Abstract
High-density whole-genome cDNA microarrays were used to investigate substrate-dependent gene expression of Methylibium petroleiphilum PM1, one of the best-characterized aerobic methyl tert-butyl ether (MTBE)-degrading bacteria. Differential gene expression profiling was conducted with PM1 grown on MTBE and ethanol as sole carbon sources. Based on microarray high scores and protein similarity analysis, an MTBE regulon located on the megaplasmid was identified for further investigation. Putative functions for enzymes encoded in this regulon are described with relevance to the predicted MTBE degradation pathway. A new unique dioxygenase enzyme system that carries out the hydroxylation of tert-butyl alcohol to 2-methyl-2-hydroxy-1-propanol in M. petroleiphilum PM1 was discovered. Hypotheses regarding the acquisition and evolution of MTBE genes as well as the involvement of IS elements in these complex processes were formulated. The pathways for toluene, phenol, and alkane oxidation via toluene monooxygenase, phenol hydroxylase, and propane monooxygenase, respectively, were upregulated in MTBE-grown cells compared to ethanol-grown cells. Four out of nine putative cyclohexanone monooxygenases were also upregulated in MTBE-grown cells. The expression data allowed prediction of several hitherto-unknown enzymes of the upper MTBE degradation pathway in M. petroleiphilum PM1 and aided our understanding of the regulation of metabolic processes that may occur in response to pollutant mixtures and perturbations in the environment.
Petroleum releases are among the most ubiquitous sources of composite organic contaminants in groundwater. The majority of petroleum-associated contaminants reach aquifers via spills or leaks from underground storage tanks at service stations (49). Over 300,000 releases from underground storage tanks have been confirmed, with more than 150,000 remediation efforts completed in the United States (32). Fuel oxygenates, such as methyl tertiary butyl ether (MTBE), often form extensive, unattenuated “plumes” in groundwater because of their high water solubility and low biodegradation rates under oxygen-limited conditions (24, 28, 34). MTBE was one of the major oxygenates incorporated into reformulated gasoline to increase the fuel's oxygen content and decrease carbon monoxide and ozone emissions. MTBE and its primary metabolite tert-butyl alcohol (TBA) are suspected and known carcinogens, respectively (1, 7, 31, 57). Recently, alternative oxygenates, such as ethanol, have been substituted for MTBE, but because of the very slow depletion of contaminant mass from spill areas under anoxic conditions, the impacts of MTBE on the subsurface will be felt for many years and likely decades to come (8, 29).
Methylibium petroleiphilum PM1 is one of the best-characterized aerobic MTBE degraders known to date, and PM1-like bacteria have been shown to be present in several MTBE-contaminated aquifers in California (19, 20, 25) and Europe (12, 33, 42). M. petroleiphilum PM1 uses MTBE as a sole carbon source, oxidizing it completely to CO2 without accumulation of TBA (16). Strain PM1 has been used successfully in two bioaugmentation field trials in gasoline-contaminated aquifers in California (43) and Montana (9). M. petroleiphilum PM1 has a broad range of novel metabolic capabilities, including heterotrophic growth under aerobic conditions on diverse carbon sources (ethanol, methanol, toluene, benzene, ethybenzene, phenol, and C4 to C12 n-alkanes (10, 37; K. R. Hristova, unpublished data). Recently, whole-genome sequencing of PM1 was conducted and revealed a ∼4-Mb circular chromosome and a ∼600-kb megaplasmid (26). Multiple operons on the chromosome were shown to code for aromatic hydrocarbon and alkane degradation, metal resistance, and methylotrophy. The megaplasmid putatively codes for important functions, including alkane degradation, and through plasmid curing experiments was shown to play an essential role in MTBE degradation (26). Impacts of interactions of MTBE and BTEX compounds (benzene, toluene, ethylbenzene, and xylenes) on biodegradation capabilities of PM1 cultures have shown inhibition of MTBE degradation in the presence of certain BTEX compounds (10, 27). However, the underlying biochemistry and complex regulation of the different pathways involved in biodegradation of these gasoline mixtures remain unknown.
To date, limited information has been available about the genetics of MTBE biodegradation. A novel ether cleavage reaction has been described as the first step in MTBE oxidation for cometabolic MTBE-degrading bacteria (23, 44, 46, 50); whether MTBE-metabolizing bacteria use a similar reaction is not yet known. Currently, there is no genetic information available concerning the identity and function of enzymes involved in MTBE and TBA oxidation in MTBE-metabolizing bacteria. However, recent studies elucidated the enzymes responsible for degradation of the MTBE metabolites 2-methyl-2-hydroxy-1-propanol (or 2-methyl-1,2-propanediol) and hydroxyisobutyraldehyde in Mycobacterium austroafricanum IFP2012 (15) and 2-hydroxyisobutyric acid (HIBA) in an environmental isolate phylogenetically similar to PM1 (42).
In this study, the M. petroleiphilum PM1 global transcriptome response in the presence of MTBE and the potential physiological stress brought about by this pollutant were evaluated for the first time. High-density oligonucleotide arrays were employed to explore the genes involved in MTBE biodegradation and to compare gene expression profiles for ethanol and MTBE as growth substrates. Results revealed links between metabolism of MTBE and metabolism of other aromatic compounds present in gasoline mixtures.
MATERIALS AND METHODS
Bacterial strain and genome sequence.
Methylibium petroleiphilum strain PM1 is a methylotroph capable of using MTBE as a sole carbon and energy source. The finished sequence of the whole genome of strain PM1 was made available though a collaborative sequencing effort between the University of California, Davis, Lawrence Livermore National Laboratory (LLNL), and the Joint Genome Institute (Walnut Creek, CA). At the time this study was initiated, a draft genome sequence of ∼8× coverage consisting of 33 contigs was available. The annotation of this draft sequence, in collaboration with Oak Ridge National Laboratory, resulted in 4,006 putative coding sequences (CDSs) that defined the genome. With completion of the genome, the number of CDSs increased to 4,479, indicating that, at the time of this expression study, our available sequence information covered nearly 90% of the genome. We are currently undertaking a follow-up study to compare gene expression of PM1 grown on MTBE, TBA, and pyruvate, utilizing the complete PM1 genome. The whole genome sequence of M. petroleiphilum PM1 is available through National Center for Biotechnology Information (NCBI), GenBank accession numbers NC_008825 for the chromosome and NC_008826 for the plasmid.
Media and growth conditions.
M. petroleiphilum PM1 was grown in liquid mineral salts medium (Tris-HCl, 0.13 M; KH2PO4, 0.023 M; K2HPO4, 0.025 M; CaCl2, 0.027 M; NaHCO3, 0.2 M; MgSO4, 0.05 M; EDTA, 0.0288 mM; and NH4Cl, 0.27 M) supplemented with trace elements (CoCl2, 0.25 μM; CuSO4, 0.3 μM; FeCl3, 40 μM; H3BO3, 50 μM; MnCl2, 10 μM; Na2MoO4, 0.1 μM; ZnSO4, 0.8 μM) and either MTBE (250 mg/liter) or ethanol (790 mg/liter) as the sole carbon source. PM1 is capable of growth on mineral salts medium with up to 1,000 mg/liter MTBE or up to 7.9 g/liter ethanol. The dimensionless Henry's constant for MTBE, 0.023, was used to calculate its solution-phase concentration. Cells were grown at 28°C in 50-ml batch cultures in 150-ml glass bottles with rotary shaking at 150 rpm. At the start of the experiment, bottles were inoculated with ∼5 ml of PM1 culture (grown in the presence of the corresponding carbon source) to achieve an optical density at 595 nm of ∼0.02. Cells from three biological replicates were harvested at mid-exponential phase after 48 h of incubation. Final optical density at 595 nm values for the ethanol- and MTBE-grown cultures were 0.6 and 0.3, respectively. Before RNA extraction, cell densities were adjusted to correspond to 5.9 × 108 and 2.5 × 108 CFU/ml for ethanol and MTBE cultures, respectively. At the time of harvesting, approximately 50% of the substrate was utilized.
RNA extraction.
Aliquots of 30 ml from liquid cultures were treated with RNAprotect to stabilize RNA (Qiagen, Valencia, CA) in a ratio of 1 part culture to 1.6 parts reagent as outlined by the manufacturer. RNA was subsequently extracted from the cells using a Gentra Purescript RNA isolation kit (Gentra Systems, MN) according to the manufacturer's protocol. A DNase treatment step was included after the RNA extraction, in which DNase I (Roche Inc., Basel, Switzerland) was added to tubes (3 U/10 μg RNA) and incubated for 30 min at 37°C, followed by enzyme inactivation at 95°C for 5 min. RNA extracts were purified with an RNeasy Mini kit and RNase-free DNase (Qiagen) according to the manufacturer's protocols. RNA was finally eluted with 50 μl RNase-free water and stored at −80°C until cDNA synthesis. Aliquots were analyzed with a Bioanalyzer (Agilent, Santa Clara, CA), which indicated minimal degradation and concentrations ranging from 409 to 620 μg/ml and A260/A280 ratios ranging from 1.8 to 2.1.
