Abstract
Staphylococcus xylosus is a commensal of the skin of humans and animals and a ubiquitous bacterium naturally present in food. It is one of the major starter cultures used for meat fermentation, but a few strains could potentially be hazardous and are related to animal opportunistic infections. To better understand the genetic diversity of S. xylosus intraspecies, suppressive and subtractive hybridization (SSH) was carried out with the S. xylosus C2a strain, a commensal of human skin, used as the driver for three tester strains, S04002 used as a starter culture, S04009 isolated from cow mastitis, and 00-1747, responsible for mouse dermatitis. SSH revealed 122 tester-specific fragments corresponding to 149 open reading frames (ORFs). A large proportion of these ORFs resembled genes involved in specific metabolisms. Analysis of the distribution of the tester-specific fragments in 20 S. xylosus strains of various origins showed that the S. xylosus species could be divided into two clusters with one composed only of potentially hazardous strains. The genetic content diversity of this species is colocalized in a region near the origin of replication of the chromosome. This region of speciation previously observed in the Staphylococcus genus corresponded in S. xylosus species to a strain-specific region potentially implicated in ecological fitness.
Staphylococcus xylosus is a commensal bacterium generally found inhabiting the skin and the mucous membranes of mammals and birds (15, 22). S. xylosus is ubiquitous, can be found in various niches (12, 25, 28), and persists in soils and on surfaces (32). In addition to its ability to form biofilms (29), the ubiquity of S. xylosus might be explained by its ability to adapt to different environments. S. xylosus is naturally present in raw meat and milk and is commonly used as a starter culture for their fermentation (14, 36). This species is virtually defined as a nonpathogenic Staphylococcus, but a few strains of S. xylosus are related to animal and human opportunistic infections (27, 33, 37, 39). Thus, most strains are commensal and useful in food while others could potentially be hazardous, showing the versatility of this species.
Genomes, and consequently phenotypes, can be highly heterogeneous in bacterial strains, even if they belong to the same species. Genomic comparative studies between all Staphylococcus sequenced genomes showed an average 78% identity (35). The 22% genetic variability observed within the Staphylococcus genus is also observed within species. Indeed, genetic diversity between 36 S. aureus strains using whole-genome DNA microarrays showed that 78% of genes were common to all strains and 22% of genes were strain specific and might play a role in adaptation to specialized niches (7). In a previous study with genomic DNA restriction and pulsed-field gel electrophoresis (PFGE) analysis of seven S. xylosus strains of various origins, we showed genomic diversity within this species, with variations of chromosome size and ribosomal operon (rrn) number (5). S. xylosus isolated from opportunistic infections seemed to have chromosomes up to 11% larger than strains isolated from the food environment (5). Genomic differences were also found between S. xylosus strains isolated from meat products by PFGE and randomly amplified polymorphic DNA analyses (5), but much remains to be learned about the genetic content differences. Strain-specific genes of S. xylosus are of major interest in explaining the versatility of this species. We expanded comparisons of S. xylosus strains by using suppressive and subtractive hybridization (SSH). This method provides comprehensive surveys of genome differences among closely related strains by facilitating the identification of DNA segments present in one genome (tester) but absent in another (driver). It has been widely used for the analysis of bacterial pathogen genomes to discover new epidemiological markers, virulence factors, and host specificity determinants (38). SSH was also successfully used to study genome plasticity or intraspecies genomic diversity (23, 30).
In this work, SSH was carried out between three S. xylosus tester strains: S. xylosus S04002, used as a starter culture, S. xylosus S04009, isolated from cow mastitis, and S. xylosus 00-1747, responsible for mouse dermatitis (39). S. xylosus C2a, a commensal of human skin, was used as a driver. The chromosomal organization of the S. xylosus C2a strain has been reported (5) and has constituted the starting point for studying the genetic diversity within this species. Specific DNA fragments of 29.7, 25.4, and 22.8 kb were isolated from the three SSH libraries obtained from S. xylosus S04002, S04009, and 00-1747, respectively. Their distributions among 20 S. xylosus strains were analyzed, and their locations in the respective chromosomes of the S. xylosus tester strains were determined.
MATERIALS AND METHODS
Strains.
The S. xylosus strains used in the study are listed in Table 1. S. xylosus C2a is derived from the type strain DSM20267 and is cured of its endogenous plasmid, pSX267 (11). The other strains are nonduplicate isolates of different origins. Bacteria were stored at −70°C. Cultures were grown in brain heart infusion broth or agar and incubated at 37°C for 16 h.
TABLE 1.
S. xylosus strains used in this study and their origins
| Strain | Origin |
|---|---|
| C2aa | Derived from DSM20267, human skin |
| 839 | Meat starter |
| 840 | Meat starter |
| S01002 | Meat starter |
| S01003 | Meat starter |
| S01004 | Meat starter |
| S01006 | Meat starter |
| S01007 | Meat starter |
| S01008 | Meat starter |
| S04002a | Meat starter |
| S01001 | Dairy starter |
| S04003 | Dairy starter |
| S04009a | Cow mastitis |
| S04010 | Cow mastitis |
| S04012 | Cow mastitis |
| S04013 | Cow mastitis |
| S04016 | Cow mastitis |
| S04017 | Cow mastitis |
| S04018 | Cow mastitis |
| S04011 | Goat mastitis |
| S04014 | Goat mastitis |
| S04019 | Goat mastitis |
| S04020 | Goat mastitis |
| 00-1747a | Mouse dermatitis |
Strain used for SSH.
Pulsed-field gel electrophoresis.
Genomic DNA of S. xylosus strains was prepared in agarose plugs as described previously (21). Plugs were restricted with the endonuclease I-CeuI (New England Biolabs, United Kingdom) or SmaI enzyme (Promega, Charbonnière, France) according to the manufacturers' instructions. Fragments were separated by PFGE in 1% agarose gels in 0.5× Tris-borate-EDTA buffer on a CHEF-DR III apparatus (Bio-Rad, Ivry, France). Electrophoretic conditions were 50 s to 100 s for 6 h and 10 s to 30 s for 18 h or 50 s to 90 s for 22 h and then 1 s to 12 s for 13 h at 14°C at a constant voltage of 6 V·cm−1 and an angle of 120° for the determination of specific subtracted fragment location. Lambda DNA concatemers were used as molecular size markers (Promega). After migration, gels were stained in ethidium bromide, digitized under UV light (Gel Doc 2000; Bio-Rad), and analyzed using the Quantity One quantitation software (Bio-Rad). Similarities between the PFGE profiles, based on band position, were derived by the Dice correlation coefficient with a maximum position tolerance of 2%. A hierarchical unweighted pair group method analysis was used to generate a dendrogram.
Chromosomal DNA and plasmid preparation.
Chromosomal DNA from S. xylosus was isolated by the method of Marmur (18). To isolate plasmid DNA from S. xylosus, cells were lysed with 16 μg/ml lysostaphin, and the QIAprep Miniprep kit (Qiagen, Courtaboeuf, France) was subsequently applied.
Suppressive-subtractive hybridizations and differential screening.
