Abstract
Macrophages and neutrophils kill the airborne fungal pathogen Aspergillus fumigatus. The dependency of this killing process on reactive oxygen intermediates (ROI) has been strongly suggested. Therefore, we investigated the enzymatic ROI detoxifying system by proteome analysis of A. fumigatus challenged by H2O2. Since many of the identified proteins and genes are apparently regulated by a putative Saccharomyces cerevisiae Yap1 homolog, the corresponding gene of A. fumigatus was identified and designated Afyap1. Nuclear localization of a functional AfYap1-eGFP fusion was stress dependent. Deletion of the Afyap1 gene led to drastically increased sensitivity of the deletion mutant against H2O2 and menadione, but not against diamide and NO radicals. Proteome analysis of the ΔAfyap1 mutant strain challenged with 2 mM H2O2 indicated that 29 proteins are controlled directly or indirectly by AfYap1, including catalase 2. Despite its importance for defense against reactive agents, the Afyap1 deletion mutant did not show attenuated virulence in a murine model of Aspergillus infection. These data challenge the hypothesis that ROI such as superoxide anions and peroxides play a direct role in killing of A. fumigatus in an immunocompromised host. This conclusion was further supported by the finding that killing of A. fumigatus wild-type and ΔAfyap1 mutant germlings by human neutrophilic granulocytes worked equally well irrespective of whether the ROI scavenger glutathione or an NADPH-oxidase inhibitor was added to the cells.
In the last few decades Aspergillus fumigatus has become the most important airborne fungal pathogen of humans. Diseases caused by A. fumigatus can be divided into three categories: allergic reactions and colonization with restricted invasiveness are observed in immunocompetent individuals, while systemic infections with high mortality rates occur in immunocompromised patients. Due to the improvement in transplant medicine and the therapy of hematological malignancies, the number of cases of invasive aspergillosis has increased. Specific diagnostics are still limited, as are the possibilities of therapeutic intervention, leading to a high mortality rate of 30 to 98% for invasive aspergillosis (8). The genome of the A. fumigatus isolate Af293 was fully sequenced. It consists of a haploid set of eight chromosomes with a total size of 29.4 Mb, of which 9,926 protein-encoding sequences were identified (44). With the genome data available the regulation of genes and the expression profile of proteins of A. fumigatus can be analyzed on a global scale, including the conditions that are related to infection.
The infectious agent of A. fumigatus are conidia, which are inhaled during routine daily activities (8). Therefore, in immunocompromised patients, the lung is the site of infection of A. fumigatus. In immunocompetent individuals, mucociliary clearance and phagocytic defense normally prevent the disease. Alveolar macrophages are the major resident cells of the lung alveoli; they, along with neutrophils (which are actively recruited during inflammation), are the major cells in the phagocytosis of A. fumigatus. Macrophages were shown to kill conidia by producing reactive oxygen intermediates (ROI) (46). Conidia escaping alveolar macrophages grow out and are attacked by neutrophils. Neutrophils adhere to the surface of the hyphae, since hyphae are too large to be engulfed. However, polymorphonuclear leukocytes (PMN) are also able to kill resting or swollen conidia (reviewed in reference 8). Contact between neutrophils and hyphae triggers a respiratory burst, secretion of ROI and hypochlorous acid, and degranulation (16, 36, 40, 52).
Several genes involved in the defense against ROI of A. fumigatus have been characterized. Although deletion of these genes increased the sensitivity to ROI of the respective mutant strains, none of them showed reduced virulence in a mouse infection model, e.g., deletion of catA (conidial catalase) resulted in increased sensitivity against H2O2 but did not affect pathogenicity (45). Mn-superoxide dismutase (SOD) and Cu/Zn-SOD were characterized but, thus far, the structural genes have not been deleted (25). Therefore, we analyzed here the proteomes of A. fumigatus wild-type strain grown without oxidative stress and in the presence of H2O2. The results obtained indicated an involvement of the key transcriptional regulator AfYap1, which was identified here, in the regulation of several defense genes against ROI. The molecular analysis of AfYap1 allowed us to study on a global scale the impact of enzymatic defense systems against ROI for virulence. Our results indicate that the AfYap1-regulated antioxidant proteins do not play a major role in the pathogenicity of A. fumigatus. Therefore, killing mechanisms other than the production of ROI by the host are presumably more important than previously thought.
MATERIALS AND METHODS
Fungal and bacterial strains, media, and growth conditions.
The A. fumigatus wild-type strain ATCC 46645 was used to generate the Afyap1 knockout (ΔAfyap1) strain (Afyap1::hph; Hygr) by using the hygromycin resistance (Hygr) gene as the selectable marker gene. The ΔAfyap1 strain was complemented with the Afyap1 gene using the phleomycin resistance gene ble as the selectable marker gene to give strain ΔAfyap1::Afyap1. Strain Afyap1-eGFP (otefp-Afyap1-eGFP; Hygr) was obtained by transformation of ATCC 46645 with an Afyap1-eGFP gene fusion controlled by the otef promoter as described below and in reference 54. As the selectable marker, the Hygr gene was used. A. fumigatus strains were cultivated at 37°C in Aspergillus minimal medium (AMM) as previously described (37). As solid medium, malt extract (2% [wt/vol] malt extract, 0.2% [wt/vol] yeast extract, 1% [wt/vol] glucose, 5 mM ammonium chloride, 1 mM dipotassium hydrogen phosphate) or AMM containing 3% (wt/vol) agar was used. Hygromycin (200 μg/ml) or phleomycin (100 μg/ml) was added to the media when required. For transformation of Escherichia coli, the strains DH5α (Bethesda Research Laboratories), XL1-Blue (Stratagene), or INVαF′ or TOP10F′ (Invitrogen, The Netherlands) were used. E. coli strains were grown at 37°C in LB medium supplemented, when required, with 50 μg of ampicillin or kanamycin per ml.
Measurement of sensitivity against reactive agents.
A total of 107 conidia of the strains tested were mixed with 2% (wt/vol) AMM agar and poured in a petri dish. A hole 1 cm in diameter was punched in the middle of the agar plate. The well was filled with 150 μl of 3% (vol/vol) H2O2, 100 μl of 0.1 M diamide solution, 100 μl of 1 mM menadione, or 100 μl of 90 mM NO donor 2,2′-(hydroxynitrosohydrazono)bis-ethanimine (DETA NO) or the corresponding DEA base. Agar plates were incubated for 16 h at 37°C. The inhibition zone of four independently incubated agar plates was measured for each tested strain.
Standard DNA techniques.
Standard techniques in the manipulation of DNA were carried out as described by Sambrook and Russell (51). Chromosomal DNA of A. fumigatus was prepared as previously described for A. nidulans (2). For Southern blot analysis, the chromosomal DNA of A. fumigatus was cut by different restriction enzymes. DNA fragments were separated on an agarose gel and blotted onto Hybond N+ nylon membranes (GE Healthcare Bio-Sciences, Germany). Labeling of the DNA probe, hybridization, and detection of DNA-DNA hybrids were performed by using the DIG HighPrime labeling and detection system (GE Healthcare Bio-Sciences) according to the manufacturer's recommendations.
Sequence analysis.
DNA was sequenced on both strands by primer walking using the BigDye terminator cycle sequencing kit (Applied Biosystems, United Kingdom). Sequencing reactions were separated on an Applied Biosystems ABI 310 sequencer. DNA sequence data were edited by using the programs Sequence Navigator and Auto Assembler (Applied Biosystems). The analysis of sequences was carried out by using GeneWorks 2.2 (IntelliGenetics, Inc.). Amino acid sequence comparisons were performed by using the BLAST 2.0 server (www.ncbi.nlm.nih.gov/BLAST). The programs PepStat and CLUSTAL W (Husar 4.0; DKFZ, Heidelberg, Germany) were used for the prediction of protein molecular masses and amino acid sequence alignments, respectively. Shading of aligned amino acid sequences was performed with MacBoxShade 2.15 (www.ch.embnet.org/software/BOX_form.html). Features and amino acid motifs of proteins were predicted by using the ISREC server (http://hits.isb-sib.ch/cgi-bin/PFSCAN).
Cloning of the A. fumigatus Afyap1 gene.
To generate an eGFP fusion of AfYap1 under the control of the otef promoter (54), cDNA was used as the template for amplification of the Afyap1 gene with the primers Yap10 (5′-ATTGGATCCGACGCGACCCATGATGTCCTC) and Yap16 (5′-ATTGGATCCCTTCGTTTTCAGTTGCCATGGCGG-3′). The PCR product generated was cloned into plasmid pCR2.1 (Invitrogen, The Netherlands). The DNA fragment encoding the Afyap1 gene was reisolated by cutting the recombinant plasmid obtained with BamHI and cloning the DNA fragment into plasmid pUCGH (34) to give plasmid pOtef-Afyap1-eGFP.
Generation of the Afyap1 knockout plasmid.
To construct the Afyap1 deletion plasmid pHyg-Δyap1, a 1.1-kbp PCR product consisting of the upstream region of the Afyap1 gene was generated by using the oligonucleotides Yap1 (5′-CACGGCCTGAGTGGCCCTAGGCCATAGGCGATTGC-3′) and Yap2 (5′-ATTGGATCCCGAAGCGGGGTATCG-3′). To amplify the 1-kbp downstream region of the Afyap1 gene, oligonucleotides Yap3 (5′-ATTAGCTTGGACATCATGGGTCGCG-3′) and Yap4 (5′-GTGGGCCATCTAGGCCCATTCGGTCCCCTTCGAC-3′) were used. Both flanking regions were cloned into plasmid pCR2.1 (Invitrogen, The Netherlands). The generated plasmid pCR2.1-right border was linearized with XhoI and XbaI. The Hygr cassette was isolated by XhoI and MluI from the plasmid pANsCos.1. The left border of the knockout construct was isolated from the plasmid pCR2.1-left border (primer Yap1 and Yap2) with MluI and XbaI. Both fragments were ligated into the plasmid pCR2.1-right border to give plasmid pHyg-Δyap1. The DNA fragment used to replace the Afyap1 gene was excised from plasmid pHyg-Δyap1 by using the restriction endonucleases SfiI and XbaI. It was used to transform the A. fumigatus wild-type strain ATCC 46645.
