Abstract
The microbial community structure and spatial distribution of microorganisms and their in situ activities in anaerobic granules were investigated by 16S rRNA gene-based molecular techniques and microsensors for CH4, H2, pH, and the oxidation-reduction potential (ORP). The 16S rRNA gene-cloning analysis revealed that the clones related to the phyla Alphaproteobacteria (detection frequency, 51%), Firmicutes (20%), Chloroflexi (9%), and Betaproteobacteria (8%) dominated the bacterial clone library, and the predominant clones in the archaeal clone library were affiliated with Methanosaeta (73%). In situ hybridization with oligonucleotide probes at the phylum level revealed that these microorganisms were numerically abundant in the granule. A layered structure of microorganisms was found in the granule, where Chloroflexi and Betaproteobacteria were present in the outer shell of the granule, Firmicutes were found in the middle layer, and aceticlastic Archaea were restricted to the inner layer. Microsensor measurements for CH4, H2, pH, and ORP revealed that acid and H2 production occurred in the upper part of the granule, below which H2 consumption and CH4 production were detected. Direct comparison of the in situ activity distribution with the spatial distribution of the microorganisms implied that Chloroflexi contributed to the degradation of complex organic compounds in the outermost layer, H2 was produced mainly by Firmicutes in the middle layer, and Methanosaeta produced CH4 in the inner layer. We determined the effective diffusion coefficient for H2 in the anaerobic granules to be 2.66 × 10−5 cm2 s−1, which was 57% in water.
Upflow anaerobic sludge blanket (UASB) reactors are commonly used in the treatment of high-strength municipal and industrial wastewaters. Their design permits the retention of a greater amount of active biomass (known as granules) than other anaerobic reactors. These anaerobic granules harbor several metabolic groups of microorganisms, including hydrolytic, fermentative, syntrophic, and methanogenic microorganisms, involved in the anaerobic degradation of complex organic compounds. These different trophic groups of anaerobes closely and coordinately interact with one another within the granules and convert complex organic compounds in wastewaters into methane (CH4) and carbon dioxide (CO2). The microbial communities of anaerobic granules used to treat different wastewaters have been investigated by culture-independent 16S rRNA gene-based molecular analyses (14, 22, 36), which allow one to obtain more-complete inventories of microorganisms in anaerobic granules. The results have shown that anaerobic granules consist of a phylogenetically diverse group of microorganisms; however, the majority of them have not yet been cultivated (14, 22, 36).
The application of fluorescence in situ hybridization (FISH) with specific oligonucleotide probes allows us to further determine the abundances and in situ spatial distributions of specific phylogenetic groups in anaerobic granules (14, 20, 35, 41). By using the FISH technique, distinct multilayered structures of different phylogenetic groups of microorganisms have been determined in different anaerobic granules cultivated on different substrates (20, 22, 35). These well-organized unique structures are thought to be a result of the sequential degradation of complex organic compounds by the different trophic groups within the granules, although this has not yet been confirmed experimentally.
In addition, the spatial distribution of the microorganisms determined by FISH analyses does not necessarily reflect microbial activity distributions. This is because the majority of microorganisms have not yet been cultivated, and phylogeny and phenotype are not necessarily congruent with physiology. The spatial distributions of in situ metabolic functions in anaerobic granules are poorly understood, mainly due to a lack of specific analytical tools with sufficient spatial resolution. Only a few studies have analyzed the distributions of glucose-degrading, fermentative, and sulfate-reducing activities in UASB granules by using microsensors for glucose, pH, and hydrogen sulfide (H2S) (13, 21, 42). In those studies, CH4 production rates were not, however, directly measured with microsensors. Santegoeds et al. (34) used CH4 biosensors and H2S microsensors to directly measure in situ methanogenic and sulfate-reducing activities in anaerobic granules and then related them to the spatial distributions of methanogenic and sulfate-reducing bacterial populations. They demonstrated a distinct layered structure of microbial activities in which sulfate reduction occurred in the outer layer, whereas CH4 production was found in the center of the granule. However, they did not investigate the in situ distributions of fermentative and syntrophic populations and their activities in the granules; hence, the overall conversion mechanism of complex organic compounds to CH4 within granules has not been elucidated.
To investigate the overall anaerobic conversion mechanism of organic compounds to CH4 within anaerobic granules, we first analyzed the bacterial and archaeal community structures by 16S rRNA gene-cloning analysis; then the spatial distributions of important phylogenetic groups in the granules were determined by FISH. Second, we applied the microsensors for CH4, H2, pH, and the oxidation-reduction potential (ORP) to determine the spatial distribution of the in situ fermentative (H+-producing), syntrophic (H2-producing), and methanogenic activities in the granules. The microbial activity distribution was directly compared with the spatial distribution of the microorganisms.
MATERIALS AND METHODS
Sludge source.