Three subtracted DNA libraries were generated following the recommendations for the Clontech PCR-Select bacterial genome subtraction kit (BD Biosciences Clontech, Erembodegem, Belgium). Genomic DNAs of S. xylosus strains S04002, S04009, and 00-1747 were used as the testers and that of S. xylosus C2a was used as a driver. To increase the proportion of chromosomal subtracted DNA, the endogenous plasmids of the tester strains were also used as drivers. Genomic and plasmid DNAs were digested by the appropriate RsaI enzyme. Suppression-subtractive hybridizations were carried out as recommended by the supplier except that hybridization temperatures were set at 61°C instead 63°C due to the low G+C content of Staphylococcus DNA. The pool of subtracted fragments was amplified by PCR using the Advantage cDNA polymerase (BD Biosciences Clontech). PCR products generated in each library were cloned into the pGEM-T Easy vector (Promega) according to the supplier's recommendations and transformed into Escherichia coli TG1 competent cells, which were then cultured on Luria-Bertani agar plates containing 100 μg/ml ampicillin, 50 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and 0.5 mM isopropyl thiogalactoside. Resulting transformants were individually grown in Luria-Bertani medium containing 100 μg/ml ampicillin in 96-well microtiter plates and stored in 20% glycerol at −70°C. Subtracted library clones were randomly selected as templates for PCR amplification using nested primers 1 and 2R, provided in the genome subtraction kit. PCR products were analyzed by agarose gel electrophoresis to verify the size of the inserts and the presence of one distinct amplified product. PCR products of each subtracted library were screened for tester-specific fragments using dot blot hybridization. They were arrayed in duplicate onto positively charged nylon membranes (Hybond N+; Amersham Biosciences, Orsay, France). One μg of RsaI-restricted genomic DNA from the driver and each tester DNA was randomly labeled with the Dig-High Prime system (Roche Applied Science, Neuilly sur Seine, France) and then used as probes to separately hybridize the blots. The digoxigenin-labeled probes were detected by the Dig color detection kit (Roche).
The distribution of tester-specific fragments was also evaluated among S. xylosus strains. Genomic DNAs from 20 strains (Table 1) were used as probes, as described above. The distribution of tester-specific fragments was analyzed by a complete linkage hierarchical cluster analysis using Microsoft Excel software with the XLStat add-in. A distance matrix was calculated by using Sokal and Sneath's (1) similarity coefficient to take into account positive (occurrence) and negative (nonoccurrence) matches.
DNA sequencing and analysis.
Plasmid DNA containing tester-specific inserts was isolated from individual clones using the NucleoSpin Plasmid QuickPure kit (Macherey-Nagel, Hoerdt, France). Sequencing reactions were performed using the BigDye terminator as recommended by the supplier (Applied Biosystems, Courtaboeuf, France) and analyzed by using an ABI 310 automated DNA sequencer. Bioinformatic analyses were performed from Web-based servers. The sequences were analyzed by BLASTX and/or BLASTN searches against the NCBI GenBank database. Based on the analysis, redundant sequences were identified and sequence assembly was done with the CAP3 program (http://bioinformatics.iastate.edu/aat/sas.html). ORF Finder and BLASTX analyses were performed on contigs. When no similarity was found, a PSI-BLAST analysis was performed.
Determination of chromosomal locations of subtracted sequences.
For Southern blotting, digested chromosomal DNAs of S. xylosus S04002, S04009, and 00-1747 strains separated by PFGE were passively transferred onto positively charged nylon membranes (Hybond-N+; Amersham Biosciences). Equal amounts of PCR amplicons of each tester-specific insert were pooled and randomly labeled using the Dig-High Prime system (Roche Applied Science). After hybridizations, detection was performed using the Dig chemiluminescence detection kit following the supplier's instructions (Roche Applied Science). To confirm the localization of tester-specific inserts, chromosomal DNA restriction fragments of interest were excised from the PFGE gel, rinsed three times for 30 min with 10 mM Tris-HCl, pH 8.5, and digested with RsaI (Promega) according to the supplier's recommendations. DNA fragments were thus extracted from agarose and purified before labeling with the AlkPhos direct labeling kit (Amersham Biosciences) and hybridized to the dot blot containing the subtracted unique sequence as described for the differential screening.
Nucleotide sequence accession numbers.
Insert sequences of the subtracted libraries have been deposited in GenBank and given accession numbers ER935832 to ER935953.
RESULTS AND DISCUSSION
PFGE analysis.
The 24 S. xylosus strains were characterized by PFGE patterns after genomic DNA restriction with the SmaI enzyme (Fig. 1). A large diversity of PFGE patterns was found between strains, as 18 distinct genetic profiles were observed. The genetic relatedness of the S. xylosus strains ranged from 10 to 100% (Fig. 1). Employing a cutoff similarity value of 70%, one cluster exclusively composed of S. xylosus strains used as a starter culture was identified, while all the other strains had their own macro-restriction profiles, except for the S01007 and S01004 strains, used as starter culture, and the strains S04016 and S04018, which were both cow mastitis isolates. Cluster analysis did not show any relationship between the origin of S. xylosus isolates and their SmaI PFGE results and confirmed the intraspecies genomic differences observed by Martin et al. (19). From our study, three strains with heterogenic profiles and of three different origins were chosen to represent S. xylosus genetic diversity: S. xylosus S04002, used as a starter culture; S04009, isolated from cow mastitis; 00-1747, responsible for mouse dermatitis.
FIG. 1.
Unweighted pair group method analysis dendrogram with Dice coefficient of PFGE profiles of 24 SmaI macro-restricted S. xylosus chromosomes.
Suppressive-subtractive hybridizations of S. xylosus C2a from S04002, S04009, or 00-1747.
Following SSH, three DNA subtraction libraries were created containing 192, 384, and 320 clones of putative specific DNAs of S. xylosus S04002, S04009, and 00-1747, respectively. The number of clones corresponded to the proportion of chromosome size differences of the three tester strains evaluated in our previous study (5). After PCR amplification of the cloned inserts, clones giving no amplicon or multiple amplicons were discarded. To check the specificity of cloned inserts of the three libraries, PCR products of the inserts were arrayed and hybridized with the genomic DNA probes of the three respective tester strains or the driver C2a strain. From S. xylosus S04002, S04009, and 00-1747 DNA subtraction libraries, a total of 75, 105, and 92 DNA fragments were found to be specific to the tester strains, respectively. All these DNA fragments were two-end sequenced, and 40, 43, and 39 different sequences were identified, respectively (Table 2). A total of 29.67, 22.78, and 25.42 kb were extracted from the S. xylosus S04002, S04009, and 00-1747 genomes, respectively. The sizes of these specific DNA fragments varied from 125 to 1,715 bp, with an average size of 700 bp. Their G+C contents were 30.3% for those of the S04002 subtracted library, 32.8% for the S04009 subtracted library, and 31.1% for the 00-1747 subtracted library. These GC percentages were at the lower limit of S. xylosus G+C content, estimated to be between 30 and 36% (31). SSH has thus been used successfully for the extraction of the differential genetic content of S. xylosus strains arising from various niches.
TABLE 2.