Complementation of the Afyap1 deletion mutant strain.
To complement the deletion of the Afyap1 mutant strain, the Afyap1 gene under the control of its own promoter was isolated from the plasmid pHyg-yap1-eGFP with PstI. The DNA fragment obtained was cloned into the plasmid pAN8.1 (accession no. Z32751), which consists of the ble gene from Streptoalloteichus hindustanus under the control of the gpdA promoter. The generated plasmid pAN8.1-yap1 was used to transform the A. fumigatus ΔAfyap1 strain. Single integration of the plasmid was checked by Southern blot analysis (data not shown).
Transformation of A. fumigatus.
Transformation of A. fumigatus was carried out using protoplasts as previously described (63).
Northern blot analysis.
RNA isolation was performed with the RNeasy plant minikit according to the manufacturer's instructions (Qiagen). 10 μg of total RNA was blotted on a Hybond N+ membrane (GE Healthcare). Hybridization and probe labeling were performed with the Dig Easy Hyb kit according to the instructions of the manufacturer (Roche Applied Sciences). The probe for cat1 (AFUA_2G00200) was amplified with the primers CAT1-F (5′-CTTCCTTGTTCCAGGGCG-3′) and CAT1-R (5′-GCGGCTGGTCATACTCC-3′); the probe for cat2 (AFUA_8G01670) was amplified with the primers CAT2-R (5′-CGATCTCCTGATCCTCACC-3′) and CAT2-R (5′-CCAGGACGATGAGATCAGC-3′).
Catalase activity assay and determination of protein concentrations.
A. fumigatus strains were grown in AMM for 24 h at 37°C. After stress induction with 2 mM H2O2 for 45 min, mycelia were harvested, frozen in liquid nitrogen, and ground with pestle and mortar. Mycelia were suspended in extraction buffer (100 mM potassium phosphate buffer [pH 6.3]) and incubated on ice for 15 min. Samples were centrifuged at 4°C with 12,000 × g for 10 min. The protein concentrations were determined according to the method of Bradford (7a). Then, 30 μg of protein extract was loaded onto an 8% polyacrylamide (wt/vol) Tris-glycine gel (Invitrogen). The catalase activity was determined directly in the gel according to the method of Goldberg and Hochman (21). The photometric determination of catalase activity was determined after addition of H2O2 by measuring the decrease in absorbance at 240 nm as described previously (7).
Isolation of PMN from peripheral blood.
PMN were isolated from the blood of healthy volunteers using polymorph-prep gradient centrifugation as described previously (49). Remaining red blood cells were lysed with ACK lysis buffer (Sigma-Aldrich, Germany). PMN purity was checked by fluorescence-activated cell sorting using anti-CD66b monoclonal antibody (BD) and was determined to be >95%. The cell number was adjusted using a hemacytometer.
Cytochrome c reduction assay.
A cytochrome c reduction assay was carried out according to the method of Babior et al. (6). A total of 106 conidia of A. fumigatus strains were incubated for 9 h at 37°C in RPMI-5% (vol/vol) fetal calf serum (FCS; Gibco, Germany). Hyphae were harvested by centrifugation, coincubated with 107 freshly isolated PMN, and resuspended in RPMI-5% (vol/vol) FCS in the presence of 75 μM cytochrome c from horse heart (Sigma-Aldrich). Samples were obtained every 30 min, cells were removed by centrifugation, and the absorbance of the supernatants was measured at 550 nm. A sample without hyphae was used as a reference control. The process was inhibited by adding horse heart SOD (Sigma-Aldrich). Three independent biological replicates were analyzed, and a standard deviation of the mean value was calculated for each time point.
Quantification of oxidative burst.
Primary human neutrophils were incubated at a multiplicity of infection (MOI) of 10 with A. fumigatus hyphae in the presence of 2.5 μM dichlorfluoresceine diacetate (DCFH-DA; Sigma-Aldrich) for 145 min at 37°C. The generation of ROI was quantified in the presence or absence of 4 mM glutathione and 16 μM diphenyleneiodonium chloride (DPI) by measuring the conversion of DCFH-DA into green fluorescent dichlorfluoresceine (49). A fluorescence reader (GENios; Tecan, Germany) which measured the relative fluorescence units in 5-min intervals (excitation, 485 nm; emission, 520 nm) was used to monitor the production of ROI.
Confrontation assay.
For plate-based inactivation assays, conidia of fungal strains were dispensed in RPMI 1640 plus 5% (vol/vol) FCS, followed by incubation overnight at room temperature in a shaker. Germ-tube formation was induced at 37°C for 4 h directly before the assay. A total of 8 × 104 PMN and 8 × 104 fungal cells were mixed in 40 μl of fresh RPMI 1640 plus 5% (vol/vol) FCS (MOI = 1), activated with 23 ng of phorbol myristate acetate/ml and incubated for different periods of time at 37°C (0, 120, 160, and 200 min). PMN were lysed by the addition of 2 ml of ice-cold distilled water. Dilutions of this suspension were plated on malt extract agar and incubated for 24 h at 37°C. When indicated, 4 mM glutathione was added to the assay to scavenge ROI or 16 μM DPI was used for the inhibition of NADPH oxidase activity. Each experiment was repeated independently with PMN from at least five different donors. Average values and standard deviations were calculated, and the significance was tested by using the Student t test.
Animal infection model.
A murine low-dose model for invasive aspergillosis was applied (37) with modifications as previously described (31). In brief, 18- to 20-g female BALB/c mice were immunosuppressed with 150 mg of cyclophosphamide (Sigma-Aldrich)/kg on days −4, −1, 2, 5, 8, 11, and 14 prior to and after infection on day 0. A single dose of cortisone acetate (200 mg/kg; Sigma-Aldrich) was injected subcutaneously on day −1. A. fumigatus conidia suspensions were harvested with phosphate-buffered saline (PBS) containing 0.1% (vol/vol) Tween 80 (Merck, Germany) and filtered twice through Miracloth (Calbiochem, Germany). Mice were anesthetized by intraperitoneal injection of 100 mg of ketamine (WDTeG, Germany) and 10 mg of xylazine (RompunR; Bayer, Germany)/kg and intranasally infected with 25 μl of a fresh suspension containing 6 × 104 conidia. Survival was monitored daily, and moribund animals were killed humanely. A control group remained uninfected (inhalation of PBS) to monitor the influence of the immunosuppression procedure on vitality. Infections were performed with a group of 10 mice for each tested strain, and the whole experiment was carried out in duplicates. Mice were cared for in accordance with the principles outlined by the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Treaty Series, no. 123 [http://conventions.coe.int/treaty/Default.asp]).
2D gel electrophoresis analysis.
Samples for two-dimensional (2D) polyacrylamide gel electrophoresis (2D-PAGE) were prepared with slight modifications as previously described (27). The pH of the samples was adjusted to 8.5 by the addition of 100 mM NaOH. Afterwards, the samples were labeled with CyDye minimal dyes according to the manufacturer's protocol (GE Healthcare Bio-Sciences). Briefly, 50 μg of protein of either sample was labeled with 300 pmol of CyDye (dissolved in dimethyl formamide). Samples from stress-induced and noninduced conditions were labeled either with Cy3 or Cy5. A pool of two samples (induced and noninduced) was prepared, labeled with Cy2, and used as an internal standard. Samples were vortex mixed and incubated for 30 min in the dark on ice. The reaction was stopped by adding 1 μl of 10 mM l-lysine. An equal volume of 4× sample buffer (composition as for the lysis buffer mentioned above, plus 3.2% [vol/vol] ampholytes and 40 mM dithiothreitol) was added. Equal amounts of each of the three label preparations were mixed and applied via anodic cup loading onto a IEF-strip.
Strips of 24 cm covering a nonlinear pH range from pH 3 to 11 or a linear pH range from pH 4 to 7 (GE Healthcare Bio-Sciences), which had been rehydrated overnight (7 M urea, 2 M thiourea, 2% [wt/vol] CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1% [wt/vol] Zwittergent 3-10, 0.002% [wt/vol] bromophenol blue, 0.5% [vol/vol] IPG buffer, 1.2% [vol/vol] De-Streak reagent [GE Healthcare Bio-Sciences]) were used for isoelectric focusing as previously described (27). Prior to the start of separation by the second dimension, the strips were equilibrated for 15 min in 10 ml of equilibration buffer containing 1% (wt/vol) dithiothreitol and subsequently for 15 min in 10 ml of equilibration buffer containing 2.5% (wt/vol) iodoacetamide. Electrophoresis was performed as described by Kniemeyer et al. (27). Proteins were visualized by analyzing the gels with a Typhoon 9410 scanner using a resolution of 100 μm. Spot detection of cropped images was performed with the DeCyder software package (version 5.0). The following parameters were applied: detection sensitivity, 2,500 spots (excluding filter set); slope, >2.1; area, <200; peak height, <600; and area, <10,000. Changes in the abundance of protein spots were regarded as significant with a threshold of two standard deviation difference. Gels of five independent experiments were analyzed with the BVA software, and average ratios as well as t test values for each spot were calculated. Only spots with a t test value below 0.05 were regarded as significant. In order to analyze differently expressed proteins by mass spectrometry (MS), separate preparative gels were run and stained with colloidal Coomassie brilliant blue (27).
MS identification.