Preparation of labeled cDNA.
cDNA production and labeling were performed by NimbleGen Systems, Inc. (Madison, WI). After thawing RNA samples on ice, 10 μg total RNA was used to perform cDNA synthesis with random hexamers and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). RNases A and H were then used to digest the RNA. The resulting single-stranded cDNA was purified by phenol extraction and precipitated after adding 10 μg glycogen (as carrier), 0.1 volume of ammonium acetate, and 2.5 volumes of 100% ethanol. The resulting pellet was dried and suspended in 30 μl water, and the cDNA yield was measured by UV/visible spectrophotometry at 260 nm. The cDNA was partially digested with DNase I (0.2 U) at 37°C for approximately 13 min, generating 50- to 200-base fragments as observed with a Bioanalyzer (Agilent). The fragmented cDNA was end labeled with biotin-N6-ddATP and terminal deoxynucleotidyl transferase (51 U) during incubation for 2 h at 37°C. The labeled product was concentrated to a 20-μl final volume using a Microcon YM-10 10,000 molecular weight cutoff filter device (Millipore, Billerica, MA) and stored at −20°C before hybridization.
Microarray design and synthesis.
Maskless, light-directed digital micromirror technology (38) was used to fabricate high-density 60-mer oligonucleotide microarrays at NimbleGen Systems, Inc. For designing oligonucleotide probes, a database of the gene sequences (CDSs) of the M. petroleiphilum PM1 genome (4,006 CDSs on 17 June 2004) was created, and a file of all possible 60-mers was generated using NimbleGen's design criteria. For each CDS, two to nine 60-base oligonucleotides (probes) were selected based on CDS length, such that each probe was at least three mismatches different than all other probes chosen. Probe sets were replicated in triplicate (representing technical replicates) on each chip. A total of 27,704 probes were designed for the genome, and these probes were randomized into a four-to-nine design on the chip (four spots with the same oligonucleotide surrounded by blank spots) to enhance sensitivity. A quality control hybridization using on-chip control oligonucleotides was performed for each array prior to hybridization with labeled cDNA from PM1.
Microarray hybridization.
The NimbleGen Systems, Inc., Hybriwheel technology was used to perform array hybridization. Briefly, arrays were prehybridized at 45°C in 50 mM 4-morpholineethanesulfonic acid buffer with 500 mM NaCl, 10 mM EDTA, and 0.005% Tween 20. Herring sperm DNA was added at 0.1 mg/ml to prevent nonspecific binding. After 15 min of prehybridization, 4 μg of labeled cDNA in hybridization buffer was added to arrays followed by incubation for 16 to 20 h at 45°C. Free probe was removed by conducting several wash steps, progressing from less to more stringent conditions. Bound probe was detected with Cy3-labeled streptavidin, with signal amplification achieved by adding biotinylated antistreptavidin goat antibody.
Data normalization and gene expression analysis.
For each experimental condition (MTBE or ethanol growth conditions), there were nine data points for each probe, representing data for three technical replicates of the entire probe set for each of three biological replicates. The arrays were analyzed using an Axon GenePix 4000B scanner (Molecular Devices Corp., Sunnyvale, CA). ImageJ software (http://rsb.info.nih.gov/ij/) was used to rotate images and double their size without interpolation. Features were extracted using GenePix 3.0 software, with a fixed feature size. The log-transformed signal (base 2) was used as the input data for analysis.
Statistical analysis.
Data analysis was performed using the R statistical package and tools available from the Bioconductor project (http://www.bioconductor.org). Data were quantile normalized (4) and background corrected and summarized using the robust multiarray average method (22). A linear model was fitted for each gene to estimate log ratios between multiple target RNA samples simultaneously by using the LIMMA package (48). The standard errors of the estimated log fold changes were moderated using empirical Bayes methods implemented in the LIMMA package, generating a moderated t statistic. P values were obtained from this moderated t statistic, after adjusting for multiple hypothesis testing using Benjamini and Hochberg's method to control the false discovery rate (14). We defined significant up- or downregulation as an expression change of ≥2-fold and P value of <0.05 (actual P values for this group were <0.01). In the text, expression values for genes in the proposed MTBE degradation pathway that fall below the twofold cutoff are indicated with an asterisk. Annotation of the significantly differentially expressed genes was derived from the Cluster of Orthologous Genes annotation for the PM1 genome. Studies of the gene ortholog neighborhood were done using the Integrated Microbial Genome database (Joint Genome Institute).
Reverse transcription-quantitative PCR analysis.
Confirmation of transcript levels for modulated genes was performed by reverse transcription-quantitative PCR (RT-qPCR) analysis of RNA samples extracted from ethanol- and MTBE-grown cultures (three replicates each). Sufficient RNA was not available from extracts used in microarray experiments, since the cDNA labeling reactions had to be repeated one or more times due to insufficient yield. Therefore, separate cultures were grown under the same conditions and extracted for RNA using the same method as described above. Total RNA (∼300 to 1,500 ng) was converted to cDNA using random hexamers and MultiScribe reverse transcriptase (Applied Biosystems, Foster City, CA). The resulting cDNA was amplified using an IQ SYBR Green RT-PCR kit (Bio-Rad, Hercules, CA) with gene-specific primers for 18 different CDSs on a MyIQ single-color real-time PCR cycler (Bio-Rad). Primers were designed using Primer Express software (Applied Biosystems) and screened for uniqueness by BLAST analysis with the PM1 genome. Calibration curves were performed with genomic DNA serially diluted over a range of 5 to 6 orders of magnitude. The PCR conditions were optimized as follows: 95°C for 5 min, and 40 cycles of 94°C for 15 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. The primers are listed in Table S1 in the supplemental material. The RNA transcript amount was normalized to the total amount of starting RNA quantified using a Bioanalyzer (Agilent). The log2 fold difference for specific transcript levels between MTBE and ethanol growth conditions was compared for RT-qPCR and microarray analyses.
Sequence analyses and generation of phylogenetic trees.
Homologs of M. petroleiphilum PM1 translated CDSs were identified using BLASTP searches against the nonredundant (nr) GenBank and SwissProt databases from NCBI. ClustalW sequence alignments as well as Protdist and neighbor-joining tree generation were carried out using the Bioedit 7.0.0 package (19).
Microarray data accession number.
Microarray data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/) under accession number GSE9381.
RESULTS AND DISCUSSION
Differential expression of genes in M. petroleiphilum PM1 grown on MTBE.
In response to growth on MTBE, 1,255 genes of the 3,941 genes represented on the arrays were differentially expressed, with 440 genes more than twofold upregulated and 815 genes more than twofold downregulated in comparison to growth on ethanol. Importantly, for genes with unknown function whose expression was altered during exposure to MTBE, our analyses identified a large number that were upregulated (172 of 440, or 39%) or downregulated (119 of 815, or 15%). In the PM1 genome, 1,559 of the 4,479 genes (35%) have not been assigned a function. The low percentage of downregulated unknowns (15%) indicates that most of the suppressed genes are known. In contrast, the high percentage of upregulated unknowns (39%) suggests that few of the genes associated with MTBE degradation have been studied to date.
MTBE pathway: gene expression.
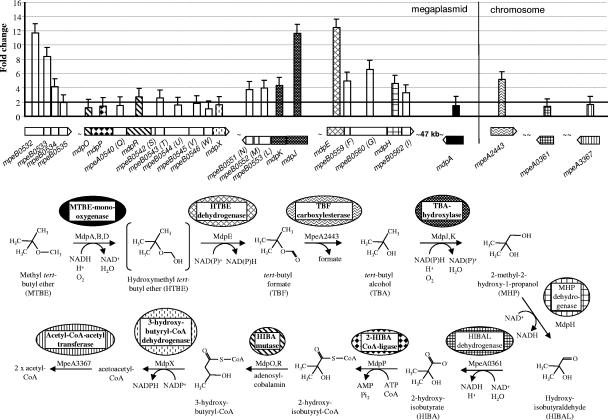
Using two independent approaches, comparative genomic hybridization and plasmid curing, we previously demonstrated that critical MTBE/TBA degradation genes are located on the PM1 plasmid (26). In this study, by comparing the whole transcriptome response of MTBE-grown and ethanol-grown cells, a large MTBE degradation regulon consisting of four major gene clusters was identified on the plasmid (Fig. 1). Genes in these clusters were designated mdp for the MTBE degradation pathway.
FIG. 1.
