Abstract
Pneumocystis jirovecii is a fungus which causes severe opportunistic infections in immunocompromised humans. The brl1 gene of P. carinii infecting rats was identified and characterized by using bioinformatics in conjunction with functional complementation in Saccharomyces cerevisiae and Schizosaccharomyces pombe. The ectopic expression of this gene rescues null alleles of essential nuclear membrane proteins of the Brr6/Brl1 family in both yeasts.
Pneumocystis spp. are extracellular opportunistic fungi that have been detected in the lungs of almost every mammalian species tested. Pneumocystis jirovecii, the species infecting human beings, causes severe, often lethal, pneumonia in immunocompromised individuals (for reviews, see references 7, 18, and 19). Because of the emergence of drug resistance in P. jirovecii, the development of new drugs is important. However, the absence of a long-term in vitro culture system for Pneumocystis organisms has impeded progress in drug design. The Pneumocystis Genome Project (http://pgp.cchmc.org) (12, 15) has completed the sequencing of a nonredundant set of 1,042 expressed sequence tags (ESTs) from RNA isolated from a single rat infected with Pneumocystis carinii (3). About 34% of the genes with the highest homology to P. carinii ESTs are found in Schizosaccharomyces pombe, the closest relative to P. carinii among fungi with sequenced genomes (6), while approximately 15% are found in the more distantly related Saccharomyces cerevisiae (3). Our strategy has been to use bioinformatics in conjunction with functional complementation in S. cerevisiae or S. pombe to assess the function of identified P. carinii genes (5). Two criteria were used to select expressed genes for analysis: (i) an essential requirement for the yeast homolog and (ii) the absence of significant homology to vertebrate genes. In this paper, we report the cloning and characterization of the P. carinii ortholog of the S. cerevisiae BRL1 and BRR6 genes and of S. pombe brl1. These genes encode integral nuclear envelope proteins that are essential and implicated in RNA export from the nucleus.
The P. carinii brl1 cDNA was isolated from the cDNA library in a Stratagene Uni-ZAP XR vector constructed by G. Smulian (University of Cincinnati). PCRs were performed using high-fidelity expand polymerase according to the manufacturer's instructions (Roche Diagnostics). For the primers used, see the supplemental material. The genomic copy of P. carinii brl1 was amplified from DNA extracted from the lungs of an infected rat (provided by A. E. Wakefield, University of Oxford).
For complementation assays in S. cerevisiae, genes were cloned into the centromeric expression vectors p416GPD (glyceraldehyde-3-phosphate dehydrogenase gene promoter) and p416TEF (translation elongation factor 1α) (9). The recombinant plasmids were introduced, using the polyethylene glycol 4000/lithium acetate technique (2), into the diploid strains Y20999, heterozygous for the brl1 null allele (Mata/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0 YHR036wΔ::kanMX4/YHR036w), and Y24614, heterozygous for the brr6 null allele (Mata/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0 YGL247wΔ::kanMX4/YGL247w) (Euroscarf, Germany). Diploid transformants were sporulated by incubation at 30°C on potassium acetate medium (2% [wt/vol] potassium acetate, 0.077% Q-Biogene dropout premix, 0.22% Difco yeast extract, 0.05% glucose, 2% Gibco agar). Tetrads were dissected on YEPD complete medium (1% [wt/vol] Difco yeast extract, 2% Difco peptone, 2% glucose) using a Zeiss Axioskop 40 microscope. Replica plating to YEPD containing 200 μg/ml Geneticin 418 (Brunschwig, Basel, Switzerland) was performed to assess the inheritance of the null allele.
The brr6 brl1 double mutant was constructed by integrative disruption (13) of BRL1 in the haploid brr6 mutant complemented with P. carinii brl1. A 616-bp internal fragment of S. cerevisiae BRL1 comprising nucleotides 542 to 1,158 of the BRL1 open reading frame (ORF) was cloned into the integrative plasmid pRS405 (14), and the recombinant plasmid was linearized by using NdeI and integrated at position 872 of BRL1.