Proteins were identified on a Bruker Ultraflex I matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)/TOF device (Bruker Daltonics, Germany) operating in reflectron mode. MS spectra were subsequently identified by searching the NCBI database using the MASCOT interface (MASCOT 2.1.03; Matrix Science, United Kingdom) with the following parameters: Cys as a S-carbamidomethyl derivative and methionine in an oxidized form (variable), one missed cleavage site, and a peptide mass tolerance of 200 ppm. Hits were considered significant according to the MASCOT score (P = 0.05). Database searches were improved by iterative recalibration and application of the peak rejection algorithm filter of the Score Booster tool implemented into the Proteinscape 1.3 database software (Protagen, Germany).
RESULTS
Proteome analysis of A. fumigatus grown under reactive oxygen stress.
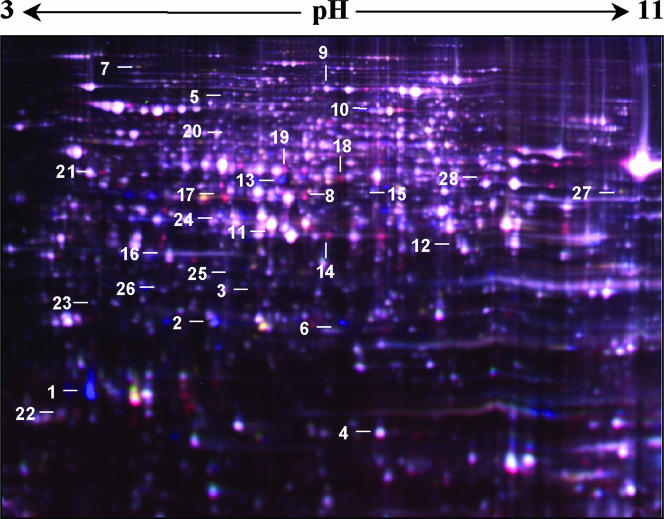
To get a comprehensive picture of the defense of A. fumigatus against ROI, we analyzed the proteomes of A. fumigatus wild-type strain grown either without oxidative stress or challenged with 2 mM H2O2 for 45 min (Fig. 1). This time point was chosen, since in a preceding enzyme assay thioredoxin reductase showed maximum activity at between 30 and 60 min after addition of H2O2 (data not shown). Furthermore, the concentration of 2 mM H2O2 did not cause any growth inhibition and did not show any effect on spore germination (data not shown). H2O2 increases intracellular peroxide (O22−) levels, which leads to the direct oxidation of the sulfur-containing amino acids and the generation of OH· radicals (22). The results of the proteome analysis are shown in Fig. 1 and Table 1. Reproducibly, 27 protein spots displayed an increase and 17 protein spots displayed a decrease in intensity of >1.5-fold (2-fold standard deviation) in extracts of H2O2 grown mycelia (see Table S1 in the supplemental material). Some proteins appeared in gels as more than one spot with the same apparent molecular mass, but with different pI values and abundance, presumably due to posttranslational modifications or isoenzyme variation, e.g., allergen AspF3 and the peroxiredoxin Prx1. Therefore, the identified 44 spots represented 28 different proteins (Table 1 and Fig. 1). The protein spot of each protein showing the highest change in abundance is depicted in Fig. 1. Several stress-associated proteins showed an increased level upon H2O2 stress (Table 1), including Cu/Zn-SOD, the mitochondrial peroxiredoxin Prx1, the allergen AspF3, and the spermidine synthase. AspF3 is a putative thioredoxin peroxidase and was the most highly induced protein after stimulation with H2O2. AspF3 is a known allergen in patients suffering from allergic bronchopulmonary aspergillosis (11). The protein could be extracted from the surface of conidia (4), although it was originally found in peroxisomal membranes (11). In Saccharomyces cerevisiae, the homolog Tsa1p functions as peroxidase, chaperone, and regulator for gene expression under stress conditions (14). It is therefore conceivable that AspF3 also functions in the detoxification of ROI. The peroxiredoxin Prx1 contains both a putative mitochondrial signal peptide and a conserved cysteine characteristic of peroxiredoxins. This finding makes it likely that this protein has thioredoxin peroxidase activity. The significant upregulation of the thioredoxin system in response to increased levels of H2O2 indicates its importance for A. fumigatus. Spermidine synthases are involved in the biosynthesis of polyamines, which play a pivotal role in defense against environmental stresses. Cryptococcus neoformans mutants with the spermidine synthase gene spe3 deleted showed reduced capsule formation, melanin production, and growth rate. A role of polyamine biosynthesis for virulence was reported (26). Several genes encoding the proteins shown in Table 1, which were upregulated under ROI stress, are known to be regulated in S. cerevisiae by the bZip-type transcription factor Yap1p. Therefore, it was likely that a homolog of Yap1 existed in A. fumigatus and, moreover, which could be of importance for defense against ROI. In the yeast S. cerevisiae, the transcriptional regulator Yap1p represents the key regulator of both the metabolic stress response and the acquisition of stress tolerance (5, 24).
FIG. 1.
2D gel electrophoresis of protein extracts of wild type (Cy3-red) and wild type challenged with 2 mM H2O2 (Cy5-blue) for 45 min. Proteins were stained with the difference in gel electrophoresis (DIGE) labeling technique. The orientation of the IEF is indicated. The numbers refer to proteins whose levels changed significantly during growth with H2O2 (see Table 1).
TABLE 1.
Differentially synthesized proteins of A. fumigatus after incubation with 2 mM H2O2
| Spot no.c | Putative function and protein namea | Ratiob
|
|
|---|---|---|---|
| 15 min | 45 min | ||
| Proteins with antioxidant properties | |||
| 1 | Allergen Asp F3 (AFUA_6G02280) | 4.3 | 10.3 |
| 2 | Mitochondrial peroxiredoxin Prx1 (AFUA_4G08580) | 2.1 | 3.7 |
| 3 | Cytochrome c peroxidase (AFUA_4G09110) | 2.0 | −2.0 |
| 4 | Cu/Zn-SOD (AFUA_5G09240) | 2.7 | 1.2 |
| 5 | Catalase Cat1 (AFUA_2G00200) | −1.3 | |
| Heat shock | |||
| 6 | 30-kDa heat shock protein (AFUA_3G14540) | 1.8 | 2.5 |
| Proteases | |||
| 7 | Zn dependent peptidase (AFUA_4G07910) | 1.2 | −2.0 |
| Protein translocation | |||
| 8 | IDI2 (AFUA_4G05830) | 1.0 | −1.6 |
| 9 | Alanyl-tRNA synthetase (AFUA_8G03880) | 1.8 | 1.4 |
| Pentose phosphate pathway | |||
| 10 | Transketolase TktA (AFUA_1G13500) | 2.1 | 2.1 |
| 11 | Transaldolase (AFUA_5G09230) | 2.3 | −1.0 |
| Glycolysis, pyruvate metabolism | |||
| 12 | Glyceraldehyde-3-phosphate-DH (AFUA_5G01970) | 1.9 | 2.3 |
| 13 | Pyruvate-DH subunit α (AFUA_1G06960) | 1.7 | 3.2 |
| Fatty acid or lipid metabolism | |||
| 14 | Zn-dependent alcohol-DH (AFUA_5G06240) | 1.0 | −2.1 |
| 15 | Acetyl coenzyme A acetyltransferase (AFUA_8G04000) | 1.2 | 1.6 |
| Polyamine pathway | |||
| 16 | Spermidine synthase (AFUA_1G13490) | 2.9 | 1.3 |
| Amino acid metabolism | |||
| 17 | Glutamine synthetase (AFUA_4G13120) | −2.0 | −2.0 |
| 18 | Carbamoylphosphate synthase (AFUA_5G06780) | −1.2 | −2.2 |
| 19 | Adenosylhomocysteinase (AFUA_1G10130) | 1.0 | 1.9 |
| Cytoskeleton | |||
| 20 | Actin interacting protein 2 (AFUA_5G02230) | −3.1 | −1.3 |
| 21 | Beta-tubulin (AAL01593) | −1.8 | 1.1 |
| 22 | Topomyosin (AFUA_7G04210) | −1.7 | −1.5 |
| Regulatory protein | |||
| 23 | 14-3-3 family protein ArtA (AFUA_2G03290) | 2.2 | 1.2 |
| Cofactor biosynthesis | |||
| 24 | Thiamine biosynthesis protein Nmt1 (AFUA_5G02470) | 1.1 | 2.0 |
| Purine and pyrimidine synthesis | |||
| 25 | Phosphoribosyltransferase (AFUA_4G04550) | 1.2 | 3.4 |
| Unclassified and proteins of unknown function | |||
| 26 | GMC-oxidoreductase (AFUA_3G01580) | 2.3 | 6.2 |
| 27 | Conserved NAD-dependent DH (AFUA_7G00350) | −1.1 | −2.1 |
| 28 | UGP1 (AFUA_7G01830) | −2.1 | −2.3 |
DH, dehydrogenase.
Average ratios extracted from statistical analysis of DIGE gels by the Decyder software programs DIA and BVA.
Spot number in Fig. 1.
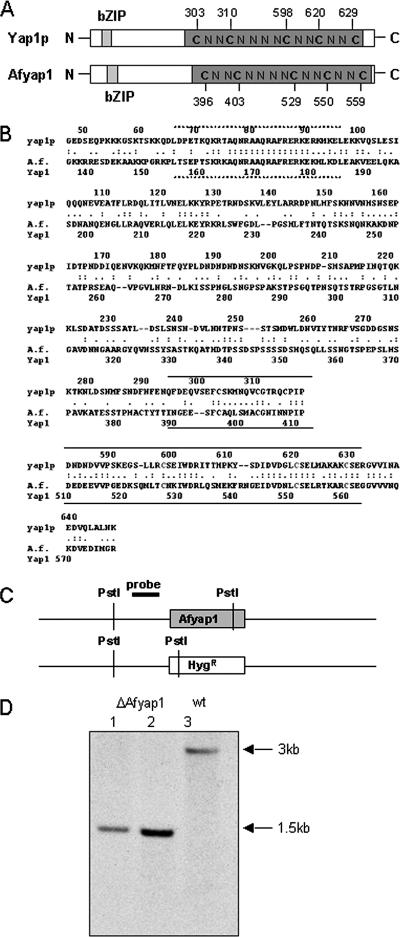
Identification and characterization of a Yap1 homolog of A. fumigatus.