Gene expression levels of the proposed MTBE degradation regulon and proposed MTBE degradation pathway in M. petroleiphilum PM1. Error bars represent 1 standard deviation. Genes within the proposed MTBE regulon whose function in the pathway has not been ascertained were left in the Mpe_BNNNN (shown as mpeBNNNN) nomenclature. Their possible positions within the mdpX nomenclature are indicated by a letter in brackets.
The relative gene expression levels in three of these clusters ranged from 2.0- to 12.4-fold. Within two clusters, Mpe_B0555 through Mpe_B0551 and Mpe_B0558 through Mpe_B0562 (upregulated 3.3- to 12-fold on MTBE), a putative iron-sulfur oxidoreductase belonging to the family of ferredoxin reductases, a hydroxylase similar to phthalate dioxygenase, and two dehydrogenase genes, mdpE (Mpe_B0558) and mdpH (Mpe_B0561), were identified.
The predicted MTBE monooxygenase gene mdpA (Mpe_B0606), 69% and 66% identical to the alkane monooxygenase AlkB of Alcanivorax borkumensis AP1 (47) and Pseudomonas putida GPo1 (carried on the OCT plasmid) (54), respectively (26), was not differentially expressed in MTBE-grown cells as determined by the microarray results (Table 1). The evidence of previous physiology studies of strain PM1, showing that two different oxygen-dependent enzymes were involved in MTBE and TBA oxidation (11; K. Hristova, unpublished data) and that an mdpA insertion mutant could not degrade MTBE to hydroxymethyl tert-butyl ether (R. Schmidt, unpublished data) strongly suggests that MdpA is the MTBE monooxygenase (Fig. 1). In addition, mdpA was 4.7-fold upregulated on ethanol-grown cells exposed to MTBE for 4 hours relative to ethanol-grown cells (microarray data not shown). This suggests that mdpA may be highly expressed early in response to the presence of MTBE. Studies with cloned alkB of Pseudomonas oleovorans showed that high expression led to physiological changes, slow growth, and eventual loss of alk+ activity (5). Our results with mdpA may reflect complex regulation of this gene in PM1. Genes for the rubredoxin (mdpB), the rubredoxin reductase (mdpD), and predicted transcriptional regulator (mdpC) were not present in the microarray. As these genes are not arranged in an operon in PM1, they may not decisively contribute to the elucidation of the MTBE pathway. For example, the rubredoxin and rubredoxin reductase genes of P. putida RR1 and Acinetobacter sp. strain M-1 are expressed constitutively (30, 52). Further studies of gene expression in response to growth on MTBE and TBA by PM1, currently under way in our laboratories, will clarify the expression profiles of these genes.
TABLE 1.
Genes involved in degradation of gasoline and aromatic compounds in M. petroleiphilum PM1
| Pathway and locus | Strand | Fold change | P value | Gene name | Predicted function |
|---|---|---|---|---|---|
| MTBE degradation | |||||
| Mpe_B0606 | − | 1.5 | 1.4E-01 | mdpA | MTBE monooxygenase |
| Mpe_B0602 | − | NDa | ND | mdpB | Rubredoxin |
| Mpe_B0597 | − | ND | ND | mdpD | Rubredoxin reductase |
| Mpe_B0601 | − | ND | ND | mdpC | ATP-dependent transcriptional regulator |
| Mpe_B0558 | + | 12.4 | 2.3E-14 | mdpE | Hydroxymethyl tert-butyl ether dehydrogenase |
| Mpe_A2443 | + | 5.2 | 2.5E-12 | tert-Butyl formate carboxylesterase | |
| Mpe_B0555 | − | 11.6 | 6.2E-11 | mdpJ | tert-Butyl alcohol hydroxylase |
| Mpe_B0554 | − | 4.3 | 1.5E-11 | mdpK | Iron-sulfur oxidoreductase |
| Mpe_B0561 | + | 4.6 | 3.9E-10 | mdpH | 2-Methyl-2-hydroxy-1-propanol dehydrogenase |
| Mpe_A0361 | − | 1.4 | 1.7E-05 | Hydroxyisobutyraldehyde dehydrogenase | |
| Mpe_B0539 | + | 1.4 | 3.4E-02 | mdpP | 2-Hydroxy-isobutyryl-CoA ligase |
| Mpe_B0541 | + | 2.7 | 2.6E-05 | mdpR | 2-Hydroxy-isobutyryl-CoA mutase |
| Mpe_B0538 | + | 1.2 | 3.3E-01 | mdpO | 2-Hydroxy-isobutyryl-CoA mutase C-terminal domain |
| Mpe_B0547 | + | 1.7 | 4.2E-04 | mdpX | 3-Hydroxybutyryl-CoA dehydrogeanse |
| Mpe_A3367 | − | 1.6 | 9.6E-05 | Acetyl-CoA acetyltransferase | |
| Phenol degradation (dmp1) | |||||
| Mpe_A2265 | − | 2.2 | 7.8E-06 | dmpI | 4-Oxalocrotonate isomerase |
| Mpe_A2266 | − | 1.5 | 3.3E-05 | dmpG | 4-Hydroxy-2-isovalerate aldolase |
| Mpe_A2267 | − | ND | ND | dmpF | Acetaldehyde dehydrogenase (acylating) |
| Mpe_A2272 | − | 1.2 | 3.3E-03 | dmpH | 4-Oxalocrotonate decarboxylase |
| Mpe_A2273 | − | 2.9 | 9.8E-09 | dmpE | 2-Hydroxypent-2,4-dienoate hydratase |
| Mpe_A2274 | − | 2.1 | 1.2E-05 | dmpD | 2-Hydroxymuconic semialdehyde hydrolase |
| Mpe_A2275 | − | 1.4 | 6.3E-06 | dmpC | 2-Hydroxymuconic semialdehyde dehydrogenase |
| Mpe_A2276 | − | 2.3 | 7.4E-09 | aphY | Conserved in some dmp-like operons |
| Mpe_A2277 | − | 2.0 | 1.2E-07 | dmpB | Catechol-2,3-dioxygenase |
| Mpe_A2278 | − | 1.8 | 1.2E-05 | dmpQ | Catechol-2,3-dioxygenase ferredoxin |
| Mpe_A2279 | + | 2.7 | 3.4E-10 | aphT | Regulator (LysR family) |
| Mpe_A2280 | − | 1.3 | 2.2E-05 | dmpP | Phenol hydrolase reductase |
| Mpe_A2281 | − | 3.1 | 3.0E-08 | dmpO | Phenol hydrolase gamma subunit |
| Mpe_A2282 | − | 2.5 | 7.3E-08 | dmpN | Phenol hydrolase alpha subunit |
| Mpe_A2283 | − | 1.3 | 1.7E-03 | dmpM | Phenol hydrolase activator |
| Mpe_A2284 | − | 1.7 | 2.5E-04 | dmpL | Phenol hydrolase beta subunit |
| Mpe_A2285 | − | 2.6 | 1.7E-06 | dmoK | Phenol hydrolase assembly |
| Mpe_A2286 | − | 1.3 | 3.7E-03 | dmpR | Regulator (HylR family) |
| Propane monooxygenase | |||||
| Mpe_A0950 | + | 1.7 | 2.2E-09 | prmA | Propane monooxygenase hydroxylase large subunit |
| Mpe_A0951 | + | 4.5 | 5.4E-15 | prmB | Propane monooxygenase reductase |
| Mpe_A0952 | + | 1.2 | 6.1E-03 | prmC | Propane monooxygenase hydroxylase small subunit |
| Mpe_A0953 | + | 1.4 | 1.7E-06 | prmD | Propane monooxygenase coupling protein |
| Cyclohexanone monooxygenases | |||||
| Mpe_B0579 | − | 1.1 | 7.0E-01 | Cyclohexanone monooxygenase | |
| Mpe_B0607 | + | −1.6 | 3.7E-07 | Cyclohexanone monooxygenase | |
| Mpe_B0610 | + | 3.1 | 3.9E-09 | Cyclohexanone monooxygenase | |
| Mpe_A0393 | + | 3.0 | 5.4E-11 | Cyclohexanone monooxygenase | |
| Mpe_A0898 | + | 1.9 | 3.3E-06 | Cyclohexanone monooxygenase | |
| Mpe_A1038 | + | 1.5 | 5.3E-06 | Cyclohexanone monooxygenase | |
| Mpe_A1351 | + | 5.7 | 2.6E-14 | Cyclohexanone monooxygenase | |
| Mpe_A2885 | + | 1.1 | 5.1E-01 | Cyclohexanone monooxygenase | |
| Mpe_A2915 | − | 1.8 | 2.3E-08 | Cyclohexanone monooxygenase | |
| Other degradation | |||||
| Mpe_A0819 | + | 2.9 | 1.2E-12 | 2-Polyprenylphenol hydroxylase | |
| Mpe_A0986 | + | 1.7 | 2.7E-08 | Phenylacetate-CoA ligase | |
| Mpe_A0987 | + | 3.0 | 8.4E-13 | Phenylacetic acid degradation protein | |
| Mpe_A0989 | + | 1.8 | 4.2E-08 | Phenylacetic acid degradation protein | |
| Mpe_A1001 | + | 2.3 | 1.9E-12 | Vanillate O-demethylase oxygenase subunit A |
ND, not determined.