For complementation assays in S. pombe, genes were cloned into the expression vector pREP41 (regulatable nmt1 promoter) (1). The diploid strain heterozygous for the null allele of brl1/SPAC8F11.06 (h+/h+ ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 SPAC8F11.06/SPAC8F11.06Δ::kanMX4) (Bioneer Corporation, Korea) was cotransformed with the recombinant plasmid and a sporulation-inducing plasmid, pON177 (16), using the lithium acetate method (8), and plated on EMM2 (8) containing appropriate supplements. Spores obtained at 29°C on EMM2 were purified (8) and plated on EMM2 medium with and without thiamine (2 μg/ml) to regulate expression from the nmt1 promoter. Replica plating to complete YE medium (8) containing 100 μg/ml geneticin 418 and 10 μg/ml phloxin B assessed whether survivors also inherited the null allele. To assess whether S. pombe brl1 is essential, tetrads from the diploid heterozygous for the br1l null allele were dissected using a Singer MSM micromanipulator, and spores were germinated at 25°C on YE.
For staining, S. pombe cells were harvested by centrifugation, fixed with 70% (vol/vol) ethanol, resuspended in phosphate-buffered saline, and stained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml) and calcofluor (10 μg/ml). Images of the S. pombe colonies on the plates were taken using a Nikon Coolpix 990 camera attached to the eyepiece of a Leitz Laborlux2 microscope equipped with a 25× Neofluor lens and 10× eyepieces. The pictures were processed with Adobe Photoshop.
Isolation of the full-length coding region of P. carinii brl1 gene.
One P. carinii EST that we identified (5) had highly significant homology with S. cerevisiae BRL1 and BRR6 and S. pombe brl1. The EST was extended by PCR from the cDNA library. The full-length P. carinii brl1 cDNA and the corresponding locus amplified from P. carinii genomic DNA were sequenced. The overall sequence identity of P. carinii Brl1p to S. cerevisiae Brl1p and Brr6p and to the S. pombe Brl1p is 23, 19, and 28%, respectively (Fig. 1).
FIG. 1.
P. carinii Brl1p (PcBrl1p) shares homology with S. cerevisiae Brl1p (ScBrl1p) and Brr6p (ScBrr6p) and with S. pombe Brl1p (SpBrl1p) (respective National Center for Research Resources Yeast Resource Center accession numbers: NP_011901, NP_011267, and SPAC8F11.06). The multiple alignment was done by using T-Coffee software and represented by using Boxshade software. Identical residues are indicated by dark areas and asterisks at the bottom of the sequence, conserved residues by gray areas and single points. The dashes within contiguous sequences are introduced by the alignment software. The black bars indicate the two putative transmembrane domains comprised within the Brl1/Brr6 homology domain (gray bar).
P. carinii brl1 functionally complements the S. cerevisiae brl1, brr6, and brr6 brl1 null mutants.
The P. carinii brl1 cDNA was cloned into the centromeric expression vector p416 under the control of the strong GPD or weaker TEF promoter and introduced into a diploid S. cerevisiae heterozygous for the brl1 or brr6 null allele. Sporulation of diploids expressing either P. carinii brl1 or S. cerevisiae BRL1 as a control gave rise to four viable haploid colonies from each tetrad dissected (n = 16 for each promoter), while only two viable kanamycin-sensitive colonies resulted from the sporulation of tetrads transformed with the empty p416GPD or p416TEF vector (Fig. 2A). The presence of the kanamycin resistance cassette and the absence of the S. cerevisiae wild-type BRL1 or BRR6 allele were confirmed by PCR analysis (Fig. 2B). The P. carinii gene was also able to complement a double brl1 brr6 mutant, indicating that it can provide the function of both genes. Green fluorescent protein tagging of P. carinii brl1 confirmed its localization in the nuclear membrane in S. cerevisiae (not shown).
FIG. 2.
P. carinii Brl1p rescues inviability of the S. cerevisiae brl1 null mutant. (A) The diploid S. cerevisiae strain heterozygous for the brl1 null allele was transformed with the indicated plasmids and sporulated, and the four spores of each tetrad were separated on rich medium (shown vertically in the picture). Two dissected tetrads for each plasmid are shown. (B) The presence of the brl1 null allele (brl1Δ) and the absence of the wild-type BRL1 gene in the haploid strain complemented with P. carinii brl1 were confirmed by PCR using locus-specific primers that are able to amplify both wild-type and brl1Δ alleles and ORF-specific primers amplifying only the wild-type allele. Lane 1, heterozygous diploid; lane 2, wild-type strain; lane 3, haploid mutant complemented by P. carinii brl1. The unrelated S. cerevisiae TAF4 genomic locus was amplified as the positive DNA control. The sizes of the PCR products of the BRL1 locus, brl1Δ, BRL1 ORF, and TAF4 were, respectively, 2,008 bp, 2,092 bp, 1,416 bp, and 2,067 bp.
