Abstract
Acidithiobacillus caldus has been proposed to play a role in the oxidation of reduced inorganic sulfur compounds (RISCs) produced in industrial biomining of sulfidic minerals. Here, we describe the regulation of a new cluster containing the gene encoding tetrathionate hydrolase (tetH), a key enzyme in the RISC metabolism of this bacterium. The cluster contains five cotranscribed genes, ISac1, rsrR, rsrS, tetH, and doxD, coding for a transposase, a two-component response regulator (RsrR and RsrS), tetrathionate hydrolase, and DoxD, respectively. As shown by quantitative PCR, rsrR, tetH, and doxD are upregulated to different degrees in the presence of tetrathionate. Western blot analysis also indicates upregulation of TetH in the presence of tetrathionate, thiosulfate, and pyrite. The tetH cluster is predicted to have two promoters, both of which are functional in Escherichia coli and one of which was mapped by primer extension. A pyrrolo-quinoline quinone binding domain in TetH was predicted by bioinformatic analysis, and the presence of an o-quinone moiety was experimentally verified, suggesting a mechanism for tetrathionate oxidation.
It is well established that consortia of bacteria and archaea play a pivotal role in the recovery of metals in industrial “biomining” operations (25, 27, 30, 32). Biomining refers to both “bioleaching,” whereby target metals are solubilized from the sulfide mineral, and “biooxidation,” where the target metal is exposed by oxidation of the sulfide mineral surrounding it. In biomining operations, the principal function of the microbial consortium is the oxidation of Fe2+ to Fe3+, whereby the microorganisms gain energy for metabolic functions and the resulting Fe3+ carries out mineral oxidation. A major limitation to the use of biomining has been the accumulation of solid elemental sulfur (S0) compounds on the surfaces of minerals (e.g., see references 14 and 42). It has been suggested that these compounds contribute to a decrease in mineral dissolution by limiting Fe3+ access to the metal sulfide bond, termed passivation (12). However, in the absence of an iron-oxidizing catalyst, the presence of S0 did not reduce leaching rates from marcasite (FeS2) and arsenopyrite (FeAsS). Possibly this was because areas of the mineral were “clean” of sulfur compounds or because the sulfur layer was permeable to the metal sulfide oxidant at the thickness observed in the study (14).
Microorganisms that use reduced inorganic sulfur compounds (RISCs) as a source of energy include archaea and bacteria and comprise acidophilic or neutrophilic photo- and chemolithotrophs that often use sulfur oxygenase (sox gene cluster) and sulfur oxygenase reductase, coded by the sor gene (16, 19, 37). However, only a few RISC-metabolizing enzymes have been characterized from industrially important acidophilic microorganisms; these include Acidithiobacillus ferrooxidans sulfur dioxygenase, which yields sulfite from S0 (33); sulfite oxidoreductase, which oxidizes sulfite to sulfate (40); a sulfide:quinone oxidoreductase which oxidizes sulfide to sulfur (41); thiosulfate oxidase, which catalyzes the oxidation of thiosulfate to tetrathionate (36); and tetrathionate hydrolase, which hydrolyzes tetrathionate to thiosulfate, sulfur, and sulfate (8). A number of enzymes and enzymatic activities have also been identified in Acidithiobacillus thiooxidans, including thiosulfate dehydrogenase (24), sulfite:ubiquinone oxidoreductase activity (38), and tetrathionate hydrolase (39).
Acidithiobacillus caldus is one of the most abundant microorganisms in industrial biomining (26, 31), where it is suggested to oxidize RISCs formed during sulfide mineral breakdown (12, 13). Elemental sulfur and tetrathionate are key intermediates in A. caldus metabolism, and tetrathionate hydrolysis yields thiosulfate, pentathionate, and eventually sulfate (6), while S0 is oxidized to sulfate via sulfite (17). The protein responsible for A. caldus tetrathionate decomposition is a periplasmic homodimer with a maximum activity at pH 3 (6). Sulfur-grown A. caldus lacks a tetrathionate-metabolizing activity, suggesting that the expression of tetrathionate hydrolase is substrate dependent and regulated on either the transcriptional or translational level (6).
