Abstract
The polyene macrolide antibiotic nystatin produced by Streptomyces noursei contains a deoxyaminosugar mycosamine moiety attached to the C-19 carbon of the macrolactone ring through the β-glycosidic bond. The nystatin biosynthetic gene cluster contains three genes, nysDI, nysDII, and nysDIII, encoding enzymes with presumed roles in mycosamine biosynthesis and attachment as glycosyltransferase, aminotransferase, and GDP-mannose dehydratase, respectively. In the present study, the functions of these three genes were analyzed. The recombinant NysDIII protein was expressed in Escherichia coli and purified, and its in vitro GDP-mannose dehydratase activity was demonstrated. The nysDI and nysDII genes were inactivated individually in S. noursei, and analyses of the resulting mutants showed that both genes produced nystatinolide and 10-deoxynystatinolide as major products. Expression of the nysDI and nysDII genes in trans in the respective mutants partially restored nystatin biosynthesis in both cases, supporting the predicted roles of these two genes in mycosamine biosynthesis and attachment. Both antifungal and hemolytic activities of the purified nystatinolides were shown to be strongly reduced compared to those of nystatin, confirming the importance of the mycosamine moiety for the biological activity of nystatin.
Nystatin is an important antifungal agent used in human therapy for treatment of certain types of mycoses. Nystatin is produced by Streptomyces noursei ATCC 11455, and its molecular structure represents a polyene macrolide with a 38-membered macrolactone ring (Fig. 1) to which deoxyaminosugar mycosamine is attached at C-19 through a β-glycosidic bond. Mycosamine moieties are also found attached to the macrolactone rings of the polyene macrolide antibiotics amphotericin (10), candicidin/FR-008 (11, 13), pimaricin (3), and rimocidin (26) which are also important antifungal agents. The mode of action of all these antibiotics is suggested to be the formation of complexes with the membrane sterols of target cells, creating hydrophilic channels (16) allowing the leakage of Na+ and K+ ions from fungal cells, eventually leading to cell death (18). For amphotericin, it has been found that each complex consists of eight antibiotic molecules, with hydrophilic polyol regions lining the aqueous central channel (4, 22). Two such complexes are assumed to align head to head to span the cell membrane, with the mycosamine moieties extending towards both the cytoplasm and the extracellular space. According to this model, the ionizable carboxyl at C-16 and the amino group of mycosamine are both involved in the interactions between the antibiotic molecules in the complex (33) and are thus presumed to be important for antifungal activity.
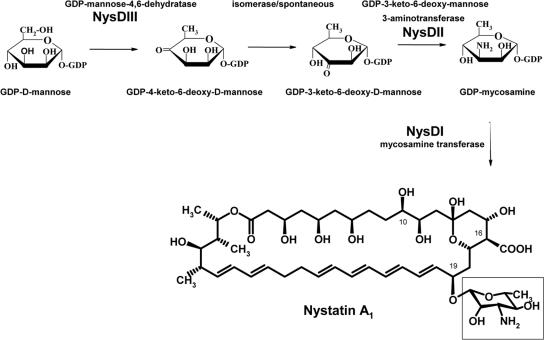
FIG. 1.
Proposed biosynthetic pathway for mycosamine, whereby the mycosamine (in the open rectangle) is shown to be attached at the C-19 position of the nystatin aglycone.
We have proposed previously that the mycosamine biosynthetic route in S. noursei is via GDP-d-mannose, which is presumably synthesized from fructose 6-phosphate by enzymes of the primary metabolism (2, 7). The polyene macrolide biosynthetic gene clusters for amphotericin, nystatin, candicidin/FR-008, and pimaricin were shown to contain genes encoding proteins (AmphDIII, NysDIII, CanM/FscMIII, and PimJ, respectively) showing homology to GDP-d-mannose 4,6-dehydratases suggested to catalyze the conversion of GDP-d-mannose to GDP-4-keto-6-deoxy-d-mannose (Fig. 1). The latter metabolite presumably undergoes a 3,4 isomerization to result in GDP-3-keto-6-deoxy-d-mannose, followed by transamination to form GDP-mycosamine. Since genes encoding GDP-4-keto-6-deoxy-d-mannose 3,4-isomerase have not been found in the polyene macrolide gene clusters, it is assumed that this isomerization is a nonenzymatic reaction (2). However, it is possible that the gene for the relevant isomerase is not physically linked to the nystatin biosynthesis gene cluster, but instead is located somewhere else in the S. noursei genome.
The polyene macrolide gene clusters of amphotericin, candicidin/FR-008, nystatin, pimaricin, and rimocidin all encode putative aminotransferases (AmphDII, CanA/FscMII, NysDII, PimC, and RimE, respectively) homologous to perosamine synthases that catalyze the conversion of GDP-4-keto-6-deoxy-d-mannose to 4-amino-4,6-dideoxy-d-mannose (GDP-perosamine) in perosamine biosynthesis (5, 24, 30). The attachment of mycosamine to the macrolactone ring by means of glycosylation is one of the post-polyketide synthase modifying steps in the polyene macrolide biosynthesis (7). Glycosyltransferases (GTases) are a diverse group of enzymes that glycosylate a wide variety of compounds. The mycosamine moiety has been shown to be important for the biological activity of some polyene macrolide antibiotics; e.g., amphotericin without mycosamine has dramatically reduced antifungal activity (9). GTases are considered to be important targets for genetic manipulation aimed at attaching alternative sugar moieties for the generation of new antibiotics (19). Genes encoding putative GTases AmphDI, NysDI, CanG/FscMI, PimK, and RimE have been identified in the biosynthetic gene clusters for amphotericin, nystatin, pimaricin, candicidin/FR-008, and rimocidin, respectively. The function of FscMI as mycosamine transferase has recently been confirmed experimentally (13).
