Abstract
The azole antifungal drugs are used to treat infections caused by Candida albicans and other fungi. These drugs interfere with the biosynthesis of ergosterol, the major sterol in fungal cells, by inhibiting an ergosterol biosynthetic enzyme, lanosterol 14 α-demethylase, encoded by the ERG11 gene. In vitro, these drugs as well as other ergosterol biosynthesis inhibitors increase ERG11 mRNA expression by activation of the ERG11 promoter. The signal for this activation most likely is the depletion of ergosterol, the end product of the pathway. To identify cis-acting regulatory elements that mediate this activation, ERG11 promoter fragments have been fused to the luciferase reporter gene from Renilla reniformis. Promoter deletions and linker scan mutations localized the region important for azole induction to a segment from bp −224 to −251 upstream of the start codon, specifically two 7-bp sequences separated by 13 bp. These sequences form an imperfect inverted repeat. The region is recognized by the transcription factor Upc2p and functions as an enhancer of transcription, as it can be placed upstream of a heterologous promoter in either direction, resulting in the azole induction of that promoter. The promoter constructs are not azole inducible in the upc2/upc2 homozygous deletion, demonstrating that Upc2p controls the azole induction of ERG11. These results identify an azole-responsive enhancer element (ARE) in the ERG11 promoter that is controlled by the Upc2p transcription factor. No other ARE is present in the promoter. Thus, this ARE and Upc2p are necessary and sufficient for azole induction of ERG11.
Candida albicans is a pathogenic yeast that causes oral, vaginal, and systemic infections (21). The current drugs used to treat candidiasis include the azoles and the polyenes, both of which target the fungal sterol ergosterol. The azoles, including imidazoles (clotrimazole [CLT], miconazole [MICA], and ketoconazole) and triazoles (fluconazole [FLC], itraconazole, and voriconazole), inhibit lanosterol 14 α-demethylase, encoded by the ERG11 gene in the ergosterol biosynthesis pathway (reviewed in reference 35). The polyenes bind to ergosterol in the fungal cell membrane and cause membrane damage. Two additional classes of antifungal drugs target other enzymes in the ergosterol pathway (35). The allylamines, such as terbinafine (TRB), inhibit the product of the ERG1 gene, which is upstream of ERG11. The morpholines, such as fenpropimorph (FEN), target the products of ERG24 and ERG2, both downstream of ERG11 (reviewed in reference 35).
Azole-resistant C. albicans and intrinsically azole-resistant species such as C. glabrata and C. krusei have emerged as serious problems in patients receiving antifungal therapy (reviewed in reference 25). Several molecular mechanisms of azole drug resistance have been identified in resistant C. albicans isolates, including overexpression of genes encoding the ATP binding cassette efflux pumps, CDR1 and CDR2 (1, 19, 20, 28, 33), and the gene encoding the major facilitator efflux pump, MDR1 (20, 28, 33). ERG11 overexpression also is associated with resistance (27, 34). In addition, point mutations and the loss of heterozygosity of ERG11 also may contribute to azole resistance (18, 34). The increased expression of several of these resistance genes has been shown to be regulated at the level of transcription by using nuclear run-on analyses (14). However, these analyses were not sensitive enough to monitor ERG11 transcription. Previous studies have shown that ERG11 transcript levels vary depending on the growth state of the culture in both Saccharomyces cerevisiae and C. albicans (9, 11, 14, 32) and that ERG11 levels respond to azole drugs (9, 30). Therefore, it is of interest to understand the regulation of expression from the ERG11 promoter.
In our previous study of transcriptional regulation of the ERG11 promoter, the full-length C. albicans ERG11 promoter was fused to the luciferase reporter gene (RLUC) from Renilla reniformis (30). RLUC has been used successfully for promoter analyses of such genes as CDR1, CDR2, MDR1, OP4, and WH11 in C. albicans (3, 7, 12, 13, 23, 31). When the ERG11 promoter was fused to RLUC, maximal induction under azole drug pressure was observed not during logarithmic growth but as cells approach stationary phase, probably because the cells have exhausted sterol stores, such as lipid droplets, at this time (30). This effect was observed for several azole drugs as well as for FEN and TRB, suggesting that it is related to ergosterol depletion more than any specific intermediate in the ergosterol pathway. The high levels of azole induction as cells approach stationary phase were observed only when the cells were grown under continuous drug pressure. In addition, the effects of pH, the carbon source, and oxygen levels were examined using the reporter gene (30). The ERG11 promoter activity was shown to parallel nascent sterol synthesis and to be inversely proportionate to total ergosterol levels. Overall, it appears that the ERG11 promoter is activated in response to sterol depletion in the cell, regardless of the mechanism of that depletion (30).
Recently, the C. albicans transcription factor Upc2p, which regulates sterol metabolism, has been identified and characterized (15, 29, 36) based on its homology to the known S. cerevisiae factors Upc2p and Ecm22p. Gene deletion of UPC2 in C. albicans results in significant increases in drug susceptibility to azoles and other ergosterol biosynthesis inhibitors (EBIs) (15, 29, 36). The Δupc2/Δupc2 homozygous deletion strain also shows reduced accumulation of radiolabeled cholesterol and reduced cellular ergosterol levels. Northern blot analysis of mRNA hybridized with several genes in the ergosterol pathway, including ERG11, demonstrated that azole induction of these genes did not occur in the Δupc2/Δupc2 strain (29). Finally, the DNA binding domain (DBD) of the C. albicans Upc2p was shown to bind to a region of the ERG2 gene containing a sequence with homology to the sterol response element (SRE) previously identified in S. cerevisiae (15).
The current analysis defines the region of the ERG11 promoter that is important for the azole regulation described above. This region has homology to the SRE from S. cerevisiae and is regulated by the transcription factor Upc2p, which has been implicated in sterol regulation in both S. cerevisiae and C. albicans. This suggests that Upc2p is an important transcription factor in the cell's response to azole drugs because of its role in the regulation of sterol metabolism.
MATERIALS AND METHODS
Strain maintenance and manipulation and chemicals.