Anaerobic granular sludge was collected from the bottom of a lab-scale UASB reactor (height, 50 cm; diameter, 5 cm) operated at 35°C. The reactor was inoculated with 0.7 liter of anaerobic granular sludge obtained from a full-scale UASB reactor treating the wastewater from an isomerized sugar-processing plant. The lab-scale reactor was fed with a synthetic medium at an average organic loading rate of 1.67 g chemical oxygen demand (COD) liter−1 day−1 and a hydraulic residence time of 8.2 h. The synthetic medium contained powdered skim milk (1,250 mg liter−1) as carbon and energy sources, NaHCO3 (1,000 mg liter−1), K2HPO4 (50 mg liter−1), and the mineral solution (4). Granule samples were obtained from the lab-scale UASB reactor after 1 year of operation. The water qualities of the effluent during the sampling period were as follows (average ± standard deviation, n = 31): COD, 30 ± 9 mg liter−1; COD removal rate, 95% ± 1%; ORP, −230 ± 20 mV; gas production rate, 0.10 ± 0.02 liter h−1; CH4 content in gas, 55 ± 15%; CO2 content in gas, 11 ± 4%; N2 content in gas, 34 ± 17%; H2 content in gas, 2,300 ± 3,300 ppm; acetate content, 165 ± 81 μM; propionate content, 77 ± 46 μM; and butyrate content, 5.5 ± 7.7 μM. Other volatile fatty acids (butyrate, isobutyrate, and formate) were sometimes detected at trace levels.
DNA extraction and PCR amplification.
The granule samples were homogenized in the sterilized, distilled water, and approximately 1.0-ml subsamples were subjected to DNA extraction. Total DNA was extracted from the homogenized granules with a FastDNA SPIN kit for soil (Bio 101; Qbiogene, Inc., Carlsbad, CA) as described in the manufacturer's instructions. 16S rRNA gene fragments were amplified from the extracted total DNA with Taq DNA polymerase (TaKaRa Bio, Inc., Ohtsu, Japan) by using a primer set of 11f (19) and 1492r (39) for the bacterial community and a primer set of A109f (40) and 1492r (39) for the archaeal community. The PCR products were electrophoresed on a 1% (wt/vol) agarose gel and purified with a Wizard PCR preps DNA purification system (Promega). To reduce the possible PCR bias, the 16S rRNA gene was amplified in 6 to 10 PCR tubes and all the tubes were combined for the next cloning step.
Cloning and sequencing of the 16S rRNA genes and phylogenetic analysis.
The purified PCR products were ligated into a qCR-XL-TOPO vector with a TOPO XL PCR cloning kit (Invitrogen, Carlsbad, CA). The ligated products were transformed into TOP10-competent Escherichia coli cells (Invitrogen). Plasmids were extracted from the cloned cells and purified with a Wizard Plus minipreps DNA purification system (Promega). Nucleotide sequencing was performed with an automatic sequencer (3100-Avant genetic analyzer; Applied Biosystems). All sequences were checked for chimeric artifacts by using a CHECK_CHIMERA program from the Ribosomal Database Project (24). Partial sequences (approximately 500 bp) were compared with similar sequences of the reference organisms by a BLAST search (1). Sequences with 97% or greater similarity were grouped into operational taxonomic units (OTUs) by a SIMILARITY_MATRIX program from the Ribosomal Database Project (24). Nearly complete sequencing of the 16S rRNA gene of each representative OTU was performed, and the sequences were aligned with a CLUSTAL W package (37). The Molecular Evolutionary Genetics Analysis program, version 3.1, was used to construct phylogenetic trees based on the neighbor-joining (33) and maximum-parsimony methods. Bootstrap resampling analysis of 1,000 replicates was performed to estimate the degree of confidence in the tree topologies.
Fixation and cryosectioning of granule samples.
Granule samples were fixed in 4% paraformaldehyde solution for 8 h at 4°C, washed three times with phosphate-buffered saline (10 mM sodium phosphate buffer, 130 mM sodium chloride [pH 7.2]), and embedded in Tissue-Tek OCT (optimal cutting temperature) compound (Sakura Finetek, Torrance, CA) overnight to infiltrate the OCT compound into the granule, as described previously (29). After rapid freezing at −21°C, 20-μm-thick slices were prepared with a cryostat (Reichert-Jung Cryocut 1800; Leica, Bensheim, Germany).
FISH.
In situ hybridization was performed according to the procedure described by Amann (2) and Okabe et al. (29). The following 16S and 23S rRNA-targeted oligonucleotide probes were used: EUB338 (3), EUB338 II (8), EUB338 III (8), ARC915 (31), ALF968 (28), BET42a (25), GNSB-941 (16), HGC69A (32), LGC354A (27), LGC354B (27), LGC354C (27), MB1174 (31), MG1200 (31), MS821 (31), and MX825 (31). The probes were labeled with fluorescein isothiocyanate (FITC) or tetramethyl rhodamine 5-isothiocyanate (TRITC). The phylum-specific probes were applied simultaneously with the ARC915 probe specific for Archaea. A model LSM510 confocal laser-scanning microscope (CLSM; Carl Zeiss, Oberkochen, Germany) equipped with an Ar ion laser (488 nm) and a HeNe laser (543 nm) was used.
Microsensor measurements.
For microsensor measurements, the granules with diameters of ca. 2 mm were selected and positioned, using five insect needles, in the flow cell reactor (4.0 liter) that was filled with the synthetic medium used for the lab-scale UASB reactor at 35°C. The medium in the flow cell reactor was kept anaerobic by adding a reducing agent (thioglycolic acid at 100 mg liter−1) and by continuous bubbling with N2, which also resulted in sufficient mixing of the medium. The ORP of the medium was ca. −200 mV. The average liquid velocity, as judged from the movement of the suspended particles, was ca. 5 mm s−1. The granules were acclimated in the medium for at least 6 h before being measured to ensure that steady-state profiles were obtained. The in situ steady-state concentration profiles of CH4, H2, pH, and ORP in the granules were measured, using microsensors as described by Okabe et al. (30). At least three concentration profiles were measured for each chemical species. For practical reasons, a concentration profile was measured only once in a granule; therefore, concentration profiles of different chemical species were measured in different granules.