In this work, we present experimental evidence for the involvement and roles of the nysDI, nysDII, and nysDIII genes in mycosamine biosynthesis and attachment in S. noursei. We also show the importance of the mycosamine moiety for both antifungal and hemolytic activities of nystatin.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. noursei strains were maintained on ISP2 agar medium (Difco), and Escherichia coli strains were grown in LB broth or LB agar (25). Conjugational plasmid transfer from E. coli ET12567 (pUZ8002) (20) and the gene replacement procedures were performed as described previously (17, 27). The transformation of E. coli was performed as described previously (15). Growth media, where appropriate, were supplemented with the following antibiotics at the indicated concentrations: chloramphenicol, 20 μg/ml; kanamycin, 25 or 50 μg/ml; ampicillin, 100 μg/ml; and apramycin, 50 μg/ml. For Flag tag expression vectors, E. coli cells were grown in LB with 0.4% glucose and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.5 mM at an optical density at 600 nm (OD600) of 0.2. Growth was continued for a further 2 to 5 h. For His tag expression vectors, IPTG was added to 1 mM at an OD600 of 0.5. Growth was continued for further 4 to 5 h.
Analyses of nystatin and nystatin analogue production in S. noursei strains.
Analyses of nystatin and nystatin analogue production were performed by liquid chromatography-mass spectrometry (LC-MS) using an Agilent 1100 high-pressure liquid chromatography (HPLC) system with both an Agilent diode array detector and an Agilent time of flight (TOF) mass-sensitive detector of dimethyl sulfoxide (DMSO) extracts of 5-day-old cultures from the microtiter well plate cultivations (8). These analyses provide both UV spectral data and mass determination of the polyene macrolides in the samples. The well plate cultivations were performed in 0.5× SAO-23 medium according to a protocol described previously (29). The production of nystatin analogues for purification was performed in 3-liter Applikon fermentors as described earlier (27).
DNA manipulation, PCR, and sequencing.
General DNA manipulations were performed as described previously (25). DNA fragments from agarose gels were purified using an QIAEX II kit (QIAGEN, Germany). Southern blot analysis was performed with the digoxigenin-11-dUTP high prime labeling kit (Roche Molecular Biochemicals) according to the manufacturer's instructions. Oligonucleotide primers were purchased from MWG Biotech (Germany). PCR was performed with the GC-rich PCR system (Roche Molecular Biochemicals) using the Eppendorf Mastercycler (Eppendorf, Germany), as described previously (6). DNA sequencing was performed at MWG Biotech (Germany).
Construction of plasmid vectors.
Information about DNA oligonucleotides is given in Table 1.
TABLE 1.
Oligonucleotides used for construction of inactivation mutants, complementation plasmids, vectors for heterologous protein expression, and DNA sequencing
| Oligonucleotide | Sequencea | Restriction site |
|---|---|---|
| For construction of mutants and complementation plasmids | ||
| DI.U.F | 5′-ctcgCTGCAGATAACTGACGAACAGG-3′ | PstI |
| DI.U.R | 5′-gatcGAATTCGACGAAGTCGGGGGTGACG-3′ | EcoRI |
| DI.D.F | 5′-ctaAAGCTTCACTGGTGGACACCGTAG-3′ | HindIII |
| DI.D.R | 5′-ggacCTGCAGCTCGACCAACCCTCCTTCAC-3′ | PstI |
| DII.U.F | 5′-gaacCTGCAGCACATAGTCGAGCTCGC-3′ | PstI |
| DII.U.R | 5′-gatcAAGCTTCTGGCGCCGACGAAGAC-3′ | HindIII |
| DII.D.F | 5′-cagcGAATTCAGGACGAGCTGTTCGGC-3′ | EcoRI |
| DII.D.R | 5′-ctacCTGCAGTTCTACGGAGTGGCATG-3′ | PstI |
| Hy.DI.Fwd.2 | 5′-catcGCATGCCGGCGGCCCGCCACATTC-3′ | SphI |
| Hy.DI.Rew | 5′-cgtgAAGCTTGTCAGTCGGTTGCCAGG-3′ | HindIII |
| DII.F.comp.Sph | 5′-catgGCATGCGGCAACCGACTGACCGAG-3′ | SphI |
| For construction of protein expression vectors | ||
| Mac.DII.Fwd | 5′-catAAGCTTATGTCCTTTACGTATCCGG-3′ | HindIII |
| Mac.DII.Rew | 5′-gtaGGTACCCATGCCACTCCGTAGAAC-3′ | KpnI |
| CTC.DIII.Fwd | 5′-catAAGCTTATGTCCAAACGAGCGCTG-3′ | HindIII |
| CTC.DIII.Rew | 5′-gatcAGATCTTGTACCAGTTGGCGGCGAGCAGC-3′ | BglII |
| DII.fwd.pqe2 | 5′-gatcCATATGTCCTTTACGTATCCGGTG-3′ | NdeI |
| DII.rew.pqe2 | 5′-gatcAAGCTTATGCCACTCCGTAGAACG-3′ | HindIII |
| DIII.fwd.pqe2 | 5′-gatcCATATGTCCAAACGAGCGCTGATC-3′ | NdeI |
| DIII.rew.pqe2 | 5′-gatcAAGCTTACCAGTTGGCGGCGAGCAG-3′ | HindIII |
Restriction enzyme sites are underlined.
nysDI deletion vector pSOK201-DIUD.
The 2.2-kb fragment, including the nysDI upstream region, was PCR amplified from recombinant phage N76 DNA (7) by using the oligonucleotides DI.U.F and DI.U.R. The 2.2-kb fragment, including the nysDI downstream region, was PCR amplified from the S. noursei total DNA by using the oligonucleotides DI.D.F and DI.D.R. The two resulting PCR products were cloned individually in pGEM3Zf(−) (Promega), further excised from the resulting constructs with EcoRI/PstI and HindIII/PstI, respectively, and ligated, together with the 3.1-kb HindIII/EcoRI fragment from pSOK201 (32), yielding the nysDI in-frame deletion plasmid pSOK201-DIUD.
nysDII deletion vector pSOK201-DIIUD.