Strains utilized in this study are summarized in Table 1. Strain CAI8 was the gift of W. A. Fonzi (4). Strains were maintained on YEPD (containing, per liter, 10 g of yeast extract, 20 g of peptone, 20 g of dextrose, with or without 15 g of Bacto agar) or on CSM-ade medium plus uridine (containing, per liter, 0.75 g CSM-ADE [Bio 101, Inc., Vista, CA], 1.7 g yeast nitrogen base without amino acids or ammonium sulfate, 5 g ammonium sulfate, 20 g dextrose, and 50 mg uridine). Strain BWP17 is a standard laboratory strain (37), and its Δupc2/Δupc2 derivative was detailed in Silver et al. (29) (Table 1). These strains were maintained on the appropriate CSM medium (Bio 101, Inc.) lacking marker amino acids as described above. When BWP17 and the Δupc2/Δupc2 strain were transformed with pCRW3 (31) that had been modified to contain the nourseothricin resistance marker SAT1 (pCRW3/SAT1), transformants were maintained on YEPD plus uridine plus nourseothricin (at 200 μg ml−1). Cells were stored at −80°C in selective medium supplemented with 10% glycerol. Overnight cultures were inoculated from a single colony from a selective agar plate into selective medium and grown overnight at 30°C, 180 rpm. These cultures then were used to inoculate cultures, usually at an optical density at 600 nm of 0.1 to 0.2, in CSM-ade medium plus uridine, with or without drug, and were grown for 48 h at 30°C and 180 rpm, after which luciferase was extracted. Medium components were obtained from Fisher Scientific (Pittsburgh, PA) or Bio 101, Inc. Chemicals were obtained from Fisher, Sigma (St. Louis, MO), or Aldrich (Milwaukee, WI).
TABLE 1.
Strains used in this study
| Designation | Archive no. | Parental strain | Relevant genotypea | Reference or source |
|---|---|---|---|---|
| SC5314 | TW00607 | Clinical isolate | Wild type | 6 |
| CAI8 | TW09007 | SC5314 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 | 4 |
| MDR1 | TW10072 | CAI8 | ADE2::(proMDR1[500]-RLUC)/ade2::hisG | 7 |
| FOR | TW13338 | CAI8 | ADE2::(proERG11[301-205] proMDR1[500]-RLUC)/ade2::hisG | This study |
| REV | TW13044 | CAI8 | ADE2::(proERG11[205-301] proMDR1[500]-RLUC)/ade2::hisG | This study |
| 1206 | TW04973 | CAI8 | ADE2::(proERG11[1206]-RLUC)/ade2::hisG | 30 |
| 806 | TW04975 | CAI8 | ADE2::(proERG11[806]-RLUC)/ade2::hisG | This study |
| 572 | TW04977 | CAI8 | ADE2::(proERG11[572]-RLUC)/ade2::hisG | This study |
| 419 | TW04978 | CAI8 | ADE2::(proERG11[419]-RLUC)/ade2::hisG | This study |
| 301 | TW04979 | CAI8 | ADE2::(proERG11[301]-RLUC)/ade2::hisG | This study |
| 276 | TW12905 | CAI8 | ADE2::(proERG11[276]-RLUC)/ade2::hisG | This study |
| 276-Ab | TW14054 | CAI8 | ADE2::(proERG11[276-A]-RLUC)/ade2::hisG | This study |
| 276-Bb | TW14051 | CAI8 | ADE2::(proERG11[276-B]-RLUC)/ade2::hisG | This study |
| 276-Cb | TW14049 | CAI8 | ADE2::(proERG11[276-C]-RLUC)/ade2::hisG | This study |
| 276-Db | TW14046 | CAI8 | ADE2::(proERG11[276-D]-RLUC)/ade2::hisG | This study |
| 276-Eb | TW14063 | CAI8 | ADE2::(proERG11[276-E]-RLUC)/ade2::hisG | This study |
| 276-Fb | TW15310 | CAI8 | ADE2::(proERG11[276-F]-RLUC)/ade2::hisG | This study |
| 276-ΔFb | TW14370 | CAI8 | ADE2::(proERG11[276-ΔF]-RLUC)/ade2::hisG | This study |
| 276-2Fb | TW12942 | CAI8 | ADE2::(proERG11[276-2F]-RLUC)/ade2::hisG | This study |
| 276-Gb | TW15319 | CAI8 | ADE2::(proERG11[276-G]-RLUC)/ade2::hisG | This study |
| 246 | TW12903 | CAI8 | ADE2::(proERG11[246]-RLUC)/ade2::hisG | This study |
| 217 | TW12901 | CAI8 | ADE2::(proERG11[217]-RLUC)/ade2::hisG | This study |
| 205 | TW04980 | CAI8 | ADE2::(proERG11[205]-RLUC)/ade2::hisG | This study |
| 99 | TW04970 | CAI8 | ADE2::(proERG11[99]-RLUC)/ade2::hisG | This study |
| BWP17 | TW14901 | SC5314 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 37 |
| Δupc2/Δupc2 | TW15343 | BWP17 | upc2::URA3/upc2::ARG4 | 29 |
| BWP17 276/ADE2/SAT1 | TW13004 | BWP17 | ADE2::(SAT1/proERG11[276]-RLUC)/ade2::hisG | This study |
| Δupc2/Δupc2 276/ADE2/SAT1 | TW13007 | Δupc2/Δupc2 | ADE2::(SAT1/proERG11[276]-RLUC)/ade2::hisG | This study |
Relative genotype is the genetic loci that differ from those of the parental strain.
The ERG11 promoter sequence for these strains is shown in Fig. 5A.
The drugs used for this study included FLC dissolved in water (stock concentration of 3.33 mg ml−1; Pfizer, Inc.), MICA in dimethyl sulfoxide (DMSO; stock concentration of 10 mg ml−1; Sigma), CLT in DMSO (stock concentration of 10 mg ml−1; Sigma), and FEN in DMSO (stock concentration of 10 mg ml−1; Sigma).