Microscale biosensors for CH4 were constructed as described by Damgaard and Revsbech (11). Because all measurements were performed under anoxic conditions, an oxygen-scavenging guard capillary (12) was not applied. Hence, the CH4 microsensor was assembled from an oxygen microsensor, a gas capillary, and a medium capillary. The tip diameters of the microsensors were 50 to 100 μm. A culture of the methane-oxidizing bacterium Methylosinus trichosporium (ATCC 49243) was injected into the medium capillary. The principle of the CH4 microsensor is based on a counter diffusion of oxygen and CH4. CH4 diffusing through the membrane of the medium capillary is consumed by the methane-oxidizing bacteria with a concomitant decrease of oxygen inside the reaction space (i.e., the medium capillary). The amount of oxygen consumed is detected by the internal oxygen microsensor and translated into a CH4 partial pressure value by calibration. Calibration was routinely performed before and after a measurement by placing a CH4 microsensor in a calibration chamber (100 ml) into which CH4 and N2 gases were continuously blown at known flow rates at 35°C. After the sensor signal stabilized, the signal was monitored and the CH4 concentration was measured by gas chromatography. The CH4 concentration was changed stepwise by changing the flow rates of CH4 and N2 gases. This procedure was repeated over the full range of 0% to 100% CH4 saturation. The response to 0% CH4 saturation (i.e., 100% N2 saturation) was typically between 70 and 90 pA, and 90% response times were ca. 200 s. The response was linear (r2 > 0.95) over the range of 0% to 100% CH4 saturation. The slope of the calibration curves was ca. 20 pA per atm of CH4 (see Fig. S1 in the supplemental material). The sensor life span was ca. 2 weeks. If the slope of the calibration curve was less than 10 pA per atm of CH4, the microsensor was discarded.
Polarographic H2 microsensors were constructed as described by Ebert and Brune (15). The tip diameters of the microsensors were ca. 10 μm, 90% response times were less than 2 s, and the detection limit was ca. 1 μM. Calibration was routinely performed by immersing a microsensor in a calibration chamber filled with the synthetic medium, which continuously bubbled with H2 and N2 gases. The H2 concentration in the medium was measured by gas chromatography. The H2 concentration was changed stepwise by changing the flow rates of H2 and N2 gases. ORP microsensors, which were made from a platinum wire coated with a glass micropipette, were constructed and calibrated as described by Jang et al. (18). All ORP data reported in this paper were the measures of the potential difference between an ORP microsensor and the Ag/AgCl reference electrode. Potentiometric pH microsensors were constructed, calibrated, and used according to the protocol described by Okabe et al. (29).
Microbial activity calculations.
Net volumetric CH4 and H2 production rates [R(CH4) and R(H2), respectively] in the granule were estimated from the steady-state concentration profiles of CH4 and H2 by using Fick's second law of diffusion as previously described by Santegoeds et al. (34) and Lorenzen et al. (23). The total H2 production rate from the granule was calculated using Fick's first law of diffusion (29). Molecular diffusion coefficients in water at 25°C (Dw 25) of 1.49 × 10−5 cm2 s−1 for CH4 and 4.50 × 10−5 cm2 s−1 for H2 were used for the calculations (7). Molecular diffusion coefficients in water at 35°C (Dw 35) were calculated according to the Stokes-Einstein relationship (7). The effective diffusion coefficients (Deff) of these compounds in the granules were estimated by correcting the Dw with the ratio of the diffusivities in granules and in water (57%), as determined with the H2 microsensor in this study (see Discussion).
Estimation of Deff.
Granules with a diameter of ca. 2 mm were selected and deactivated in pure chloroform for 10 min. After being rinsed, a deactivated granule was mounted in the flow cell reactor containing the synthetic medium at 35°C for microsensor measurements, and a H2 microsensor was positioned in the center of the granule with a micromanipulator. After a preincubation of at least 30 min, H2 gas was bubbled. The evolution of the H2 concentration in the center of the granule was monitored over time in triplicate, and the average Deff was calculated according to the protocol described by Cronenberg and van den Heuvel (6).
Chemical analyses.
The concentrations of COD were determined according to standard methods (5). Volatile fatty acid concentrations were determined by a high-performance liquid chromatography system (LC-10AD system; Shimadzu Co., Kyoto, Japan) equipped with a Shimadzu Shim-pack SCR-102H column (0.8 by 30 cm) after filtration with 0.2-μm-pore-size membranes (Advantec Co., Ltd., Tokyo, Japan). The levels of CH4, H2, and CO2 in gaseous samples were determined by a gas chromatography system (GC-14B; Shimadzu Co., Kyoto, Japan) equipped with a thermal-conductivity detector and a Shincarbon-ST column (Shimadzu Co., Kyoto, Japan). The ORP and pH were directly determined by using an ORP and a pH electrode, respectively.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of the clones used for the phylogenetic analysis are AB329636 to AB329664.
RESULTS
Bacterial and archaeal community structures.