This report presents a novel gene cluster containing the gene coding for the A. caldus tetrathionate hydrolase, shows its differential expression by using quantitative PCR (Q-PCR) and Western blot analysis, maps the promoter regions, and demonstrates the presence of an o-quinone cofactor in tetrathionate hydrolase (TetH). Knowledge of RISC oxidation regulation in this industrially important bacterium could contribute to the development of new approaches in biomining.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. caldus strain KU (ATCC 51756 and DSM 8584) was cultivated in mineral salts medium (MSM) (acidified to pH 2.5 with H2SO4) containing trace elements (12). The MSM was heated at 121°C for 15 min before filter-sterilized (0.22-μm PES membrane filter; Millipore) trace elements were added, and either sterile-filtered tetrathionate (5 mM) or 0.5% (wt/vol) S0 (sterilized at 105°C for 24 h) was added as the energy source. Cultivation with 10 mM thiosulfate as the substrate was performed in a 500-ml continuous culture vessel (dilution rate = 0.006 h−1; Watson Marlow 205S pump) to minimize acid degradation of the thiosulfate. The culture vessel was stirred (300 rpm) and aerated with 300 ml CO2-enriched air min−1 (2%, vol/vol) at 45°C. Cultivation of “Ferroplasma acidarmanus” (proposed name) Fer1 (10, 15) was carried out in MSM (pH 2) with 0.02% (wt/vol) yeast extract (autoclaved at 121°C for 15 min) at 37°C with sparging with 300 ml air min−1 and 3% (wt/vol) pyrite. The pyrite concentrate has been previously described (11). The mixed culture of “F. acidarmanus” and A. caldus was grown as described for “F. acidarmanus” except that the yeast extract was omitted. Escherichia coli strains DH5α and BL21(DE3) were grown in Luria-Bertani medium with the addition of an appropriate antibiotic.
Bioinformatic analysis of the tetH gene cluster.
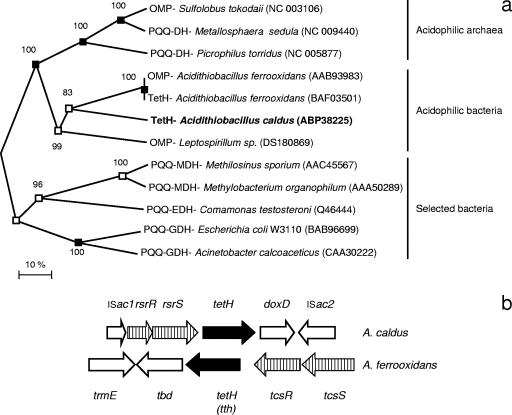
Candidate protein-coding genes of the partial genome sequence of A. caldus KU were predicted using GLIMMER (9) and CRITICA (3). The gene prediction results were combined, and the corresponding amino acid sequences were compared against GenBank's nonredundant database using BLASTP (1). The alignments of the N terminus of each gene model versus the best match were used to select the preferred gene model. The revised genes were compared against the GenBank nonredundant database and the Swissprot, Pfam, Tigrfam, PROSITE, PRINTS, and COGS databases. The annotated sequences were displayed in Artemis (5) to facilitate further functional curation. Promoter prediction was performed using programs available at www.fruitfly.org/seq_tools/promoter.html and www.softberry.com and a combined Hidden Markov Model/Neural network program developed by David Holmes's laboratory (unpublished). The TetH alignment and construction of the phylogenetic tree were carried out using MEGA version 3.1 (20). Three separate phylogenetic trees were created by distance and neighbor joining, parsimony, and minimum evolution methods, and the neighbor joining tree is presented (see Fig. 2). Those nodes supported by all three trees and by two trees have been indicated.
FIG. 2.
(a) Unrooted neighbor joining tree of the A. caldus tetH (in bold) alignment with closest relatives from the NCBI database containing a PQQ binding domain and selected neutrophilic dehydrogenases also containing a PQQ binding domain. Phylogenetic analysis was carried out by the minimum evolution, distance neighbor joining, and maximum parsimony methods in MEGA; the nodes supported by all three trees (filled boxes) and by two trees (open boxes) have been marked, and the values by the nodes are bootstrap values of 1,000 runs. Accession numbers are given in parentheses. The scale bar represents 10% sequence similarity. (b) Gene block comparison of the A. caldus and A. ferrooxidans gene clusters. Vertical lines represent the two-component regulation system, and solid black denotes the tetrathionate hydrolase gene (termed tth in A. ferrooxidans).
N-terminal sequencing of the tetrathionate hydrolase.
To ensure that the tetH gene sequence coded for the purified tetrathionate hydrolase protein (6), N-terminal and internal amino acid sequences were determined by Edman degradation and in-gel trypsin digestion (carried out by the Protein Analysis Center, Karolinska Institute, Sweden).
DNA manipulation and sequencing.
A. caldus cells were cultured as described above and harvested at 10,000 × g for 10 min, washed twice in 10 mM Tris HCl buffer (pH 8), and lysed by the addition of 100 mg lysozyme ml−1 for 10 min at room temperature followed by a final concentration of 0.5% (wt/vol) sodium dodecyl sulfate (SDS) for 30 min. Genomic DNA was prepared from the cell lysate using phenol:chloroform:isoamyl alcohol extraction and isopropanol precipitation (34). Established methods were used for restriction enzyme digestion, ligation, transformation, and plasmid purification.