Constructs containing ERG11 promoter elements fused with RLUC.
Seventeen ERG11 promoter fragments of various lengths were PCR amplified using plasmid pCaERG1200 (30) as the template and the oligonucleotides listed in Table 2. Oligonucleotides were synthesized with PstI and KpnI sites on the 5′ ends (Sigma-Aldrich). Seven of the fragments were modified by altering the sequence of the oligonucleotide used to amplify the fragment (276A to 276G). Products then were cloned into pCR-TOPO2.1, transformed into Escherichia coli TOP10 cells (Invitrogen), and screened for inserts of the appropriate size by PstI and KpnI digestion. All ERG11 promoter fragments were excised from the pCR-TOPO2.1 vector with PstI and KpnI, gel purified, and ligated to the PstI and KpnI sites in the multiple-cloning site immediately upstream of RLUC in pCRW3. The plasmid pCRW3 was a gift of David Soll (31). Resulting plasmids containing inserts of the appropriate sizes were sequenced by an ABI automated sequencer using Taq dye terminator and primer chemistries (Applied Biosystems, Foster City, CA) in order to ensure the correct promoter sequence identity and fusion.
TABLE 2.
Oligonucleotides used in the analyses
| Namea | Sequenceb |
|---|---|
| Actin 50-mer | 5′ GATTTAGGTTTGGAAGCTGCTGGTATTGACCAAACCACTTTCAACTCC 3′ |
| Ade2-11545 | 5′ CAGTTAAATAGTCTTCATATC 3′ |
| Ade2-11487R | 5′ CGTTTACTTGTTTAATATGC 3′ |
| ERG1206 | 5′ GGCGGTACCAGGAAAATGAAAGGGAC 3′ |
| ERG806 | 5′ GGCGGTACCTTTCCATTCTGTTGTCTC 3′ |
| ERG572 | 5′ GGCGGTACCCAACAACAATAATAATGAAA 3′ |
| ERG419 | 5′ CCAGGTACCGCCACACGACAACTTTC 3′ |
| ERG301 | 5′ CCAGGTACCTGTGTGAGAATTTGATAAAG 3′ |
| ERG276 | 5′ AGTCGGTACCGAAATATTGGGTTTTGCTTGTATTCAATATCGTACCCG 3′ |
| ERG276-A | 5′ ATATGGTACCACCCGCTTGGGTTTTGCTTGTATTCAATATCGTACCCG 3′ |
| ERG276-B | 5′ ATCGGGTACCGAAATAGGAAAGTTTGCTTGTATTCAATATCGTACCCG 3′ |
| ERG276-C | 5′ ATCGGGTACCGAAATATTGGGTGGGATGTgTATTCAATATCGTACCCG 3′ |
| ERG276-D | 5′ ATCGGGTACCGAAATATTGGGTGGGATGGAGCGGCAATATCGTACCCG 3′ |
| ERG276-E | 5′ ATCGGGTACCGAAATATTGGGTTTTGCTTGTATTTCCGCGCGTACCCGATTATGTCG 3′ |
| ERG276-F | 5′ ATCGGGTACCGAAATATTGGGTTTTGCTTGTATTCAATATCGTACTTACGGCTGTCGTATATTCTTTTTTCAATGTCAATTTG 3′ |
| ERG276-2Fa | 5′ CAATATCGTACTTACGGCTTACGGCTGTCGTATATTC 3′ |
| ERG276-2Fb | 5′ GAATATACGACAGCCGTAAGCCGTAAGTACGATATTG 3′ |
| ERG276-ΔFa | 5′ GCTTGTATTCAATATCGTAC*TGTCGTATATTC 3′ |
| ERG276-ΔFb | 5′ GAATATACGACA*GTACGATATTGAATACAAGC 3′ |
| ERG276-G | 5′ ATCGGGTACCGAAATATTGGGTTTTGCTTGTATTCAATATCGTACCCGATTATGGTAGCGCTTCTTTTTTCAATGTCAATTTG3′ |
| ERG246 | 5′ AgtcGGTACCCGTACCCGATTATGTCG 3′ |
| ERG217 | 5′ AgtcGGTACCCAATGTCAATTTGAGAACG 3′ |
| ERG205 | 5′ CCAGGTACCGAGAACGAGAACGAAAAC 3′ |
| ERG99 | 5′ CCAGGTACCATAGACAAAGAAAGGGAAT 3′ |
| pSP73-2358 | 5′ CAGATTGTACTGAGAGTG 3′ |
| PstERG-3′ | 5′ ATACTGCAGATTGAGTTATGATCTTCTTG 3′ |
| RLUC | 5′ CACCACTGCGGACCAGTTATCATCCGTTTCC 3′ |
| 300-200a | 5′ ATCGATGAATTTGATAAAGAGAAAAAAGAAATATTGGGTTTTGCTTGTATTCAATATCGTAC 3′ |
| 300-200b | 5′ ATCGATCAAATTGACATTGAAAAAAGAATATACGACATAATCGGGTACGATATTGAATACAA 3′ |
| SAT1-5DR | 5′ AGTTGGTACCGTTTTCCCAGTCACGACG 3′ |
| SAT1-3DR | 5′ ATGTGGTACCTTGTGGATTGTGAGCGC 3′ |
ERG numbers refer to the position upstream of the ATG start codon of ERG11.
Appropriate restriction enzymes sites were included at the 5′ end of the oligonucleotides for the purpose of directional cloning of PCR products. The PstI site is italicized, and KpnI sites are underlined. Bases in boldface represent linker-scanning mutations introduced in those constructs. An asterisk indicates the location of the deletion of F nucleotides.
Two promoter constructs (ΔF and 2F) were prepared by site-directed mutagenesis of existing promoter elements using Stratagene QuikChange II with the oligonucleotides listed in Table 2 per the manufacturer's instructions. Fusions between the ERG11 promoter and the MDR1 promoter were prepared by PCR amplification and cloning of the bp 200 to 300 fragment from the ERG11 promoter (with oligonucleotides 300-200a and 300-200b) and inserting the fragment at the KpnI site of the MDR1 construct 500 (7).