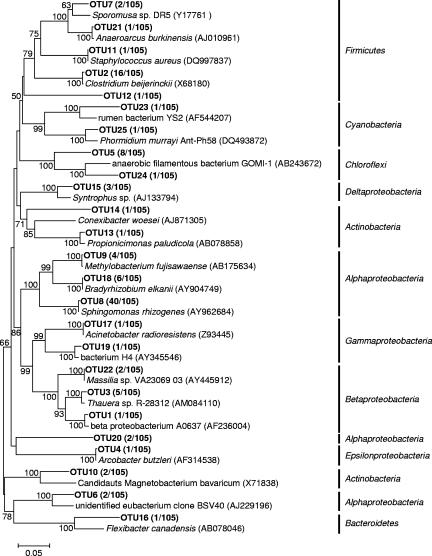
16S rRNA gene clone libraries of Bacteria and Archaea were constructed from the anaerobic granule samples taken from the UASB reactor. From the Bacteria clone library, 105 clones were randomly selected and partial sequences of about 500 bp were analyzed. In total, the clones were grouped into 25 OTUs on the basis of more than 97% sequence similarity within an OTU. Then a nearly complete 16S rRNA gene sequence of a representative clone of each OTU was analyzed. The phylogenetic trees were constructed by the neighbor-joining and maximum-parsimony methods, and both methods resulted in essentially the same topology. The phylogenetic tree inferred from the neighbor-joining method is shown in Fig. 1. The distribution of cloned sequences among phyla is as follows: Alphaproteobacteria, 51%; Firmicutes, 20%; Chloroflexi, 9%; Betaproteobacteria, 8%; Actinobacteria, 4%; Deltaproteobacteria, 3%; Cyanobacteria, 2%; Gammaproteobacteria, 2%; Epsilonproteobacteria, 1%; and Bacteroidetes, 1%. The most frequently detected clones (40 out of 105 clones), which were represented by OTU8, were closely related to “Sphingomonas rhizogenes” (99.8% sequence similarity).
FIG. 1.
Phylogenetic tree representing affiliation of 16S rRNA clone sequences of Bacteria retrieved from granule samples (OTU numbers). The tree was generated by using nearly full-length 16S rRNA gene sequences and the neighbor-joining method. The scale bar represents 5% estimated divergence. The numbers at the nodes are bootstrap values (1,000 replicates) with more than 50% bootstrap support.
For the Archaea clone library, 48 clones were randomly selected and partial sequences of about 500 bp were analyzed. The clones were grouped into four OTUs. The clones belonging to OTU1 and OTU4 were related to Methanosaeta, whereas the clones belonging to the other OTUs (OTU2 and OTU3) were related to Methanomicrobiales. Their representative sequences were used for phylogenetic analysis (Fig. 2). The most frequently detected clones (33 out of 48 clones; detection frequency, 69%) represented by OTU1 were affiliated with Methanosaeta concilii, with a 98.3% sequence similarity. The clones represented by OTU4 (2 of 48; detection frequency, 4%) were affiliated with Methanosaeta harundinacea, with a 99.6% sequence similarity. The clones represented by OTU2 (9 of 48; detection frequency, 19%) and OTU3 (4 of 48; detection frequency, 8%) were related to the order Methanomicrobiales, with 96.2% and 97.8% sequence similarities, respectively. These results indicate that the diversity of the Archaea clone library is less than that of the Bacteria.
FIG. 2.
Phylogenetic tree representing affiliation of 16S rRNA clone sequences of Archaea retrieved from granule samples (OTU numbers). The tree was generated by using nearly full-length 16S rRNA gene sequences and the neighbor-joining method. The scale bar represents 5% estimated divergence. The numbers at the nodes are bootstrap values (1,000 replicates) with more than 50% bootstrap support.
Spatial distribution of microorganisms in granules.
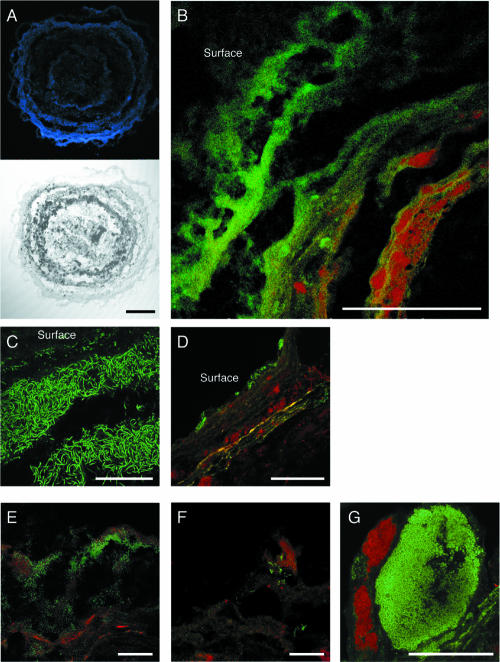
Cross-sectional differential interference contrast images of the granules showed that the granules had a multilayered structure consisting of biomass and interstitial voids (Fig. 3A). Analysis of the granules after staining with DAPI (4′,6′-diamidino-2-phenylindole) showed that the microorganisms were present predominantly in the outer 400 μm (Fig. 3A). It is most likely that the dark, nonstaining center consisted of inert matter and dormant microbial cells, which can probably be attributed to substrate limitation in the centers of the granules due to the relatively low COD loading rate in the reactor from which the granule samples were taken. The nonstaining center was always observed in the granules with diameters exceeding about 1,000 μm. FISH, using a FITC-labeled EUB338-mixed probe and a TRITC-labeled ARC915 probe, revealed that the outer layer (ca. 250 μm thick) was dominated by bacterial cells whereas the inner layer (below 250 μm from the surface) was occupied mainly by archaeal cells (Fig. 3B). Archaeal and bacterial signals were low in the granule interior (below ca. 400 μm from the surface). This layered structure was repeatedly observed in all of the granular sections analyzed. Filamentous cells were observed in the uppermost layer of the granules, and these cells were hybridized with the probe GNSB-941, which is specific for almost all members of the phylum Chloroflexi (Fig. 3C). The BET42a-stained cells were also present in the outer shell of the granule (Fig. 3D). The cells hybridized with probe LGC354, specific for Firmicutes, were numerically important Bacteria in the inner layer of the granule (Fig. 3E). The abundance and fluorescence intensity of the ALF968-stained cells were low (Fig. 3F), although members of Alphaproteobacteria were predominant in the bacterial clone library (Fig. 1). The MX825-stained cells surrounded the dense spherical microcolonies that were composed of a number of coccoid cells stained with the probe HGC69A (i.e., Actinobacteria) in the middle layer (at a depth of ca. 200 μm) of the granule (Fig. 3G). Among archaeal cells, the MX825-stained cells (i.e., Methanosaeta) predominated while the remaining portion of archaeal cells could not be hybridized with the MG1200, MB1174, and MS821 probes (data not shown).