We hypothesize that the gene mdpE (Mpe_B0558), 12-fold upregulated on MTBE and coding for a dehydrogenase, may be involved in the production of tert-butyl formate (TBF) (Fig. 1). The conserved motif (230-GQHKGSA-236) and the conserved residue E319 of MdpE clearly identify it as a member of a recently described family of (S)-2-hydroxyacid dehydrogenases that bind NADP/NADPH as cofactors in a novel, non-Rossman fold (21, 35, 36). With one exception, these functionally diverse enzymes act on 2-oxo or 2-hydroxy acids (35, 36, 58). Sequence alignment and phylogenetic analysis of MdpE with proteins belonging to the seven proposed clades of (S)-2-hydroxyacid dehydrogenases (35, 36) suggest that this enzyme is deeply branching. Therefore, it is not possible to assign this enzyme into any of the described groups, as it may represent a separate clade. However, based on the functionality of the MdpE enzyme and its 12-fold increase in expression on MTBE, we propose this enzyme to be the dehydrogenase required for complete conversion of MTBE to the intermediate TBF.
It has been demonstrated that the hydrolysis of TBF to TBA occurs spontaneously and rapidly under low-pH conditions (6, 45). However, on the basis of growth in a buffered mineral medium used in this study, as well as physiology studies in other organisms (45), it seems most probable that TBF hydrolysis in PM1 is an esterase-catalyzed process. A gene for an esterase (Mpe_B0604) is located downstream of mdpA on the megaplasmid, but our analyses provide evidence that preclude its involvement in TBF hydrolysis. This esterase gene was not significantly differentially expressed on MTBE (it was downregulated 1.2-fold), which may be the result of interruption by an ISmp1 element. In addition, an Mpe_B0604 homolog is lacking in PM1-like MTBE-degrading environmental isolates that also lack the ISmp1 element (26), suggesting the involvement of another esterase.
No other prospective esterases were found on the megaplasmid; however, a 5.2-fold-upregulated gene for a possible TBF esterase was found on the main chromosome (Mpe_A2443). The Mpe_A2443 protein belongs to the hormone-sensitive lipase family. The bacterial members of this family are known to act on short-chain (C4 to C8) carboxylic esters, but their physiological function is largely unknown (17). The Mpe_A2443 protein is most closely related (53% identity) to a putative esterase from Rhodococcus sp. strain RHA1 (GenBank accession no. YP_706618) and contains the conserved active site and G-D/E-S-A-G motif of acetyl esterases, such as Aes of Escherichia coli (17). Further physiological and genetic studies are required to clarify whether the Mpe_A2443 protein functions as an esterase and whether it is responsible for TBF hydrolysis in PM1.
The monooxygenase enzyme alkane hydroxylase (alkB) was suggested to be responsible for TBA oxidation in M. austroafricanum strains (15), as well as in cometabolic oxidation of MTBE and TBA in Mycobacterium vaccae JOB5 (45). However, based on the microarray analyses, sequence comparisons, and protein homology modeling, we propose that a new Rieske non-heme iron subunit (mdpJ; 11.7-fold upregulated on MTBE) of a multicomponent enzyme system and an associated Fe-S reductase (mdpK; 4.3-fold upregulated on MTBE) are involved in TBA oxidation in PM1 (Fig. 1).
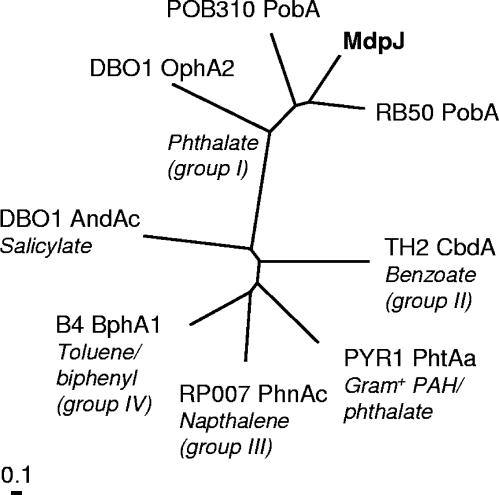
A more detailed sequence analysis of MdpJ was performed due to its high upregulation (11.7-fold) on MTBE. The analysis identified an N-terminal Rieske-type [2Fe-2S] domain (C85-X-H-X16-C-X2-H107) and a conserved C-terminal mononuclear, non-heme iron-binding motif (D/E190-X3-D-X2-H-X4-H202) typical of Rieske non-heme iron dioxygenases. This class of enzymes uses molecular oxygen, adding both atoms of O2 to the aromatic ring of the substrate, including aromatic and polycyclic aromatic hydrocarbons and chlorinated aromatic, nitroaromatic, aminoaromatic, and heterocyclic aromatic compounds. Enzymes in this family are also involved in benzylic and methyl group hydroxylation, desaturation, sulfoxidation, and dealkylation reactions (39). A phylogenetic comparison showed MdpJ belongs to the phthalate group (group I) of dioxygenases as described by Parales and Resnick (39) (Fig. 2). This grouping was of particular interest, since some enzymes of the phthalate family function as monooxygenases and not dioxygenases with their native substrates (39). Those best studied are toluene and naphthalene dioxygenases, and the change in functionality in each case is probably a result of positioning of the compound in the active site (40, 41). Further study is necessary to determine if TBA is the native substrate for MdpJ. However, together with the high expression value of 11.7-fold upregulation in MTBE-grown cells, MdpJ could carry out the hydroxylation of TBA to 2-methyl-2-hydroxy-1-propanol (Fig. 1) in M. petroleiphilum PM1.
FIG. 2.
MdpJ fits in the phthalate (group 1) of aromatic ring hydroxylating dioxygenases. Sequences were aligned, and the tree was generated using the Bioedit 7.0.0 package (19). Representative α subunit sequences are from the following organisms/dioxygenase systems: MdpJ, proposed TBA hydroxylase M. petroleiphilum PM1 (ZP_00241560); B4 BphA1, BPDO, Pseudomonas sp. strain B4 (AJ251217); DBO1 OphA2, phthalate dioxygenase, B. cepacia DBO1 (AF095748); POB310 PobA, phenoxybenzoate dioxygenase, P. pseudoalcaligenes POB310 (X78823); PYR-1 PhtAa, phthalate dioxygenase, Mycobacterium vanbaalenii PYR-1 (AY365117); RB50_PobA, iron-sulfur protein, Bordetella bronchiseptica RB50 (CAE34653); RP007 PhnAc, polycyclic aromatic hydrocarbon dioxygenase: Burkholderia sp. strain RP007 (AF061751); TH2 CbdA, 2-halobenzoate dioxygenase, Burkholderia sp. strain TH2 (AB035324).
The protein product of the gene immediately downstream of mdpJ, mdpK, shares 39% identity with PobB, a reductase component of phenoxybenzoate dioxygenase. In addition, MdpK contains domains typically conserved in class IA oxygenase ferredoxin reductases: a flavin mononucleotide isoalloxazine-binding domain (61RxYSL65), an NAD ribose-binding domain (125GGIGITP131), and a [2Fe-2S] ferredoxin-binding domain (288Cx4Cx2Cx29C324) situated at the C terminus (55). The MdpK protein therefore most likely represents the ferredoxin reductase component of the predicted TBA hydroxylase. The presence of a specific and unique TBA oxidation enzyme system in PM1, responsible for the oxidation of TBA to 2-methyl-2-hydroxy-1-propanol, may help explain the unique capability of PM1 to efficiently degrade TBA without substantial accumulation of the intermediate.
The conversion of 2-methyl-2-hydroxy-1-propanol (MHP) to HIBA was recently hypothesized to be a two-step process involving alcohol dehydrogenase (MpdB) and aldehyde dehydrogenase (MpdC) in M. austroafricanum IFP 2012 (15). A BLAST search with the M. austroafricanum IFP 2012-predicted amino acid sequence of the mpd cluster genes mpdB and mpdC retrieved from the NCBI nr database against the PM1 whole-genome sequence database showed the highest identities to MpeA0945 (33%) and MpeA1909 (40%), respectively (26). Both genes are located on the chromosome, and the expression levels of Mpe_A0945 and Mpe_A1909 were neutral and decreased 1.7-fold, respectively, on MTBE in comparison to ethanol, making their involvement in MTBE degradation unlikely. Based on microarray data, we propose that in PM1 a putative plasmid-encoded dehydrogenase, mdpH (Mpe_B0561), upregulated 4.6-fold on MTBE, acts as the MHP dehydrogenase in PM1 (Fig. 1). Interestingly, this gene shows 32% identity to human 3-HIBA dehydrogenase.