The 2.2-kb fragment, including the nysDII upstream region, was PCR amplified from the plasmid pL7611B DNA (7) by using the oligonucleotides DII.U.F and DII.U.R. The 2.2-kb fragment, including nysDII downstream region, was PCR amplified from S. noursei total DNA by using the oligonucleotides DII.D.F and DII.D.R. The resulting two PCR products were cloned individually in pGEM3Zf(−), further excised from the resulting constructs with HindIII/PstI and EcoRI/PstI, respectively, and ligated, together with the 3.1-kb HindIII/EcoRI fragment from pSOK201, yielding the nysDII in-frame deletion plasmid pSOK201-DIIUD.
Plasmids pSOK804-ErmDI and pSOK804-ErmDI.2 for complementation of the nysDI deletion mutant.
The 1.5-kb fragment encompassing the nysDI coding sequence was PCR amplified from N76 DNA (7) with the oligonucleotides Hy.DI.Fwd.2 and Hy.DI.Rew. The PCR product was cloned in pGEM11Zf(+) (Promega), further excised from the resulting construct with SphI/HindIII, and ligated, together with the 0.3-kb EcoRI/SphI fragment (ermE*p promoter) of pGEM7ZfErmE*Li (from C. R. Hutchinson), into the HindIII/EcoRI sites of the integrative vector pSOK804 (28), yielding plasmid pSOK804-ErmDI. The 0.96-kb fragment from pL76B11 was excised with SpeI/NsiI and cloned into the corresponding SphI/NsiI sites of pSOK804-ErmDI, yielding plasmid pSOK804-ErmDI.2. The resulting two constructs have short and long versions of the nysDI coding sequence (see Results) under transcriptional control of the ermE*p promoter.
Plasmid pSOK804-ErmDII for complementation of the nysDII deletion mutant.
The 1.1-kb fragment containing the nysDII coding sequence was PCR amplified from N76 DNA (7) with the oligonucleotides DII.F.comp.Sph and DII.R-comp.H. The obtained PCR product was cloned in pGEM11Zf(+), further excised from the resulting construct with SphI/EcoRI, and ligated, together with the 0.3-kb EcoRI/SphI fragment of pGEM7ZfErmE*Li, into the HindIII/EcoRI-digested vector pSOK804, yielding plasmid pSOK804-ErmDII.
Vectors for heterologous expression of NysDII and NysDIII.
Two commercial expression vector systems were used for the expression of NysDII and NysDIII proteins in E. coli: Flag protein expression systems (Sigma-Aldrich) using vectors pFLAG-MAC (Flag tag, N terminal) and pFLAG-CTC (Flag tag, C terminal) and QIAexpress (QIAGEN, Germany) using vector pQE2 (His tag, N terminal).
DNA oligonucleotides used in vector construction are listed in Table 1. The 1.1-kb nysDII and 1-kb nysDIII gene coding fragments were amplified by PCR, introducing appropriate digestion sites for cloning into the respective expression vectors. PCR amplification of nysDII and nysDIII for cloning into Flag tag expression vectors was performed with the oligonucleotides Mac.DII.Fwd and Mac.DII.Rew; PCR amplification of nysDII and nysDIII for cloning into the His tag expression vectors was performed with the oligonucleotides DII.fwd.pqe2, DII.rew.pqe2, DIII.fwd.pqe2, and DIII.rew.pqe2. The resulting constructs were named DII-Mac, DIII-CTC, and DII- and DIII-pQE2.
DNA sequencing.
All constructs for gene replacement, complementation, and heterologous expression were verified by DNA sequencing. In addition, the absence of unwanted mutations in the deletion mutants was verified by PCR amplification of chromosomal DNA fragments, cloning, and sequencing.
Preparative LC-MS purification of nystatinolides.
Samples for purification of nystatinolides were made from methanol extracts of the whole-cell cultures. The extracts were dried under vacuum and dissolved in DMSO. Purification of the nystatinolides from the resulting DMSO stock was performed on an Agilent Prep-C18 LC column (50 by 250 mm, 10-μm particles) run at an ambient temperature and a flow of 85 ml/min. The mobile phase was 41% acetonitrile in 10 mM ammonium acetate, pH 4.0, and the hardware was an Agilent 1100 preparative HPLC system equipped with an active splitter and an Agilent single quadrupole mass selective detector. The LC fractions containing the nystatinolides were subsequently reduced under vacuum to remove the organic solvent; thereafter, the precipitated polyene was washed with distilled water and freeze-dried.
Bioassay for antifungal activity.
The test organism used for polyene macrolide bioactivity assays was Candida albicans ATCC 10231, grown in a total volume of 100 μl with standard M19 medium without NaCl (including an inoculum of 1,000 CFU per well and the antibiotics). Stock solutions of the polyene macrolides (5 mg/ml in DMSO) were diluted to different concentrations in the M19 medium. The DMSO concentration was adjusted to 2% in all cultures independently of the antibiotic concentration. The test organism cultures with and without antibiotics were then incubated in 96-well microtiter plates at 34°C without shaking in the robot TECAN Genesis RSP 200, and after 6, 8, 10, 12, 14, 16 and 18 h, growth was measured as the OD660. The data were plotted against the antibiotic concentration, and the MICs resulting in the MIC50 were estimated from the regression curves.
In vitro measurement of hemolytic activity.
