Abstract
The existence of specific respiratory supercomplexes in mitochondria of most organisms has gained much momentum. However, its functional significance is still poorly understood. The availability of many deletion mutants in complex I (NADH:ubiquinone oxidoreductase) of Neurospora crassa, distinctly affected in the assembly process, offers unique opportunities to analyze the biogenesis of respiratory supercomplexes. Herein, we describe the role of complex I in assembly of respiratory complexes and supercomplexes as suggested by blue and colorless native polyacrylamide gel electrophoresis and mass spectrometry analyses of mildly solubilized mitochondria from the wild type and eight deletion mutants. As an important refinement of the fungal respirasome model, we found that the standard respiratory chain of N. crassa comprises putative complex I dimers in addition to I-III-IV and III-IV supercomplexes. Three Neurospora mutants able to assemble a complete complex I, lacking only the disrupted subunit, have respiratory supercomplexes, in particular I-III-IV supercomplexes and complex I dimers, like the wild-type strain. Furthermore, we were able to detect the I-III-IV supercomplexes in the nuo51 mutant with no overall enzymatic activity, representing the first example of inactive respirasomes. In addition, III-IV supercomplexes were also present in strains lacking an assembled complex I, namely, in four membrane arm subunit mutants as well as in the peripheral arm nuo30.4 mutant. In membrane arm mutants, high-molecular-mass species of the 30.4-kDa peripheral arm subunit comigrating with III-IV supercomplexes and/or the prohibitin complex were detected. The data presented herein suggest that the biogenesis of complex I is linked with its assembly into supercomplexes.
The filamentous fungus Neurospora crassa, with a sequenced 40-Mb genome (31, 52), is an outstanding eukaryotic model for all aspects of the biosciences, with numerous molecular genetics tools available (19). So-called RIPing (repeat-induced point mutation), which is an effective defense against duplicated sequences, is commonly used to achieve gene disruption (8, 74). Overall, N. crassa has been vastly used in mitochondria research, such as studies of the protein import machinery (2, 43, 75).
The standard oxidative phosphorylation (OXPHOS) system is composed of the four major respiratory chain complexes and the FOF1-ATP synthase (complex V) and is responsible for most of the cellular ATP production. Research on complex I (NADH:ubiquinone oxidoreductase), the largest and most intricate OXPHOS component, has taken on greater significance since many human mitochondrial diseases involve structural and functional defects of this enzyme complex (reviewed in references 30 and 42). Contradicting the predominant view (35), the respirasome model was introduced based on the results by blue native polyacrylamide gel electrophoresis (BN-PAGE) of efficiently but mildly solubilized bovine heart mitochondria that led to the separation of high yields of stoichiometric respiratory supercomplexes and of ATP synthase dimers (67, 68). In line with the ancient “solid-state model” (13), this model postulates the quantitative assembly of the I, III, and IV respiratory complexes into two different supercomplexes, I1III2IV4 and III2IV4, occurring in a 2:1 ratio. Furthermore, the complexes I, III, and IV are present in a 1:3:6 stoichiometry (37, 68). In fact, the analysis of digitonin-solubilized mitochondria, isolated from fresh bovine heart, by a particular gentle colorless native PAGE (CN-PAGE), recovered nearly all of the I, III, and IV complexes as supercomplexes and most of the ATP synthases as dimers and higher oligomers (46, 48). Likewise, specific respiratory supercomplexes were detected by similar approaches in mitochondria of various eukaryotes (15, 26, 27, 45, 47, 61, 62, 64, 71, 94, 97, 98), including the filamentous fungus Podospora anserina (45), as well as in bacteria (79). Recently, the first single particle structures of respiratory supercomplexes were determined (24, 39, 66), and several other important studies by the bioenergetics/mitochondrial community have supported the respirasome model (1, 6, 10, 18, 21, 49, 56, 62, 85, 86, 88, 96). However, the functional significance of respiratory supercomplexes, including their enzymatic advantages, is still poorly documented. Since these supercomplexes represent assemblies of sequential enzymes, substrate channeling has been proposed (67), which is plausible in theory but difficult to prove experimentally (77). Accumulating evidence from the investigation of mutants and in human patients, with specific assembly defects of a single respiratory complex, suggests that complexes III and IV are involved to various extents in the assembly/stabilization of complex I in mammals (1, 7, 18, 21, 56, 71). In contrast, the absence of complex I in mitochondria of human patients (71) and plants (60, 62) was found not to reduce the abundance of the other OXPHOS complexes. However, site-directed mutations of the NDUFS2 (49-kDa subunit) and NDUFS4 (18-kDa AQDQ; homologue of the 21-kDa subunit of N. crassa) genes in human patients (85) and the gene encoding the 51-kDa subunit homologue in Caenorhabditis elegans (32, 33) were reported to reduce the amounts of both assembled complex I and of complex III (85) or complex IV (32, 33), respectively.