The identity of the hydroxyisobutyraldehyde (HIBAL) dehydrogenase is less clear. In total, there are 11 genes belonging to the aldehyde dehydrogenase superfamily in the PM1 genome, but none showed significant upregulation when grown on MTBE. Based on predicted function, gene arrangement, and 1.4-fold upregulation, we propose Mpe_A0361 as the most likely candidate gene for HIBAL dehydrogenase in PM1. The chromosome-encoded Mpe_A0361 shows 33% identity to MpdC of M. austroafricanum IFP 2012. More significantly, the peptide shows 58% identity to AldA of E. coli, an enzyme active on a number of small α-hydroxyaldehyde substrates, including lactaldehyde, glycolaldehyde, and methylglyoxal (3, 13, 18).
Based on comparative sequence analysis, the products of mdpP (Mpe_B0539), mdpX (Mpe_B0547), and mdpO/R (Mpe_B0538/Mpe_B0541) were annotated as 2-HIBA coenzyme A (CoA) ligase, 3-hydroxybutyryl-CoA dehydrogenase, and a two-component HIBA mutase, respectively (26). These genes were upregulated 1.4-, 1.6-, and 1.2-/2.7-fold, respectively, on MTBE-grown relative to ethanol-grown cells (Fig. 1). The function and the expression of MdpX and MdpO/R are in agreement with a recently proposed pathway of HIBA degradation by a cobalamin-dependent mutase (42). In addition, we propose MdpP as the 2-HIBA-CoA ligase. A neighboring gene product, the Mpe_B0543 protein, is a putative ATP-binding cobalamin adenosyl transferase.
The product of 3-hydroxybutyryl-CoA dehydrogenase, acetoacetyl-CoA, is potentially converted by a predicted acetyl-CoA acetyltransferase (Mpe_A3367) to two acetyl-CoA molecules (Fig. 1), which can feed into the tricarboxylic acid cycle. Mpe_A3367 is upregulated 1.6-fold on MTBE. This enzyme can also catalyze the reverse reaction as the first step in synthesis of polyhydroxybutyrate, which may be used as a carbon and energy reserve when nitrogen and/or phosphorus are limiting.
The microarray expression data also revealed significant upregulation of genes with unknown function belonging to the cluster Mpe_B0532 through Mpe_B0535 (Fig. 1). These genes do not show significant similarity to any known proteins in gene and protein database searches. We cannot formulate hypotheses for the role of the cluster at this point, except to note that they are highly expressed in MTBE-grown cells and could be involved in degradation of MTBE or aromatic compounds in M. petroleiphilum PM1.
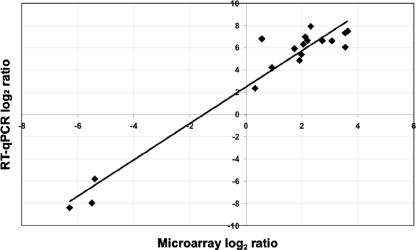
Evaluation of the expression ratios by quantitative RT-PCR.
RT-qPCR was employed to confirm the trends observed in the expression data. Eighteen genes, chosen based on genomic location and differential expression, were compared using the two techniques. In general, the RT-qPCR results, expressed as the difference between MTBE- and ethanol-grown cells, showed the same trends as the log differences for the same treatments for the microarray data (Fig. 3); the data were well correlated (r2, ∼0.94). For some of the CDSs, including Mpe_B0606 and Mpe_B0559, the RT-qPCR log difference was considerably greater than the microarray difference for MTBE versus ethanol (1.5-fold and 0.7-fold for the microarray analysis and 110- and 240-fold for the RT-qPCR analysis for Mpe_B0606 and Mpe_B0559, respectively), which caused the slope to deviate from 1. Attempts were made to reproduce growth conditions, since the same extracts used in the microarray analysis were not available for the RT-qPCR analysis; thus, it was likely that slight variations in culture conditions were present. Because of these slight variations in culturing, the variability between analyses was likely higher, but the trends seen in the microarray analysis are consistent with those observed with the RT-qPCR analysis. The smaller differences observed with the microarray data may be caused in part by data normalization, which tends to compress the microarray data.
FIG. 3.
Correlation between MTBE upregulation, determined by RT-qPCR analysis versus microarray analysis and expressed as log2 ratios for the following locus tags: Mpe_A0474, Mpe_A0475, Mpe_A0476, Mpe_B0532, Mpe_B0533, Mpe_B0534, Mpe_B0535, Mpe_B0548, Mpe_B0551, Mpe_B0553, Mpe_B0554, Mpe_B0555, Mpe_B0558, Mpe_B0559, Mpe_B0560, Mpe_B0561, Mpe_B0562, and Mpe_B0606. The linear relationship is expressed by the equation y = 1.64x + 2.47, and r2 = 0.94. The absolute standard deviations for the RT-qPCR log2 fold differences ranged from 0.30 to 1.2 and averaged 0.55.
Ethanol oxidation in PM1: the QEDH regulon.
While the primary focus of this study was the elucidation of genes involved in MTBE degradation, a converse analysis of the data provided information for genes upregulated in response to ethanol and provided validation of the microarray data set. In response to growth on ethanol, the most significantly upregulated gene cluster (2.7- to 79.6-fold) in PM1 is the quinoprotein ethanol dehydrogenase (QEDH) cluster (Table 2), compared with growth on MTBE. The QEDH regulon extends from Mpe_A0473 to Mpe_A0481 and includes the quinoprotein ethanol dehydrogenase genes exaA1 (Mpe_A0476) and exaA2 (Mpe_A0473) and two copies of the cytochrome c550 precursor gene, exaB1 (Mpe_A0480) and exaB2 (Mpe_A0474). Quinoprotein alcohol dehydrogenases are a family of proteins found in methylotrophic and autotrophic bacteria that use pyrroloquinoline quinone as their prosthetic group and contain a C-terminal cytochrome c domain (http://www.bioconductor.org). A two-component regulatory system consisting of a sensor histidine kinase gene, exaD (Mpe_A0477), and a response regulator gene, exaE (Mpe_A478), is present in the PM1 operon, like that of the ethanol oxidation regulon in Pseudomonas aeruginosa ATCC 17933 (16). The QEDH regulon also contains a gene with unknown function (Mpe_A475) that was highly upregulated in cells grown on ethanol (45-fold).
TABLE 2.
Genes involved in ethanol oxidation in M. petroleiphilum PM1
| Locus | Strand | Fold change | P value | Gene name | Predicted function |
|---|---|---|---|---|---|
| Mpe_A0473 | + | −3.0 | 1.2E-07 | exaA2 | Quinoprotein ethanol dehydrogenase |
| Mpe_A0474 | − | −41.7 | 1.6E-17 | exaB2 | Cytochrome c550 |
| Mpe_A0475 | − | −45.1 | 5.8E-17 | Pentapeptide repeat family protein | |
| Mpe_A0476 | − | −79.6 | 9.0E-20 | exaA1 | Quinoprotein alcohol dehydrogenase |
| Mpe_A0477 | + | −1.9 | 6.5E-04 | exaD | Two-component sensor histidine kinase |
| Mpe_A0478 | + | NDa | ND | exaE | Two-component response regulator |
| Mpe_A0479 | − | −4.2 | 2.5E-10 | Sigma-54-dependent transcriptional regulator, Fis family | |
| Mpe_A0480 | + | ND | ND | exaB1 | Cytochrome c550 |
| Mpe_A0481 | + | −8.2 | 4.9E-17 | Hypothetical | |
| Mpe_A0599 | − | −11.2 | 7.1E-11 | exaC | Acetaldehyde dehydrogenase (NAD+) |
| Mpe_A3829 | ND | ND | pqqA | Pyrroloquinoline quinone biosynthesis protein A | |
| Mpe_A2585 | + | −2.7 | 5.6E-10 | pqqB | Pyrroloquinoline quinone biosynthesis protein B |
| Mpe_A2586 | + | ND | ND | pqqC | Coenzyme PQQ synthesis protein C |
| Mpe_A2587 | + | ND | ND | pqqD | Pyrroloquinoline quinone biosynthesis protein D |
| Mpe_A2588 | + | −2.9 | 6.6E-15 | pqqE | Coenzyme PQQ synthesis protein E |
ND, not determined.