FIG. 3.
Confocal laser scanning microscope images of thin sections of the anaerobic granules showing the in situ spatial organization of bacteria and archaea. (A) DAPI-stained and differential interference contrast images. (B) FISH with TRITC-labeled probe ARC915 and the FITC-labeled EUB338-mixed probe. (C) FISH with FITC-labeled probe GNSB-941. (D) FISH with TRITC-labeled probe ARC915 and FITC-labeled probe BET42a. (E) FISH with TRITC-labeled probe ARC915 and FITC-labeled probe LGC354. (F) FISH with TRITC-labeled probe ARC915 and FITC-labeled probe ALF968. (G) FISH with TRITC-labeled probe MX825 and FITC-labeled probe HGC69A. Scale bars indicate 200 μm (panels A and B) and 50 μm (panels C through G). For panels B through G, TRITC-labeled probes are red and FITC-labeled probes are green.
Concentration profiles and spatial distribution of microbial activities.
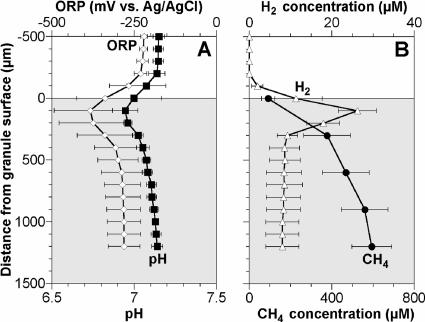
Steady-state concentration profiles of the ORP, pH, H2, and CH4 in the granules are shown in Fig. 4. A decrease in pH (i.e., acid production) was found in the upper parts of the granules, below which pH increased (Fig. 4A). When H2 profiles were measured in medium without NaHCO3, the H2 profile showed a peak of 26 μM at a depth of 100 μm and the H2 concentration readily decreased in the inner layer of the granule (Fig. 4B). The addition of 12 mM of NaHCO3 stimulated H2 consumption activity, and the H2 concentration in the granule decreased to under the detection limit (1 μM) (data not shown). The CH4 concentration gradually increased throughout the granule, and its gradient was steeper in the outer layer of the granule (Fig. 4B).
FIG. 4.
Concentration profiles of pH, ORP, CH4, and H2 in the anaerobic granule. Each profile value is the average of three measurements, and the error bars represent the standard deviations of triplicate measurements. Zero on the vertical axis corresponds to the surface of the granule.
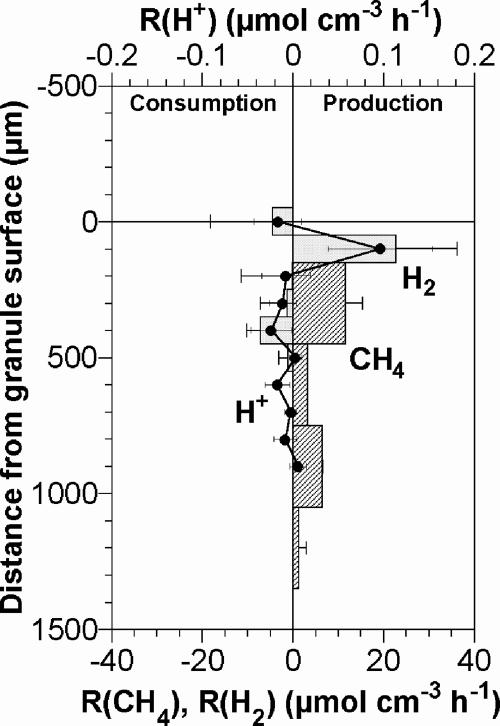
The spatial distributions of R(CH4) and R(H2) were calculated on the basis of the measured profiles. Figure 5 shows that anaerobic processes occurred in distinctly different layers within the granule. H2 production was detected exclusively at a depth of 100 μm. The H2 produced was partly consumed below 300 μm from the surface and was emitted from the granule at a total production rate of 2.3 ± 4.3 μmol cm−2 h−1. CH4 was produced mainly in the inner layer (below ca. 300 μm from the surface) at a maximum rate of 11.5 ± 3.9 μmol cm−3 h−1 at a depth of 300 μm.