S. xylosus S04002-, S04009-, and 00-1747-specific sequences with significant protein matches in the GenBank database
| Function group and predicted encoded protein | Fragment | E value | % Identity | Organism | Accession no. | Homology found
|
||
|---|---|---|---|---|---|---|---|---|
| S04002 | S04009 | 00-1747 | ||||||
| Metabolism | ||||||||
| Biotin metabolism | ||||||||
| Biotin synthase BioB | E5 | 2.00E-44 | 85 | Staphylococcus aureus COL | ER935916 | + | ||
| E129 | 2.00E-34 | 80 | S. aureus COL | ER935940 | + | |||
| Adenosylmethionine-8-amino-7-oxononanoate aminotransferase BioA BioD | E5 | 1.00E-17 | 62 | S. aureus Mu50 | ER935916 | + | ||
| D128 | 6.00E-125 | 82 | S. aureus COL | ER935891 | + | |||
| D128 | 4.00E-26 | 63 | Staphylococcus epidermidis RP62A | ER935891 | + | |||
| Myo-inositol catabolism | ||||||||
| IolA | Mb262 | 5.00E-133 | 74 | Bacillus licheniformis ATCC 14580 | ER935857 | + | + | |
| D76 | 6.00E-40 | 65 | Geobacillus kaustophilus HTA426 | ER935886 | + | |||
| IolB | Mb11 | 1.00E-61 | 52 | B. licheniformis ATCC 14580 | ER935832 | + | + | |
| IolC | Mb11 | 7.00E-59 | 63 | Bacillus halodurans C-125 | ER935832 | + | + | |
| IolC (putative) | M141 | 1.00E-30 | 66 | B. licheniformis ATCC 14580 | ER935866 | + | + | |
| IolD | M141 | 8.00E-71 | 70 | B. halodurans C-125 | ER935866 | + | + | |
| IolG | E126 | 2.00E-64 | 50 | Bacillus cereus E33L | ER935938 | + | ||
| IolE | Mb118 | 2.00E-79 | 63 | Clostridium tetani E88 | ER935843 | + | + | |
| Pentose and glucuronate interconversion | ||||||||
| d-Arabino-3-hexulose-6-phosphate formaldehyde lyase | E51 | 2.00E-14 | 95 | Staphylococcus saprophyticus ATCC 15305 | ER935923 | + | + | |
| Putative hexulose-6-phosphate synthase | E28 | 4.00E-75 | 95 | S. saprophyticus ATCC 15305 | ER935918 | + | + | |
| Putative 6-phospho-3-hexuloisomerase | E28 | 9.00E-77 | 83 | S. saprophyticus ATCC 15305 | ER935918 | + | + | |
| UTP-glucose-1-phosphate uridyltransferase | E36 | 7.00E-75 | 90 | S. saprophyticus ATCC 15305 | ER935921 | + | ||
| 2-dehydro-3-deoxyphosphogluconate aldolase | Mb275 | 3.00E-78 | 63 | Bacillus thuringiensis serovar Israelensis ATCC 35646 | ER935858 | + | + | |
| 2-dehydro-3-deoxygluconokinase PfkB | D151 | 5.00E-63 | 55 | Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 | ER935896 | + | ||
| D242 | 6.00E-29 | 49 | Bacillus weihenstephanensis KBAB4 | ER935906 | + | + | ||
| Alpha/beta hydrolase fold family protein | Mb59 | 3.00E-27 | 52 | Flavobacteriales bacterium HTCC2170 | ER935840 | + | ||
| Fructose and mannose metabolism | ||||||||
| Phosphomannomutase | E127 | 4.00E-83 | 65 | S. epidermidis RP62A | ER935939 | + | ||
| Hexitol dehydrogenase | Mb27 | 4.00E-94 | 69 | S. aureus COL | ER935835 | + | + | |
| M139 | 1.00E-14 | 72 | S. aureus COL | ER935864 | + | + | ||
| Sorbitol dehydrogenase | M139 | 3.00E-10 | 62 | S. aureus COL | ER935864 | + | + | |
| Fermentative metabolism | ||||||||
| Acetoin (diacetyl) reductase ButA | E129 | 9.00E-57 | 81 | S. epidermidis ATCC 12228 | ER935940 | + | ||
| α-Acetolactate synthase AlsS | E190 | 0.0 | 97 | S. saprophyticus ATCC 15305 | ER935953 | + | ||
| Amino acid metabolism | ||||||||
| Saccharopine dehydrogenase-related protein | D85 | 2.00E-04 | 54 | Lactococcus lactis subsp. cremoris SK11 | ER935887 | + | ||
| D218 | 2.00E-35 | 64 | L. lactis subsp. cremoris SK11 | ER935905 | + | |||
| Sarcosine oxidase alpha subunit | D129 | 5.00E-77 | 51 | B. thuringiensis serovar Konkukian 97-27 | ER935892 | + | ||
| Sarcosine oxidase beta subunit | D298 | 4.00E-45 | 41 | B. cereus ATCC 14579 | ER935912 | + | ||
| Betaine aldehyde dehydrogenase | D298 | 9.00E-25 | 63 | B. weihenstephanensis KBAB4 | ER935912 | + | ||
| Dihydrodipicolinate synthase | D176 | 2.00E-06 | 42 | Rickettsia felis URRWXCal2 | ER935899 | + | ||
| Other | ||||||||
| Lipase | E179 | 4.00E-92 | 92 | S. saprophyticus | ER935951 | + | ||
| Putative lipase | D218 | 2.00E-88 | 83 | S. saprophyticus ATCC 15305 | ER935905 | + | ||
| UDP-glucose 4-epimerase | D44 | 2.00E-20 | 59 | G. kaustophilus | ER935881 | + | ||
| Dihydroorotase | D23 | 1.00E-71 | 56 | B. cereus G9241 | ER935879 | + | + | |
| Putative nucleoside diphosphate-sugar epimerase DTDP-D-glucose 4,6-dehydratase | D46 | 1.00E-55 | 82 | S. saprophyticus ATCC 15305 | ER935882 | + | + | |
| COG1670, putative acetyltransferase | D96 | 5.00E-45 | 54 | S. saprophyticus ATCC 15305 | ER935888 | + | + | |
| Aldehyde dehydrogenase AldA | D106 | 4.00E-55 | 69 | Bacillus sp. B14905 | ER935889 | + | ||
| Riboflavin kinase RibR | D213 | 9.00E-10 | 40 | B. subtilis subsp. subtilis 168 | ER935904 | + | + | + |
| COG2936, putative peptidase S9, prolyl oligopeptidase precursor | D273 | 5.00E-50 | 81 | S. aureus RF122 | ER935909 | + | ||
| Similar to oxidoreductase | Mb127 | 8.00E-16 | 36 | S. aureus Mu50 | ER935844 | + | + | |
| COG0655, putative iron-sulfur flavoprotein (hypothetical protein SAS0063) | E184 | 1.00E-16 | 53 | S. aureus MSSA476 | ER935952 | + | ||
| Short-chain alcohol dehydrogenase | E139 | 9.00E-73 | 98 | S. saprophyticus ATCC 15305 | ER935942 | + | ||
| COG4221, putative short-chain alcohol dehydrogenase of unknown specificity | E51 | 2.00E-42 | 100 | S. saprophyticus ATCC 15305 | ER935923 | + | ||
| COG1063, alcohol dehydrogenase | M65 | 2.00E-141 | 97 | S. saprophyticus ATCC 15305 | ER935860 | + | + | |
| Regulatory | ||||||||
| Putative regulator of HxlAB (hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase) | E111 | 1.00E-35 | 98 | S. saprophyticus ATCC 15305 | ER935932 | + | ||
| Transcriptional regulator, LysR family probable AlsR regulator | E173 | 4.00E-37 | 33 | Bacillus clausii KSM-K16 | ER935950 | + | ||
| Putative regulator of sorbitol operon | D7 | 4.00E-18 | 28 | Clostridium beijerincki NCIMB 8052 | ER935875 | + | ||
| Sorbitol operon activator | D254 | 2.00E-24 | 39 | C. beijerincki KSM-K16 | ER935908 | + | ||
| Transcriptional regulator putative hydrogen peroxide-inducible genes activator | E85 | 3.00E-81 | 99 | S. saprophyticus ATCC 15305 | ER935928 | + | + | |
| PTS multidomain regulator BglG | Mb48 | 6.00E-41 | 44 | S. aureus MSSA476 | ER935839 | + | ||
| D63 | 2.00E-16 | 45 | S. aureus COL | ER935884 | + | + | ||
| Putative PTS multidomain regulator EIIA putative fructose/mannitol specific | Mb147 | 6.00E-45 | 45 | S. aureus MSSA476 | ER935850 | + | + | |
| PTS system enzyme II B component putative sorbitol specific | M149 | 6.00E-35 | 87 | S. aureus Mu50 | ER935867 | + | + | |