The two putative quinoprotein ethanol dehydrogenases, the Mpe_A0476 and Mpe_A0473 proteins, share a 52% identity. The Mpe_A0476 protein showed a significantly higher identity to the ExaA from Pseudomonas aeruginosa (70%) compared to Mpe_A0473 (53%). In addition, the expression level of Mpe_A0476 was much higher than that of Mpe_A0473 (∼80-fold versus 3-fold). The role of the Mpe_A0473 dehydrogenase gene has not been clearly elucidated; however, an exaA knockout in P. aeruginosa did not eliminate ethanol oxidation, suggesting metabolic redundancy and a role for a second dehydrogenase (57a). Based on the ethanol degradation pathway in E. coli, it is likely that Mpe_A0476 codes for an alcohol dehydrogenase that converts ethanol to acetaldehyde. This product is likely converted to acetyl-CoA (33) by the second NADH dehydrogenase (acetaldehyde dehydrogenase, ExaC; Mpe_A0599), which shows 71% identity to ExaC (from the ExaABC cluster in P. aeruginosa) and is suspected to play a role in ethanol oxidation (51). In addition, the gene Mpe_A0599 is upregulated 11-fold on ethanol. It is not known if one or both putative cytochromes c550 exaB1 (Mpe_A0480) and exaB2 (Mpe_A0474) function in electron transfer during ethanol degradation in strain PM1.
PM1 biodegradation capacity for pollutants.
M. petroleiphilum PM1 grows on phenol, and two distinct clusters of dimethylphenol (dmp)-like genes are present on the chromosome (Mpe_A2265 to Mpe_A2267, Mpe_A2272 to Mpe_A2286, Mpe_A3305 to Mpe_A3313, and Mpe_A3321 to Mpe_A3325). Compared to growth on ethanol, in MTBE-grown cells, significant upregulation of structural genes in the Dmp pathway (P < 0.05) was observed for dmp operon I (Table 1), but not in dmp operon II, except for dmpH. Genetic analysis of the dmp operon II suggested it was not functional, since it lacks DmpP (phenol hydroxylase reductase) and DmpC (2-hydroxymuconic semialdehyde dehydrogenase). Additionally, upregulation of the genes in the tbu toluene degradation operons I and II (P < 0.5) was observed when cells were grown on MTBE. A definite conclusion on the expression patterns of the tbu operons is not possible, because only two genes from operon I showed >2-fold upregulation on MTBE, while six genes from operon II were not available in the microarray. The two available structural tbu operon II genes showed 2.5- and 4.1-fold upregulation, respectively.
Of interest was the differential expression of the regulators of the tbu (toluene) and dmp (phenol) degradation operons. Both tbu operons of PM1 have a two-component sensor-regulator gene pair located immediately downstream of the operon. These regulators are divergently expressed but showed less than twofold expression increases in the presence of MTBE. Only dmp operon I showed significant upregulation (2.0- to 3.1-fold). Included among the upregulated genes was a LysR family type regulator encoded by Mpe_A2279, which is most similar to aphT of Commamonas testosteroni (1). AphT is related to regulators of pathways for ortho cleavage of catechol or chlorinated catechols. The Fis family regulator gene Mpe_A2286, closely related to the phenol regulator gene dmpR (GenBank accession no. CAA48174), did not show differential expression under our test conditions.
Several genes coding for enzymes involved in degradation of aromatic compounds, including phenylacetic acid degradation proteins (Mpe_A0987 and Mpe_A0989 proteins), phenylpropionate dioxygenase (Mpe_A1001), and 2-polyprenylphenol hydroxylase (Mpe_A0819), were also upregulated in cells grown on MTBE.
M. petroleiphilum PM1 contains an alkane monooxygenase pathway on its plasmid and a propane monooxygenase pathway on its chromosome, which may facilitate its growth on n-alkanes. The alkane monooxygenase was already discussed in the context of the MTBE degradation pathway. The propane monooxygenase (pmo) reductase (Mpe_A0951) was upregulated approximately 4.4-fold in MTBE-grown relative to ethanol-grown cells. It is not currently known whether PM1 can grow on propane or whether the Pmo pathway is functional.
The PM1 genome has nine CDSs with similarity to cyclohexanone monooxygenases (CHMOs) (31). In MTBE-grown cells, there was greater-than-twofold upregulation of three CHMO genes (Mpe_B0610, Mpe_A0393, and Mpe_A1351) and downregulation of one CHMO gene (Mpe_A0607) (Table 1).
Previous physiology studies have shown a complex pattern of interactions between MTBE and individual BTEX compounds in PM1 cultures and raised the question of whether these interactions are regulatory or mechanistic in nature (10, 27). The MTBE-facilitated upregulation of several enzymes involved in BTEX degradation as well as a number of other environmental contaminants suggests the pattern stems from a regulatory network. As previous studies have suggested a possible hierarchy to such a network (e.g., benzene used preferentially to MTBE) (10), we hope to compare MTBE and BTEX expression data in future studies to further elucidate the regulatory response to complex mixtures of environmental contaminants.
MTBE pathway: gene arrangement and mobilization.
Genes specifying the biodegradation of recalcitrant compounds are usually clustered on the same genomic locus, although degradative genes can also be widely separated. Examples of the latter arrangement include the dioxin/dibenzofuran pathway of Sphingomonas sp. strain RW1, whose degradative genes were found to be scattered around the chromosome (2), or the chromosomally encoded naphthalene conversion to salicylate and plasmid-encoded salicylate degradation in P. putida PMD-1 (59). In PM1, the majority of the MTBE pathway genes appear to be localized to a main cluster (Mpe_B0538 to Mpe_B0562), but several genes are found on the plasmid outside of this locus, and at least two of the predicted genes are on the chromosome.
Often, degradative genes or gene clusters are flanked by insertion sequences forming degradative transposons. This allows for the shuttling of catabolic genes and entire gene clusters between different replicons (53). The PM1 genome has a number of complex repetitive elements, including eight families of insertion sequences (ISmp1 to -8), and two large genomic segments that appear to have undergone recent duplications, including the plasmid-based 29-kb phosphonate transport/cobalamin biosynthesis direct repeat found in tandem and flanked by ISmp8 elements and a 40-kb duplication found on both the chromosome and the plasmid, where it appears to have integrated and interrupted the deoxycytidine deaminase gene Mpe_B0168/Mpe_B0202 (26).
The presence of many IS elements in the vicinity of the main MTBE pathway gene cluster may enable mobilization (as a composite transposon) and confer selective advantage if retained. The ISmp8 element is restricted to only one strand on the plasmid (five copies) and thus has the potential for deletions or to transpose larger segments as a transposon. In fact, the majority of the MTBE gene cluster is flanked by two ISmp8 transposases (Mpe_B0489 and Mpe_B0570) that are 79 kb apart. Similarly, ISmp7 copies are found within (Mpe_B0549/Mpe_B0550, Mpe_B0572/Mpe_B0571, and Mpe_B0586/Mpe_B0587) and flanking (Mpe_B0004/Mpe_B0005 and Mpe_B0070/Mpe_B0071) the MTBE gene cluster and could be involved in gene rearrangement, deletion, and mobilization. In addition, IS elements may influence transcription (e.g., a divergent ISmp4-like IS element may interrupt the promoter of TBA hydroxylase mdpJ) and contribute coding sequence (e.g., ISmp4 sequence may add to the 3′ end of the Mpe_A2443 esterase gene) of key MTBE degradative enzymes. Interestingly, ISmp1 (3 copies), ISmp7 (7 copies), and ISmp4 (12 copies) were among the 5% of genes showing highest expression on both ethanol and MTBE. This high expression was observed even though multiple copies of each IS (except for ISmp1) were present in the microarray and shows evidence that the IS elements continue to shape the PM1 genome.
The G+C content of the Mpe_A0375 to Mpe_A0384 IS locus downstream of the predicted HIBAL dehydrogenase Mpe_A0361is 65.0%, compared with an average of 69.2% for the chromosome and 66.0% for the plasmid; in addition, it encodes several hypothetical proteins and lies within a region rich in hypothetical genes. While these represent the only three ISmp2s in the genome, the ISmp1 in this region is identical to the one located on the plasmid (Mpe_B0605), which we predict disrupts the esterase Mpe_B0604 and lies downstream of mdpA (Mpe_B0606).
Several of the PM1 IS elements are also similar to ones found associated with catabolic transposons in other environmental bacteria. The ISmp1 transposase is similar to a transposase from a catabolic transposon carrying tfd genes for 2,4-dichlorophenoxyacetic acid degradation on pEST4011 of Achromobacter denitrificans (56). The unique Mpe_B0528 and Mpe_B0529 transposases (part of a predicted composite transposon consisting of genes Mpe_B0527 to Mpe_B0529 and located just upstream of the main MTBE gene cluster) are similar to a transposase associated with catechol 1,2-dioxygenase in Burkholderia sp. strain TH2 (51). Finally, a transposase associated with the p-toluenesulfonate degradation transposon of pTSA in Comamonas testosteroni T-2 is similar to the ISmp8 transposase. The possible involvement of IS elements in mobilization of MTBE genes onto or from the plasmid, as well as in disrupting functions and/or regulation of expression, is intriguing from the standpoint of genome and metabolic pathway evolution. Further research is required in order to answer these and other pertinent questions.