FIG. 5.
Spatial distributions and magnitudes of the net volumetric production rates of CH4 and H2. The rates were calculated based on the corresponding concentration profiles shown in Fig. 4. Each profile value is the average of three measurements, and the error bars represent the standard deviations of triplicate measurements. Negative values indicate consumption rates. Zero on the vertical axis corresponds to the surface of the granule.
Transient measurements for determination of Deff.
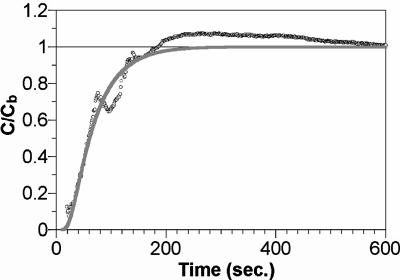
For the determination of the Deff, H2 concentrations were continuously monitored at the center of the granule that was deactivated by pure chloroform. The ratio of the H2 concentration at the granule center to that in the bulk liquid (C/Cb) was calculated, and a representative profile of C/Cb transience is shown in Fig. 6. A mathematical model that describes C/Cb transience in the granule provided a good fit to the profile measured (correlation coefficient, 0.98). Based on the model, the Deff for H2 in the granule was determined to be 2.66 ± 0.13 × 10−5 cm2 s−1 (average ± standard deviation, n = 3).
FIG. 6.
A typical transient H2 concentration profile measured at the center of the granule inhibited by chloroform. Points (○) indicate H2 concentrations measured, and the solid line is a theoretical curve.
DISCUSSION
The microsensor measurements indicated a distinct layered structure of the microbial activities in the anaerobic granule, with net acid (H+) and H2 production (i.e., fermentative and syntrophic activities) at a depth of 100 μm and net H2 consumption and CH4 production (i.e., methanogenesis) below 300 μm from the surface (Fig. 5). Because anaerobic degradation of organic compounds is a multistep process, a layered structure was developed. The layers consist of bacteria that hydrolyze complex organic compounds in wastewater to fundamental structural building blocks (e.g., glucose and amino acids) at the granule surface, fermentative bacteria that ferment these products to fatty acids and, subsequently, syntrophic bacteria that oxidize fatty acids and alcohols to H2 and acetate in the middle layer, and methane-producing archaea in the inner layer. Our knowledge of the spatial distribution of microbial activities, especially the CH4-producing activity in anaerobic granules, is very limited because in situ activity measurements require specific analytical tools (e.g., microsensors). CH4 microprofiles and the distributions of CH4 production activity in a sewage biofilm (10), a rice paddy soil (12), and a lake sediment (9) have been measured with micrometer resolution by using CH4 microsensors. Only one study indicated that there were CH4 microprofiles in anaerobic granules (34). This study demonstrated a similar, layered structure of the microbial activities in methanogenic-sulfidogenic aggregates, in which sulfidogenic activity was found in the outer layer and CH4 production started at from 300 μm onward inside the aggregate. In this study, sulfate reduction could be ignored because the sulfate concentration was less than 2 μM in the synthetic medium for cultivation and microsensor measurements.
In situ hybridization results showed that the outer layer (ca. 250-μm thick) of the granule was dominated by Bacteria whereas the inner layer (below 250 μm from the surface) was dominated by Archaea (Fig. 3B). Similar layered structures of microorganisms in anaerobic granules have been reported elsewhere (20, 35). Direct comparison of the microsensor results with FISH results revealed that H2 might be produced at a depth of 100 μm mainly by members of Firmicutes and that Archaea affiliated with Methanosaeta produced CH4 below 300 μm. Phylogenetic analysis revealed that members of the phyla Alphaproteobacteria, Firmicutes, Chloroflexi, and Betaproteobacteria dominated the bacterial clone library. This result was in good agreement with the FISH results. Chloroflexi were detected mainly in the granule surface (Fig. 3C), indicating that they contributed to the hydrolysis of complex organic compounds. Ariesyady et al. (4) analyzed the in situ function (i.e., glucose-, propionate-, butyrate-, and acetate-degrading activities) of bacteria in a full-scale anaerobic sludge digester by using a microautoradiography-FISH technique and revealed that Chloroflexi were one of the numerically dominant glucose-degrading bacterial groups. The filamentous Chloroflexi-like bacteria also contributed to the formation and maintenance of dense and rigid granule structures by wrapping up other microorganisms. Bacteria belonging to Firmicutes have been detected in methanogenic granules (14, 22). These bacteria can anaerobically utilize glucose, propionate, butyrate, and acetate (4) and produce H2 (17). In contrast, Ariesyady et al. (4) demonstrated that the glucose-utilizing rate of Betaproteobacteria was very low and that Alphaproteobacteria did not utilize all of the substrates tested (i.e., glucose, propionate, butyrate, and acetate) in an anaerobic sludge digester, indicating that the contributions of these bacteria to the degradation of organic compounds in the granules was relatively low.