| D53 | 2.00E-36 | 84 | S. aureus Mu50 | ER935883 | + | + | ||
| PTS transport system IIA component putative galactitol specific | D53 | 1.00E-07 | 44 | S. aureus MRSA252 | ER935883 | + | + | |
| Accessory gene regulator C agrC | E154 | 5.00E-110 | 83 | S. saprophyticus ATCC 15305 | ER935944 | + | ||
| Protein SSP2020 putative ebsC transcription regulation protein | M160 | 6.00E-40 | 61 | S. saprophyticus ATCC 15305 | ER935868 | + | + | |
| NmrA-like transcriptional regulator | D11 | 8.00E-22 | 38 | Salinispora tropica CNB-440 | ER935876 | + | ||
| Transcriptional regulator, LysR family domain protein | E184 | 4.00E-56 | 66 | S. aureus MSSA476 | ER935952 | + | ||
| E116 | 8.00E-13 | 46 | S. aureus USA300 | ER935933 | + | |||
| Transcriptional regulator, GntR family/aminotransferase, class I | E3 | 1.00E-53 | 57 | S. aureus COL | ER935914 | + | + | |
| Mb46 | 1.00E-49 | 59 | S. aureus COL | ER935838 | + | |||
| Mb145 | 9.00E-20 | 81 | S. aureus COL | ER935849 | + | |||
| COG1167, putative transcriptional regulator, GntR family | E100 | 2.00E-165 | 98 | S. saprophyticus ATCC 15305 | ER935929 | + | ||
| Transcriptional regulator, AraC/XylS family | D141 | 2.00E-43 | 58 | Oceanobacillus iheyensis HTE831 | ER935895 | + | + | |
| COG1733, putative transcriptional regulator | E139 | 3.00E-17 | 93 | S. saprophyticus ATCC 15305 | ER935942 | + | + | |
| Cell membrane/surface structure | ||||||||
| Putative autolysin (mannosyl-glycoprotein endo-β-N-acetylglucosamidase) | E73 | 3.00E-55 | 99 | S. saprophyticus ATCC 15305 | ER935926 | + | + | + |
| E130 | 2.00E-178 | 97 | S. saprophyticus ATCC 15305 | ER935941 | + | |||
| Autolysin (mannosyl-glycoprotein endo-β-N-acetylglucosamidase) | D294 | 5.00E-07 | 66 | S. epidermidis ATCC 12228 | ER935911 | + | + | + |
| 67-kDa myosin-cross-reactive streptococcal antigen-like protein | E110 | 3.00E-26 | 94 | S. epidermidis | ER935931 | + | ||
| smart00255, putative Toll-interleukin receptor | D157 | 1.00E-35 | 69 | S. aureus MSSA476 | ER935897 | + | ||
| Glycosyl transferase, family 2, CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase | E121 | 1.00E-32 | 40 | Clostridium phytofermentans ISDg | ER935935 | + | ||
| Lipopolysaccharide modification acyltransferase | Mb127 | 8.00E-19 | 78 | S. epidermidis ATCC 1228 | ER935844 | + | + | |
| Biofilm-associated protein Bap | D137 | 1.00E-18 | 28 | Staphylococcus hyicus | ER935894 | + | ||
| LPXTG cell wall surface anchor family protein, hypothetical protein SSPP105 | E25 | 1.00E-27 | 98 | S. saprophyticus ATCC 15305 | ER935917 | + | ||
| Cell wall surface anchor family protein, hypothetical protein SSPP105 | E34 | 2.00E-114 | 98 | S. saprophyticus ATCC 15305 | ER935920 | + | ||
| Putative membrane protein of unknown function | D157 | 3.00E-05 | 36 | S. saprophyticus ATCC 15305 | ER935897 | + | ||
| COG3152, predicted membrane protein | D158 | 0.056 | 31 | Staphylococcus haemolyticus | ER935898 | + | ||
| COG3949, SSP0396 uncharacterized membrane protein | D294 | 9.00E-24 | 39 | S. saprophyticus ATCC 15305 | + | + | + | |
| Resistance | ||||||||
| Methicillin resistance protein MecA1 | Mb201 | 4.00E-38 | 52 | Staphylococcus sciuri | ER935854 | + | ||
| MecR1, methicillin resistance regulatory protein | M116 | 5.00E-73 | 58 | S. aureus | ER935862 | + | ||
| Metallothiol transferase FosB (fosfomycin resistance protein) | M130 | 3.00E-69 | 89 | S. haemolyticus JCSC1435 | ER935863 | + | ||
| MphBM macrolide 2′-phosphotransferase II | M180 | 1.00E-92 | 86 | S. aureus | ER935869 | + | + | |
| Micrococcin P1 peptide synthetase | Mb107 | 6.00E-123 | 58 | Staphylococcus equorum | ER935842 | + | + | |
| Putative tetronasin resistance transmembrane protein | D178 | 3.00E-18 | 36 | B. halodurans C-125 | ER935901 | + | + | |
| Aminoglycoside adenyltransferase | D121 | 7.00E-51 | 79 | S. saprophyticus ATCC 15305 | ER935890 | + | ||
| Arsenate reductase | E125 | 3,00E-73 | 100 | S. xylosus plasmid Psx267 | ER935937 | + | + | + |
| Arsenic efflux pump protein | E125 | 2.00E-61 | 100 | S. aureus | ER935937 | + | + | + |
| Arsenic efflux pump membrane protein | D284 | 4.00E-19 | 100 | S. xylosus(Psx267) | ER935910 | + | + | + |
| Arsenic resistance operon repressor | D284 | 4.00E-49 | 89 | S. aureus N315 | ER935910 | + | + | + |
| Membrane protein TerC, possibly involved in tellurium resistance | D16 | 1.00E-11 | 78 | S. saprophyticus ATCC 15305 | ER935877 | + | ||
| Probable drug resistance transporter of EmrB/QacA family | Mb12 | 4.00E-19 | 46 | Listeria monocytogenes 4b F2365 | ER935833 | + | + | |
| Putative drug exporter of RND superfamily | E4 | 8.00E-91 | 74 | S. saprophyticus ATCC 15305 | ER935915 | + | ||
| COG2329, putative antibiotic biosynthesis monooxygenase | M266 | 2.00E-27 | 55 | S. aureus JH1 | ER935873 | + | + | |
| Transport | ||||||||
| l-lactate permease | E117 | 2.00E-47 | 92 | S. saprophyticus ATCC 15305 | ER935934 | + | ||
| Na+:myo-inositol cotransporter | Mb42 | 1,00E-56 | 70 | Bacillus clausii KSM-K16 | ER935837 | + | + | |
| Amino acid permease-associated region lysine-specific permease precursor | D132 | 4.00E-58 | 53 | Pseudomonas fluorescens PfO-1 | ER935893 | + | ||
| PTS enzyme IIC component putative galactitol specific I | M283 | 9.00E-34 | 57 | Salmonella enterica serovar Typhimurium LT2 | ER935874 | + | ||
| Ferrous iron transport protein B | Mb142 | 8.00E-42 | 76 | S. epidermidis RP62A | ER935848 | + | + | |
| ATPase copper transport | E158 | 4.00E-62 | 86 | S. aureus USA300 | ER935947 | + | ||
| D182 | 4.00E-110 | 85 | S. aureus USA300 | ER935902 | + | |||
| Putative permease, putative role in amino acid adenylation | E157 | 2.00E-51 | 81 | S. aureus USA300 | ER935946 | + | + | |
| Putative permease of drug and metabolite transporter superfamily | E38 | 6.00E-122 | 98 | S. saprophyticus ATCC 15305 | ER935922 | + | + | |
| Putative transporter, EamA family | E77 | 8.00E-50 | 53 | S. aureus Mu50 | ER935927 | + | ||
| Putative ABC-type nitrate sulfonate taurine bicarbonate transport system ATPase component | M237 | 3.00E-65 | 89 | S. saprophyticus ATCC 15305 | ER935871 | + | + | |
| TRAP-T family transporter, putative tripartite tricarboxylate transporter | D177 | 2.00E-17 | 49 | Rhodobacter sphaeroides 2.4.1 | ER935900 | + | + | |
| ABC transporter, ATP-binding/permease protein | D19 | 6.00E-18 | 73 | S. aureus COL | ER935878 | + | ||
| Putative ABC transporter permease | E61 | 1.00E-10 | 89 | S. saprophyticus ATCC 15305 | ER935925 | + | ||