RNA preparation, RT-PCR, and Q-PCR.
RNA was prepared from exponential-phase cells grown on tetrathionate or S0 by using an RNeasy mini kit (QIAGEN) with subsequent treatment with a DNA-free kit (Ambion), according to the manufacturers' recommendations. The RNA concentrations were quantified using a NanoDrop ND-1000 spectrophotometer (Saveen Werner). Reverse transcription-PCR (RT-PCR) was performed in two steps. First, cDNA was produced from total RNA by using a RevertAid first strand cDNA synthesis kit (Fermentas) and random hexamer primers. In the second step, PCR was carried out with sequence-specific primers (RT-PCR oligonucleotides shown in Table 1) and PuRe Taq ready-to-go PCR beads (GE Healthcare). To ensure that the RT-PCR was functioning correctly, positive controls using convergent primers within tetH and doxD were also carried out. Controls using the same primers to test for DNA contamination in the RNA preparations were all negative. Q-PCR was also prepared in a two-step manner with the cDNA produced from RNA using a RevertAid first strand cDNA synthesis kit (Fermentas) and random hexamer primers. The second step was performed with sequence-specific primers (Q-PCR primers shown in Table 1) and iTaq SYBR green supermix (Bio-Rad) using a Bio-Rad iCycler iQ multicolor real-time PCR detection system. The expression of the analyzed genes was calculated using iCycler iQ optical system software version 3.1 (Bio-Rad). All Q-PCR experiments were carried out in triplicate from two independent experiments.
TABLE 1.
PCR, RT-PCR, and Q-PCR oligonucleotides used in this study
| Name | Sequence (5′ to 3′) | Orientation | Application |
|---|---|---|---|
| a | AAGAATTCCGTGACCATAGCTTTTGCCAT | Forward | P0 promoter/lac operon fusions |
| b | AAGGATCCCCGTTGCTGATCAGATCCAA | Reverse | P0 promoter/lac operon fusions |
| c | TTTGAATTCAACGCACGTAAATTGTA | Forward | P1 promoter/lac operon fusions |
| d | TTGGATCCAGGGTTGCGGTAACGGCAA | Reverse | P1 promoter/lac operon fusions |
| e | ACAGATTGCCAGGCACCTTGTA | Forward | RT-PCR |
| f | GATCCATCCATATGCGAGCAGAT | Reverse | RT-PCR, primer extension |
| g | CCAAAATGTGTCTCTTGGTCAT | Forward | RT-PCR |
| h | TTCATACATATTTCTGGCTCATCG | Reverse | RT-PCR, Q-PCR |
| i | CGCCGACGATTACCTATCAAA | Forward | RT-PCR, Q-PCR |
| j | GTCATCGCATCATGGCATTGCACA | Forward | RT-PCR |
| k | TGGGGTAGGTATAAACGCGCAA | Reverse | RT-PCR, primer extension |
| l | TTGCGCGTTTATACCTACCCCA | Forward | RT-PCR |
| m | CGTATATGAATCGCCTGGATCC | Reverse | RT-PCR |
| n | TTGCGCGTTTGTACCTACC | Forward | Q-PCR |
| o | GCCGTCTACTTGAGCTCC | Reverse | Q-PCR |
| p | CTCTAATTTCGCTATGGGGATCAC | Forward | Q-PCR |
Protein purification, antibody preparation, and Western blotting.
TetH was purified from A. caldus cells according to the method of Bugaytsova and Lindström (6), with an additional purification step of polyacrylamide gel electrophoresis for the enzyme prior to use for antibody generation. Polyclonal antibodies against TetH were raised using a series of four immunizations in a rabbit (AgriSera AB, Vännäs, Sweden). Western blotting detection of TetH (34) was carried out using anti-tetrathionate hydrolase primary and horseradish peroxidase-labeled secondary antibodies. The Western blot was visualized by using an ECL-PLUS detection kit (Amersham Pharmacia), and images were captured with a GelDoc XR system equipped with a charge-coupled-device camera and Quantity One software version 4.6.1 (Bio-Rad). The Western blot image was processed using Adobe Photoshop.
Promoter fusion and β-galactosidase assays.
DNA fragments containing predicted promoters P0 and P1 (Fig. 1) were amplified using primer pairs a and b and c and d (Table 1), digested with EcoRI and BamHI, and cloned upstream from the promoterless lac operon in the multiple cloning site on the broad-host-range plasmid pRW2 (21). β-Galactosidase activity measurements were performed in E. coli strain DH5α (22). β-Galactosidase measurements were carried out in triplicate, and the means ± standard deviations are presented below.