Hemolytic assay for the polyene macrolides was performed by monitoring the ability of the macrolides to cause lysis of erythrocytes in defibrinated horse blood under buffered conditions, essentially as described previously by Seco et al. (26). From stock solutions of the purified polyenes (5 mg/ml in DMSO), samples (0.1 ml) with different polyene macrolide concentrations (10 to 500 μg/ml in DMSO) were prepared. The diluted samples were mixed with 0.9 ml horse blood-phosphate-buffered saline (PBS), which is PBS buffer containing 2.5% defibrinated horse blood (Statens Serum Institut, Copenhagen, Denmark [www.ssi.dk]), and incubated in a water bath at 37°C. Samples were collected at different time points (30 and 60 min), centrifuged at 5,000 rpm for 5 min, and the OD545 was measured. One hundred percent hemolysis was defined as the OD545 value obtained from a suspension of 2.5% horse blood in distilled water, and 0.9 ml horse blood-PBS with the addition of 0.1 ml DMSO was used as a “blank.” Each experiment was performed in three parallels, and samples containing nystatin were used as standards.
Recombinant protein detection and purification. (i) Flag tag system.
Pre- and postinduction samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting by using the Anti-Flag M2 monoclonal antibody (Sigma-Aldrich). Purification was performed according to the protocol for Flag protein expression systems by using Anti-Flag M2 Affinity gel, 0.1 M glycine-HCl (pH 3.5), and Tris-buffered saline. In the final purification step, the bound Flag fusion protein was eluted into vials containing 50 μl Tris-HCl (1 M, pH 8.0).
(ii) His tag system.
Pre- and postinduction samples were analyzed by SDS-PAGE and Western blotting using the Penta-His monoclonal antibody (QIAGEN, Germany). Purification was performed according to the protocol of QIAexpress, under native conditions with washing and denaturation solution containing 10 mM HEPES. In a final purification step, the imidazole was removed by dialysis by using buffer containing 20 mM HEPES and 1% glycerol.
SDS-PAGE and Western blotting were performed as described previously (25). Anti-Flag M2 (1 μg/ml) and Penta-His (0.2 μg/ml) antibodies were used for detecting Flag- and His-tagged recombinant proteins.
Enzymatic assays for NysDIII (GDP-d-mannose-4,6-dehydratase) and NysDII (aminotransferase).
The GDP-d-mannose-dehydratase activity was assayed with a procedure similar to that of Markovitz (21). The assay mixture contained 0.25 M KH2PO4, 0.25 M Na2HPO4 (final pH 7.5), 4 mM dithiothreitol, 12 mM EDTA, 1.3 mM GDP-d-mannose, and different amounts of purified NysDIII (1 to 10 μl) expressed from Flag-CTC (ca 0.06 to 0.6 μg) and pQE2 (ca. 0.17 to 1.7 μg) in a total volume of 75 μl. The reaction was performed at 37°C for 30 min, followed by heat denaturation; 700 μl 0.1 M NaOH was added, and the mixture was incubated for 20 min at 37°C. The absorption was measured at 318 nm (due to the formation of enolate anion of GDP-4-keto-6-d-mannose) by using the assay mixture without heat-denatured enzyme as a “blank.” A coupled assay was performed for both purified NysDIII (expressed from Flag-CTC and pQE2) and NysDII (expressed from Flag-MAC and pQE2) by adding 50 μM pyridoxal 5-phosphate, 4 mM sodium-l-glutamate to the assay mixture described above and NysDIII and NysDII in different amounts (NysDIII as above and 0.1 to 1.0 μg Flag tag expressed and 0.02 to 0.2 μg His-tag-expressed NysDII). The reaction was performed at 37°C for 30 min. For the HPLC analysis, the samples were boiled for 1 min and the proteins were precipitated by centrifugation.
HPLC analysis of NysDIII and NysDII enzyme activity.
GDP-d-mannose and GDP-4-keto-6-deoxy-d-mannose were analyzed by HPLC and detected by UV spectrophotometry in the wavelength range of 190 to 500 nm. A reversed-phase column, Waters Nova-Pak C18 (2.1 by 150 mm), was used as stationary phase. Thirty millimolars of phosphate buffer, pH 6.0, containing 5 mM tetrabutylammonium hydrogensulfate and 2% acetonitrile was used as a mobile phase at 0.3 ml/min (1). The analysis was performed on an Agilent 1100 HPLC system.
RESULTS
Heterologously expressed NysDIII protein exhibits GDP-d-mannose 4,6-dehydratase activity in vitro.
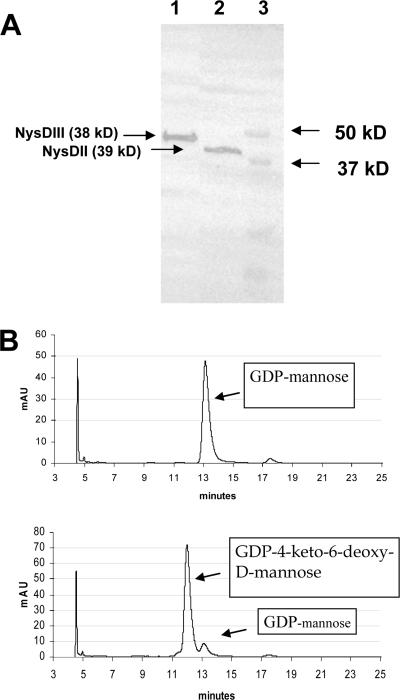
In order to confirm the proposed GDP-d-mannose 4,6-dehydratase function of NysDIII, the protein was expressed in E. coli by using the vectors DIII-pQE2 (His tag, N terminal) and DIII-CTC (Flag tag, C terminal) (Fig. 2A) as described in Materials and Methods. NysDIII expressed from both vectors was shown to be soluble and was purified successfully for enzymatic assays. The activity of purified NysDIII expressed with both His and Flag tags was monitored spectroscopically by absorption at 318 nm (see Materials and Methods for details) in order to detect the enolate anion of GDP-4-keto-6-d-mannose (23). The activity of the Flag-tagged enzyme was measured to be ca 0.18 ΔA·min−1·μg−1 protein and that for the His-tagged enzyme was ca 0.005 ΔA·min−1·μg−1 protein. The reaction products in the conversion of GDP-mannose to GDP-4-keto-6-deoxy-d-mannose by NysDIII were detected by HPLC for Flag-CTC (C-terminal-located Flag peptide) expressed protein (Fig. 2B).