| Major facilitator family transporter | E142 | 3.00E-32 | 54 | Listeria innocua Clip11262 | ER935943 | + | + | |
| Putative permease of the major facilitator superfamily | D301 | 4.00E-20 | 96 | S. saprophyticus ATCC 15305 | ER935913 | + | ||
| Phage-related and IS elements | ||||||||
| IS605/IS200-like transposase | Mb168 | 3.00E-86 | 89 | S. epidermidis ATCC 1228 | ER935851 | + | + | |
| Recombinase Sin | E110 | 3.00E-75 | 94 | S. epidermidis ATCC 12228 | ER935931 | + | ||
| Integrase | Mb194 | 7.00E-66 | 64 | S. aureus RF122 | ER935852 | + | ||
| Phage infection protein | E168 | 0.0 | 99 | S. epidermidis RP62A | ER935949 | + | + | |
| Phage terminase large subunit, 77ORF003 | Mb33 | 5.00E-68 | 62 | Bacteriophage 77 | ER935836 | + | ||
| ORF001 | Mb66 | 3.00E-11 | 60 | Bacteriophage 2638A | ER935841 | + | ||
| ORF094 | Mb66 | 7.00E-13 | 79 | Bacteriophage 2638A | ER935841 | + | ||
| ORF028 | Mb66 | 3.00E-06 | 77 | Bacteriophage 2638A | ER935841 | + | ||
| Conserved phage protein probable ORF029 of bacteriophage 37 | Mb131 | 1.00E-16 | 56 | S. haemolyticus JCSC1435 | ER935845 | + | ||
| phiPVL ORF050-like protein | Mb133 | 3.00E-06 | 36 | S. aureus USA300 | ER935846 | + | ||
| ORF025 | Mb137 | 3.00E-08 | 51 | Bacteriophage EW | ER935847 | + | ||
| ORF60 | Mb215 | 2.00E-06 | 46 | S. phage 187 | ER935856 | + | ||
| ORF077 | Mb215 | 0.012 | 41 | Bacteriophage EW | ER935856 | + | ||
| Capsid protein, 77ORF006 | M85 | 7.00E-55 | 80 | Bacteriophage 77 | ER935861 | + | ||
| 77ORF042 | M85 | 1.00E-12 | 47 | Bacteriophage 77 | ER935861 | + | ||
| ORF001 | M256 | 3.00E-59 | 59 | Bacteriophage 2638A | ER935872 | + | ||
| Phage helicase, C terminal | M200 | 2.00E-22 | 31 | Clostridium thermocellum ATCC 27405 | ER935870 | + | ||
| Integrase, ORF010 | D65 | 2.00E-30 | 56 | Bacteriophage 52A | ER935885 | + | ||
| Hypothetical protein SSPP103, phage infection protein | E160 | 3.00E-11 | S. saprophyticus ATCC 15305 | ER935948 | + | + | + | |
| Hypothetical protein SAR0381, probable phage protein | Mb279 | 5.00E-09 | 38 | S. aureus MRSA252 | ER935859 | + | ||
| Unknown function | ||||||||
| COG3877, unnamed protein product | E31 | 7.00E-37 | 79 | S. haemolyticus JCSC1435 | ER935919 | + | ||
| Hypothetical protein SSP0169 | E55 | 2.00E-09 | 85 | S. saprophyticus ATCC 15305 | ER935924 | + | ||
| Similar to unknown protein SH2317 | E108 | 7.00E-09 | 60 | S. haemolyticus | ER935930 | + | ||
| Hypothetical protein SE2200 | E123 | 3.00E-09 | 89 | S. epidermidis ATCC 12228 | ER935936 | + | + | |
| Hypothetical protein SH0055 | E156 | 1.00E-34 | 70 | S. haemolyticus JCSC1435 | ER935945 | + | ||
| COG1284, uncharacterized conserved protein SSP1949 | Mb26 | 2.00E-36 | 84 | S. saprophyticus ATCC 15305 | ER935834 | + | + | |
| Hypothetical protein cn33 | Mb198 | 2.00E-19 | 37 | S. epidermidis bacteriophage CNPH82 | ER935853 | + | ||
| Hypothetical protein SH1793 | Mb215 | 0.001 | 54 | S. haemolyticus JCSC1435 | ER935856 | + | ||
| Hypothetical protein, no similarity found | Mb213 | ER935855 | + | |||||
| Hypothetical protein SAB0188 | M139 | 7.00E-07 | 64 | S. aureus RF122 | ER935864 | + | + | |
| Hypothetical protein Mbar_A2529 | M266 | 2.00E-12 | 30 | Methanosarcina barkeri strain Fusaro | ER935873 | + | + | |
| Hypothetical protein | D39 | 8.00E-12 | 45 | S. aureus Mu50 | ER935880 | + | ||
| Truncated hypothetical protein | D121 | 1.00E-10 | 83 | S. saprophyticus ATCC 15305 | ER935890 | + | ||
| Hypothetical protein SSP2368 | D197 | 4.00E-05 | 49 | S. saprophyticus ATCC 15305 | ER935903 | + | + | |
| No significant similarity found | M140 | ER935865 | + | |||||
| No significant similarity found | D246 | ER935907 | + | |||||
Identification of specific sequences of S. xylosus S04002, S04009, and 00-1747 and a large proportion of specific metabolism sequences.
The 122 tester-specific fragments were analyzed by BLASTX and PSI-BLAST searches and led to the identification of 149 open reading frames (ORFs), as shown in Table 2. These ORFs were assigned to one of seven classes: metabolism, regulatory, cell membrane/surface structure, resistance, transport, phage-related structure, and IS elements and unknown functions. Fourteen ORFs resembled hypothetical bacterial genes of unknown function, while two fragments had no significant matches with known sequences. The other ORFs had significant similarity to sequences with known functions largely described in the Staphylococcus genus. However, for the S. xylosus 00-1747 strain, 20 of its 47 detected ORFs corresponded to genes from other bacterial genera (Table 2). A majority of ORFs seemed to be related to specific metabolisms as biotin, myo-inositol, and sorbitol, to amino acid and fermentative metabolisms, and to pentose and glucuronate interconversions (Table 2).
In the S. xylosus S04009 strain genome, fragments corresponding to five genes involved in myo-inositol metabolism as well as the Na+:myo-inositol cotransporter were subtracted (Table 2). Myo-inositol is a hexitol which is a constituent of virtually every living cell; it is synthesized by plants and is essential for humans and animals. Different bacterial species (Klebsiella spp., Salmonella spp., Pseudomonas spp., Bacillus subtilis, etc.) are able to use myo-inositol as carbohydrate source, but genes implicated in this catabolism have been described in few species (16). In Bacillus subtilis, two iol operons were described, including an operon of 10 genes, iolABCDEFGHIJ, and an operon of two genes, iolRS (40). Until now, these genes had never been described in a Staphylococcus genus.
Five ORFs of the S. xylosus 00-1747 strain were related to lysine and glycine metabolism genes, none of which corresponded to staphylococcal genes (Table 2). Sarcosine oxidase and betaine aldehyde dehydrogenase were shown to be implicated in metabolism of glycine betaine, an effective osmoprotectant (1, 4). Only betaine aldehyde dehydrogenase and dihydropicolinate synthase were identified in actual staphylococcal sequenced genomes, but these genes showed no significant match at the DNA level with these ORFs. Sarcosine oxidase and saccharopine dehydrogenase have never been observed in staphylococcal genomes. The fact that the S. xylosus 00-1747 strain contained numerous nonstaphylococcal homology ORFs might be explained by its origin. This strain was isolated from mouse dermatitis in Korea. The particular origin of this strain suggests that ecological and/or geographical divergences in S. xylosus species may exist.