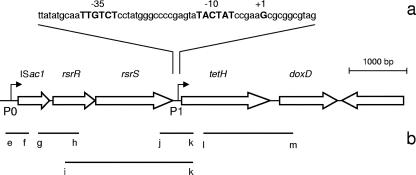
FIG. 1.
(a) A. caldus tetH gene cluster with the expanded portion describing the P1 promoter region showing the −10 and −35 sequences and transcription start site (all in bold caps). (b) Positive transcripts detected by RT-PCR represented by lines under the corresponding genes (to scale; see Table 1 for primers). The second transposase was not labeled as it is coded on the opposite strand.
Primer extension.
Primer extension was carried out according to Balsalobre et al. (4), except that Moloney murine leukemia virus reverse transcriptase (SuperScript II reverse transcriptase; Invitrogen) instead of avian myeloblastosis virus reverse transcriptase was used with the supplied buffer in the extension reaction mixture. Primers f and k were used for P0 and P1, respectively (Table 1). The primer extension image was processed using Adobe Photoshop.
Measurement of the TetH PQQ content.
Purified TetH (1.6 mg) was lyophilized, and the dried pellet extracted with 2 ml methanol. After incubation at 37°C for 30 min, the suspension was centrifuged at 10,000 × g for 10 min and the supernatant evaporated. The dry material was resuspended in 5 mM Tris-HCl buffer (pH 8.0) and centrifuged at 10,000 × g for 10 min, and the supernatant used as an extract. The pyrrolo-quinoline quinone (PQQ) content was measured enzymatically using the recombinant soluble apoenzyme from Acinetobacter calcoaceticus LMD 79.41 (28). Fifty microliters of the extract and 10 μl of 0.4 M CaCl2 were added to 10 μl of apo-glucose dehydrogenase (5 mg/ml). The mixture was incubated at 30°C for 30 min (5 mM Tris-HCl buffer [pH 8.0] was used instead of extract in the control experiments). The glucose dehydrogenase activity was assayed by using a dye-linked system containing 20 mM glucose, 50 μM phenazine methosulfate, and 100 μM 2,6-dichlorophenol indophenol (DCPIP; total volume 1 ml) at 30°C. One unit of enzyme activity was defined as the amount of enzyme that catalyzes the reduction of 1 μmol DCPIP min−1. Experiments were carried out in triplicate, and the means ± standard deviations are presented below.
Staining for quinoproteins.
Quinoproteins were detected by staining with nitro blue tetrazolium (NBT; 0.24 mM in 2 M potassium glycinate, pH 10) (29). The protein samples were applied to the nitrocellulose filter, dried at room temperature, immersed in the glycinate-NBT solution for 45 min in the dark, and then dipped in 0.1 M sodium borate, pH 10. Quinoproteins were specifically stained as purple-blue bands due to NBT reduction to formazan.
Nucleotide sequence accession number.
The DNA sequence of the tetH gene cluster is available under the GenBank accession number EF460464.
RESULTS AND DISCUSSION
TetH sequencing and bioinformatic analysis of the gene cluster containing tetH.
Trypsin digestion and N-terminal sequencing of the tetrathionate hydrolase protein provided three peptide sequences (SITPVLQPGNPFDPDSPFARLYLPQNA, GVQWNFP, and GEIPGAVNTG) that were used to confirm the corresponding gene in the genome sequence of A. caldus. TetH (GenBank accession number ABP38225) is predicted to be a 503-amino-acid-long protein containing a 24-amino-acid Sec signal peptide with a signal peptide cleavage site. It is predicted to be a periplasmic exposed protein with four transmembrane spanning loops, two nonoverlapping WD-40 repeats, and a quinone binding domain (Table 2). tetH was linked to the upstream genes rsrR and rsrS, predicted to encode a two-component response regulator belonging to the osmoregulatory family, and to a downstream gene, doxD, predicted to encode a subunit of thiosulfate:quinol oxidoreductase. The cluster is flanked by two predicted transposases (Table 2; Fig. 1a).
TABLE 2.
Cluster of genes containing tetH and flanked by two insertion sequences
| Gene name | Suggested function | Best hit in NCBI database | Scored | E valued | % Sa | No. of TMb | Domain(s) and motif(s)c |
|---|---|---|---|---|---|---|---|
| ISac1 | Transposase (IS family 256) | Transposase (Azoarcus sp. strain EbN1) | 131 | 3e−29 | 72 | None | PF00872 |
| rsrR | Two-component transcriptional regulator system | Response regulator receiver (Acidovorax sp. strain JS42) | 195 | 1e−48 | 67 | None | PD000039, PF00072, D000329, PF00486, SSF52172, S50110, SM00448 |
| rsrS | Two-component transcriptional regulator system | Periplasmic sensor signal transduction histidine kinase (Shewanella baltica OS195) | 119 | 4e−25 | 46 | 4 | PF02518, SM00387, SSF55874, PF00672, PS50885, PF00512 |
| tetH | PQQ binding dehydrogenase or kinase | Tetrathionate hydrolase (A. ferrooxidans) | 506 | 1e−141 | 71 | 4 | PF01011, SM00564, SSf50998 |
| doxD | Quinol oxidase subunit (DoxD) | DoxD subunit (Gluconobacter oxydans 621H) | 307 | 5e−82 | 67 | 5 | PF04173 |
| ISac2 | Transposase (IS family 630) | Transposase (Burkholderia vietnamiensis) | 413 | 7e−114 | 76 | None | SSF46689, SSF53098 |
S, similarity of the alignment between the gene and the top hit from the NCBI database using BLASTP.