FIG. 2.
(A) Western blot demonstrating the expression of NysDIII (38 kDa) expressed in Flag-CTC vector and NysDII (37 kDa) expressed in Flag-MAC vector (anti-Flag). Lane 1, NysDIII expressed in DIII-CTC; lane 2, NysDII expressed in DII-MAC; lane 3, molecular mass marker. (B) HPLC chromatograms confirming the GDP-d-mannose 4,6-dehydratase activity of NysDIII. The top panel shows the substrate GDP-mannose. The bottom panel shows the conversion of GDP-mannose to GDP-4-keto-6-deoxy-d-mannose after incubation with recombinant NysDIII.
In an attempt to confirm the proposed aminotransferase function of NysDII, the protein was expressed using vectors DII-pQE2 and DII-MAC as described in Materials and Methods. NysDII recombinant proteins expressed from both vectors were shown to be soluble. The recombinant proteins were purified and used in coupled enzyme assays, together with recombinant NysDIII (see Materials and Methods). However, the expected reaction product (GDP-mycosamine) in this coupled reaction with NysDIII could not be detected by HPLC analysis using either Flag- or His-tagged NysDII. There might be several reasons behind this negative result: inactivation of NysDII by the tag, suboptimal assay conditions, or the requirement of isomerization to supply NysDII with its proposed substrate GDP-3-keto-6-deoxy-d-mannose.
Inactivation of nysDI and nysDII genes in S. noursei leads to the production of nystatinolides.
To confirm the roles of NysDII and NysDI in mycosamine biosynthesis, the in-frame deletion mutants of the corresponding genes were constructed by double homologous recombination in S. noursei. Several attempts (using ca 1.5-kb flanking regions for the gene replacement used routinely for such purposes in S. noursei) were made for the construction of such mutants without success (6, 28). By extending the flanking regions to about 2.2 kb, both in-frame deletion mutants were constructed successfully. The deletions within nysDII and nysDI genes in the corresponding mutants, designated NDD2 and NDD1, respectively, were confirmed by Southern blot analysis (data not shown). The deletion in the nysDI gene eliminated the central region of the gene, thereby removing 413 out of 506 amino acids from the NysDI protein. In the case of nysDII, the deletion also affected the central part of the gene, removing 327 out of 352 amino acids from the NysDII protein. The chromosomal DNA regions flanking the deletions in both mutants were PCR amplified and sequenced in order to verify that no unexpected mutations were introduced during the gene replacement procedure (data not shown).
To analyze polyene macrolide production by the NDD2 and NDD1 mutants, the strains were incubated for 5 days in 0.5× SAO-23 medium and culture extracts were analyzed for the presence of nystatin-related polyene macrolides by using LC-MS-TOF. Several independent clones for each mutant were used in this analysis.
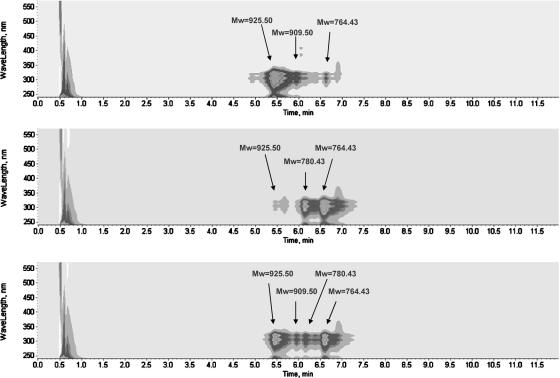
Based on UV spectra and molecular masses, the NDD1 mutant was shown to produce the major polyenes nystatinolide at a level of 28% (±2%) and 10-deoxynystatinolide at 63% (±4%) relative to the level of wild-type nystatin production. In addition, the mutant produced nystatin at a level of 1.2% (±0.2%) relative to the level of nystatin produced in the wild type and 10-deoxynystatin was produced in trace amounts (Table 2; Fig. 3).
TABLE 2.
Nystatin and its analogues produced in S. noursei wild type, NDD1, NDD1 C, NDD2, and NDD2 Ca
| S. noursei strain | % Production for antibioticb
|
|||
|---|---|---|---|---|
| Nystatin | Nystatinolide | 10-Deoxynystatinolide | 10-Deoxynystatin | |
| Wild type | 100 ± 12 | 1.0 ± 0.3 | 4.0 ± 0.5 | 2.2 ± 0.5 |
| NDD1 | 1.2 ± 0.2 | 28 ± 2 | 63 ± 4 | <0.05 |
| NDD1 C | 55 ± 5 | 10 ± 3 | 27 ± 8 | 1.7 ± 0.3 |
| NDD2 | 0.05 ± 0.02 | 0.31 ± 0.15 | 0.09 ± 0.04 | |
| NDD2 C | 0.26 ± 0.12 | 0.03 ± 0.01 | ||
NDD1 C is NDD1 complemented with nysDI (pSOK804-ErmDI.2), and NDD2 C is NDD2 complemented with nysDII (pSOK804-ErmDII).
The production levels are given in the percentage relative to nystatin production in the wild type (1.5 g/liter is set to 100%) and are average numbers from at least three parallels. The Mw was 926 for nystatin, 780 for nystatinolide, 764 for 10-deoxynystatinolide, and 910 for 10-deoxynystatin.
FIG. 3.