Clones affiliated with aceticlastic Methanosaeta were frequently detected in the archaeal clone library (Fig. 2), and cells hybridized with probe MX825, specific for the genus Methanosaeta, were the dominant members of Archaea in the granule. In general, aceticlastic methanogens are more abundant than hydrogenotrophic ones in methanogenic consortia (14, 35). Although the hydrogenotrophic methanogens affiliated with Methanomicrobiales were identified by phylogenetic analysis, the number of hydrogenotrophic methanogens hybridized with probes MG1200 and MB1174 was under the detection limit in this study. The probe MG1200 is specific for Methanomicrobiales, whereas the probe MB1174 is specific for Methanobacteriales. Moreover, the 16S rRNA gene sequence of the clones belonging to OTU2 in the Archaea clone library, which were more frequently detected than those belonging to OTU3, had two mismatches with the sequence of probe MG1200. Therefore, further studies are urgently needed to identify the hydrogenotrophic methanogens with additional oligonucleotide probes specific for them and to evaluate their significance in methane production.
We determined the Deff for H2 in the anaerobic granules with H2 microsensors. In this study, the Deff for H2 in the granules at 35°C was 2.66 × 10−5 ± 0.13 × 10−5 cm2 s−1, which was 57% ± 3% of the Dw for H2 in water (Dw 35; 4.65 × 10−5 cm2 s−1). Similar ratios were determined for glucose by applying microsensors to anaerobic granules (21, 34). Based on these results, the Deff values for CH4 and H2 were assumed to be 57% of their D in water, and these values were used to calculate the levels of microbial activities (R) in the granules (Fig. 5). The calculated CH4 production rates in the granule (11.5 μmol cm−3 h−1) were comparable to those in the methanogenic aggregates (ca. 15 μmol cm−3 h−1) (34) and were two orders of magnitude higher than those in a sewage biofilm (ca. 0.18 μmol cm−3 h−1) (10). The high level of CH4 production activity of the anaerobic granules might be attributed to the higher abundance of methanogens.
The maximum H2 concentration (26 μM) in the granule (Fig. 4B) was one order of magnitude higher than that in anaerobic aggregates (less than 5 μM) (34). By contrast, these values were lower than those for the hindgut of a wood-feeding lower termite (ca. 50 μM) (15). In methanogenic consortia, H2 produced by syntrophic bacteria should be efficiently consumed, because the oxidation of intermediary metabolites (e.g., fatty acids and alcohols) into H2 is thermodynamically feasible only at very low H2 partial pressures (e.g., between 10−4 and 10−6 atm for anaerobic propionate degradation) (26). To investigate whether H2 concentrations in the granules analyzed in this study were low enough to carry out the syntrophic H2-producing reaction (e.g., propionate oxidation), the relationship between the H2 concentration and the Gibbs free energy change (ΔG) for propionate oxidation was calculated according to the equation ΔG = ΔG0′ + RT ln {([H2]3 [Ace])/[Pro])}, where ΔG is the actual free energy, ΔG0′ is the standard free energy (76.5 kJ mol−1), R is the ideal gas constant, T is the temperature, and [H2], [Ace], and [Pro] are the molar H2, acetate, and propionate concentrations, respectively (38). The average concentrations of propionate (77 μM) and acetate (165 μM) and the temperature (35°C) in the lab-scale UASB reactor from which the granule samples were taken were used for the calculation. The H2 concentration for ΔG = 0 was calculated to be 36.8 μM, indicating that the measured H2 concentration in the granule (<26 μM) was low enough for the syntrophic propionate oxidation to proceed.
There was a difficulty with the CH4 microsensor measurements. It is curious that low-level but significant CH4-producing activities were detected in the centers of granules in which the density of Archaea was low. A possible explanation could be compression of the granules by insertion of the CH4 microsensor, because the diameter of its tip was relatively large (50 to 100 μm) and conical compared to those of other microsensors. Therefore, CH4 concentration profiles were monitored by advancing the microsensor in steps of 300 μm (Fig. 4B).
In conclusion, combining 16S rRNA gene-based molecular techniques with microsensors provided direct information about the phylogenetic diversities, spatial distributions, and activities of Bacteria and Archaea in anaerobic granules. The spatial distributions of the microorganisms and their in situ activities in the granules were characterized by a distinct layered structure. Acid and H2 production occurred in the upper part of the granule, below which H2 consumption and CH4 production were found. Chloroflexi might contribute to the hydrolysis of complex organic compounds in the outer shell of the granule, H2 was produced mainly by members of Firmicutes in the middle layer, and Archaea affiliated with Methanosaeta were produced CH4 in the inner layer. Further studies with the microsensor for volatile fatty acids and additional oligonucleotide probes for syntrophic bacteria and hydrogenotrophic methanogens are needed for better understanding of the overall degradation mechanism of complex organic matter in anaerobic granules.
Supplementary Material
Acknowledgments
This study was carried out as a part of the Project for Development of Technologies for Analyzing and Controlling the Mechanism of Biodegrading and Processing, a project of the New Energy and Industrial Technology Development Organization (NEDO), Japan.
This work was partially supported by a Grant-in-Aid for Developmental Scientific Research (grant number 18710013) from the Ministry of Education, Science and Culture of Japan and by funding from the Maeda Engineering Foundation.
We thank Tsukasa Ito and Tomonori Kindaichi for valuable assistance and discussions during preparation of the manuscript.
Footnotes
Published ahead of print on 28 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkerman, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariesyady, H. D., T. Ito, and S. Okabe. 2007. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 41:1554-1568. [DOI] [PubMed] [Google Scholar]
- 5.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC.