TM, transmembrane domain; predicted by the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html).
Domains were identified in the proteins by using InterProScan (http://www.ebi.ac.uk/InterProScan/).
Statistics derived from BLASTP (NCBI database).
The predicted PQQ binding domain is a propeller structure found in dehydrogenases that use PQQ as a cofactor (2). Bioinformatic analysis showed that TetH was similar to several other membrane-bound PQQ dehydrogenases, and the TetH amino acid sequence formed a clade with A. ferrooxidans tetrathionate hydrolase and other PQQ binding domain-containing proteins from acidophilic bacteria and archaea that were distinct from neutrophilic quinone dehydrogenases (Fig. 2a). The proteins from acidophilic microorganisms were suggested to be membrane-bound dehydrogenases that agreed with the suggested localization of A. caldus TetH. The abundance of this gene group among acidophilic bacteria and archaea suggests its specificity and importance at low pH. The A. caldus tetH gene cluster context was compared to that found in A. ferrooxidans (Fig. 2b), and both clusters contain a two-component regulatory system. The A. ferrooxidans TcsRS system was most similar to the σ54-dependent ZraRS-like two-component systems, whereas the A. caldus RsrRS was similar to OmpRS-like systems.
RT-PCR analysis of the tetH cluster.
Amplification products were obtained by RT-PCR experiments between primer pairs g and h, i and k, j and k, and l and m, indicating that ISac1, rsrR, rsrS, tetH, and doxD are cotranscribed. ISac2 is not part of the operon as it is oriented in the opposite direction (Fig. 1b).
Q-PCR analysis of tetH expression.
Two-step Q-PCR was performed with RNA samples prepared from A. caldus cultures grown utilizing either S0 or tetrathionate as the growth substrate. All three tested genes (rsrR, tetH, and doxD) were upregulated with tetrathionate as the substrate in comparison to their levels with growth on S0, by 6.5-fold ± 5.6-fold, 233.5-fold ± 134.0-fold, and 25.3-fold ± 20.2-fold, respectively (for all, n [number of replicate experiments] = 6). This suggests that internal promoters may be present within the gene cluster, that posttranscriptional processing of mRNA takes place, or that there are different levels of primer binding due to mRNA structure.
Western blotting shows high levels of TetH in tetrathionate-grown A. caldus.
Western blots identified bands corresponding to TetH in cells grown on tetrathionate, thiosulfate, and a mixture of tetrathionate and S0 (Fig. 3). The positive result with cells grown on thiosulfate may be explained by the fact that the products of thiosulfate oxidation include tetrathionate (17). A weaker band was found in A. caldus and “F. acidarmanus” cells grown in mixed culture on pyrite that is oxidized to Fe2+ and thiosulfate (35) but not in “F. acidarmanus” protein alone as it does not oxidize RISCs (10). The weak band may be explained by a standard concentration of protein being loaded onto the gel that would have been derived from both species. The expression of TetH during bioleaching suggests its potential importance, as the end point of RISC metabolism is sulfuric acid and this produces the acidic environment necessary for the growth of A. caldus (and other biomining microorganisms). An industrial aim within bioleaching is the ability to control the oxidation of RISCs, and this study is the first step in understanding its regulation. The TetH enzyme was not observed when 50 μg total cell protein from S0-grown A. caldus was loaded on the gel. However, with 500 μg total protein, a faint band was observed (data not shown). This result correlates with the Q-PCR data, where it was shown that the expression of the tetH gene was strongly upregulated during cultivation on tetrathionate compared to its expression during cultivation on S0, and with previous data demonstrating lack of TetH activity in A. caldus cultured with S0 (6). The expression of tetH in cells cultured on tetrathionate and S0 suggests that its expression was induced by the presence of tetrathionate rather than repression by S0 and that it was regulated at the transcriptional level, depending on the growth conditions.
FIG. 3.