HPLC-diode array detector-MS analysis of nystatin and its analogue production in S. noursei wild type (top), NDD1 (middle), and NDD1 complemented with nysDI (bottom). Nystatin, Mw 925.50; nystatinolide, Mw 780.43; 10-deoxynystatinolide, Mw 764.43; and 10-deoxynystatin, Mw 909.50.
The NDD2 mutant was shown to produce, based on UV spectra and molecular masses, both nystatinolide and 10-deoxynystatinolide in very small amounts. The production levels were 0.31% (±0.15) for nystatinolide and 0.09% (±0.04) for 10-deoxynystatinolide relative to the wild-type nystatin production levels (Table 2). The production of nystatin was also detected in the NDD2 mutants at a level of 0.05% (± 0.02). In addition, low but significant amounts of tetraenes with molecular masses corresponding to the noncarboxylated nystatin analogues were detected in the NDD2 mutant. The determination of accurate mass by using LC-MS-TOF strongly suggested that these tetratenes were represented by 16-decarboxy-16-methylnystatin and 16-decarboxy-16-methylnystatinolide with a mycarose moiety at C-35, which have been described previously (8). These analogues were not found in the complemented mutant NDD2 C (see below).
nysDI and nysDII genes expressed in trans can complement the respective mutants.
Next, the NDD1 and NDD2 mutants were complemented with the respective genes cloned under control of the ermE*p promoter (9) into a pSOK804 vector (pSOK804-ErmDI, pSOK804-ErmDI.2, and pSOK804-ErmDII) that integrates site specifically into the genome of S. noursei (28). As we were not certain about the location of the start codon for the nysDI gene, two alternative complementation constructs were made (see Materials and Methods). Eight to 10 clones for each complemented mutant were then analyzed for the production of polyene macrolides in parallel with the parent and wild-type strains.
Complementation of the NDD1 mutant with the construct pSOK804-ErmDI did not have any effect on metabolite profile, while complementation with pSOK804-ErmDI.2 led to a restoration of nystatin production to a level of 55% (±5) relative to that of the wild type (Table 2; Fig. 3). Simultaneously, complemented NDD1 mutant produced nystatinolide at a level of 10% (±3), 10-deoxynystatinolide at 27% (±8), and 10-deoxynystatin at 1.7% (±0.3) relative to the levels produced by the wild type. These results support the proposed function of NysDI as a glycosyltransferase attaching mycosamine to the nystatin aglycone and also confirm the correct translation start of the nysDI gene located 43 codons upstream of the start proposed originally (7).
Complementation of the NDD2 mutant with the construct pSOK804-ErmDII led to a restoration of nystatin production to only 0.26% (±0.12) of the wild-type level. Simultaneously, the level of nystatinolide was reduced to 0.03% (±0.01). Although the amount of nystatin produced was still low, the complementation clearly increased the nystatin yield, thus confirming the proposed involvement of NysDII in mycosamine biosynthesis. Neither 16-decarboxy-16-methylnystatin nor 16-decarboxy-16-methylnystatinolide with a mycarose moiety at C-35 was identified in the extracts of the complemented nysDII mutant (see Discussion).
Nystatinolides show some residual antifungal activity, but no hemolytic activity.
To test whether the mycosamine group is significant for the antifungal activity of nystatin, nystatinolide and 10-deoxynystatinolide were purified from the NDD1 mutant and their MIC50 values were determined by using C. albicans as a test organism. The MIC50 for nystatinolide was 60 μg/ml, that for 10-deoxynystatinolide was 20 μg/ml, and the MIC50 value for nystatin was 1.2 μg/ml in the same experiment. These results clearly demonstrate the significance of the mycosamine moiety for the antifungal activity of nystatin.
In order to test the in vitro toxicity of the nystatinolides, we carried out determinations of their hemolytic activity in comparison with that of nystatin (see Materials and Methods). The results of these experiments showed that hemolytic activities of both nystatinolide and 10-deoxynystatinolide were more than 90% lower compared to that of nystatin (data not shown). These results demonstrate that the mycosamine sugar moiety contributes significantly to the hemolytic activity of antibiotic nystatin in vitro.
DISCUSSION
Sugar moieties are crucial for the biological activity of many antibiotics, and an understanding of their biosynthesis and attachment may open new possibilities for combinatorial biosynthesis (31). The majority of polyene macrolide antibiotics contain one single deoxyaminosugar, mycosamine, which so far has not been found in any other natural products (2). For the polyene macrolide antibiotic nystatin, mycosamine has been presumed, but until now not proven experimentally, to be important for its antifungal activity. Thereby, mycosamine is potentially an interesting moiety of the nystatin that can be modified or substituted using genetic engineering, leading to the production of novel nystatin analogues. From the suggested mycosamine biosynthetic route (7) and organization of the polyene macrolide biosynthetic gene clusters, it was suggested that the enzymes appearing early in the pathway are most likely to be recruited from the primary metabolism, i.e., from the pathway responsible for the biosynthesis of lipopolysaccharides. This in turn suggests that the three genes identified within the nystatin cluster must play a pivotal role in mycosamine formation, performing the final steps in GDP-mycosamine biosynthesis and its attachment to the nystatin aglycone. The homologue of the nysDIII gene in the amphotericin biosynthetic gene cluster of S. nodosus, amphDIII, has been deleted, resulting in the production of amphoteronolides and therefore demonstrating its requirement for mycosamine biosynthesis (9). In this study, we investigated the function of nysDIII through recombinant expression and enzymatic assays, which confirmed its function as GDP-d-mannose 4,6-dehydratase. Our coupled NysDII-NysDIII enzyme assay failed to confirm the proposed aminotransferase activity of NysDII, presumably due to suboptimal assay conditions or negative influence of the tag on the activity of the recombinant protein. Importantly, however, we managed to express soluble recombinant proteins involved in mycosamine biosynthesis, which can potentially be used in the future for in vitro synthesis of deoxyaminosugars.