From the S. xylosus meat starter S04002 subtraction, fragments corresponding to fermentative metabolism were found: an acetolactate synthase which transforms pyruvate in acetolactate, its regulator AlsR, and an acetoin dehydrogenase which transforms acetoin into diacetyl, as well as a lactate permease also implicated in anaerobic metabolism (Table 2). Acetoin and diacetyl are aroma compounds. Their production in fermented meat products has been characterized with a dairy product odor (20). Some S. xylosus strains produce these aroma compounds more than other staphylococcus species (34). This property is variable between S. xylosus strains, and a correlation between strain persistence in ripened meat and the capacity of strains to produce acetoin has been shown (9). In lactic acid bacteria isolated from fermented sausages, the production of acetoin is stimulated by a low pH and a low sugar concentration, conditions that could be found at the end of the ripening process (9). Recently, Fuchs et al. (8) showed that in S. aureus the production of acetoin and lactate was stimulated by anaerobic growth conditions.
The large number of differential genes predicted to be involved in metabolism strongly suggested a wide adaptation of S. xylosus strains to their environment and to the availability of the different substrates.
Horizontal transfer in S. xylosus S04009.
The S. xylosus S04009 strain isolated from cow mastitis showed 12 specific subtracted fragments corresponding to phage-related sequences (Table 2). The presence of at least three different bacteriophages, 77, 2638A, and EW, was deduced, and an IS605/IS200 transposase was also detected. Three phage-related proteins were found in S. xylosus S04002 and one in 00-1747 subtracted fragments. Phages in S. aureus are well studied: 37 phage genomes are reported in the GenBank database (3, 17). To our knowledge, there are few studies of phages from coagulase-negative staphylococci, and only two phages from Staphylococcus epidermidis have been completely sequenced and molecularly characterized (3). Other putative evidence of horizontal transfer in S. xylosus S04009 is the presence of specific sequences corresponding to antibiotic resistance as methicillin, fosfomycin, and macrolide resistance proteins (Table 2).
Particular S. xylosus surface structures.
Specific surface structures were found in the S. xylosus tester strains. The meat starter strain S. xylosus S04002 possessed a specific fragment corresponding to a 67-kDa myosin-cross-reactive streptococcal antigen-like protein. This protein family is thought to have structural features in common with the beta chain of the class II antigens. In Streptococcus pyogenes it reacts with antimyosin antibodies and plays a role in the pathogenesis of streptococcal infections (13). This gene is found in other Staphylococcus sequenced genomes, such as S. aureus, S. epidermidis, or Staphylococcus haemolyticus, but its function in those bacteria is still unknown. The S. xylosus 00-1747 strain implicated in mouse dermatitis contained a specific fragment resembling a Toll-interleukin receptor (TIR). Such a eukaryotic signal-mimicking molecule has been described in Salmonella enterica serovar Enteriditis (24). It was demonstrated that it is an important virulence factor in vivo by modulating host defense mechanisms involved in regulation of NF-κB and caspase activation. Experiments in mice infected with a Salmonella strain deficient in TIR showed reduced lethality. Predicted TIR-like domains are widespread and show an unusual distribution in bacterial species. A lateral transfer of this gene has been supposed (24).
Specific fragments shared by S. xylosus S04002, S04009, and 00-1747.
The presence of common fragments in the three S. xylosus tester strain chromosomes was detected by dot blot hybridization using genomic DNA of the three strains as a probe. The three strains shared only six specific fragments. Those six common fragments corresponded to arsenic resistance genes with the arsenate reductase and the arsenical efflux pump membrane protein, a riboflavin kinase, a hypothetical phage infection protein, and two fragments which were similar to an autolysin, a mannosyl-glycoprotein endo-β-N-acetylglucosamidase (Table 2).
Among the 122 specific fragments, 66 hybridized with S. xylosus S04009 genomic DNA. This mastitis isolate contained the highest number of specific fragments in its genome.
The S. xylosus strains S04009 and 00-1747 isolated from mastitis and dermatitis, respectively, shared the highest number of specific fragments, with 29 in common. They had specific sequences similar to a hexitol-specific PTS system, probably sorbitol specific. Indeed, other fragments related to sorbitol assimilation were isolated, such as a sorbitol dehydrogenase and the putative regulator of the sorbitol operon. Those two commensal strains also shared numerous specific DNA sequences implicated in resistance to antibiotics and other toxic components, such as tellurium, but also antibiotic synthesis-related sequences, such as a monooxygenase and a lipopeptide, micrococcin P1. These two strains also had in common the feoB gene, implicated in ferrous ion transport. This gene was identified in S. aureus and S. epidermidis genomes but had not been characterized (10). The ability to acquire iron (Fe) in a host is known to be an essential attribute of most pathogenic bacteria (2).
The meat starter strain S04002 and the strain S04009 isolated from mastitis shared 20 fragments. They have in common the iolABCDE myo-inositol genes but not the iolG gene, which was present in S04002 but absent in S04009. IolG corresponds to a myo-inositol 2-dehydrogenase, which is the first enzyme of myo-inositol catabolism, degrading the incoming myo-inositol into 2-keto-myo-inositol. The iolFHIJ and iolRS genes described in B. subtilis were not detected in the subtracted libraries (40). The gene iolR corresponds to a negative regulator of the iol operon, iolF is related to a myo-inositol transporter, and iolE codes for a 2-keto-myo-inositol dehydratase, while the functions of the other iol genes are still unclear in B. subtilis (40).
The meat starter S. xylosus S04002 had only nine fragments in common with the mouse dermatitis strain 00-1747. The biotin synthesis-related sequences obtained after genomic subtraction of these two strains seemed to be drastically different at the DNA level, and none of these fragments hybridized with the two genomic DNAs.
Conservation of tester-specific fragments within S. xylosus strains.
The extent of polymorphism among S. xylosus strains for the specific fragments was assessed. The RsaI-digested genomic DNAs of 20 different S. xylosus strains, 10 used as starters in the food industry and 10 isolated from cow or goat mastitis (Table 1), were hybridized with the 122 specific fragment sequences of the three tester strains. Four specific fragments (D39, D129, D137, and D158) from the S. xylosus 00-1747 library and four (Mb33, Mb66, M85, and M130) from the S. xylosus S04009 library were absent in the genomic DNA of the 20 S. xylosus strains. No specific fragment was present in all the S. xylosus genomes. The subtracted fragment most commonly found, which hybridized with 21 of the 23 S. xylosus strains (including the three tester strains), was fragment D294, corresponding to a hypothetical membrane protein and a CHAP domain corresponding to an amidase domain frequently found in autolysin precursor. A hierarchic ascendant classification gathered the strains into two clusters named I and II (Fig. 2).
FIG. 2.
Distribution of tester-specific fragments within S. xylosus strains.
Cluster I comprised five strains, all isolated from animal-associated opportunistic infections, sharing eight fragments: Mb26, Mb48, Mb147, M139, M149, D63, D96, and D213 (Table 2). These fragments corresponded to metabolic genes as PTS system specific to hexitol and its associated antiterminator Bgl, a sorbitol dehydrogenase, an acetyltransferase of unknown specificity, and a riboflavin kinase. Three of these fragments (Mb147, M139, and M149) corresponding to a hexitol assimilation system were absent in cluster II except one strain. The cluster could be divided into two subgroups named Ia and Ib. The Ia subgroup included the tester strain S. xylosus S04009 and the strain S04013, both isolated from cow mastitis. These two strains had 43 of the 122 specific fragments in common, 25 of which were from the S. xylosus S04009 library, showing a highly related genetic content between these two strains. Subgroup Ib comprised the tester strain S. xylosus 00-1747, isolated from mouse dermatitis, and two strains isolated from goat mastitis, S04019 and S04020. The genomes of these three strains shared 20 fragments, 9 of which were common to the genome of the tester strain S04009 of subgroup Ia.
Cluster II comprised 18 strains that shared a few fragments with cluster I, and they shared the E154 fragment, which was absent in cluster I except for one strain. This fragment corresponded to the accessory gene regulator C (agrC), which is a staphylococcal conserved regulator. It has been observed that there is a high genetic variability of the agr locus in Staphylococcus species and particularly in the agrC sequence (26). This variability was observed not only between Staphylococcus species but also between subpopulations of a single species (6). In S. aureus, agrD and agrC polymorphism permitted definition of a group broadly correlated with genotype which was correlated with pathotype (26). The existence of at least two different genotypic clusters (I and II) within the S. xylosus species was confirmed by the argC polymorphism distribution observed.