Western spot immunodetection of A. caldus TetH. Lane 1, A. caldus cells grown in mixed culture with “F. acidarmanus” on pyrite; lane 2, negative control with “F. acidarmanus” grown on pyrite; lane 3, A. caldus grown on a mixture of S0 and tetrathionate; lane 4, A. caldus grown on S0; lane 5, A. caldus grown on tetrathionate; and lane 6, A. caldus grown on thiosulfate. All lanes except lane 6 (100 μg) were loaded with 50 μg total protein. The dashed line indicates where the image of the single gel was cropped.
Investigation of the tetH operon promoters.
The two predicted promoters within the tetH cluster (P0 and P1) (Fig. 1) were cloned upstream of the pRW2 promoterless lac operon in E. coli strain DH5α, and the β-galactosidase activities were measured. The β-galactosidase activities were 42 ± 15, 103 ± 29, and 33,011 ± 4,675 Miller units for the vector control, P0, and P1 promoters, respectively (for all, n = 3). This gives 2.4- and 786-fold higher β-galactosidase expression from the P0 and P1 promoters, respectively, than from the vector control, suggesting that the promoters were active.
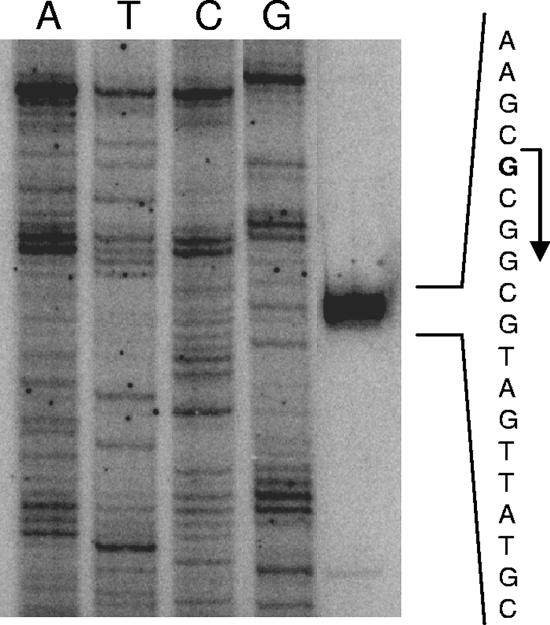
Primer extensions using total RNA from A. caldus identified a transcription start site located between rsrR and tetH for promoter P1 (Fig. 4). However, no band was detected in association with the P0 promoter (data not shown). This was probably due to the low expression level from this promoter, as shown by Q-PCR with A. caldus and by lac promoter fusion experiments with E. coli. Therefore, tetH regulation may occur via the rsrRS system at the P1 promoter, and future work will aim to elucidate possible transcriptional regulation by several predicted transcriptional factor (Fnr, ArcA, OmpR, and GcvA) binding sites. Interestingly, the A. ferrooxidans tetrathionate hydrolase gene (tth) also has a two-component regulatory system directly upstream, although, unlike the A. caldus rsrRS, the A. ferrooxidans system belongs to the σ54-specific family.
FIG. 4.
Primer extension analysis of the A. caldus P1 promoter during growth on tetrathionate. The predicted transcription start site is marked in bold with a bent arrow.
Identification of a TetH-bound quinoid compound.
The prediction of a putative PQQ binding domain in TetH was surprising, as previous analyses had not suggested a dehydrogenase activity (6, 17). Therefore, it was tested whether a TetH methanol extract restored the function of an apo form of the recombinant PQQ-dependent glucose dehydrogenase. The negative controls (lacking TetH extract, apoenzyme, or glucose; for all, n = 3) all gave a glucose dehydrogenase activity of ≤0.09 U ml−1 ± 0.03 U ml−1, whereas the positive control with added PQQ gave 86.2 U ml−1 ± 0.7 U ml−1. In the presence of TetH methanol extract, the activity was 0.09 U ml−1 ± 0.03 U ml−1, suggesting that TetH does not contain PQQ (data not shown). However, native TetH was suggested to contain a quinoid compound by NBT-glycinate staining after direct blotting on a nitrocellulose filter (Fig. 5) and sodium dodecyl sulfate electrophoresis with subsequent blotting (data not shown). However, it was not possible to identify the cofactor, as the staining does not discriminate between different quinoid compounds. The predicted quinone binding domain and the positive o-quinone staining suggest that TetH is involved in quinone turnover. However, the fact that the TetH extract did not restore the function of the apo form of glucose dehydrogenase suggests that this cofactor is not PQQ.
FIG. 5.
o-Quinone staining of two different amounts of native TetH (a) and TetH boiled and precipitated with trichloroacetic acid before blotting and staining (b). The amounts of TetH were 15 and 80 μg protein in lanes 1 and 2, respectively.
Conclusions.
