas. 2007. Combinatorial mutasynthesis of flavonoid analogues from acrylic acids in microorganisms. Org. Lett. 9:1855-1858. [DOI] [PubMed] [Google Scholar]
- 10.Draths, K. M., D. R. Knop, and J. W. Frost. 1999. Shikimic acid and quinic acid: replacing isolation from plant sources with recombinant microbial biocatalysis. J. Am. Chem. Soc. 121:1603-1604. [Google Scholar]
- 11.Halkier, B. A., H. L. Nielsen, B. Koch, and B. L. Moller. 1995. Purification and characterization of recombinant cytochrome P450TYR expressed at high levels in Escherichia coli. Arch. Biochem. Biophys. 322:369-377. [DOI] [PubMed] [Google Scholar]
- 12.Hotze, M., G. Schroder, and J. Schroder. 1995. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett. 374:345-350. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Q., C. A. Roessner, R. Croteau, and A. I. Scott. 2001. Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg. Med. Chem. 9:2237-2242. [DOI] [PubMed] [Google Scholar]
- 14.Joo, H., Z. Lin, and F. H. Arnold. 1999. Laboratory evolution of peroxide-mediated cytochrome P450 hydroxylation. Nature 399:670-673. [DOI] [PubMed] [Google Scholar]
- 15.Jung, W., O. Yu, S. M. Lau, D. P. O'Keefe, J. Odell, G. Fader, and B. McGonigle. 2000. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18:208-212. [DOI] [PubMed] [Google Scholar]
- 16.Koepp, A. E., M. Hezari, J. Zajicek, B. S. Vogel, R. E. LaFever, N. G. Lewis, and R. Croteau. 1995. Cyclization of geranylgeranyl diphosphate to taxa-4(5),11(12)-diene is the committed step of taxol biosynthesis in Pacific yew. J. Biol. Chem. 270:8686-8690. [DOI] [PubMed] [Google Scholar]
- 17.Leonard, E., K. H. Lim, P. N. Saw, and M. A. Koffas. 2007. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl. Environ. Microbiol. 73:3877-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard, E., Y. Yan, and M. A. Koffas. 2006. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab. Eng. 8:172-181. [DOI] [PubMed] [Google Scholar]
- 19.Leonard, E., Y. Yan, K. H. Lim, and M. A. Koffas. 2005. Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:8241-8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, C. J., J. W. Blount, C. L. Steele, and R. A. Dixon. 2002. Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc. Natl. Acad. Sci. USA 99:14578-14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, V. J. J., D. J. Pitera, S. T. Withers, J. D. Newman, and J. D. Keasling. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796-802. [DOI] [PubMed] [Google Scholar]
- 22.Miura, Y., and A. J. Fulco. 1974. (ω-2) hydroxylation of fatty acids by a soluble system from Bacillus megaterium. J. Biol. Chem. 249:1880-1888. [PubMed] [Google Scholar]
- 23.Nielsen, J. S., and B. L. Moller. 2000. Cloning and expression of cytochrome P450 enzymes catalyzing the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of cyanogenic glucosides in Triglochin maritima. Plant Physiol. 122:1311-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otey, C. R., M. Landwehr, J. B. Endelman, K. Hiraga, J. D. Bloom, and F. H. Arnold. 2006. Structure-guided recombination creates an artificial family of cytochromes P450. PLoS Biol. 4:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paterson, I., and E. A. Anderson. 2005. Chemistry. The renaissance of natural products as drug candidates. Science 310:451-453. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer, B. A., S. J. Admiraal, H. Gramajo, D. E. Cane, and C. Khosla. 2001. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790-1792. [DOI] [PubMed] [Google Scholar]
- 27.Sevrioukova, I. F., H. Li, H. Zhang, J. A. Peterson, and T. L. Poulos. 1999. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. USA 96:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steele, C. L., M. Gijzen, D. Qutob, and R. A. Dixon. 1999. Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch. Biochem. Biophys. 367:146-150. [DOI] [PubMed] [Google Scholar]
- 29.Williams, P. A., J. Cosme, V. Sridhar, E. F. Johnson, and D. E. McRee. 2000. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell 5:121-131. [DOI] [PubMed] [Google Scholar]
- 30.Williams, P. A., J. Cosme, V. Sridhar, E. F. Johnson, and D. E. McRee. 2000. Microsomal cytochrome P450 2C5: comparison to microbial P450s and unique features. J. Inorg. Biochem. 81:183-190. [DOI] [PubMed] [Google Scholar]
- 31.Yamazaki, S., K. Sato, K. Suhara, M. Sakaguchi, K. Mihara, and T. Omura. 1993. Importance of the proline-rich region following signal-anchor sequence in the formation of correct conformation of microsomal cytochrome P-450s. J. Biochem. (Tokyo) 114:652-657. [DOI] [PubMed] [Google Scholar]
- 32.Yu, O., J. Shi, A. O. Hession, C. A. Maxwell, B. McGonigle, and J. T. Odell. 2003. Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63:753-763. [DOI] [PubMed] [Google Scholar]