We analyzed the A. fumigatus genome for the presence of a putative S. cerevisiae yap1 homologue. On chromosome 6, a single gene was identified that we designated Afyap1 (Fig. 2). Mutational analyses had established that a set of cysteine residues located in the carboxy-terminal cysteine-rich domain (c-CRD) region of both S. cerevisiae Yap1 and Cap1, the Yap1 homolog in C. albicans (1), is required for the export of Yap1 and Cap1 from the nucleus to the cytoplasm by the export protein Crm1 under nonstress conditions. Upon oxidant exposure, which changes the conformation of the c-CRD and masks the nuclear export signal (NES), Yap1 and Cap1 accumulate in the nucleus (10, 30, 64, 66). As shown in Fig. 2A, the five cysteine residues required for nuclear maintenance upon oxidation of Yap1p are conserved in AfYap1. In addition, putative homologs for the yeast gene gpx3, whose protein product was shown to oxidize Yap1 upon challenge with ROI (13), and the exportin gene crm1, were identified in the A. fumigatus genome as AFUA_3G12270 and AFUA_1G08790, respectively. The deduced A. fumigatus open reading frames show sequence similarity to the respective S. cerevisiae proteins of 59 and 54%, respectively.
FIG. 2.
A. fumigatus Yap1. (A) Schematic view of characteristic motifs of yeast protein Yap1p and A. fumigatus protein AfYap1. Numbers indicate the position of conserved cysteine residues in the NES. (B) Partial sequence alignment of AfYap1 from A. fumigatus (AFUA6G09930) with Yap1p of S. cerevisiae (YML007W) with CLUSTAL W. Identical amino acids between the different amino acid sequences at the same position are marked with colons. The numbers above the sequence indicate the positions of amino acids shown in the respective proteins. DNA interaction domain (bZIP domain) is marked with a dashed line, and the CRDs are marked with a contin- uous line. (C) Deletion of A. fumigatus Afyap1 gene. Schematic representation of the chromosomal Afyap1 locus of the wild type and the ΔAfyap1 deletion mutants. The PstI cleavage sites and the position to which the probe hybridizes are indicated. (D) Southern blot analysis of the Afyap1 deletion strains. Chromosomal DNA of the parental wild-type strain ATCC 46645 (lane 3) and the mutant ΔAfyap1 strain (lanes 1 and 2) was cut by PstI. An 830-bp Afyap1 flanking PCR fragment was used as a probe. In the ΔAfyap1 mutant strain, the band characteristic of the wild type (lane 3) had disappeared. Instead, bands characteristic for a gene replacement at the Afyap1 locus were detected.
Deletion of the Afyap1 gene and analysis of its importance for defense against H2O2, menadione, diamide, and NO radicals.
To analyze the importance of AfYap1 for the defense of A. fumigatus against reactive agents, we deleted the Afyap1 gene in the A. fumigatus wild-type strain ATCC 46645. For this purpose, plasmid Δyap1-Hyg-TOPO was generated (see Materials and Methods). As the selection marker, the hygromycin B resistance gene was used. After transformation, deletion of the Afyap1 gene in the resulting transformants was detected by Southern blot analysis (Fig. 2C and D). The band characteristic for the Afyap1 gene of the wild type (3 kbp) had disappeared in the ΔAfyap1 mutant strains. Instead, the band characteristic of gene replacement of the Afyap1 gene of 1.5 kbp was detected. The ΔAfyap1 strain was viable and showed slightly reduced growth on malt agar plates (data not shown). As a control, the ΔAfyap1 deletion mutant was complemented using the wild-type Afyap1 gene under the control of its own promoter and using phleomycin as the selection marker gene (data not shown). To verify a single ectopic integration, the complementation was checked by Southern blotting with an internal Afyap1 probe.
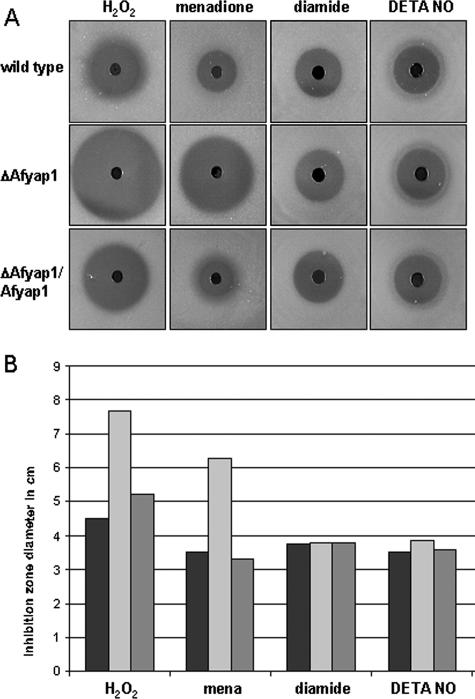
To determine whether AfYap1 was required for the resistance of A. fumigatus against reactive agents, the wild type, the ΔAfyap1 mutant strain, and a complemented mutant strain (ΔAfyap1/Afyap1) were placed on AMM agar plates containing a concentration gradient of H2O2, menadione, diamide, or the NO donor DETA NO. Among the stress-inducing agents selected, menadione generates superoxide anions (O2−) in a redox cycle, which inactivates 4Fe-4S cluster-containing proteins. Menadione may also affect the reduced glutathione (GSH) pool directly via a detoxification reaction catalyzed by glutathione S-transferase (48). Diamide is a thiol-oxidizing agent resulting in fast oxidation of GSH to oxidized glutathione (GSSG), resulting in a GSH/GSSG redox imbalance (57), and proteins with the possibility to form disulfide bonds become oxidized. DETA NO is a compound leading to the release of NO· (17). The NO radicals interact with, e.g., superoxide O2− anions, and form the more reactive peroxynitrite. However, other oxidation states of NO· and the formation of other reactive nitrogen intermediates (RNI) are also possible. The MIC at which 50% of C. albicans isolates are inhibited by DETA NO was determined to be 0.25 to 1 mg/ml (39). RNI have been shown to inhibit mitochondrial respiration, causing DNA damage and leading to nitrosylation of thiol groups (17).
As shown in Fig. 3, the Afyap1 deletion mutant was much more susceptible to H2O2 and menadione than was the wild type. Expression of the wild-type Afyap1 gene complemented the hypersensitivity of the ΔAfyap1 strain against H2O2 and menadione to an extent similar to that of the wild-type strain. Interestingly, the ΔAfyap1 mutant strain did not exhibit increased sensitivity to diamide and DETA NO. The phenotypes observed suggest that AfYap1 is a key regulator of antioxidant defense of A. fumigatus directed toward peroxides and superoxide anions. In contrast, AfYap1 is not involved in the detoxification of NO radicals and diamide. Diamide oxidizes sulfhydryl groups of proteins and leads to Yap1p nuclear localization in S. cerevisiae (58). A deletion of yap1 leads in contrast to A. fumigatus to an increased sensitivity to diamide (30).
FIG. 3.
Sensitivity of A. fumigatus strains against different reactive agents. (A) Inhibition zone assay. A total number of 107 conidia were mixed with 2% (wt/vol) AMM top agar. In the middle of the agar plate a well was filled either with H2O2, menadione, diamide, or DETA NO. The inhibition zones were measured 16 h after incubation at 37°C. (B) Schematic view of sensitivity assay. Inhibition zone diameters of the wild-type strain (black bar), ΔAfyap1 strain (light gray bar), and complemented ΔAfyap1/Afyap1 strain (dark gray bar) are indicated in centimeters. The ROI-producing agents H2O2, menadione (mena), diamide, and DETA NO are indicated on the left. The base DETA that corresponds to DETA NO was also tested, and no difference in sensitivity was observed for the ΔAfyap1 and the wild-type strain (data not shown).
Identification of putative AfYap1 targets.
To analyze possible targets of AfYap1-dependent regulation, we examined the global effect of Afyap1 deletion at the proteomic level. For this purpose, protein extracts of the wild-type and ΔAfyap1 strains challenged with H2O2 were compared by 2D-PAGE. Reproducibly, 21 protein spots displayed an increase and 36 protein spots displayed a decrease in intensity of more than 1.3-fold (2-fold standard deviation) in ΔAfyap1 extracts (see Table S2 in the supplemental material). Some proteins appeared in gels as more than one spot with a different pI, e.g., methyltranferase, transketolase TktA, and UGP 1. In all, 29 protein spots, including catalases, showed a decreased level in the ΔAfyap1 mutant compared to the wild-type strain after exposure to 2 mM H2O2 and therefore present putative AfYap1 targets (Table 2).
TABLE 2.