The nysDII and nysDI genes were investigated through gene inactivation, complementation, and phenotypic analysis of the resulting recombinant strains. The nysDI mutants produced nystatinolide and 10-deoxynystatinolide in significantly higher amounts than did the wild-type strain as well as trace amounts of nystatin. The latter is surprising, considering that the nysDI gene was deleted from the chromosome, and can be explained only by the existence of an alternative glycosyltransferase capable of attaching mycosamine to nystatinolide with low efficiency. By reintroduction (complementation) of the nysDI gene in the mutant, the nystatin biosynthesis was restored to a level of ca. 55% compared to that of the wild-type strain, unequivocally confirming the involvement of this gene in mycosamine attachment.
Inactivation of the nysDII gene yielded a mutant producing nystatinolide at a very low yield, and the reason for the latter remains unclear. We PCR amplified and sequenced the region surrounding the deletion in the nysDII mutant and could not identify any mutation that might have affected other nystatin biosynthetic genes. Still, complementation of this mutant with nysDII resulted in increased nystatin production, with a concomitant decrease in production of nystatinolide, thus confirming the role of nysDII in mycosamine biosynthesis. From the suggested function of NysDII (Fig. 1), the inactivation of its gene could lead to the production of nystatin with deaminated sugar attached. This was the case when the homologous amphDII gene was deleted in Streptomyces nodosus, resulting in the attachment of a neutral deoxyhexose to amphotericin aglycone (12). In S. noursei, however, the situation was different, as the nysDII mutant produced nystatinolide as the major product. Based on these results, we suggest that the NysDI glycosyltransferase has a strong preference for GDP-mycosamine as a substrate, while its counterpart in S. nodosus, AmphDI, has some sugar substrate flexibility. The amphotericin analogue with deoxyhexose displayed low antifungal activity, but in this strain, the P450 monooxygenase amphN gene was deleted as well, resulting in the replacement of the exocyclic carboxyl group with a methyl group (12). Both the amino sugar and the exocyclic carboxy group are supposed to be important for channel formation by the antibiotic molecules and thereby for antifungal activity (14).
In addition to nystatinolide, we observed the production of putative 16-decarboxy-16-methylnystatin and 16-decarboxy-16-methylnystatinolide with a mycarose moiety on C-35 in trace amounts by the nysDII mutant. Considering the very low production level for nystatinolides in NDD2, these results were rather surprising, as the NysDII was presumed to be an aminotransferase involved in mycosamine biosynthesis and its absence will not affect other post-polyketide synthase modifications, such as oxidation of the C-16 methyl group. Chen et al. detected noncarboxylated analogues of the polyene macrolide FR-008 (candicidin) in Streptomyces sp. strain FR-008 mutants deficient in either the mycosamynil transferase FscMI or the putative aminotransferase FscMII, the homologues of NysDI and NysDII, respectively (13). The authors suggested that the nonglycosylated analogues may inhibit an enzyme responsible for the oxidation of the exocyclic methyl group. This is probably not the case for S. noursei, since noncarboxylated nystatinolides could no longer be detected upon complementation of the nysDII mutant. Currently, we do not have enough data to suggest a plausible explanation for this phenomenon.
Finally, in order to confirm the importance of the mycosamine moiety for antifungal activity of nystatin, we measured MIC50 values for both nystatinolide and 10-deoxynystatinolide by using C. albicans as a test organism. Apparently, the antifungal activity of both nystatinolides was reduced significantly compared to that of nystatin, confirming the importance of mycosamine for biological activity. Interestingly, the antifungal activity of 10-deoxynystatinolide was higher than that of nystatinolide, suggesting that the mechanism behind antifungal activity of nystatin aglycones might differ from that of nystatin. The fact that both nystatin aglycones displayed greatly reduced hemolytic activity compared to nystatin suggests that these molecules fail to form stable channels in the eukaryotic cell membranes. The latter is probably due to the lack of efficient interaction between the polyene antibiotic molecules, which is suggested to be ensured through the formation of hydrogen bonds between carboxy and amino groups of the neighboring molecules (4).
Acknowledgments
This work was supported by research grants from the Norwegian University of Science and Technology (no. 8115710) and the Research Council of Norway (no. 146610/431).