Concluding remarks.
In this study, high-density, whole-genome cDNA microarrays were used to investigate differential gene expression when M. petroleiphilum PM1 was grown on MTBE and ethanol as sole carbon sources. This is the first time that experimental evidence has been presented that links all the enzymatic steps of the upper MTBE degradation pathway from TBF dehydrogenase to MHP dehydrogenase with candidate genes. The microarray studies conducted thus far have led to interesting and testable hypotheses concerning plasmid- and chromosome-encoded genes that may function in each step of the MTBE degradation pathway (Fig. 1) and have led to interesting hypotheses regarding the acquisition and evolution of MTBE genes as well as the involvement of IS elements in these complex processes. To further elucidate the function of the PM1 TBA hydroxylase enzyme system, we are currently performing whole-genome microarray studies with MTBE- and TBA-grown cells, using the completed genome annotation. In addition, gene knockout experiments are being focused on mdpA, mdpJ, and mdpK to test the hypotheses developed from microarray, comparative genomic, and proteomic analyses.
Overall expression results confirm the upregulation of more genes in total, as well as higher expression levels for energy metabolism and housekeeping genes in the presence of the higher-energy-yielding and less-recalcitrant substrate, ethanol (data not shown). In spite of this clear trend, the higher number of unknown genes expressed in the presence of MTBE points to a wealth of untapped information related to bacterial survival in the presence of a recalcitrant, toxic carbon source.
M. petroleiphilum PM1 is known to be a member of subsurface microbial communities at several gasoline-contaminated sites. Given that many contaminated sites have mixtures of organic contaminants including BTEX compounds and fuel oxygenates, it is ideal that bioremediation technologies would utilize microorganisms capable of metabolizing the target contaminant, as well as other contaminants present. In PM1, the exposure to MTBE induces pathways for degradation of a spectrum of aromatic compounds, such as benzene, toluene, xylene (BTX), phenolic compounds, alkanes, and alicyclic, aliphatic, or aryl ketones. This result suggests that PM1 could coexpress pathways for biodegradation of BTX and fuel oxygenates in the bioremediation of gasoline-contaminated aquifers.
Supplementary Material
Acknowledgments
We thank Binyam Gebreyesus at University of California, Davis (UCD), for assistance with RNA extractions, and we thank Rebecca Parales (UCD) and Harry Beller (LLNL) for helpful comments on the manuscript. In addition, we thank the reviewers for their insightful comments that strengthened the manuscript.
We acknowledge the University of California Office of the President for funding through the Campus-Laboratory Collaboration Program as well as the LLNL Laboratory-Directed Research and Development Program. This research was also supported by grant number 5 P42 ES04699-16 from the National Institute of Environmental Health Sciences (NIEHS), NIH. This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract W-7405-Eng-48.
This report's contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS.
Footnotes
Published ahead of print on 21 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmed, F. E. 2001. Toxicology and human health effects following exposure to oxygenated or reformulated gasoline. Toxicol. Lett. 123:89-113. [DOI] [PubMed] [Google Scholar]
- 2.Armengaud, J., K. N. Timmis, and R.-M. Wittich. 1999. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J. Bacteriol. 181:3452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldoma, L., and J. Aguilar. 1987. Involvement of lactaldehyde dehydrogenase in several metabolic pathways of Escherichia coli K12. J. Biol. Chem. 262:13991-13996. [PubMed] [Google Scholar]
- 4.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Q., D. B. Janssen, and B. Witholt. 1996. Physiological changes and alk gene instability in Pseudomonas oleovorans during induction and expression of alk genes. J. Bacteriol. 178:5508-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church, C. D., J. F. Pankow, and P. G. Tratnyek. 1999. Hydrolysis of tert-butyl formate: kinetics, products and implication of the environmental impact of methyl tert-butyl ether. Environ. Toxicol. Chem. 18:2789-2796. [Google Scholar]
- 7.Cirvello, J. D., A. Radovsky, J. E. Heath, D. R. Farnell, and C. Lindamood. 1995. Toxicity and carcinogenicity of t-butyl alcohol in rats and mice following chronic exposure in drinking-water. Toxicol. Ind. Health 11:151-165. [DOI] [PubMed] [Google Scholar]
- 8.Dakhel, N., G. Pasteris, D. Werner, and P. Hohener. 2003. Small-volume releases of gasoline in the vadose zone: impact of the additives MTBE and ethanol on groundwater quality. Environ. Sci. Technol. 37:2127-2133. [DOI] [PubMed] [Google Scholar]
- 9.Davis-Hoover, W. J., S. A. Stavnes, J. J. Fleischman, S. C. Hunt, J. Goetz, M. Keper, M. Roulier, K. Hristova, K. Scow, K. Knutson, W. Mahaffee, and D. J. Slomczynski. 2003. BTEX, MTBE bioremediation: bionets containing PM1, SOS or air, p. E-25. In V. S. Magar and M. E. Kelley (ed.), Seventh International In Situ and On-Site Bioremediation Symposium. Battelle Press, Columbus, OH.
- 10.Deeb, R. A., H.-Y. Hu, J. R. Hanson, K. M. Scow, and L. Alvarez-Cohen. 2001. Substrate interactions in BTEX and MTBE mixtures by an MTBE-degrading isolate. Environ. Sci. Technol. 35:312-317. [DOI] [PubMed] [Google Scholar]
- 11.Deeb, R. A., S. Nishino, J. Spain, H. Y. Hu, K. M. Scow, and L. Alvarez-Cohen. 2000. MTBE and benzene biodegradation by PM1 via two independent monooxygenase-initiated pathways. Abstr. Pap. Am. Chem. Soc. 219:ENVR 228. [Google Scholar]
- 12.De Marco, P., C. C. Pacheco, A. R. Figueiredo, and P. Moradas-Ferreira. 2004. Novel pollutant-resistant methylotrophic bacteria for use in bioremediation. FEMS Microbiol. Lett. 234:75-80. [DOI] [PubMed] [Google Scholar]
- 13.Di Costanzo, L., G. A. Gomez, and D. W. Christianson. 2007. Crystal structure of lactaldehyde dehydrogenase from Escherichia coli and inferences regarding substrate and cofactor specificity. J. Mol. Biol. 366:481-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudoit, S., J. Shaffer, and J. Boldrick. 2003. Multiple hypothesis testing in microarray experiments. Stat. Sci. 18:71-103. [Google Scholar]
- 15.Ferreira, N. L., D. Labbe, F. Monot, F. Fayolle-Guichard, and C. W. Greer. 2006. Genes involved in the methyl tert-butyl ether (MTBE) metabolic pathway of Mycobacterium austroafricanum IFP 2012. Microbiology 152:1361-1374. [DOI] [PubMed] [Google Scholar]
- 16.Hanson, J. R., C. E. Ackerman, and K. M. Scow. 1999. Biodegradation of methyl tert-butyl ether by a bacterial pure culture. Appl. Environ. Microbiol. 65:4788-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haruki, M., Y. Oohashi, S. Mizuguchi, Y. Matsuo, M. Morikawa, and S. Kanaya. 1999. Identification of catalytically essential residues in Escherichia coli esterase by site-directed mutagenesis. FEBS Lett. 454:262-266. [DOI] [PubMed] [Google Scholar]
- 18.Hidalgo, E., Y. M. Chen, E. C. C. Lin, and J. Aguilar. 1991. Molecular cloning and DNA sequencing of the Escherichia coli K-12 Ald gene encoding aldehyde dehydrogenase. J. Bacteriol. 173:6118-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hristova, K. R., B. Gerbreyesus, D. Mackay, and K. M. Scow. 2003. Naturally occurring bacteria similar to the methyl tert-butyl ether (MTBE)-degrading strain PM1 are present in MTBE-contaminated groundwater. Appl. Environ. Microbiol. 69:2616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hristova, K. R., C. M. Lutenegger, and K. M. Scow. 2001. Detection and quantification of MTBE-degrading strain PM1 by real-time TaqMan PCR. Appl. Environ. Microbiol. 67:5154-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irimia, A., D. Madern, G. Zaccai, and F. M. D. Vellieux. 2004. Methanoarchaeal sulfolactate dehydrogenase: prototype of a new family of NADH-dependent enzymes. EMBO J. 23:1234-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:31-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, E. L., C. A. Smith, K. T. O'Reilly, and M. R. Hyman. 2004. Induction of methyl tertiary butyl ether (MTBE)-oxidizing activity in Mycobacterium vaccae JOB5 by MTBE. Appl. Environ. Microbiol. 70:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, R., J. Pankow, D. Bender, C. Price, and J. Zogorski. 2000. MTBE: to what extent will past releases contaminate community water supply wells? Environ. Sci. Technol. 34:210A. [DOI] [PubMed] [Google Scholar]
- 25.Kane, S. R., H. R. Beller, T. C. Legler, C. J. Koester, H. C. Pinkart, R. U. Halden, and A. M. Happel. 2001. Aerobic biodegradation of methyl tert-butyl ether by aquifer bacteria from leaking underground storage tank sites. Appl. Environ. Microbiol. 67:5824-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane, S. R., A. Y. Chakicherla, P. S. G. Chain, R. Schmidt, M. W. Shin, T. C. Legler, K. M. Scow, F. W. Larimer, S. M. Lucas, P. M. Richardson, and K. R. Hristova. 2007. Whole-genome analysis of methyl tert-butyl ether -degrading beta-proteobacterium Methylibium petroleiphilum PM1. J. Bacteriol. 189:1931-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane, S. R., T. C. Legler, L. M. Balser, and K. T. O'Reilly. 2003. Aerobic biodegradation of MTBE by aquifer bacteria from LUFT sites, p. E-12. In V. S. Magar and M. E. Kelley (ed.), Seventh International In Situ and On-Site Bioremediation Symposium. Battelle Press, Columbus, OH.