Decreased protein spots (and therefore putative AfYap1 targets) in A. fumigatus ΔAfyap1 strain compared to wild type after exposure to 2 mM H2O2 for 45 min (pH gradient gel 4 to 7)
| Function and protein namea | Ratiob |
|---|---|
| Proteins with antioxidant properties | |
| Allergen Asp F3 (AFUA_6G02280) | −2.81 |
| Mitochondrial peroxiredoxin Prx1 (AFUA_8G07130) | −2.31 |
| Cytochrome c peroxidase (AFUA_4G09110) | −1.43 |
| Catalase Cat1 (AFUA_2G00200) | −2.01 |
| Catalase/peroxidase Cat2 (AFUA_8G01670) | −2.02 |
| Heat shock | |
| Chaperone HSP70 HscA (AFUA_8G03930) | −1.75 |
| HSP70 chaperone Hsp88 (AFUA_1G12610) | −1.68 |
| 30-kDa heat shock protein (AFUA_3G14540) | −1.67 |
| Heat shock protein class 1 (AFUA_6G06470) | −2.01 |
| Proteases/protein degradation | |
| Ubiquitin UbiA (AFUA_1G04040) | −2.46 |
| Protein synthesis | |
| Elongation factor EF-3 (AFUA_7G05660) | −2.22 |
| Elongation factor-1γ (AFUA_1G17120) | −4.79 |
| Eukaryotic initiation factor 5A (AFUA_1G04070) | −2.91 |
| Glycolysis | |
| Pyruvate-DH subunit α (AFUA_1G06960) | −1.71 |
| Pentose phosphate pathway | |
| Transketolase TktA (AFUA_1G13500) | −2.04 |
| Phosphoglucomutase PgmA (AFUA_3G11830) | −1.67 |
| Purine and pyrimidine synthesis | |
| GMP synthase (AFUA_3G01110) | −1.51 |
| Unclassified and proteins of unknown function | |
| Methionine synthase MetH/D (AFUA_4G07360) | −2.39 |
| Aminotransferase (AFUA_2G12470) | −1.82 |
| Peptidyl cis-trans isomerase (AFUA_3G07430) | −1.85 |
| Histone demethylase Aof2 (AFUA_4G13000) | −2.02 |
| Septin AspB (AFUA_7G05370) | −3.03 |
| Hypothetical protein AFUA_5G14680 | −2.01 |
| UGP 1 (AFUA_7G01830) | −2.11 |
| Hypothetical protein AFUA_3G00730 | −1.54 |
| p-Nitroreductase family protein (AFUA_5G09910) | −2.25 |
| PH domain protein (AFUA_4G12450) | −1.79 |
| Hypothetical protein AFUA_3G11400 | −1.57 |
| Hypothetical protein AFUA_1G09890 | −1.46 |
Name and function are derived from GenBank (National Center for Biotechnology Information). DH, dehydrogenase.
Average ratios are extracted from statistical tables of the Decyder software after DIA and BVA analysis.
Catalases regulated by AfYap1.
The proteome analysis suggested that catalases are regulated by AfYap1. By measuring catalase activity, we examined which of the catalases was controlled by AfYap1. For this purpose, the wild-type strain and the Afyap1 deletion strain were challenged with H2O2 for 45 min. Protein extracts were generated, and equal amounts of protein were loaded onto a native polyacrylamide gel. The observed, negative-stained bands were assigned according to Paris et al. (45). As shown in Fig. 4A, the activity of catalase Cat2 was completely absent in the ΔAfyap1 mutant strain, and the activity of catalase Cat1 was reduced. Inspection of the putative promoter regions of the two catalase-encoding genes in A. fumigatus suggests the presence of two putative AfYap1 binding sites [TTA(G/C/T)TAA] in the Cat2 promoter and none in the Cat1 promoter (29). In addition, low activity of the conidial CatA was detected (Fig. 4). Catalase activity was also determined photometrically, and the decreased activity in the ΔAfyap1 strain confirmed the results obtained by the activity staining after native PAGE. Protein extracts derived from wild-type mycelium showed an increase in catalase/peroxidase activity from 2.6 to 4.4 μmol min−1 mg of protein after the addition of 2 mM H2O2, whereas in the ΔAfyap1 strain the activity increased only from 0.6 to 2.6 μmol min−1. 2D gels from proteins extracted from the cytosol showed a similar regulation of Cat1 and Cat2, as observed in the catalase activity assays (Fig. 4B). Both catalases were also detectable in supernatants of culture medium (data not shown). To confirm the data obtained by both 2D gel electrophoresis and activity staining on the transcriptional level, we carried out Northern blot analysis. The steady-state mRNA level of catalase cat1 in A. fumigatus was only slightly H2O2 stress or AfYap1 dependent (Fig. 4C). The transcript level of catalase cat2 was highly upregulated 15 min after the addition of H2O2 and slightly reduced after 45 min (Fig. 4C). In the Afyap1 deletion strain no induction of cat2 transcript levels was detectable, suggesting that the Yap1 response elements (YREs) at positions −140 and −605 in the promoter region of cat2 are presumably functional.
FIG. 4.
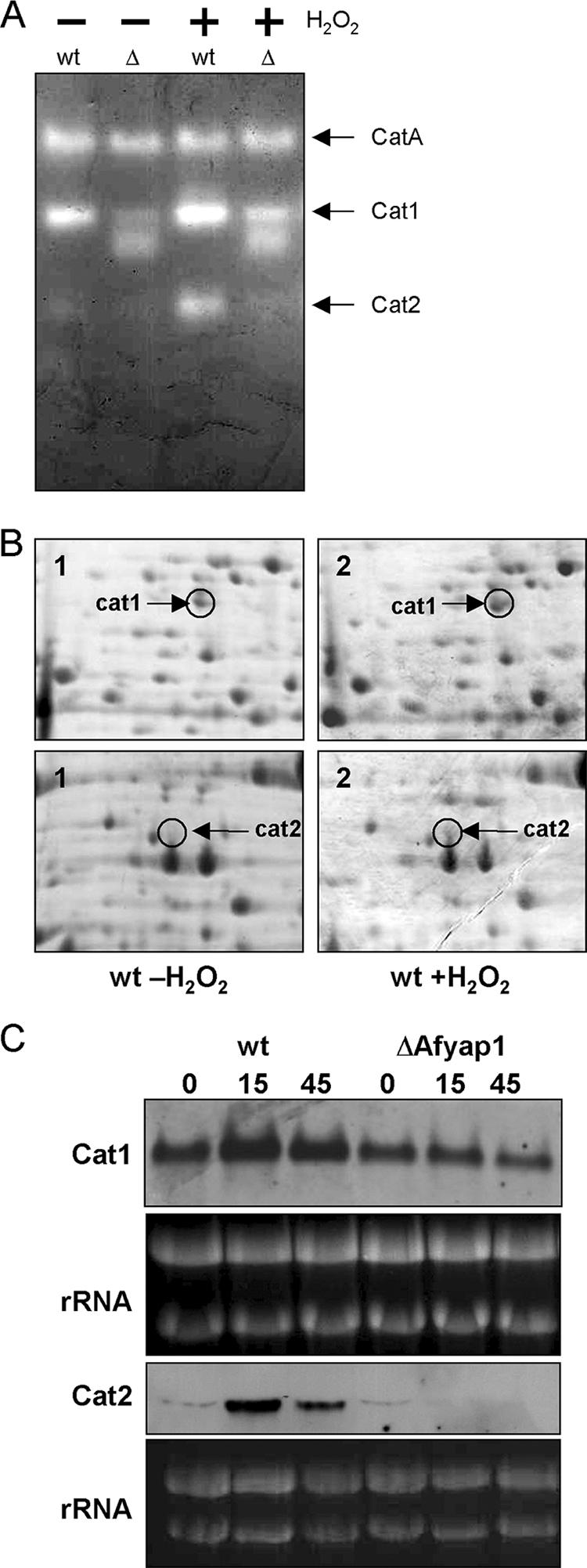
Regulation of catalases 1 and 2 by oxidative stress. (A) Catalase activity staining modified according to the method of Goldberg and Hochman (21). Overnight cultures of wild type (wt) and ΔAfyap1mutant strain (Δ) were incubated with (+) or without (−) 2 mM H2O2 for 45 min before protein extracts were isolated. The negatively stained bands were allocated to the three different catalases CatA, Cat1, and Cat 2 as described previously (45). (B) Sections of Coomassie blue-stained 2D gels from A. fumigatus wild-type mycelium grown with (panels 1) or without (panels 2) 2 mM H2O2 for 45 min. Catalases 1 and 2 were identified by MALDI-TOF MS and marked with arrows. (C) Northern blot analysis of the Afyap1-dependent expression of catalase cat1 and cat2 under oxidative stress conditions. RNA was isolated from H2O2 stressed mycelia of A. fumigatus wild-type and ΔAfyap1 strains after 0, 15, and 45 min of stress induction. rRNA bands are shown as controls.
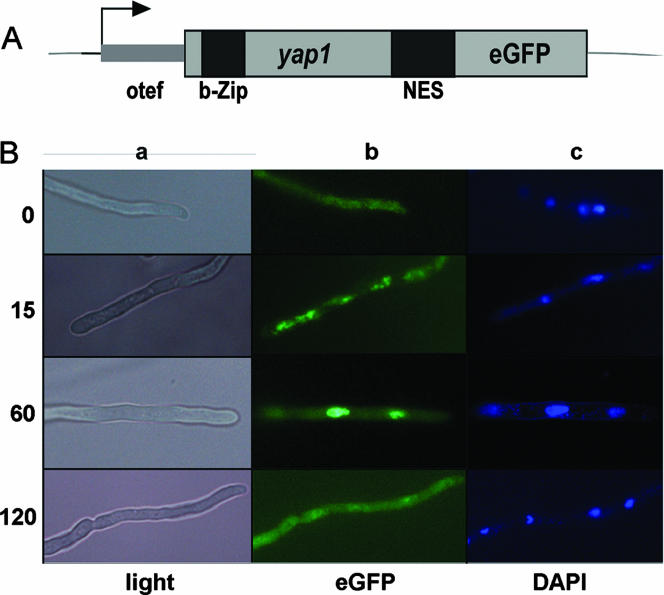
Nuclear localization of AfYap1 is stress dependent.