P. carinii brl1 functionally complements the S. pombe brl1 null mutant.
Blast searches in the S. pombe genome revealed only a single ORF (SPAC8F11.06) with significant homology to the Brr6/Brl1 gene family (Fig. 1). The diploid heterozygous for the null allele of this ORF was induced to sporulate, and the tetrads were dissected. Only two of four spores formed colonies in each of 17 tetrads. Examination of the plates revealed that the two other spores from each tetrad germinated and arrested as single, elongated cells (Fig. 3A). This phenotype is characteristic of a cell cycle mutant (10).
FIG. 3.
P. carinii Brl1p rescues the cell cycle arrest phenotype of the S. pombe brl1 null mutant. (A) The S. pombe diploid brl1 null mutant was sporulated, the four spores of each tetrad were separated on rich medium, and the phenotypes of the haploid cells arising from the dissected tetrads were examined. Two spores from each tetrad gave rise to wild-type, kanamycin-sensitive colonies, and two spores from each tetrad gave rise to a single elongated cell. (B) The presence of the brl1 null allele (brl1Δ) in the haploid strain complemented with P. carinii brl1 was confirmed by PCR using locus-specific primers that are able to amplify both wild-type and brl1Δ alleles. Lane 1, wild-type strain; lane 2, heterozygous diploid; lane 3, null mutant complemented by S. pombe brl1; and lane 4, null mutant complemented by P. carinii brl1. As predicted, the observed sizes of the amplified fragments from the S. pombe brl1 and brl1Δ alleles were 1,220 bp and 1,731 bp, respectively. (C) The diploid S. pombe brl1 null mutant was transformed with the indicated plasmids and sporulated, and the phenotypes of the haploid cells arising from the dissected tetrads were examined. The arrows indicate cells in which the mitotically unstable plasmid has probably been lost. ON and OFF indicate medium without and with thiamine, respectively, which shuts off the nmt1 promoter and, therefore, P. carinii or S. pombe brl1 expression. Scale bars, 10 μm.
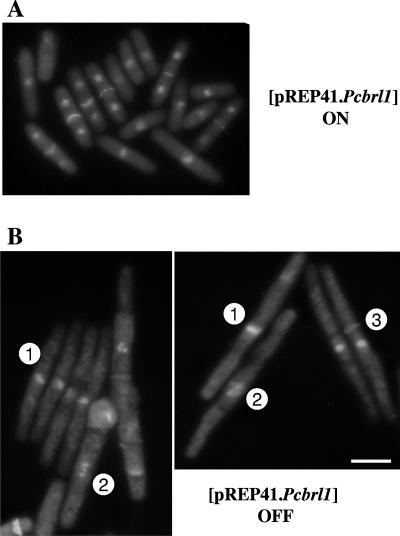
P. carinii brl1 was cloned into the expression vector pREP41 under the control of the regulatable nmt1 promoter and introduced into a diploid S. pombe heterozygous for the brl1 null allele. Spores were prepared, and replica plating on relevant media established that viable haploid colonies carrying the brl1 null allele were recovered from diploids expressing either P. carinii brl1 or S. pombe brl1 as a control, but not with the empty pREP41 vector. PCR analysis confirmed the absence of the S. pombe brl1 wild-type allele in these colonies (Fig. 3B). Examination of the edges of rescued colonies revealed elongated cells (Fig. 3C, arrows), likely due to loss of the plasmid. To confirm that inactivation of Brl1p in exponentially growing cells gives rise to the same phenotype, we repressed the expression of the P. carinii brl1 gene in the haploid brl1 null mutant by replica plating to medium containing thiamine to inactivate the nmt1 promoter. The cells stopped dividing and became highly elongated (Fig. 3C). DAPI staining of fixed cells revealed that the elongated cells had a single nucleus, which is consistent with a cell cycle arrest (Fig. 4). Some cells also showed condensed chromosomes and septa. These latter cells may result from entry into mitosis with insufficient Brl1p, which may indicate a role for Brl1p during mitosis.
FIG. 4.