RT-PCR results show that tetH is cotranscribed with doxD. Although the role of doxD in A. caldus has not been defined, it is similar to a gene from the acidophilic thermophile Acidianus ambivalens that encodes a subunit of thiosulfate:caldariella quinone oxidoreductase and to a gene encoding one of the components of a predicted thiosulfate:quinol oxidoreductase in A. ferrooxidans. However, A. ferrooxidans contains further copies of doxDA, possibly used for thiosulfate metabolism (23). It has been previously reported that uncouplers and inhibitors of electron transport, such as carbonyl cyanide m-chlorophenyl-hydrazone and 2,4-dinitrophenol suppress tetrathionate hydrolysis of A. caldus (17). Moreover, bioinformatic analysis suggests that tetH is similar to PQQ binding dehydrogenases that normally transfer electrons to the quinol oxidase or bc1 complex via ubiquinone as a primary electron acceptor (2, 7). Based on this data, we propose that TetH is connected to the respiratory chain through an o-quinone cofactor. Although the A. caldus and A. ferrooxidans tetrathionate hydrolases had 71% similarity, the latter is not inhibited by 2,4-dinitrophenol (18) and doxD genes are not located in the neighborhood of tetH on the A. ferrooxidans chromosome (Fig. 2b), suggesting that the proteins may have different mechanisms for transferring electrons to the respiratory chain.
Acknowledgments
We acknowledge Anna Åberg and Claudia Muller for advice on the primer extension and Siv Sääf for technical assistance. Plasmid pRW2 was generously provided by Victoria Schingler, Umeå University, and the pyrite was kindly supplied by Boliden Mineral AB.
This work was carried out in the frame of the European Commission project BioMinE under the sixth framework program for research and development (European project contract NMP1-CT-500329-1). D.H. acknowledges Fondecyt grant 1050063, DI-UNAB grant 34-06, and a Microsoft-sponsored research award.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, C. 2004. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 428:2-9. [DOI] [PubMed] [Google Scholar]
- 3.Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512-524. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre, C., J. Morschhäuser, J. Jass, J. Hacker, and B. E. Uhlin. 2003. Transcriptional analysis of the sfa determinant revealing multiple mRNA processing events in the biogenesis of S fimbriae in pathogenic Escherichia coli. J. Bacteriol. 185:620-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berriman, M., and K. Rutherford. 2003. Viewing and annotating sequence data with Artemis. Brief. Bioinform. 4:124-132. [DOI] [PubMed] [Google Scholar]
- 6.Bugaytsova, Z., and E. B. Lindström. 2004. Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 271:272-280. [DOI] [PubMed] [Google Scholar]
- 7.Davidson, V. L. 2004. Electron transfer in quinoproteins. Arch. Biochem. Biophys. 428:32-40. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, G. A. H., W. Hazeu, P. Bos, and J. G. Kuenen. 1997. Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 143:499-504. [DOI] [PubMed] [Google Scholar]
- 9.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dopson, M., C. Baker-Austin, A. Hind, J. P. Bowman, and P. L. Bond. 2004. Characterization of Ferroplasma isolates and Ferroplasma acidarmanus sp. nov., extreme acidophiles from acid mine drainage and industrial bioleaching environments. Appl. Environ. Microbiol. 70:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dopson, M., and E. B. Lindström. 2004. Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite and chalcopyrite. Microb. Ecol. 48:19-28. [DOI] [PubMed] [Google Scholar]
- 12.Dopson, M., and E. B. Lindström. 1999. Potential role of Thiobacillus caldus in arsenopyrite bioleaching. Appl. Environ. Microbiol. 65:36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, K. J., P. L. Bond, and J. F. Banfield. 2000. Characteristics of attachment and growth of Thiobacillus caldus on sulphide minerals: a chemotactic response to sulphur minerals? Environ. Microbiol. 2:324-332. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, K. J., P. L. Bond, G. K. Druschel, M. M. McGuire, R. J. Hamers, and J. F. Banfield. 2000. Geochemical and biological aspects of sulfide mineral dissolution: lessons from Iron Mountain, California. Chem. Geol. 169:383-397. [Google Scholar]
- 15.Edwards, K. J., P. L. Bond, T. M. Gihring, and J. F. Banfield. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796-1799. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39:235-289. [DOI] [PubMed] [Google Scholar]
- 17.Hallberg, K. B., M. Dopson, and E. B. Lindstrom. 1996. Reduced sulfur compound oxidation by Thiobacillus caldus. J. Bacteriol. 178:6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazeu, W., W. H. Batenburg-van der Vegte, P. Bos, R. K. van der Pas, and J. G. Kuenen. 1988. The production and utilization of intermediary elemental sulfur during the oxidation of reduced sulfur compounds by Thiobacillus ferrooxidans. Arch. Microbiol. 150:574-579. [Google Scholar]
- 19.Kelly, D. P., J. K. Shergill, W. P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenoek 71:95-107. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Lodge, J., R. Williams, A. Bell, B. Chan, and S. Busby. 1990. Comparison of promoter activities in Escherichia coli and Pseudomonas aeruginosa: use of a new broad-host-range promoter-probe plasmid. FEMS Microbiol. Lett. 67:221-225. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. 1972. Experiments in molecular genetics, 1st ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Muller, F. H., T. M. Bandeiras, T. Urich, M. Teixeira, C. M. Gomes, and A. Kletzin. 2004. Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate: quinone oxidoreductase. Mol. Microbiol. 53:1147-1160. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, K., M. Nakamura, H. Yoshikawa, and Y. Amano. 2001. Purification and properties of thiosulfate dehydrogenase from Acidithiobacillus thiooxidans JCM7814. Biosci. Biotechnol. Biochem. 65:102-108. [DOI] [PubMed] [Google Scholar]
- 25.Norris, P. R. 2007. Acidophile diversity in mineral sulfide oxidation, p. 199-235. In D. E. Rawlings and D. B. Johnson (ed.), Biomining. Springer-Verlag, Berlin, Germany.