Constructs containing the SAT1 selectable marker.
To clone the nourseothricin resistance gene SAT1 into the ERG11 constructs, PCR was performed using plasmid pA83 containing the SAT1 gene (24) (a gift of J. Morschhauser) with primers SAT1-5DR and SAT1-3DR (Table 2), which include KpnI sites at the 5′ end and which include homology to the 5DR and 3DR regions of pA83. The resulting fragments containing SAT1 were cloned into pCR-TOPO2.1, transformed into TOP10 cells, and screened for the presence of inserts of the appropriate sizes. The DNA fragment containing SAT1 was excised from the plasmid with KpnI, gel purified, and ligated at the KpnI site of ERG11 construct 276. Before ligation, KpnI-digested construct 276 was phosphatased using shrimp alkaline phosphatase by following the manufacturer's instructions (Promega, Madison, WI).
Candida transformants.
NsiI was used to linearize all pCRW3-based constructs within the ADE2 gene. The digested plasmids were phosphatased as described above to prevent end-to-end joining. Approximately 10 μg of digested and phosphatased plasmid DNA was added to aliquots of competent cells.
Transformation by electroporation was used for ADE2 selections. Electrocompetent cells were generated as described previously (7, 30), although all cells were freshly prepared. Conditions for electroporation were 1.6 kV, 25 μF, and 200 Ω. All transformations were conducted with a Bio-Rad Gene Pulser (Hercules, CA). One milliliter of 1 M sorbitol was added to the cuvette, and cells were plated onto the appropriate selective medium and grown at 30°C for 2 to 5 days.
Transformation by lithium acetate/heat shock (5) was used for SAT1 selection. This only reflects a general shift to lithium acetate/heat shock as the preferred transformation protocol in the laboratory. After heat shock, cells were pelleted and resuspended in 1 ml of YEPD in the absence of drug and were grown at 30°C and 180 rpm for 4 h. Cells then were plated on YEPD agar supplemented with nourseothricin (200 μg/ml).
Transformed colonies from either method then were restreaked three times on selective medium to ensure the presence of single colonies that retained the selectable marker. Each transformant was characterized by PCR and Southern blot analysis to ensure a single integration at the ADE2 locus.
PCR and Southern blot analysis.
Genomic DNA was prepared as previously described (10). For the PCR screens, pSP73-2358, Ade2-11545, and Ade2-11487R oligonucleotides (Table 2) were used to determine the correct gene integrations. PCR-positive transformants were further characterized by Southern blot analysis. Five milligrams of genomic DNA was digested separately with PstI, EcoRV, and XhoI (Promega) and was resolved by electrophoresis in 0.8% agarose gels. A 32P-kinase-labeled RLUC oligonucleotide was hybridized to Southern blots to determine the location and copy number of the inserted construct. Standard techniques were used for the Southern blot analyses (17, 26).
In vitro assay of RLUC activity.
Luciferase assays were performed as previously described (30). Specific activity was defined as the relative luminescence per 10 s per μg of protein extract. Protein concentrations were determined by the Bradford assay (Bio-Rad Laboratories). Azole induction (n-fold induction) is expressed as the ratio of luciferase activity in the presence of drug to the luciferase activity in the absence of drug.
Luciferase experiments were repeated at least three times. However, absolute levels of luciferase vary from experiment to experiment based on the activity of the stock solution freshly prepared for each experiment. As azole induction is a ratio of normalized values, standard deviation calculations among different experiments with different absolute levels are not valid. The data presented in the figures represent the results from representative experiments. Variation within a single experiment has been repeatedly determined by testing luciferase at the same time from three independent cultures prepared in parallel (7, 30). When these experiments are performed, the results differ by less than 10%, most often less than 5%.
RESULTS
Promoter deletion construction.
The intergenic region upstream of ERG11 (open reading frame [ORF] 19.922) and downstream of ORF 19.921 is 1,469 bp in length. Previous studies used a 1,206-bp fragment (originally referred to as CaErg1200, here termed the 1206 construct), which spans a significant part of this region, as a representation of the full-length ERG11 promoter (30). The 1206 construct and a full-length 1,469-bp construct behave similarly with regard to azole induction (data not shown).
To understand the transcriptional regulation of this promoter, nested deletions in the 1,206-bp region were constructed to include regions of the promoter 1,206, 805, 572, 419, 301, or 205 bp upstream of the start codon ATG. Each promoter region was amplified by PCR and cloned upstream of the RLUC reporter gene in plasmid pCRW3 containing the ADE2 gene. The plasmids were transformed into C. albicans strain CAI8 (Table 1), and ADE2 prototrophs were selected. Strains were screened for a single integration of the plasmid at the ADE2 locus using PCR and genomic Southern analysis. Strains with correct single integrations at the ADE2 locus were designated based on the length of the promoter fragment: 1,206, 805, 572, 419, 301, and 205 bp.
A 99-bp construct also was created, although the start of transcription previously has been localized at −127 to −129 (8). Subsequent analysis showed that the 99-bp construct had no significant promoter activity above that of the vector control, and it was not considered further.
Deletion analysis of basal transcription.
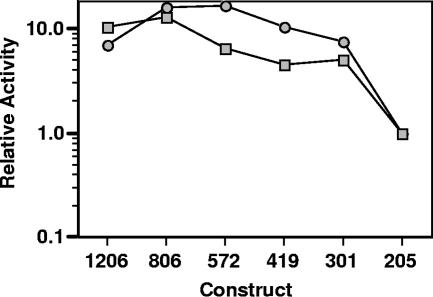
Previous work (30) detected expression from the 1206 promoter in logarithmic (6 h), late logarithmic (24 h), and postdiauxic (48 h) growth, while the most significant azole induction was observed during postdiauxic growth. The deletion constructs first were analyzed for basal transcription (Fig. 1). Activity was measured for each of the constructs during logarithmic growth and postdiauxic growth. In both growth conditions, the 205 construct demonstrated activity that was at least 20-fold above that of the empty vector control (data not shown). All other constructs, 301 to 1206, had significantly high levels of transcriptional activity. This suggests that the region between bp 205 and 301 upstream of the ATG is important for basal transcription.