We are grateful to Randi Aune for assisting with fermentations.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Albermann, C., and W. Piepersberg. 2001. Expression and identification of the RfbE protein from Vibrio cholerae O1 and its use for the enzymatic synthesis of GDP-D-perosamine. Glycobiology 11:655-661. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, J. F., P. Caffrey, J. A. Gil, and S. B. Zotchev. 2003. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 61:179-188. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, J. F., R. Fouces, M. V. Mendes, N. Olivera, and J. F. Martin. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 7:895-905. [DOI] [PubMed] [Google Scholar]
- 4.Baginski, M., H. Resat, and J. A. McCammon. 1997. Molecular properties of amphotericin B membrane channel: a molecular dynamics simulation. Mol. Pharmacol. 52:560-570. [DOI] [PubMed] [Google Scholar]
- 5.Bilge, S. S., J. C. Vary, Jr., S. F. Dowell, and P. I. Tarr. 1996. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect. Immun. 64:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brautaset, T., P. Bruheim, H. Sletta, L. Hagen, T. E. Ellingsen, A. R. Strom, S. Valla, and S. B. Zotchev. 2002. Hexaene derivatives of nystatin produced as a result of an induced rearrangement within the nysC polyketide synthase gene in S. noursei ATCC 11455. Chem. Biol. 9:367-373. [DOI] [PubMed] [Google Scholar]
- 7.Brautaset, T., O. N. Sekurova, H. Sletta, T. E. Ellingsen, A. R. Strom, S. Valla, and S. B. Zotchev. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395-403. [DOI] [PubMed] [Google Scholar]
- 8.Bruheim, P., S. E. Borgos, P. Tsan, H. Sletta, T. E. Ellingsen, J. M. Lancelin, and S. B. Zotchev. 2004. Chemical diversity of polyene macrolides produced by Streptomyces noursei ATCC 11455 and recombinant strain ERD44 with genetically altered polyketide synthase NysC. Antimicrob. Agents Chemother. 48:4120-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrne, B., M. Carmody, E. Gibson, B. Rawlings, and P. Caffrey. 2003. Biosynthesis of deoxyamphotericins and deoxyamphoteronolides by engineered strains of Streptomyces nodosus. Chem. Biol. 10:1215-1224. [DOI] [PubMed] [Google Scholar]
- 10.Caffrey, P., S. Lynch, E. Flood, S. Finnan, and M. Oliynyk. 2001. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem. Biol. 8:713-723. [DOI] [PubMed] [Google Scholar]
- 11.Campelo, A. B., and J. A. Gil. 2002. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 148:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Carmody, M., B. Murphy, B. Byrne, P. Power, D. Rai, B. Rawlings, and P. Caffrey. 2005. Biosynthesis of amphotericin derivatives lacking exocyclic carboxyl groups. J. Biol. Chem. 280:34420-34426. [DOI] [PubMed] [Google Scholar]
- 13.Chen, S., X. Huang, X. Zhou, L. Bai, J. He, K. J. Jeong, S. Y. Lee, and Z. Deng. 2003. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem. Biol. 10:1065-1076. [DOI] [PubMed] [Google Scholar]
- 14.Chéron, M., B. Cybulska, J. Mazerski, J. Grzybowska, A. Czerwinski, and E. Borowski. 1988. Quantitative structure-activity relationships in amphotericin B derivatives. Biochem. Pharmacol. 37:827-836. [DOI] [PubMed] [Google Scholar]
- 15.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Kruijff, B., and R. A. Demel. 1974. Polyene antibiotic-sterol interactions in membranes of Acholeplasma laidlawii cells and lecithin liposomes. III. Molecular structure of the polyene antibiotic-cholesterol complexes. Biochim. Biophys. Acta 339:57-70. [DOI] [PubMed] [Google Scholar]
- 17.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 18.Hammond, S. M. 1977. Biological activity of polyene antibiotics. Prog. Med. Chem. 14:105-179. [DOI] [PubMed] [Google Scholar]
- 19.Hu, Y., and S. Walker. 2002. Remarkable structural similarities between diverse glycosyltransferases. Chem. Biol. 9:1287-1296. [DOI] [PubMed] [Google Scholar]
- 20.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 21.Markovitz, A. 1964. Biosynthesis of guanosine diphosphate D-rhamnose and guanosine diphosphate D-talomethylose from guanosine diphosphate alpha-D-mannose. J. Biol. Chem. 239:2091-2098. [PubMed] [Google Scholar]
- 22.Matsumori, N., Y. Sawada, and M. Murata. 2005. Mycosamine orientation of amphotericin B controlling interaction with ergosterol: sterol-dependent activity of conformation-restricted derivatives with an amino-carbonyl bridge. J. Am. Chem. Soc. 127:10667-10675. [DOI] [PubMed] [Google Scholar]
- 23.Oths, P. J., R. M. Mayer, and H. G. Floss. 1990. Stereochemistry and mechanism of the GDP-mannose dehydratase reaction. Carbohydr. Res. 198:91-100. [DOI] [PubMed] [Google Scholar]
- 24.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Seco, E. M., T. Cuesta, S. Fotso, H. Laatsch, and F. Malpartida. 2005. Two polyene amides produced by genetically modified Streptomyces diastaticus var. 108. Chem. Biol. 12:535-543. [DOI] [PubMed] [Google Scholar]
- 27.Sekurova, O., H. Sletta, T. E. Ellingsen, S. Valla, and S. Zotchev. 1999. Molecular cloning and analysis of a pleiotropic regulatory gene locus from the nystatin producer Streptomyces noursei ATCC 11455. FEMS Microbiol. Lett. 177:297-304. [DOI] [PubMed] [Google Scholar]
- 28.Sekurova, O. N., T. Brautaset, H. Sletta, S. E. Borgos, M. O. Jakobsen, T. E. Ellingsen, A. R. Strom, S. Valla, and S. B. Zotchev. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 186:1345-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sletta, H., S. E. Borgos, P. Bruheim, O. N. Sekurova, H. Grasdalen, R. Aune, T. E. Ellingsen, and S. B. Zotchev. 2005. Nystatin biosynthesis and transport: nysH and nysG genes encoding a putative ABC transporter system in Streptomyces noursei ATCC 11455 are required for efficient conversion of 10-deoxynystatin to nystatin. Antimicrob. Agents Chemother. 49:4576-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trefzer, A., J. A. Salas, and A. Bechthold. 1999. Genes and enzymes involved in deoxysugar biosynthesis in bacteria. Nat. Prod. Rep. 16:283-299. [DOI] [PubMed] [Google Scholar]
- 31.Walsh, C., C. L. Freel Meyers, and H. C. Losey. 2003. Antibiotic glycosyltransferases: antibiotic maturation and prospects for reprogramming. J. Med. Chem. 46:3425-3436. [DOI] [PubMed] [Google Scholar]
- 32.Zotchev, S., K. Haugan, O. Sekurova, H. Sletta, T. E. Ellingsen, and S. Valla. 2000. Identification of a gene cluster for antibacterial polyketide-derived antibiotic biosynthesis in the nystatin producer Streptomyces noursei ATCC 11455. Microbiology 146:611-619. [DOI] [PubMed] [Google Scholar]
- 33.Zotchev, S. B. 2003. Polyene macrolide antibiotics and their applications in human therapy. Curr. Med. Chem. 10:211-223. [DOI] [PubMed] [Google Scholar]