- 6.Cronenberg, C., and J. C. van den Heuvel. 1991. Determination of glucose diffusion coefficients in biofilms with microsensors. Biosens. Bioelectronics 6:255-262. [DOI] [PubMed] [Google Scholar]
- 7.Cussler, E. L. 1997. Diffusion mass transfer in fluid systems, 2nd ed. Cambridge University Press, Cambridge, England.
- 8.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 9.Damgaard, L. R., L. H. Larsen, and N. P. Revsbech. 1995. Microscale biosensors for environmental monitoring. Trends Anal. Chem. 14:300-303. [Google Scholar]
- 10.Damgaard, L. R., L. P. Nielsen, and N. P. Revsbech. 2001. Methane microprofiles in a sewage biofilm determined with a microscale biosensor. Water Res. 35:1379-1386. [DOI] [PubMed] [Google Scholar]
- 11.Damgaard, L. R., and N. P. Revsbech. 1997. A microscale biosensor for methane containing methanotrophic bacteria and an internal oxygen reservoir. Anal. Chem. 69:2262-2267. [DOI] [PubMed] [Google Scholar]
- 12.Damgaard, L. R., N. P. Revsbech, and W. Reichardt. 1998. Use of an oxygen-insensitive microscale biosensor for methane to measure methane concentration profiles in a rice paddy. Appl. Environ. Microbiol. 64:864-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Beer, D., J. W. Huisman, J. C. Van den Heuvel, and S. P. P. Ottengraf. 1992. The effect of pH profiles in methanogenic aggregates on the kinetics of acetate conversion. Water Res. 26:1329-1336. [Google Scholar]
- 14.Díaz, E. E., A. J. M. Stams, R. Amils, and J. L. Sanz. 2006. Phenotypic properties and microbial diversity of methanogenic granules from a full-scale upflow anaerobic sludge bed reactor treating brewery wastewater. Appl. Environ. Microbiol. 72:4942-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebert, A., and A. Brune. 1997. Hydrogen concentration profiles at the oxic-anoxic interface: a microsensor study of the hindgut of the wood-feeding lower termite Reticulitermes flavipes (Kollar). Appl. Environ. Microbiol. 63:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 17.Girbal, L., G. von Abendroth, M. Winkler, P. M. Benton, I. Meynial-Salles, C. Croux, J. W. Peters, T. Happe, and P. Soucaille. 2005. Homologous and heterologous overexpression in Clostridium acetobutylicum and characterization of purified clostridial and algal Fe-only hydrogenases with high specific activities. Appl. Environ. Microbiol. 71:2777-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang, A., J.-H. Lee, P. R. Bhadri, S. A. Kumar, W. Timmons, F. R. Beyette, Jr., I. Papautsky, and P. L. Bishop. 2005. Miniaturized redox potential probe for in situ environmental monitoring. Environ. Sci. Technol. 39:6191-6197. [DOI] [PubMed] [Google Scholar]
- 19.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanthier, M., B. Tartakovsky, R. Villemur, G. DeLuca, and S. R. Guiot. 2002. Microstructure of anaerobic granules bioaugmented with Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 68:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lens, P. N., D. de Beer, C. C. Cronenberg, F. P. Houwen, S. P. Ottengraf, and W. H. Verstraete. 1993. Heterogeneous distribution of microbial activity in methanogenic aggregates: pH and glucose microprofiles. Appl. Environ. Microbiol. 59:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, W. T., O. C. Chan, and H. H. P. Fang. 2002. Characterization of microbial community in granular sludge treating brewery wastewater. Water Res. 36:1767-1775. [DOI] [PubMed] [Google Scholar]
- 23.Lorenzen, J., L. H. Larsen, T. Kjar, and N. P. Revsbech. 1998. Biosensor detection of the microscale distribution of nitrate, nitrate assimilation, nitrification, and denitrification in a diatom-inhabited freshwater sediment. Appl. Environ. Microbiol. 64:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maidak, B. L., G. L. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 26.McCarty, P. L., and D. P. Smith. 1986. Anaerobic wastewater treatment. Environ. Sci. Technol. 20:1200-1206. [Google Scholar]
- 27.Meier, H., R. Amann, W. Ludwig, and K.-H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 28.Neef, A. 1997. Anwendung der in situ-Einzelzell-Identifizierung von bakterin zur populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 29.Okabe, S., H. Satoh, and Y. Watanabe. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 34.Santegoeds, C. M., L. R. Damgaard, G. Hesselink, J. Zopfi, P. Lens, G. Muyzer, and D. de Beer. 1999. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl. Environ. Microbiol. 65:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 1999. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekiguchi, Y., Y. Kamagata, K. Syutubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voolapalli, R. K., and D. C. Stuckey. 1999. Relative importance of trophic group concentrations during anaerobic degradation of volatile fatty acids. Appl. Environ. Microbiol. 65:5009-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitehead, T. R., and M. A. Cotta. 1999. Phylogenetic diversity of methanogenic Archaea in swine waste storage pits. FEMS Microbiol. Lett. 179:223-226. [DOI] [PubMed] [Google Scholar]
- 41.Yamada, T., Y. Sekiguchi, H. Imachi, Y. Kamagata, A. Ohashi, and H. Harada. 2005. Diversity, localization, and physiological properties of filamentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl. Environ. Microbiol. 71:7493-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi, T., S. Yamazaki, S. Uemura, I. C. Tseng, A. Ohashi, and H. Harada. 2001. Microbial-ecological significance of sulfide precipitation within anaerobic granular sludge revealed by microsensors study. Water Res. 35:3411-3417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.