FIG. 1.
Promoter deletion analyses of basal transcription. C. albicans strains containing the indicated luciferase deletion constructs were grown to mid-log-phase growth (optimal density of 1) (squares) and postdiauxic growth (48 h) (circles), and the relative specific activities were determined. The relative activity is the specific activity of each promoter construct divided by the specific activity of construct 205. The specific activity of the 205 construct was routinely 20-fold above that of the background or empty vector control, but the mid-log-phase and postdiauxic growth needs to be compared to that of one active construct, such as 205, rather than to that of the background. These results are from one representative experiment (see Materials and Methods).
Deletion analysis of azole induction.
In a previous work (30), azole induction of the full-length promoter was analyzed for timing and drug concentrations. It was shown that azole induction is maximal after 24 to 48 h, presumably as ergosterol is exhausted from the cells. Azole induction also is maximal above the MIC, which would be predicted as concentrations below the MIC do not significantly affect the growth of the cells. The induction conditions with the full-length promoter were used in an analysis of the deletion promoter constructs described above.
Azole induction was measured for each of the promoter constructs (Fig. 2). The results show that azoles induce the ERG11 promoter if the promoter includes at least 301 bp upstream of the ATG, and there is no obvious additional effect if larger regions of the promoter are included. The same pattern of induction is observed for three different azoles, FLC, MICA, and CLT. A similar pattern of induction also was observed for TRB (data not shown) and FEN (Fig. 2), even though they target the pathway up- and downstream of the azole target Erg11p. These results demonstrate that the region between bp 205 and 301 upstream of the ATG (termed the 205-301 region) is important for induction by azoles and other ergosterol biosynthesis inhibitors, probably as a result of sterol depletion. It is important to stress that azole induction is observed above the basal level of transcription of these promoter fragments (data not shown). As all azoles and other ergosterol biosynthesis inhibitors behave in the same way with the full-length ERG11 promoter (30) and with the promoter deletions (Fig. 2), further analyses of azole induction were studied using a single azole, MICA, which has the highest induction level of the azoles (and is readily available at an affordable price).
FIG. 2.
Promoter deletion analyses of azole induction. C. albicans constructs containing the indicated luciferase deletion constructs were grown in the presence and absence of EBIs for 48 h. EBIs include CLT and MICA (squares and circles, respectively; concentration, 10 μg ml−1) and FLC and FEN (diamonds and triangles, respectively; concentration, 100 μg ml−1). Induction (n-fold) is expressed as the ratio of luciferase activity in the presence of drug to the luciferase activity in the absence of drug for each construct. These results are from one representative experiment (see Materials and Methods).
The 205-301 region acts as an enhancer.
To determine if the 205-301 region of the promoter is a region important for basal transcription or if it is an enhancer of transcription, the 205-301 region was inserted upstream of a promoter that is transcriptionally active but not azole inducible. When the promoter of the MDR1 gene was fused to RLUC as a reporter, the promoter had basal activity and responded to benomyl but not to other drugs, such as the azoles (7). The 205-301 region from the ERG11 promoter was inserted in both orientations upstream of this 500-bp MDR1 promoter (Fig. 3A). The 500-bp MDR1 promoter alone was not affected by azoles, while the constructs containing the 301-205 region in either orientation were azole inducible (Fig. 3B). These data suggest that the 301-205 azole-inducible region can act as a transcriptional enhancer, since an enhancer upregulates transcription and has effects over long distances and in either orientation.
FIG. 3.
ERG11/MDR1 chimeric luciferase reporter gene constructs. (A) The non-azole-inducible MDR1 promoter (MDR1-500) (7) was inserted upstream of the luciferase reporter gene (RLUC). The azole-inducible ERG11 region, identified in Fig. 1 and located at bp 301 to 205 upstream of the start of translation (region 301-205), was ligated upstream of the MDR1 promoter in its native, forward orientation (FOR). It also was ligated upstream of the MDR1 promoter in the reverse orientation (REV). (B) ERG11/MDR1 chimeric and reverse chimeric strains were grown in the presence and absence of 10 μg ml−1 MICA for 48 h, and luciferase assays were performed. Induction (n-fold) is expressed as the ratio of luciferase activity in the presence of drug to the luciferase activity in the absence of drug for each construct. These results are from one representative experiment (see Materials and Methods).
Focused deletion analysis of azole induction.
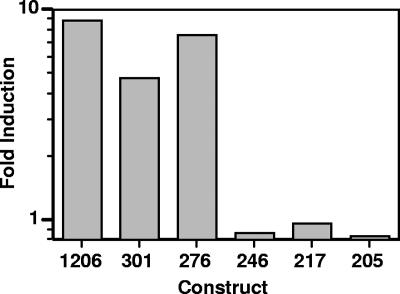
To refine the analysis of the 205-301 region as it relates to azole induction, additional deletion constructs were generated (276, 246, and 217 bp extending to the ATG; termed constructs 276, 246, and 217, respectively) and tested for azole induction (Fig. 4). The 246 and 217 constructs are not azole inducible, while the 276 construct is inducible at levels similar to that of the 301 or 1206 construct. This suggests that the region of the promoter important for azole induction is localized at or near the region between bp 246 and 276 (termed the 246-276 region).
FIG. 4.
Azole-inducible region of the ERG11 promoter is localized between bp 276 and 246 upstream of the translational start site. Strains containing the deletion constructs indicated were grown in the presence or absence of 10 μg ml−1 MICA for 48 h, and luciferase assays were performed. Induction (n-fold) is expressed as the ratio of luciferase activity in the presence of drug to the luciferase activity in the absence of drug for each construct. These results are from one representative experiment (see Materials and Methods).