Previous work on both S. cerevisiae and C. albicans demonstrated that regulation of the Yap1 transcription factors involved oxidant-dependent nuclear localization of the protein (30, 66). As shown in Fig. 2A, the five cysteine residues required for nuclear maintenance upon oxidation of Yap1 are conserved in AfYap1. To analyze whether AfYap1 is also regulated by subcellular localization, the Afyap1 gene was fused in frame with the gene encoding the enhanced green fluorescence protein (egfp) (Fig. 5). After transformation of A. fumigatus with plasmid otefp-Afyap1-eGFP, the cellular localization of the AfYap1-eGFP fusion was analyzed in the transformant strains. As shown in Fig. 5, in the absence of H2O2 the protein fusion exhibited diffuse cytoplasmic fluorescence. At 15 min after treatment with H2O2, the protein fusion began to localize in the nucleus, as seen by coincidence with the DAPI (4′,6′-diamidino-2-phenylindole) stain. After 60 min of H2O2 treatment, the AfYap1-eGFP signal fully accumulated in the nucleus. This accumulation was reversible, and after 120 min a diffuse cytoplasmic fluorescence was observed again. Hence, the cellular localization of the AfYap1-eGFP fusion protein is indicative of H2O2 stress to the fungal cell.
FIG. 5.
Oxidative stress-regulated nuclear localization of AfYap1-eGFP. (A) Schematic representation of otefp-Afyap1-eGFP fusion construct. The putative bZIP domain and NES of AfYap1 are indicated. (B) Localization of AfYap1-eGFP. Overnight culture of strain otef-AfYap1-eGFP in AMM was challenged with 2 mM H2O2 for 0, 15, 60, and 120 min. Samples were analyzed by light microscopy (a) or fluorescence microscopy (b and c). Panel b shows the green fluorescent protein fluorescence signal. Nuclei were stained with DAPI (c).
In the ΔAfyap1 mutant, the presence of the GFP fusion protein containing wild-type AfYap1 exhibited H2O2 resistance that was indistinguishable from that of the wild-type strain (data not shown). Consistently, when the AfYap1-eGFP fusion protein was produced in the wild-type strain, the respective mutant strain showed elevated H2O2 tolerance above that of the wild-type strain, indicating the functionality of the protein fusion (data not shown).
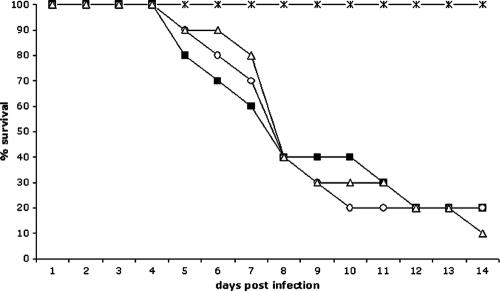
Despite the importance of AfYap1 for defense against ROI, deletion of Afyap1 did not result in attenuated virulence in a mouse infection model.
To assess the role of AfYap1 in pathogenesis, the corresponding deletion mutant was tested in an animal infection model of invasive aspergillosis. Groups of 10 immunosuppressed, neutropenic mice were infected by inhalation with 6 × 104 conidia of the different strains. The results are shown in Fig. 6. In the groups infected with wild-type conidia (strain ATCC 46645), the mortality was 90% after 17 days. Similarly, when mice were infected with conidia of the ΔAfyap1 mutant strain, the mortality was almost the same as that observed for the wild-type strain (Fig. 6). As a control, mice were infected with conidia of the complemented Afyap1 mutant (ΔAfyap1/Afyap1), which showed the same virulence as wild-type conidia (Fig. 6). In the uninfected control group (PBS), mortality was 0%.
FIG. 6.
Virulence of an Afyap1 deletion strain. Virulence of A. fumigatus wild-type ATCC 46645 (▪), ΔAfyap1 (○), and complemented ΔAfyap1 (▵) strains in mice. Groups of 10 BALB/c mice were infected with 6 × 104 A. fumigatus conidia each, as indicated, by nasal inhalation. Survival was monitored for 14 days. A group of mice which inhaled PBS served as control (×).
Generation of ROI by PMN during contact with conidia of the different strains.
To study whether the generation of ROI by PMN depends on the different A. fumigatus mutant strains analyzed, a cytochrome c reduction assay (61) for measuring the presence of peroxides O2− was applied. Thus, freshly isolated PMN from healthy human donors were coincubated with wild-type, ΔAfyap1, and ΔAfyap1/Afyap1 hyphae, and the reduction of cytochrome c was measured over time at 550 nm (data not shown). No significant difference in the amount of cytochrome c reduction was observed for the tested strains, excluding that the amount of ROI generated by PMN differs between the strains analyzed.
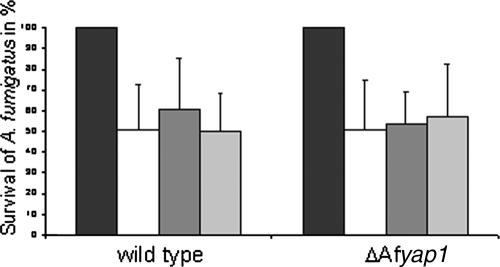
Survival of A. fumigatus wild type and ΔAfyap1 mutant confronted with PMN.
To test whether ROIs are involved in the killing of hyphae by human PMN, A. fumigatus germ tubes of the wild-type strain, the ΔAfyap1 mutant strain, and the complemented ΔAfyap1/Afyap1 mutant strain (data not shown) were coincubated for 160 min with freshly isolated human PMN. In preliminary experiments with the wild-type strain, ca. 50% killing was achieved after 160 min of coincubation. No phagocytosis was seen, but attachment of PMN to the germ tubes was observed. As shown in Fig. 7, there was no significant difference in survival rate irrespective of whether the wild type or the Afyap1 deletion mutant was tested. The same was also true for the complemented Afyap1 deletion mutant (ΔAfyap1/Afyap1) (data not shown). The addition of glutathione to the assay did not display any effect on killing of A. fumigatus hyphae by PMN, further supporting that it is unlikely that ROI are important for killing of A. fumigatus hyphae by PMN under the conditions tested. At this concentration, glutathione was able to scavenge ca. 90% of ROI produced by human PMNs after stimulation with A. fumigatus hyphae in preliminary experiments using DCFH-DA as an indicator for ROI (data not shown). In control experiments, 4 mM glutathione did not affect fungal viability (J. Loeffler, unpublished data). To further substantiate these findings, NADPH oxidase activity in PMNs was inhibited by 5 μg of DPI/ml, which corresponds to a concentration of 16 μM. This concentration reduced the amount of ROI produced by PMN after contact with A. fumigatus hyphae to ca. 30% and did not significantly affect fungal viability (data not shown). In former studies, 10 μM DPI was shown to inhibit the rate of killing of the bacterial pathogen Staphylococcus aureus by 77% (23). Based on control experiments, higher concentrations of DPI were not used due to adverse effects on fungal growth. When added to the assay, the survival rate of hyphae of the wild type did not differ from that of the ΔAfyap1 strain in the presence of PMN at any time point with or without ROI inhibitors (Fig. 7).
FIG. 7.
Survival rate of A. fumigatus hyphae of the wild-type (ATCC 46645) and ΔAfyap1 strains after confrontation with human PMN. Freshly isolated human PMN were stimulated for 30 min with phorbol myristate acetate and coincubated for 160 min with young hyphae from the wild type (left) or the ΔAfyap1 mutant strain (right) at an MOI of 1. Viable hyphae were quantified by plating after osmotic lysis of the PMN. White bars represent the percentage of viable wild-type or mutant hyphae in relation to controls without PMN (black bars). The addition of 4 mM GSH (dark gray bars) or 16 μM DPI (light gray bars) showed no effect on the survival of A. fumigatus.
DISCUSSION
A central aim of this study was the characterization of the stress response of A. fumigatus to ROI. The mechanisms of cellular redox homeostasis and the ROI metabolism are of particular interest, because ROI were shown to be important for macrophages in defense against the fungal pathogen A. fumigatus (46). We report here the identification of 27 proteins with increased levels and 17 protein spots with decreased levels upon exposure to 2 mM H2O2 by proteome analysis. In S. cerevisiae, this H2O2 concentration was found to induce stress defense at the molecular level (12). Consistently, as shown here, the Afyap1-eGFP was located in the nucleus at this H2O2 concentration, showing that AfYap1 was activated. Based on the proteome data a distinct picture of cellular processes involved in the adaptation to higher oxygen peroxide levels was obtained. Primarily affected are proteins required for antioxidant defense; heat shock proteins; the protein translation apparatus; proteins of central metabolic pathways such as glycolysis, the tricarboxylic acid cycle, and the pentose phosphate cycle; proteins of the amino acid and trehalose metabolism; and proteins of the cytoskeleton. Thus, the antioxidant response to ROI in A. fumigatus resembles the protection mechanism in S. cerevisiae (19, 20), Schizosaccharomyces pombe (62), and Candida albicans (32). However, as also found here, the response of proteins of the cytoskeleton to ROI seems to be stronger in filamentous fungi (47). Moreover, the upregulation of the thioredoxin system clearly shows its importance in the protection of A. fumigatus against H2O2, whereas the upregulation of catalases was only moderate and, interestingly, proteins of the glutathione system were not detected at all. The importance of the thioredoxin system for redox regulation has also been shown for A. nidulans (55).