- 28.Klinger, J., C. Stieler, F. Sacher, and H. J. Branch. 2002. MTBE (methyl tertiary-butyl ether) in groundwaters: monitoring results from Germany. J. Environ. Monit. 4:276-279. [DOI] [PubMed] [Google Scholar]
- 29.Mackay, D., N. de Sieyes, M. Einarson, K. Feris, A. Pappas, I. Wood, L. Jacobsen, L. Justice, M. Noske, J. Wilson, C. Adair, and K. Scow. 2007. Impact of ethanol on the natural attenuation of MTBE in a normally sulfate-reducing aquifer. Environ. Sci. Technol. 41:2015-2021. [DOI] [PubMed] [Google Scholar]
- 30.Marin, M. M., L. Yuste, and F. Rojo. 2003. Differential expression of the components of the two alkane hydroxylases from Pseudomonas aeruginosa. J. Bacteriol. 185:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGregor, D. B., G. Cruzan, R. D. Callander, K. May, and M. Banton. 2005. The mutagenicity testing of tertiary-butyl alcohol, tertiary-butyl acetate (TM) and methyl tertiary-butyl ether in Salmonella typhimurium. Mutat. Res. 565:181-189. [DOI] [PubMed] [Google Scholar]
- 32.Moran, M. J., J. S. Zogorski, and P. J. Squillace. 2005. MTBE and gasoline hydrocarbons in ground water of the United States. Ground Water 43:615-627. [DOI] [PubMed] [Google Scholar]
- 33.Moreels, D., L. Bastiaens, F. Ollevier, R. Merckx, L. Diels, and D. Springael. 2004. Effect of in situ parameters on the enrichment process of MTBE degrading organisms. Commun. Agric. Appl. Biol. Sci. 69:3-6. [PubMed] [Google Scholar]
- 34.Moyer, E. E. 2003. Introduction, p. 3-9. In E. E. Moyer and P. T. Kostecki (ed.), MTBE remediation handbook. Amherst Scientific Publishers, Amherst, MA.
- 35.Muramatsu, H., H. Mihara, M. Goto, I. Miyahara, K. Hirotsu, T. Kurihara, and N. Esaki. 2005. A new family of NAD(P)H-dependent oxidoreductases distinct from conventional Rossmann-fold proteins. J. Biosci. Bioeng. 99:541-547. [DOI] [PubMed] [Google Scholar]
- 36.Muramatsu, H., H. Mihara, R. Kakutani, M. Yasuda, M. Ueda, T. Kurihara, and N. Esaki. 2005. The putative malate/lactate dehydrogenase from Pseudomonas putida is an NADPH-dependent Δ1-piperideine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductase involved in the catabolism of d-lysine and d-proline. J. Biol. Chem. 280:5329-5335. [DOI] [PubMed] [Google Scholar]
- 37.Nakatsu, C. H., K. R. Hristova, S. Hanada, X.-Y. Meng, J. R. Hanson, K. M. Scow, and Y. Kamagata. 2006. Methylibium petrolelphilum gen. nov., sp nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int. J. Sys. Evol. Microbiol. 56:983-989. [DOI] [PubMed] [Google Scholar]
- 38.Nuwaysir, E. F., W. Huang, T. J. Albert, J. Singh, K. Nuwaysir, A. Pitas, T. Richmond, T. Gorski, J. P. Berg, J. Ballin, M. McCormick, J. Norton, T. Pollock, T. Sumwalt, L. Butcher, D. Porter, M. Molla, C. Hall, F. Blattner, M. R. Sussman, R. L. Wallace, F. Cerrina, and R. D. Green. 2002. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 12:1749-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parales, R. E., and S. M. Resnick. 2006. Aromatic ring hydroxylating dioxygenases, p. 287-340. In J.-L. Ramos and R. C. Levesque (ed.), Pseudomonas, vol. 4. Springer Netherlands, Dordrecht, The Netherlands. [Google Scholar]
- 40.Resnick, S. M., K. Lee, and D. T. Gibson. 1996. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J. Ind. Microbiol. Biotechnol. 17:438-457. [Google Scholar]
- 41.Robertson, J. B., J. C. Spain, J. D. Haddock, and D. T. Gibson. 1992. Oxidation of nitrotoluenes by toluene dioxygenase: evidence for a monooxygenase reaction. Appl. Environ. Microbiol. 58:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohwerder, T., U. Breuer, D. Benndorf, U. Lechner, and R. H. Muller. 2006. The alkyl tert-butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl. Environ. Microbiol. 72:4128-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, A. E., K. Hristova, I. Wood, D. M. Mackay, E. Lory, D. Lorenzana, and K. M. Scow. 2005. Comparison of biostimulation versus bioaugmentation with bacterial strain PM1 for treatment of groundwater contaminated with methyl tertiary butyl ether (MTBE). Environ. Health Persp. 113:317-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, C. A., and M. R. Hyman. 2004. Oxidation of methyl tert-butyl ether by alkane hydroxylase in dicyclopropylketone-induced and n-octane-grown Pseudomonas putida GPo1. Appl. Environ. Microbiol. 70:4544-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, C. A., K. T. O'Reilly, and M. R. Hyman. 2003. Characterization of the initial reactions during the cometabolic oxidation of methyl tert-butyl ether by propane-grown Mycobacterium vaccae JOB5. Appl. Environ. Microbiol. 69:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, C. A., K. T. O'Reilly, and M. R. Hyman. 2003. Cometabolism of methyl tertiary butyl ether and gaseous n-alkanes by Pseudomonas mendocina KR-1 grown on C5 to C8 n-alkanes. Appl. Environ. Microbiol. 69:7385-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smits, T. H. M., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed]
- 49.Squillace, P. J., J. S. Zogorski, W. G. Wilber, and C. V. Price. 1996. Preliminary assessment of the occurrence and possible sources of MTBE in groundwater in the United States, 1993-1994. Environ. Sci. Technol. 30:1721-1730. [Google Scholar]
- 50.Steffan, R. J., K. McClay, S. Vainberg, C. W. Condee, and D. Zhang. 1997. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki, K., A. Ichimura, N. Ogawa, A. Hasebe, and K. Miyashita. 2002. Differential expression of two catechol 1,2-dioxygenases in Burkholderia sp. strain TH2. J. Bacteriol. 184:5714-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tani, A., T. Ishige, Y. Sakai, and N. Kato. 2001. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 183:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262-269. [DOI] [PubMed] [Google Scholar]
- 54.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Rothlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 55.van der Geize, R., G. I. Hessels, and L. Dijkhuizen. 2002. Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid Δ1-dehydrogenase isoenzyme. Microbiology 148:3285-3292. [DOI] [PubMed] [Google Scholar]
- 56.Vedler, E., M. Vahter, and A. Heinaru. 2004. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vosahlikova, M., T. Cajthaml, K. Demnerova, and J. Pazlarova. 2006. Effect of methyl tert-butyl ether in standard tests for mutagenicity and environmental toxicity. Environ. Toxicol. 21:599-605. [DOI] [PubMed] [Google Scholar]
- 57a.Vrionis, H. A., A. J. Daugulis, and A. M. Kropinski. 2002. Identification and characterization of the AgmR regulator of Pseudomonas putida: role in alcohol utilization. Appl. Microbiol. Biotechnol. 58:469-475. [DOI] [PubMed] [Google Scholar]
- 58.Yew, W. S., and J. A. Gerlt. 2002. Utilization of l-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 184:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuniga, M. C., D. R. Durham, and R. A. Welch. 1981. Plasmid-mediated and chromosome-mediated dissimilation of naphthalene and salicylate in Pseudomonas putida Pmd-1. J. Bacteriol. 147:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.