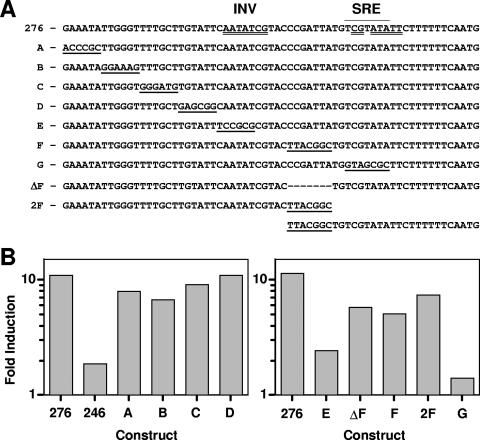
Scanning mutations in the 246-276 region.
Further definition of the 246-276 region was performed by constructing linker-scanning mutations of six bases, each using the following substitution rules: G to A, A to C, C to T, and T to G (Fig. 5A). Four 6-bp linker-scanning mutations, designated A to D, were analyzed for azole induction (Fig. 5B). These constructs all were azole inducible to levels similar to those of the 276 construct. A fifth construct, E (Fig. 5C), is significantly reduced in azole induction at levels approaching that of construct 246, suggesting that the azole-inducible region is localized to the 6-bp region at positions 252 to 247.
FIG. 5.
Scanning mutations across the azole-inducible region. (A) The region from bp 276 to 212 is shown. An SRE was identified by homology to Saccharomyces and is overlined (region 232-226). A potential inverted repeat sequence at the SRE also was identified (double-underlined sequence spanning region 251-224). The two halves of the inverted repeat include a region coincident with the SRE (region 231-224) and a second, inverted region (INV; region 251-245). Rows A to G indicate constructs with scanning mutations (underlined) not found in the wild-type promoter. ΔF has a deletion of 7 bp (indicated with dashes). 2F has a duplication of 7 bp. To align the sequences, the 2F construct is continued on a second line at the duplication. (B) Promoter constructs were grown in the absence or presence of MICA (10 μg ml−1) for 48 h, and luciferase assays were performed. Induction (n-fold) is expressed as the ratio of luciferase activity in the presence of drug to the luciferase activity in the absence of drug for each construct. These results are from one representative experiment (see Materials and Methods). Left panel, promoter constructs 276 and 246 and linker scan mutations A to D. Right panel, promoter construct 276 and linker scan mutations E, F, and G, and two mutations designed to test spacing. ΔF, deletion of the 7-bp mutation in F; 2F, duplication of the 7-bp mutation in F.
Azole induction linked to an SRE.
Previous work (15, 29) has shown that the Candida transcriptional regulator Upc2p is homologous to the Saccharomyces Upc2p factor. Upc2p in Saccharomyces recognizes an 11-bp recognition element with a 7-bp conserved core sequence, 5′ TCGTATA 3′. Silver et al. (29) located this 7-bp core at positions 232 to 226 in the C. albicans ERG11 promoter (Fig. 5A). This SRE and the E mutation (positions 252 to 247) that eliminates azole induction appear to form an imperfect inverted repeat. The E region with limited inverse homology to the SRE is designated INV. At least one group has hypothesized that the Saccharomyces Upc2p functions as a dimer (2). Therefore, it is hypothesized that the two regions of the ERG11 promoter, SRE and INV, are two halves of a dimer recognition sequence. Therefore, two additional linker mutations were constructed: construct F contained a 7-bp mutation centered between the SRE and INV, and construct G was a 7-bp mutation of the SRE (Fig. 5A). When tested for azole induction (Fig. 5C), construct G was not azole inducible, while construct F, which maintains the distance between the SRE and INV, showed significant azole induction. This adds support to the idea that the SRE and INV are two halves of a dimer recognition sequence and that the spacing, but not the sequence between the two recognition sites, is important for azole induction. Together, the SRE and INV halves of this DNA element appear to form an azole-responsive element (ARE).
To further characterize the spacing between the INV and SRE, two additional mutations were constructed. ΔF is a construct in which the 7 bp that was mutated in construct F was deleted. 2F is a construct in which the 7 bp that was mutated in construct F was duplicated. Both constructs ΔF and 2F show azole induction comparable to that of construct F (Fig. 5C), suggesting that the sequence between E and G is a spacer or filler between the SRE and INV sequences.
Effect of Δupc2/Δupc2 deletion on the ERG11 promoter.
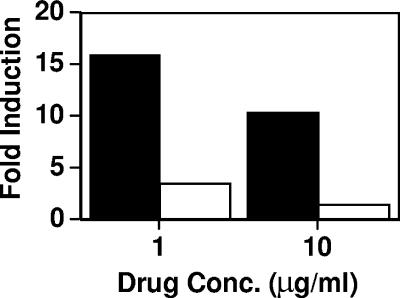
To demonstrate that the Upc2p transcription factor was responsible for azole induction of the ERG11 promoter at the ARE, the 276 fragment linked to the reporter gene was tested in the Δupc2/Δupc2 strain (29). To achieve this, a plasmid was created that contained (i) the 276 promoter fragment linked to the luciferase reporter gene, (ii) the ADE2 gene as a site for integration, and (iii) the nourseothricin resistance gene SAT1 as a selectable marker. The SAT1 selectable marker is required, as the Δupc2/Δupc2 strain is not an ade2 mutant auxotroph. The 276/ADE2/SAT1 construct was integrated at the ADE2 locus in the Δupc2/Δupc2 deletion strain and the parental strain, BWP17. Azole induction was observed for the parental strain at two different MICA drug concentrations, 1 and 10 μg/ml (Fig. 6). Two different drug concentrations were tested, as the Δupc2/Δupc2 deletion strain is 8- to 11-fold more susceptible to azoles than the parental strain. The Δupc2/Δupc2 deletion strain at either concentration has limited azole induction compared to that of the parental strain at either concentration. This is important, as the two strains differ in their azole susceptibilities.
FIG. 6.