In the present study, the Yap1 homologue of A. fumigatus was identified and designated AfYap1. The genes of several H2O2-induced proteins in A. fumigatus contained putative YREs in their promoter regions, such as allergen Asp F3, peroxiredoxin Prx1, and transaldolase. This finding suggests that the genes are direct targets of AfYap1, provided the identified YREs are functional. In contrast, it is also possible that the genes are only indirectly regulated by AfYap1 or that AfYap1 regulates the genes in cooperation with other transcription factors such as Skn7 and TFIIA, as known for genes in S. cerevisiae (28, 42). While the two transcription factors Yap1p and AfYap1 exhibit a high degree of sequence similarity, little information was available on the regulation of Yap1p homologs in filamentous fungi. We provide several lines of evidence that the bZip transcription factor AfYap1 functions in a similar way. First, the AfYap1-eGFP fusion protein showed nuclear localization under H2O2 stress (Fig. 5) similar to Yap1 accumulation in both S. cerevisiae and C. albicans (30, 66). Second, deletion strains of Afyap1 in A. fumigatus exhibited increased sensitivity against H2O2 and menadione on agar plates similar to the S. cerevisiae Δyap1 and C. albicans Δcap1 mutants (60, 66). Third, complementation of the Afyap1 deletion mutant by ectopic integration of the Afyap1 gene restored the wild-type phenotype (Fig. 3). In contrast to S. cerevisiae (30), deletion of Afyap1 did not lead to increased sensitivity to diamide. A similar phenotype has also been shown for A. nidulans (3). Because wild-type and Afyap1 deletion strains did not exhibit increased sensitivity against the NO· donor agent DETA NO and the corresponding base DEA on agar plates, this led us to conclude that AfYap1 does not play a role in NO· detoxification in A. fumigatus. However, this finding does not exclude that an overlap between antioxidant and antinitrosative defense exists, because both ROI and RNI target thiols or iron-sulfur clusters (18). Consistently, a Cryptococcus neoformans mutant with a thiol peroxidase gene (tsa1) deleted was sensitive to both nitrosative and oxidative stress (41).
To characterize the targets of AfYap1 in the filamentous fungus A. fumigatus, we compared the proteome of the wild type and the ΔAfyap1 deletion strain upon exposure to H2O2. The peroxiredoxin Prx1 did not show different levels in dependence of the Afyap1 deletion upon challenge with H2O2 and is presumably not a target of AfYap1. Other known Yap1 targets followed characteristic AfYap regulation: thioredoxin peroxidase (AspF3), cytochrome peroxidase, and the protein AFUA_3G00730, which appears to belong to the glutathione S-transferase family. Proteins of the protein degradation pathway were regulated by AfYap1 as well, which is also known for yeast species (19, 60). In addition, a new putative AfYap1 target was identified with the p-nitroreductase family protein AFUA_5G09910. The members of this large family of eubacterial conserved proteins catalyze the reduction of nitro-substituted compounds using FMN or FAD as a prosthetic group and NAD(P)H as a reducing agent. The biological functions of these enzymes have not been elucidated, but a putative role in maintaining the oxidative and/or nitrosative balance within cells was discussed (15).
Because of the important function of catalases for scavenging ROI and their possible regulation by AfYap1, these enzymes were investigated in more detail. Three catalases were described in A. fumigatus: the conidial catalase CatA and the two mycelial catalases Cat1 and Cat2. Deletion of cat1 in A. fumigatus increased the sensitivity of the deletion mutant to ROI but had no effect on pathogenicity (9, 45). Only a cat1/cat2 double deletion in A. fumigatus increased the sensitivity of the respective mutant to ROI and caused a delayed infection in a murine infection model (45). Here, we identified both mycelial catalases Cat1 and Cat2 by 2D gel analysis of H2O2 challenged mycelia and found that the activity of Cat2 but not of Cat1 clearly depends on the presence of ROI and AfYap1. We therefore conclude that catalase 2 is controlled by AfYap1. A Yap1-dependent regulation was also shown for catalase A in S. cerevisiae (19) and C. albicans (32). However, in S. cerevisiae in cooperation with the transcription factor Skn7, Yap1 controls several genes of the oxidative stress response, such as thioredoxin, thioredoxin reductase, and the catalase T (35). Such a regulation is also likely to occur for some genes in A. fumigatus, such as the catalase Cat 1. Also, an Skn7 ortholog is present in A. fumigatus (33).
The role of ROI for host resistance during microbial infections is a matter of debate. As reported here, the Afyap1 deletion in A. fumigatus exhibited drastically increased sensitivity to H2O2 and menadione (Fig. 3) but did not show reduced pathogenicity in an in vitro killing assay with human PMN and in an infection model applying immunosuppressed, neutropenic mice (Fig. 6). The same was shown for a putative interaction partner of AfYap1, the response regulator Skn7. Deletion of the skn7 homolog AfSKN7 in A. fumigatus led to increased sensitivity against peroxides, but virulence was not affected. In contrast to the ΔAfyap1 strain, the ΔAfskn7 mutant exhibited no enhanced sensitivity against menadione (33). Therefore, the outcome of our study implies that superoxide and peroxide detoxification mechanisms of A. fumigatus do not play an important role during infection in immunocompromised mice or in the extracellular killing of A. fumigatus germ-tubes by neutrophils. Consequently, killing mechanisms other than the production of ROI by the host appear to be more relevant. Alternatively, it cannot be ruled out that the protective role of AfYap1 against ROI is dispensable in vivo because other mechanisms take over this task. Nevertheless, our results are in agreement with recent findings that the granule proteins in neutrophils play a major role in the killing process of microbes and ROI only function as activators of vacuolar enzymes (50, 53). This conclusion was also supported by the finding that knockout mice lacking the ability to produce cathepsin G and elastase were found to be susceptible to Aspergillus infection (56). Granulocytes are also able to produce RNI and hypochlorous acid to kill microbes (16, 17). The production of both RNI and ROI in granulocytes is regulated by gamma interferon. The production of ROI is therefore always coupled to the production of RNI. Hence, RNI may play a more relevant role in the killing of fungi than originally anticipated. However, NO synthase deficiency is not always detrimental (38), and macrophages generally produce considerably more RNI than neutrophils (43). In the same way, macrophages from iNO synthase-deficient mice did not show reduced in vitro killing of A. fumigatus conidia (46). Alternatively, the myeloperoxidase-H2O2-halide system may also be involved in the damage of hyphae by neutrophils (16), but a direct role for this system remains to be demonstrated. In addition, ROI are required as a substrate for the production of hypochlorous acid (53). Hence, the mechanism for how the innate immune system kills and digests spores and germlings of A. fumigatus needs to be further elucidated with a focus on non-ROI-mediated killing. One candidate for such non-ROI-mediated killing is represented by the recently found extracellular fibers called neutrophil extracellular traps. They may mediate the extracellular killing of hyphae of A. fumigatus, since this has been shown for both the yeast-form and hyphal cells of C. albicans (59). The transferrin family protein lactoferrin with a high sequestration activity for iron is another recently found factor secreted by PMN, which mediates the inhibition of conidial growth of A. fumigatus independently of ROI production (65).
In summary, in the present study we show that the regulatory response upon oxidative stress of A. fumigatus is to a large extent regulated by AfYap1. For the first time, the ROI detoxifying system of A. fumigatus against H2O2 was characterized in detail. Taken as a whole, the present study strongly argues that the function and regulation during oxidative stress mediated by AfYap1 is not of significant importance for pathogenicity in an immunocompromised host. Consequently, our data challenge the idea that the innate immune system, in particular PMN, kills A. fumigatus mycelium during infection by the production of peroxides and superoxide anions.
Supplementary Material
Acknowledgments
We thank Olaf Scheibner and Robert Winkler for MS analysis, Marcel Thoen for providing the thioredoxin reductase assay, and Sven Krappmann for the gift of plasmid pAN8.1. We thank Matthias Brock, Christian Fleck, and Thorsten Heinekamp for helpful discussions and Silke Steinbach for technical assistance.
This study was supported by the Deutsche Forschungsgemeinschaft Priority Program 1160 (Colonization and Infection by Human-Pathogenic Fungi). Work in the laboratories of O.K. and J.L. was financed by a grant from the Interdisciplinary Forum for Clinical Research at the University of Wuerzburg (IZKF-A50).
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrianopoulos, A., and M. J. Hynes. 1988. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol. Cell. Biol. 8:3532-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano, Y., D. Hagiwara, T. Yamashino, and T. Mizuno. 2007. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71:1800-1803. [DOI] [PubMed] [Google Scholar]
- 4.Asif, A. R., M. Oellerich, V. W. Amstrong, B. Riemenschneider, M. Monod, and U. Reichard. 2006. Proteome of conidial surface associated proteins of Aspergillus fumigatus reflecting potential vaccine candidates and allergens. J. Proteome Res. 5:954-962. [DOI] [PubMed] [Google Scholar]
- 5.Azevedo, D., F. Tacnet, A. Delaunay, C. Rodrigues-Pousada, and M. B. Toledano. 2003. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35:889-900. [DOI] [PubMed] [Google Scholar]
- 6.Babior, B. M., R. S. Kipnes, and J. T. Curnutte. 1973. Biological defense mechanisms. The production by leukocytes of superoxide, a potential bactericidal agent. J. Clin. Investig. 52:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beers, R. F., Jr., and I. W. Sizer. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133-140. [PubMed] [Google Scholar]
- 7a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brakhage, A. A. 2005. Systemic fungal infections caused by Aspergillus species: epidemiology, infection process, and virulence determinants. Curr. Drug Targets 6:875-886. [DOI] [PubMed] [Google Scholar]
- 9.Calera, J. A., S. Paris, M. Monod, A. J. Hamilton, J. P. Debeaupuis, M. Diaquin, R. Lopez-Medrano, F. Leal, and J. P. Latge. 1997. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect. Immun. 65:4718-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, S. T., E. A. Epping, S. M. Steggerda, and W. S. Moye-Rowley. 1999. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 19:8302-8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crameri, R. 1999. Epidemiology and molecular basis of the involvement of Aspergillus fumigatus in allergic diseases. Contrib. Microbiol. 2:44-56. [DOI] [PubMed] [Google Scholar]
- 12.Delaunay, A., A. D. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaunay, A., D. Pflieger, M. B. Barrault, J. Vinh, and M. B. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471-481. [DOI] [PubMed] [Google Scholar]
- 14.Demasi, A. P., G. A. Pereira, and L. E. Netto. 2006. Yeast oxidative stress response: influences of cytosolic thioredoxin peroxidase I and of the mitochondrial functional state. FEBS J. 273:805-816. [DOI] [PubMed] [Google Scholar]
- 15.de Oliveira, I. M., J. A. Henriques, and D. Bonatto. 2007. In silico identification of a new group of specific bacterial and fungal nitroreductases-like proteins. Biochem. Biophys. Res. Commun. 355:919-925. [DOI] [PubMed] [Google Scholar]
- 16.Diamond, R. D., and R. A. Clark. 1982. Damage to Aspergillus fumigatus and Rhizopus oryzae hyphae by oxidative and nonoxidative microbicidal products of human neutrophils in vitro. Infect. Immun. 38:487-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, F. C. 1997. Perspectives series: host/pathogen interactions: mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Investig. 99:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820-832. [DOI] [PubMed] [Google Scholar]
- 19.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godon, C., G. Lagniel, J. Lee, J. M. Buhler, S. Kieffer, M. Perrot, H. Boucherie, M. B. Toledano, and J. Labarre. 1998. The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273:22480-22489. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg, I., and A. Hochman. 1989. Purification and characterization of a novel type of catalase from the bacterium Klebsiella pneumoniae. Biochim. Biophys. Acta 991:330-336. [DOI] [PubMed] [Google Scholar]
- 22.Halliwell, B., and J. M. C. Gutteride. 1999. Free radicals in biology and medicine. Oxford Science Publications, Oxford, England.