Cluster II could be divided into two subgroups named IIa and IIb. Subgroup IIa comprised the tester strain S04002 and the S01001 and S01008 strains of meat origin and six other strains isolated from mastitis (Fig. 2; Table 1). Within this heterogeneous subgroup, only two fragments were found to be in common: D294, described previously to be the most shared fragment between the 23 S. xylosus strains, and the fragment E154, already mentioned above. Subgroup IIb comprised eight S. xylosus strains used as starters (Fig. 2; Table 1) and one S. xylosus isolated from cow mastitis S04016. Twenty-one fragments were common between all these strains. They corresponded to the six myo-inositol metabolism genes (iolABCDEG), all the fermentative metabolism genes (als, acetoin reductase, and lactate permease genes), and the lipase gene (E179). The presence of iol genes in the majority of starter S. xylosus strains and their absence in the actual staphylococcal sequenced genomes indicate a specific and fortuitous acquisition of these genes by some S. xylosus strains. The presence of fermentative metabolism may be an adaptation to anaerobic growth conditions. In S. aureus, the overexpression of the genes implicated in butanediol fermentation and lactate production were recently observed under fermentative conditions (8). Adaptation to anaerobic conditions and lipase activity could procure a technological advantage for S. xylosus used in food processing.
Location of tester-specific fragments on their respective chromosomes.
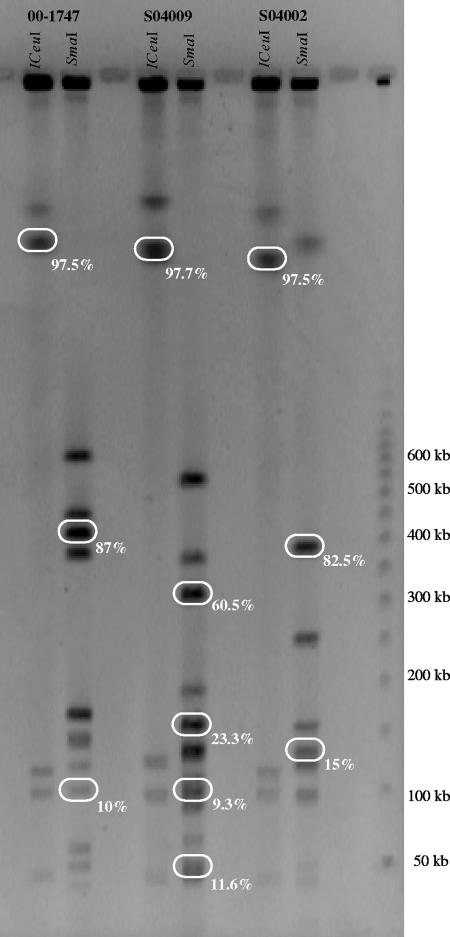
The location of each specific fragment from the three DNA subtraction libraries was analyzed by Southern hybridization. Genomic DNAs of the S. xylosus S04002, S04009, and 00-1747 strains were digested with I-CeuI or SmaI, resolved by PFGE (Fig. 3), and blotted for Southern hybridization with a labeled pool of the specific fragments of each library. The specific fragments were located on the ∼1,100-kb I-CeuI fragment of each tester strain chromosome and on a few SmaI fragments. The specific fragments from the S. xylosus S04002 library were located on the 375-kb and 125-kb SmaI fragments, those of the S. xylosus S04009 were mainly on the 300-kb but also on the 150-kb, 100-kb, and 50-kb SmaI fragments, and the fragments of S. xylosus 00-1747 were located on the 400-kb and 100-kb SmaI fragments (Fig. 3). To confirm the probable colocalization of the SSH library-specific fragments, a second hybridization was performed using PFGE macro-restricted fragments as probes. The distribution of all the specific fragments from the three libraries is shown in Fig. 3. For each of the S. xylosus S04002, S04009, and 00-1747 subtracted libraries, more than 97.5% of inserts were located in the ∼1,100 kb I-CeuI fragment. Most of these fragments were located on the 300-/400-kb SmaI fragments on each S. xylosus tester strain, confirming that numerous specific fragments were colocalized on the S. xylosus S04002, S04009, and 00-1747 chromosomes. In a previous study we demonstrated by positioning the dnaA and gyrA genes, which are often found near the chromosome origin of replication of bacteria (oriC), that in S. xylosus C2a the origin of replication oriC was in the 1,138-kb I-CeuI and 365-kb SmaI fragments (5). To check if the colocalization region corresponded to the oriC of the three tester strain chromosomes, we hybridized dnaA and gyrA probes on I-CeuI or SmaI restriction fragments of S. xylosus S04002, S04009, and 00-1747 strains (data not shown). The genes dnaA and gyrA were located on the ∼1,100-kb I-CeuI fragment and on 300-/400-kb SmaI fragments of the three tester strains. Takeuchi et al. (35) observed in a comparative analysis of the S. haemolyticus genome with the seven S. aureus and the two S. epidermidis sequenced genomes that a region named the “oriC environ” emerged as a chromosomal region unique to each staphylococcal species. It dictates many biological features that characterize each staphylococcal species. For example, in the oriC environ of S. aureus, important virulence determinants such as protein A (spa), coagulase (coa), and the capsule locus [cap5(8)A to -P] were present. This oriC environ region was defined in the downstream region 30 kb from the oriC. It contained the staphylococcal cassette chromosomes, which might serve as a vehicle for introduction of exogenous genes into this region as well as insertion sequence elements and other recombinases serving to excise unused genes. A species-specific upstream region was also observed in some Staphylococcus strains. The authors' theory was that certain useful foreign genes were preserved by being transferred upstream of the oriC, protecting them from insertion sequence-mediated deletion occurring in the oriC environ. It was observed that the oriC environ was similar within a species but diverse among the different species, and the seven S. aureus sequenced strains had the downstream part of the oriC environ in common (35). In this study, we showed that the polymorphism of S. xylosus strains was principally centered on this oriC environ region, a species-specific region and also a strain-specific region in S. xylosus. Analysis of the differential genetic content of different S. xylosus demonstrated that this region is implicated in staphylococcal speciation but also actively participates in species evolution by the acquisition of new metabolism functions, like assimilation of new carbohydrate sources, antibiotic resistance, or factors possibly implicated in virulence. These strain-specific genes may be used as markers in epidemiological or evolutionary studies. The genomic evolution capacity is essential for ubiquitous species such as S. xylosus. Sequencing of this strain-specific “oriC environ” region in numerous Staphylococcus species and numerous strains within a species should provide a better understanding of staphylococcal genome plasticity and the genetic fitness of this genus in response to specialized niches.
FIG. 3.
PFGE patterns of the S. xylosus 00-1747, S04009, and S04002 tester strains. The percentage of specific fragments that hybridized with restriction fragments is detailed for each restriction pattern.
Acknowledgments
We thank Young-Suk Won for giving us the strain implicated in mouse dermatitis and Pascal Rainard for the strains isolated from mastitis. We thank Isabelle Lebert for helpful discussions, Sébastien Masseglia for technical assistance, Brigitte Duclos for secretarial assistance, and David Marsh for revision of the English.
Emilie Dordet-Frisoni is the recipient of a fellowship from the French Ministry of “Education Nationale et Recherche.” This work was financially supported by the ANR project “Genoferment” ANR-05-PNRA-020.
Footnotes
Published ahead of print on 21 September 2007.