Azole induction of the ERG11 promoter in Δupc2/Δupc2 mutant strains. The inducible 276-bp construct, tagged with the SAT1 marker, was introduced into wild-type BWP17 (black bars) and the upc2-homozygous gene deletion (white bars). Strains were grown in the presence or absence of 10 and 1.0 μg ml−1 MICA for 48 h, and luciferase assays were performed. Induction (n-fold) is expressed as the ratio of luciferase activity in the presence of drug to the luciferase activity in the absence of drug for each construct. These results are from one representative experiment (see Materials and Methods). Basal levels were essentially equivalent in the absence of drug for these strains.
DISCUSSION
This study has identified an ARE, a specific region of the ERG11 promoter that is necessary and sufficient for the upregulation of ERG11 transcription under drug pressure from azoles and other EBIs. This ARE can act as an enhancer when placed upstream of a non-azole-inducible promoter and is regulated by the transcription factor Upc2p.
ERG11 basal transcription showed similar overall activities in logarithmic and postdiauxic growth (Fig. 1). Basal promoter activity was observed for the 205 construct (20-fold above that of the vector control), but significantly more promoter activity was observed for constructs containing more than 301 bp of the promoter. Levels of azole induction are equivalent for constructs 301 to 1206, which contain the ARE (Fig. 2).
The ARE is induced by several EBIs, including azoles, FEN, and TRB (Fig. 2 and data not shown). This suggests that the induction of ERG11 at the ARE is mediated by a lack of ergosterol, the end product of the pathway. A lack of ergosterol is likely to activate Upc2p, and potentially other transcription factors, through unknown mechanisms (36). This activation will increase the expression of genes throughout the pathway, as has been observed for mRNA expression for several ERG genes (29). This is consistent with the Upc2p regulation of the ARE (Fig. 6).
The ARE acts as an enhancer, since it can be placed 500 bp upstream of a non-azole-inducible promoter in either orientation and can induce transcription of the promoter in the presence of azoles (Fig. 3). The azole induction is not the result of endogenous promoter activity from within the ARE, as it is observed in both orientations, and the 500-bp MDR1 promoter contains five potential ATG start sites that would interfere with the correct translation of luciferase if the mRNA initiated from within the ARE.
Small deletions and linker-scanning mutations were used to define the regions of the ARE (Fig. 4 and 5). Summarizing these results, the ARE is composed of two 7-bp regions (INV and SRE) of the ERG11 promoter separated by 13 bp. Construct 276F demonstrates that the region between the INV and SRE can be mutated without significant loss of azole induction. Constructs ΔF and 2F demonstrate that the F region is indeed a spacer that can be deleted or expanded without appreciable loss of azole induction. The alterations in spacing (±7 bp) will alter the face of the DNA to which Upc2p would bind. The fact that ΔF and 2F are both azole inducible argues that the transcription complex that recognizes INV and SRE is adaptable to significant changes in the spacer region.
The C. albicans ERG11 SRE is homologous to the core SRE as defined for Saccharomyces. However, the C. albicans ERG11 INV is not homologous to the SRE from Saccharomyces, as it is an imperfect inverted repeat, but the data from 276E and 246 clearly show that it is essential for azole induction. The requirement for both the INV and SRE suggests that the Upc2p transcription factor works as a dimer, although currently there is no definitive evidence for either Candida or Saccharomyces. Further work is necessary to clarify the role of the 27 bp of the ARE, including the INV, the SRE, and the spacer.
A similar ARE, with two elements separated by a spacer, may exist within the C. albicans ERG2 promoter. SREs have been identified in many ERG genes, either defined by homology to the 7-bp SRE core (29) or by similarity to the larger 11-bp sequence (15). In ERG2, one SRE at −217 to −210 is defined by both methods. A second SRE exists between positions −239 and −232. The two elements in ERG2 are in inverted orientation and are separated by 14 bp. The two ERG2 elements are not identical, differing by 1 bp within the 7-bp core. Thus, both ERG11 and ERG2 have a region that includes two SRE-like elements that differ in sequence, separated by 13 or 14 bp.
Previous work has identified additional SREs in other ERG genes (15, 29), although preliminary surveys have not identified other inverted repeats (data not shown). There are additional SREs farther upstream in ERG11 (15, 29), but there is no indication that these elements have an effect on the azole induction of the promoter (Fig. 2). It is clear that experiments are necessary to more clearly define SREs and INV elements in other ERG genes.
Electrophoretic mobility shift assays (EMSAs) are routinely used to identify interactions between proteins and DNA. EMSA has been used to show that the DBD of C. albicans Upc2p binds to three SREs within the C. albicans ERG2 promoter, including the −217 to −210 and −239 to −232 sequences (15) that make up the inverted repeat. The core region of the ERG2 −217 to −210 sequence is identical to the SRE of ERG11, so it is highly likely that the Upc2p DBD also will bind to the ERG11 SRE. Since this Upc2p DBD binds separately to each half of the ERG2 inverted repeat, EMSA analysis alone will not be sufficient to explain the requirement for both halves of the ERG11 inverted repeat in azole induction. One possibility is that a single Upc2p can bind to either half of the inverted repeat, but the dimer must bind to both elements to activate transcription. Further work is necessary to determine the interactions between the ARE, Upc2p, and other potential transcription factors.
The ARE is not functional in the Δupc2/Δupc2 strain at either of two drug concentrations. The use of two different drug concentrations is necessary, as the Δupc2/Δupc2 strain is 10-fold more susceptible to azole drugs (i.e., a lower MIC) than the wild-type strain (29). Azole induction is significantly reduced at both drug concentrations, suggesting that the reduction in promoter activity is not a pleiotrophic effect of strain susceptibility but a direct effect of the transcription factor on the promoter.
Based on the results of the present study and previous research, it is hypothesized that azole induction of ERG11 transcription is a response to the depletion of ergosterol or another downstream product of the ergosterol biosynthetic pathway. Cells require ergosterol when they are actively dividing during logarithmic growth; therefore, ergosterol is both being used by the cells and being inhibited by the azoles during growth in the presence of drugs. Growth in the presence of azoles does occur, as the azoles are fungistatic and inhibit growth but do not kill cells. The lack of ergosterol likely activates Upc2p, which in turn increases the expression of ERG11 and other genes. Attempts to abolish azole induction with exogenous ergosterol have been unsuccessful (data not shown), most likely because of an inability of Candida cells to import ergosterol under aerobic conditions, as has been observed similarly for S. cerevisiae (22).