- 23.Hampton, M. B., and C. C. Winterbourn. 1995. Modification of neutrophil oxidant production with diphenyleneiodonium and its effect on bacterial killing. Free Radic. Biol. Med. 18:633-639. [DOI] [PubMed] [Google Scholar]
- 24.Harshman, K. D., W. S. Moye-Rowley, and C. S. Parker. 1988. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell 53:321-330. [DOI] [PubMed] [Google Scholar]
- 25.Holdom, M. D., B. Lechenne, R. J. Hay, A. J. Hamilton, and M. Monod. 2000. Production and characterization of recombinant Aspergillus fumigatus Cu, Zn superoxide dismutase and its recognition by immune human sera. J. Clin. Microbiol. 38:558-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingsbury, J. M., Z. Yang, T. M. Ganous, G. M. Cox, and J. H. McCusker. 2004. Novel chimeric spermidine synthase-saccharopine dehydrogenase gene (SPE3-LYS9) in the human pathogen Cryptococcus neoformans. Eukaryot. Cell 3:752-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kniemeyer, O., F. Lessing, O. Scheibner, C. Hertweck, and A. A. Brakhage. 2006. Optimisation of a 2-D gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus. Curr. Genet. 49:178-189. [DOI] [PubMed] [Google Scholar]
- 28.Kraemer, S. M., D. A. Goldstrohm, A. Berger, S. Hankey, S. A. Rovinsky, W. Scott Moye-Rowley, and L. A. Stargell. 2006. TFIIA plays a role in the response to oxidative stress. Eukaryot. Cell 5:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuge, S., and N. Jones. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kupfahl, C., T. Heinekamp, G. Geginat, T. Ruppert, A. Hartl, H. Hof, and A. A. Brakhage. 2006. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol. Microbiol. 62:292-302. [DOI] [PubMed] [Google Scholar]
- 32.Kusch, H., S. Engelmann, D. Albrecht, J. Morschhauser, and M. Hecker. 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7:686-697. [DOI] [PubMed] [Google Scholar]
- 33.Lamarre, C., O. Ibrahim-Granet, C. Du, R. Calderone, and J. P. Latge. 2007. Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet. Biol. 44:682-690. [DOI] [PubMed] [Google Scholar]
- 34.Langfelder, K., B. Philippe, B. Jahn, J. P. Latge, and A. A. Brakhage. 2001. Differential expression of the Aspergillus fumigatus pksP gene detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 69:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 36.Levitz, S. M., and T. P. Farrell. 1990. Human neutrophil degranulation stimulated by Aspergillus fumigatus. J. Leukoc. Biol. 47:170-175. [DOI] [PubMed] [Google Scholar]
- 37.Liebmann, B., M. Muller, A. Braun, and A. A. Brakhage. 2004. The cyclic AMP-dependent protein kinase A network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72:5193-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacMicking, J., Q. W. Xie, and C. Nathan. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323-350. [DOI] [PubMed] [Google Scholar]
- 39.McElhaney-Feser, G. E., R. E. Raulli, and R. L. Cihlar. 1998. Synergy of nitric oxide and azoles against Candida species in vitro. Antimicrob. Agents Chemother. 42:2342-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meshulam, T., S. M. Levitz, L. Christin, and R. D. Diamond. 1995. A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanilide (XTT). J. Infect. Dis. 172:1153-1156. [DOI] [PubMed] [Google Scholar]
- 41.Missall, T. A., M. E. Pusateri, and J. K. Lodge. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51:1447-1458. [DOI] [PubMed] [Google Scholar]
- 42.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 45.Paris, S., D. Wysong, J. P. Debeaupuis, K. Shibuya, B. Philippe, R. D. Diamond, and J. P. Latge. 2003. Catalases of Aspergillus fumigatus. Infect. Immun. 71:3551-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Philippe, B., O. Ibrahim-Granet, M. C. Prevost, M. A. Gougerot-Pocidalo, M. Sanchez Perez, A. Van der Meeren, and J. P. Latge. 2003. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect. Immun. 71:3034-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pocsi, I., M. Miskei, Z. Karanyi, T. Emri, P. Ayoubi, T. Pusztahelyi, G. Balla, and R. A. Prade. 2005. Comparison of gene expression signatures of diamide, H2O2 and menadione exposed Aspergillus nidulans cultures-linking genome-wide transcriptional changes to cellular physiology. BMC Genomics 6:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pocsi, I., R. A. Prade, and M. J. Penninckx. 2004. Glutathione, altruistic metabolite in fungi. Adv. Microb. Physiol. 49:1-76. [DOI] [PubMed] [Google Scholar]
- 49.Radsak, M. P., H. R. Salih, H. G. Rammensee, and H. Schild. 2004. Triggering receptor expressed on myeloid cells-1 in neutrophil inflammatory responses: differential regulation of activation and survival. J. Immunol. 172:4956-4963. [DOI] [PubMed] [Google Scholar]
- 50.Reeves, E. P., H. Lu, H. L. Jacobs, C. G. Messina, S. Bolsover, G. Gabella, E. O. Potma, A. Warley, J. Roes, and A. W. Segal. 2002. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416:291-297. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. Russel. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Schaffner, A., C. E. Davis, T. Schaffner, M. Markert, H. Douglas, and A. I. Braude. 1986. In vitro susceptibility of fungi to killing by neutrophil granulocytes discriminates between primary pathogenicity and opportunism. J. Clin. Investig. 78:511-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segal, A. W. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23:197-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spellig, T., A. Bottin, and R. Kahmann. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503-509. [DOI] [PubMed] [Google Scholar]
- 55.Thon, M., Q. Al-Abdallah, P. Hortschansky, and A. A. Brakhage. 2007. The thioredoxin system of the filamentous fungus Aspergillus nidulans: impact on development and oxidative stress response. J. Biol. Chem. 282:27259-27269. [DOI] [PubMed] [Google Scholar]
- 56.Tkalcevic, J., M. Novelli, M. Phylactides, J. P. Iredale, A. W. Segal, and J. Roes. 2000. Impaired immunity and enhanced resistance to endotoxin in the absence of neutrophil elastase and cathepsin G. Immunity 12:201-210. [DOI] [PubMed] [Google Scholar]
- 57.Toledano, M. B., A. Delaunay, B. Biteau, D. Spector, and D. Azevedo. 2003. Oxidative stress responses in yeast, p. 305-387. In S. Hohman and W. H. Mager (ed.), Yeast stress responses. Springer, Berlin, Germany.
- 58.Toone, W. M., B. A. Morgan, and N. Jones. 2001. Redox control of AP-1-like factors in yeast and beyond. Oncogene 20:2336-2346. [DOI] [PubMed] [Google Scholar]
- 59.Urban, C. F., U. Reichard, V. Brinkmann, and A. Zychlinsky. 2006. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 8:668-676. [DOI] [PubMed] [Google Scholar]
- 60.Wang, Y., Y. Y. Cao, X. M. Jia, Y. B. Cao, P. H. Gao, X. P. Fu, K. Ying, W. S. Chen, and Y. Y. Jiang. 2006. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic. Biol. Med. 40:1201-1209. [DOI] [PubMed] [Google Scholar]
- 61.Watson, F., J. Robinson, and S. W. Edwards. 1991. Protein kinase C-dependent and -independent activation of the NADPH oxidase of human neutrophils. J. Biol. Chem. 266:7432-7439. [PubMed] [Google Scholar]
- 62.Weeks, M. E., J. Sinclair, A. Butt, Y. L. Chung, J. L. Worthington, C. R. Wilkinson, J. Griffiths, N. Jones, M. D. Waterfield, and J. F. Timms. 2006. A parallel proteomic and metabolomic analysis of the hydrogen peroxide- and Sty1p-dependent stress response in Schizosaccharomyces pombe. Proteomics 6:2772-2796. [DOI] [PubMed] [Google Scholar]
- 63.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]
- 64.Yan, C., L. H. Lee, and L. I. Davis. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zarember, K. A., J. A. Sugui, Y. C. Chang, K. J. Kwon-Chung, and J. I. Gallin. 2007. Human polymorphonuclear leukocytes inhibit Aspergillus fumigatus conidial growth by lactoferrin-mediated iron depletion. J. Immunol. 178:6367-6373. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, X., M. De Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36:618-629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.