Inactivation of S. pombe brl1 causes a delay in mitotic entry and progression. Haploid S. pombe brl1Δ cells covered by pREP41.Pcbrl1 cells were grown to early exponential phase in supplemented minimal medium, thiamine was added to half the culture, and the cells were grown at 25°C for 16 h. Cells were harvested by centrifugation, fixed with ethanol, and stained with DAPI and Calcofluor. (A) Induced control. (B) Cells from cultures to which thiamine had been added. Note that these latter cells are elongated compared to the induced control, which resembles the wild type. 1, cell with a single nucleus arrested in interphase; 2, cell with condensed chromosomes; 3, cell with a septum. Scale bar, 10 μm.
We have demonstrated that the sole identifiable S. pombe member of the Brr6/Brl1 gene family is essential. The null mutant dies in the first cell cycle after germination as a single, elongated, mononucleate cell. This does not merely reflect a requirement for brl1 during spore germination, as inactivation of the protein during exponential growth gives rise to the same phenotype. The finding is somewhat unexpected, given the apparent involvement of S. cerevisiae Brl1p and Brr6p in mRNA transport (4, 11). However, it is consistent with the suggestion of a role for these proteins in nuclear membrane formation (11). Since the S. pombe gene complements both the S. cerevisiae brl1 and brr6 mutants (11), it is likely that they perform similar functions in the cell. It is noteworthy that the published thermosensitive brl1 mutants of S. cerevisiae (11) arrest with a large-budded phenotype, characteristic of cells arrested before nuclear division. This may indicate a conserved function for Brl1p in regulating entry into mitosis. It is possible that a subset of mRNAs affected by BRL1 may be important for cell cycle progression in S. pombe. Alternatively, the S. pombe brl1 protein may have additional functions unrelated to mRNA transport that are also important for cell cycle progression and which have not been observed in studies of the S. cerevisiae mutants. In this context, it is relevant that a recent screen for extragenic suppressors of mutants in the S. pombe cut12 protein, which is a spindle-pole-body protein that participates in determining the timing of mitotic entry, identified components of the translation and transcription machinery (17).
Inspection of the phylogenetic relationships within the fungal and protozoan kingdoms (6) suggests that BRL1 and BRR6 are paralogs that arose from a genome duplication event specific to the Saccharomycotina subphylum of the Ascomycota. Two Brl1/Brr6 family members have been identified within the genomes of S. cerevisiae, Candida glabrata, Ashbya gossypii, Kluyveromyces lactis, Debaryomyces hansenii and Yarrowia lipolitica, while among the Brl1/Brr6 family members so far identified in diverse fungal and protozoan species lying outside of the Saccharomycotina, no more than a single ortholog has been detected in any one species. Thus, the conserved function of the Brr6/Brl1 family is likely to have been originally conveyed by a single-copy gene. Both the S. pombe and S. cerevisiae Brr6/Brl1 family null mutants are rescued by the sole identifiable P. carinii Brr6/Brl1 family gene, indicating that P. carinii Brl1p fulfils the essential roles of these proteins in both S. pombe and S. cerevisiae. However, the roles of S. cerevisiae BRR6 and BRL1 may have diverged (4, 11), and dissection of the family's conserved function may be easier to study in a species that harbors only a single ortholog. Since the conserved Brr6/Brl1 family genes are essential in both species where their function has been examined, it is tempting to speculate that the ortholog will also be essential in P. carinii. Unfortunately, the inability to genetically manipulate P. carinii in vitro precludes a direct test of this question at present.
The P. carinii brl1 gene is present in the P. carinii EST library, which indicates that it is expressed during infection of rat lung tissue. It appears to be conserved in all organisms presumed to have a closed mitosis (in which the nuclear membrane does not break down), and lacks any strong homology to any vertebrate proteins. Finally, searches in the databases identify clear orthologs in other fungal and protozoan pathogens (not shown). These features suggest that the Brl1/Brr6 protein family may provide a useful target for the development of new drugs to combat Pneumocystis infections, as well as a wide range of other organisms. The inhibition of the ability of heterologous brl1 to rescue the distinctive phenotype of the S. pombe mutant could be used to screen libraries of potential inhibitors.
Nucleotide sequence accession numbers.
The GenBank accession numbers of P. carinii brl1 cDNA and genomic DNA sequences are EF547361 and EF547362, respectively.
Supplementary Material
Acknowledgments
We are indebted to M. T. Cushion for providing helpful information on the Pneumocystis genomic databases, to G. Smulian for the gift of the P. carinii cDNA library, and to C. Sibley and I. Macreadie for providing plasmids.
This study was supported by Swiss National Science Foundation grant 310000-112360.