- 26.Okibe, N., M. Gericke, K. B. Hallberg, and D. B. Johnson. 2003. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred-tank bioleaching operation. Appl. Environ. Microbiol. 69:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson, G. J., J. A. Brierley, and C. L. Brierley. 2003. Bioleaching review part B. Progress in bioleaching: applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 63:249-257. [DOI] [PubMed] [Google Scholar]
- 28.Olsthoorn, A. J., and J. A. Duine. 1996. Production, characterization, and reconstitution of recombinant quinoprotein glucose dehydrogenase (soluble type; EC 1.1.99.17) apoenzyme of Acinetobacter calcoaceticus. Arch. Biochem. Biophys. 336:42-48. [DOI] [PubMed] [Google Scholar]
- 29.Paz, M. A., R. Fluckiger, A. Boak, H. M. Kagan, and P. M. Gallop. 1991. Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 266:689-692. [PubMed] [Google Scholar]
- 30.Rawlings, D. E. 2002. Heavy metal mining using microbes. Annu. Rev. Microbiol. 56:65-91. [DOI] [PubMed] [Google Scholar]
- 31.Rawlings, D. E., N. J. Coram, and M. N. D. Gardner. 1999. Thiobacillus caldus and Leptospirillium ferrooxidans are widely distributed in continuous flow biooxidation tanks used to treat a variety of metal containing ores and concentrates, p. 777-786. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment: toward the mining of the 21st century, vol. A. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 32.Rohwerder, T., T. Gehrke, K. Kinzler, and W. Sand. 2003. Bioleaching review part A. Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63:239-248. [DOI] [PubMed] [Google Scholar]
- 33.Rohwerder, T., and W. Sand. 2003. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149:1699-1709. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Schippers, A., and W. Sand. 1999. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Appl. Environ. Microbiol. 65:319-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silver, M., and D. G. Lundgren. 1968. The thiosulfate oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 46:1215-1220. [DOI] [PubMed] [Google Scholar]
- 37.Sorokin, D. Y., and J. G. Kuenen. 2005. Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes. FEMS Microbiol. Rev. 29:685-702. [DOI] [PubMed] [Google Scholar]
- 38.Sugio, T., T. Hisazumi, T. Kanao, K. Kamimura, F. Takeuchi, and A. Negishi. 2006. Existence of aa3-type ubiquinol oxidase as a terminal oxidase in sulfite oxidation of Acidithiobacillus thiooxidans. Biosci. Biotechnol. Biochem. 70:1584-1591. [DOI] [PubMed] [Google Scholar]
- 39.Tano, T., H. Kitaguchi, K. Harada, T. Nagasawa, and T. Sugio. 1996. Purification and some properties of a tetrathionate decomposing enzyme from Thiobacillus thiooxidans. Biosci. Biotechnol. Biochem. 60:224-227. [DOI] [PubMed] [Google Scholar]
- 40.Vestal, J. R., and D. G. Lundgren. 1971. The sulfite oxidase of Thiobacillus ferrooxidans (Ferrobacillus ferrooxidans). Can. J. Biochem. 49:1125-1130. [DOI] [PubMed] [Google Scholar]
- 41.Wakai, S., M. Kikumoto, T. Kanao, and K. Kamimura. 2004. Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 68:2519-2528. [DOI] [PubMed] [Google Scholar]
- 42.Watling, H. R. 2006. The bioleaching of sulphide minerals with emphasis on copper sulphides: a review. Hydrometallurgy 84:81-108. [Google Scholar]