Upc2p is a member of the Zn2Cys6 (class III) type of zinc finger proteins (16). Many Zn2Cys6 transcription factors recognize binding elements that include CGG or CCG. However, it is not unusual for some of these factors to bind unrelated sequences, such as the TCGTATA sequence that has been suggested for Upc2p in C. albicans and S. cerevisiae (16). Many Zn2Cys6 factors bind to promoters as dimers, either homo- or heterodimers binding to two elements, in an inverted, everted, or direct orientation. Thus, the SREs and INV elements that are present in ERG11 as inverted imperfect repeats are consistent with the binding of Upc2p as a homo- or heterodimer. The identity of the factor that binds to the INV sequence (Upc2p or another transcription factor) awaits further analyses. Finally, the spacing between the SRE and INV is not inconsistent with the spacing of other inverted repeats recognized by Zn2Cys6 factors (16).
This study has clearly defined the ARE in the ERG11 promoter that is directly responsible for the azole induction of the promoter. This element functions as an enhancer, as it can be placed upstream of other promoters to confer azole inducibility. The increased expression of the ERG11 promoter is dependent on the Upc2p transcription factor, and with data showing that the Upc2p DBD interacts with an identical SRE in ERG2 (15), it is likely that this is a direct effect. This study contributes to the basic understanding of the ERG11 promoter, which is paramount in understanding azole drug interactions and resistance in C. albicans.
Acknowledgments
We thank William Fonzi (Georgetown University, Washington, DC) for providing us with the CAI8 strain, David Soll (University of Iowa, Iowa City, IA) for the Renilla luciferase reporter RLUC pCRW3 plasmid, and Joachim Morschhauser for the nourseothricin-selectable marker SAT1 plasmid pA83. We thank members of the White laboratory for their valuable comments and support on the manuscript.
This research was funded by NIH NIDR grants R01 DE11367 and R01 DE14161. J.L.S. and B.G.O. were supported by NIH pathobiology training grant T32 AI 07509.
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowley, J. H., F. W. Leak, Jr., K. V. Shianna, S. Tove, and L. W. Parks. 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180:4177-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 4.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 6.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 7.Harry, J. B., B. G. Oliver, J. L. Song, P. M. Silver, J. T. Little, J. Choiniere, and T. C. White. 2005. Drug-induced regulation of the MDR1 promoter in Candida albicans. Antimicrob. Agents Chemother. 49:2785-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harry, J. B., J. L. Song, C. N. Lyons, and T. C. White. 2002. Transcription initiation of genes associated with azole resistance in Candida albicans. Med. Mycol. 40:73-81. [DOI] [PubMed] [Google Scholar]
- 9.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 11.Krishnamurthy, S., V. Gupta, R. Prasad, S. L. Panwar, and R. Prasad. 1998. Expression of CDR1, a multidrug resistance gene of Candida albicans: transcriptional activation by heat shock, drugs and human steroid hormones. FEMS Microbiol. Lett. 160:191-197. [DOI] [PubMed] [Google Scholar]
- 12.Kvaal, C. A., T. Srikantha, and D. R. Soll. 1997. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect. Immun. 65:4468-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lockhart, S. R., M. Nguyen, T. Srikantha, and D. R. Soll. 1998. A MADS box protein consensus binding site is necessary and sufficient for activation of the opaque-phase-specific gene OP4 of Candida albicans. J. Bacteriol. 180:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyons, C. N., and T. C. White. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacPherson, S., B. Akache, S. Weber, X. De Deken, M. Raymond, and B. Turcotte. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49:1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacPherson, S., M. Larochelle, and B. Turcotte. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70:583-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. S. Ramaekers, F. C. Odds, and H. Vanden Bossche. 1999. Contribution of mutations in the cytochrome P450 14 alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 10:2701-2713. [DOI] [PubMed] [Google Scholar]
- 19.Marr, K. A., C. N. Lyons, T. R. Rustad, R. A. Bowden, and T. C. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niimi, M., and R. Cannon. 1997. Multidrug resistance genes in Candida albicans. Jpn. J. Med. Mycol. 38:297-302. [Google Scholar]
- 21.Odds, F. C. 1988. Candida and candidosis: a review and bibliography. Bailliere Tindall, London, United Kingdom.
- 22.Parks, L. W., and W. M. Casey. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49:95-116. [DOI] [PubMed] [Google Scholar]
- 23.Puri, N., S. Krishnamurthy, S. Habib, S. E. Hasnain, S. K. Goswami, and K. Prasad. 1999. CDR1, a multidrug resistance gene from Candida albicans, contains multiple regulatory domains in its promoter and the distal AP-1 element mediates its induction by miconazole. FEMS Microbiol. Lett. 180:213-219. [DOI] [PubMed] [Google Scholar]
- 24.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 25.Rogers, T. R. 2006. Antifungal drug resistance: limited data, dramatic impact? Int. J. Antimicrob. Agents 27(Suppl. 1):7-11. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14 α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silver, P. M., B. G. Oliver, and T. C. White. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song, J. L., J. B. Harry, R. T. Eastman, B. G. Oliver, and T. C. White. 2004. The Candida albicans lanosterol 14-α-demethylase (ERG11) gene promoter is maximally induced after prolonged growth with antifungal drugs. Antimicrob. Agents Chemother. 48:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srikantha, T., A. Klapach, W. W. Lorenz, L. K. Tsai, L. A. Laughlin, J. A. Gorman, and D. R. Soll. 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 178:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turi, T. G., and J. C. Loper. 1992. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11). J. Biol. Chem. 267:2046-2056. [PubMed] [Google Scholar]
- 33.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14 α-demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White, T. C., and P. M. Silver. 2005. Regulation of sterol metabolism in Candida albicans by the UPC2 gene. Biochem. Soc. Trans. 33:1215-1218. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]