Footnotes
Published ahead of print on 9 November 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 2.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 3.Cushion, M. T., A. G. Smulian, B. E. Slaven, T. Sesterhenn, J. Arnold, C. Staben, A. Porollo, R. Adamczak, and J. Meller. 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS ONE 2:e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bruyn Kops, A., and C. Guthrie. 2001. An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J. 20:4183-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser, P. M., L. Lo Presti, M. Cockell, L. Cerutti, and V. Simanis. 2006. Analysis of Pneumocystis carinii gene function by complementation in yeast mutants. J. Eukaryot. Microbiol. 53:S149-S150. [DOI] [PubMed] [Google Scholar]
- 6.James, T. Y., F. Kauff, C. L. Schoch, P. B. Matheny, V. Hofstetter, C. J. Cox, G. Celio, C. Gueidan, E. Fraker, J. Miadlikowska, H. T. Lumbsch, A. Rauhut, V. Reeb, A. E. Arnold, A. Amtoft, J. E. Stajich, K. Hosaka, G. H. Sung, D. Johnson, B. O'Rourke, M. Crockett, M. Binder, J. M. Curtis, J. C. Slot, Z. Wang, A. W. Wilson, A. Schussler, J. E. Longcore, K. O'Donnell, M. S. Ozley-Standridge, D. Porter, P. M. Letcher, M. J. Powell, J. W. Taylor, M. M. White, G. W. Griffith, D. R. Davies, R. A. Humber, J. B. Morton, S. J. Ugiyama, A. Y. Rossman, J. D. Rogers, D. H. Pfister, D. Hewitt, K. Hansen, S. Hambleton, R. A. Shoemaker, J. Kohlmeyer, B. Volkmann-Kohlmeyer, R. A. Spotts, M. Serdani, P. W. Crous, K. W. Hughes, K. Matsuura, E. Langer, G. Langer, W. A. Untereiner, R. Lucking, B. Budel, D. M. Geiser, A. Aptroot, P. Diederich, I. Schmitt, M. Schultz, R. Yahr, D. S. Hibbett, F. Lutzoni, D. J. McLaughlin, J. W. Spatafora, and R. Vilgalys. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818-822. [DOI] [PubMed] [Google Scholar]
- 7.Kovacs, J. A., V. J. Gill, S. Meshnick, and H. Masur. 2001. New insights into transmission, diagnosis, and drug treatment of Pneumocystis carinii pneumonia. JAMA 286:2450-2460. [DOI] [PubMed] [Google Scholar]
- 8.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 9.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 10.Nurse, P., P. Thuriaux, and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167-178. [DOI] [PubMed] [Google Scholar]
- 11.Saitoh, Y. H., K. Ogawa, and T. Nishimoto. 2005. Brl1p: a novel nuclear envelope protein required for nuclear transport. Traffic 6:502-517. [DOI] [PubMed] [Google Scholar]
- 12.Sesterhenn, M. T., B. E. Slaven, A. G. Smulian, and M. T. Cushion. 2003. Generation of sequencing libraries for the Pneumocystis Genome Project. J. Eukaryot. Microbiol. 50:S663-S665. [DOI] [PubMed] [Google Scholar]
- 13.Shortle, D., J. E. Haber, and D. Botstein. 1982. Lethal disruption of the yeast actin gene by integrative DNA transformation. Science 217:371-373. [DOI] [PubMed] [Google Scholar]
- 14.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slaven, B. E., J. Meller, A. Porollo, T. Sesterhenn, A. G. Smulian, and M. T. Cushion. 2006. Draft assembly and annotation of the Pneumocystis carinii genome. J. Eukaryot. Microbiol. 53:S89-S91. [DOI] [PubMed] [Google Scholar]
- 16.Styrkarsdottir, U., R. Egel, and O. Nielsen. 1993. The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr. Genet. 23:184-186. [DOI] [PubMed] [Google Scholar]
- 17.Tallada, V. A., A. J. Bridge, P. A Emery, and I. M. Hagan. 2007. Suppression of the Schizosaccharomyces pombe cut12.1 cell-cycle defect by mutations in cdc25 and genes involved in transcriptional and translational control. Genetics 176:73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas, C. F., and A. H. Limper. 2004. Pneumocystis pneumonia. N. Engl. J. Med. 350:2487-2498. [DOI] [PubMed] [Google Scholar]
- 19.Thomas, C. F., and A. H. Limper. 2007. Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat. Rev. Microbiol. 5:298-308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.