REFERENCES
- 1.Boch, J., B. Kempf, and E. Bremer. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartron, M. L., S. Maddocks, P. Gillingham, C. J. Craven, and S. C. Andrews. 2006. Feo transport of ferrous iron into bacteria. Biometals 19:143-157. [DOI] [PubMed] [Google Scholar]
- 3.Daniel, A., P. E. Bonnen, and V. A. Fischetti. 2007. First complete genome sequence of two Staphylococcus epidermidis bacteriophages. J. Bacteriol. 189:2086-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diab, F., T. Bernard, A. Bazire, D. Haras, C. Blanco, and M. Jebbar. 2006. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 152:1395-1406. [DOI] [PubMed] [Google Scholar]
- 5.Dordet-Frisoni, E., R. Talon, and S. Leroy. 2007. Physical and genetic map of the Staphylococcus xylosus C2a chromosome. FEMS Microbiol. Lett. 266:184-193. [DOI] [PubMed] [Google Scholar]
- 6.Dufour, P., S. Jarraud, F. Vandenesch, T. Greenland, R. P. Novick, M. Bes, J. Etienne, and G. Lina. 2002. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald, J. R., S. D. Reid, E. Ruotsalainen, T. J. Tripp, M. Liu, R. Cole, P. Kuusela, P. M. Schlievert, A. Jarvinen, and J. M. Musser. 2003. Genome diversification in Staphylococcus aureus: molecular evolution of a highly variable chromosomal region encoding the staphylococcal exotoxin-like family of proteins. Infect. Immun. 71:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, S., J. Pane-Farre, C. Kohler, M. Hecker, and S. Engelmann. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189:4275-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Varona, M., E. M. Santos, I. Jaime, and J. Rovira. 2000. Characterisation of Micrococcaceae isolated from different varieties of chorizo. Food Microbiol. 54:189-195. [DOI] [PubMed] [Google Scholar]
- 10.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotz, F., J. Zabielski, L. Philipson, and M. Lindberg. 1983. DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid 9:126-137. [DOI] [PubMed] [Google Scholar]
- 12.Kessie, G., M. Ettayebi, A. M. Haddad, A. M. Shibl, F. J. al-Shammary, A. F. Tawfik, and M. N. al-Ahdal. 1998. Plasmid profile and antibiotic resistance in coagulase-negative staphylococci isolated from polluted water. J. Appl. Microbiol. 84:417-422. [DOI] [PubMed] [Google Scholar]
- 13.Kil, K. S., M. W. Cunningham, and L. A. Barnett. 1994. Cloning and sequence analysis of a gene encoding a 67-kilodalton myosin-cross-reactive antigen of Streptococcus pyogenes reveals its similarity with class II major histocompatibility antigens. Infect. Immun. 62:2440-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloos, W. E., and K. H. Schleifer. 1986. Genus IV. Staphylococcus, p. 1013-1035. In Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD.
- 15.Kloos, W. E., R. J. Zimmerman, and R. F. Smith. 1976. Preliminary studies on the characterization and distribution of Staphylococcus and Micrococcus species on animal skin. Appl. Environ. Microbiol. 31:53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krings, E., K. Krumbach, B. Bathe, R. Kelle, V. F. Wendisch, H. Sahm, and L. Eggeling. 2006. Characterization of myo-inositol utilization by Corynebacterium glutamicum: the stimulon, identification of transporters, and influence on l-lysine formation. J. Bacteriol. 188:8054-8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. USA 102:5174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganism. J. Mol. Biol. 3:208-218. [Google Scholar]
- 19.Martin, B., M. Garriga, M. Hugas, S. Bover-Cid, M. T. Veciana-Nogues, and T. Aymerich. 2006. Molecular, technological and safety characterization of gram-positive catalase-positive cocci from slightly fermented sausages. Int. J. Food Microbiol. 107:148-158. [DOI] [PubMed] [Google Scholar]
- 20.Montel, M. C., J. Reitz, R. Talon, J. L. Berdagué, and S. Rousset-Akrim. 1996. Biochemical activities of Micrococcaceae and their effects on the aromatic profiles and odours of a dry sausage model. Food Microbiol. 13:489-499. [Google Scholar]
- 21.Morot-Bizot, S., R. Talon, and S. Leroy-Setrin. 2003. Development of specific PCR primers for a rapid and accurate identification of Staphylococcus xylosus, a species used in food fermentation. J. Microbiol. Methods 55:279-286. [DOI] [PubMed] [Google Scholar]
- 22.Nagase, N., A. Sasaki, K. Yamashita, A. Shimizu, Y. Wakita, S. Kitai, and J. Kawano. 2002. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 64:245-250. [DOI] [PubMed] [Google Scholar]
- 23.Nesbo, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman, R. M., P. Salunkhe, A. Godzik, and J. C. Reed. 2006. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun. 74:594-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nimrat, S., S. Siriboonlamom, S. Zhang, Y. Xu, and V. Vuthiphandchai. 2006. Chilled storage of white shrimp (Litopenaeus vannamei) spermatophores. Aquaculture 261:944-951. [Google Scholar]
- 26.Novick, R. 2006. Staphylococcal pathogenesis and pathogenicity factors: genetics and regulation, p. 496-516. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnog, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 27.Pinna, A., S. Zanetti, M. Sotgiu, L. A. Sechi, G. Fadda, and F. Carta. 1999. Identification and antibiotic susceptibility of coagulase negative staphylococci isolated in corneal/external infections. Br. J. Ophthalmol. 83:771-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pioch, G., H. Heyne, and W. Witte. 1988. Coagulase-negative Staphylococcus species in mixed fodder and on grain. Zentralbl. Mikrobiol. 143:157-171. [PubMed] [Google Scholar]
- 29.Planchon, S., B. Gaillard-Martinie, E. Dordet-Frisoni, M. N. Bellon-Fontaine, S. Leroy, J. Labadie, M. Hebraud, and R. Talon. 2006. Formation of biofilm by Staphylococcus xylosus. Int. J. Food Microbiol. 109:88-96. [DOI] [PubMed] [Google Scholar]
- 30.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 31.Schleifer, K. H., and W. E. Kloos. 1975. Isolation and characterization of staphylococci from human skin. I. Amended descriptions of Staphylococcus epidermidis and Staphylococcus saprophyticus and descriptions of three new species: Staphylococcus cohnii, Staphylococcus haemolyticus, and Staphylococcus xylosus. Int. J. Syst. Bacteriol. 25:50-61. [Google Scholar]
- 32.Shale, K., J. F. R. Lues, P. Venter, and E. M. Buys. 2005. The distribution of Staphylococcus sp. on bovine meat from abattoir deboning rooms. Food Microbiol. 22:433-438. [Google Scholar]
- 33.Siqueira, J. F., Jr., and K. C. Lima. 2002. Staphylococcus epidermidis and Staphylococcus xylosus in a secondary root canal infection with persistent symptoms: a case report. Aust. Endod. J. 28:61-63. [DOI] [PubMed] [Google Scholar]
- 34.Sondergaard, A. K., and L. Stahnke. 2002. Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum: a comparative study in model systems. Int. J. Food Microbiol. 75:99-109. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talon, R., S. Leroy-Sétrin, and S. Fadda. 2002. Bacterial starters involved in the quality of fermented meat products, p. 175-191. In F. Toldra (ed.), Handbook of research advances in quality of meat and meat products. Research Signpost, Kerala, India.
- 37.Tompkins, J. C., J. Figueroa, and R. W. Steele. 2004. Occult bacteremia. Infect. Med. 21:68-72. [Google Scholar]
- 38.Winstanley, C. 2002. Spot the difference: applications of subtractive hybridization to the study of bacterial pathogens. J. Med. Microbiol. 51:459-467. [DOI] [PubMed] [Google Scholar]
- 39.Won, Y. S., H. J. Kwon, G. T. Oh, B. H. Kim, C. H. Lee, Y. H. Park, B. H. Hyun, and Y. K. Choi. 2002. Identification of Staphylococcus xylosus isolated from C57BL/6J-Nos2tm1Lau mice with dermatitis. Microbiol. Immunol. 46:629-632. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida, K. I., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]