The molecular details of complex I biogenesis are far from elucidated. However, and in contrast to human complex I (3, 86, 91), a modular assembly pathway of the peripheral and membrane arms has been described in N. crassa (84). This is at least partially due to the existence of a comprehensive collection of complex I mutants in N. crassa (reviewed in references 55 and 89), which only exists because the fungus possesses several alternative NADH dehydrogenases (12, 55, 89). To unravel the molecular details underlying the formation of respiratory supercomplexes and their functional significance, we analyzed the steady-state supramolecular organization of the OXPHOS machinery of wild-type and eight complex I deletion mutants of N. crassa using BN- and CN-PAGE. For the first time, putative complex I dimers could be detected as components of the standard respiratory chain in a filamentous fungus. In addition, we provide evidence showing that I-III-IV supercomplexes have nonrespiratory relevance, since the inactive complex I of nuo51 is assembled into supercomplexes like those found in wild-type mitochondria as well as in the nuo21 and nuo29.9 mutants. Furthermore, digitonin-stable ATP synthase dimers and III-IV supercomplexes were also present in all strains studied. In two-dimensional (2D) BN-sodium dodecyl sulfate (SDS) gels of complex I deletion mutants lacking a membrane arm subunit, considerable amounts of total 30.4-kDa subunit comigrated with III-IV supercomplexes and/or the prohibitin complex, suggesting a putative interaction with them or other nonidentified assembly factors.
MATERIALS AND METHODS
Strains and isolation of crude mitochondria.
The wild type, 74-OR23-1A (74A), and eight mutant strains of N. crassa (nuo 9.8, nuo11.5, nuo14, nuo20.8, nuo21, nuo29.9, nuo30.4, and nuo51) were investigated in this study. Crude mitochondria of these strains were prepared as described elsewhere (93) using slight alterations. Mycelium was grown in 1× liquid Vogel's medium supplemented with 1.5% of sucrose for 16 to 18 h, whereas some mutant strains required 20 to 24 h of growth at 26°C with shaking. The mycelium was separated from the culture medium by filtration through a layer of cheesecloth. All steps described afterwards were done at 4°C. The mycelia were disrupted mechanically in a buffer containing 0.44 M sucrose, 30 mM Tris-HCl (pH 7.4), 2 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride. Then, cell debris, nuclei, and mycelia were removed by centrifuging twice at 2,000 × g for 5 min. The pellet was discarded, while the supernatant was centrifuged at 20,000 × g for 45 min. The crude mitochondrial pellet obtained was suspended in the same washing buffer and centrifuged at 31,000 × g for 20 min. Finally, the mitochondrial pellet was suspended in the same buffer. After isolation, the crude mitochondria were frozen as aliquots in liquid nitrogen and stored at −80°C until use.
Electrophoretic techniques.
Digitonin and n-dodecylmaltoside (DDM) were of a high-purity grade from Calbiochem Merck Biosciences GmbH, and Triton X-100 of high-purity grade was purchased from Roche Diagnostics GmbH.
Mitochondria were thawed on ice and centrifuged at 10,000 × g for 5 min. The pellet was suspended in the solubilization buffer containing 50 mM NaCl, 50 mM imidazole-HCl (pH 7.0), 10% glycerol, and 5 mM 6-aminocaproic acid (final concentration). Mitochondria from N. crassa strains were solubilized with digitonin using a detergent/protein ratio of 3.5 g/g or with Triton X-100 or DDM using a detergent/protein ratio of 1.5 to 3 g/g at a final detergent concentration of 1% by adding a freshly prepared 10% detergent solution. The samples were incubated for 30 min on ice followed by centrifugation at 20,800 × g for 10 min. Each lane was loaded with the extract from mitochondria containing 150 μg (for in-gel activity), 300 μg (for 2D SDS-PAGE), or 400 μg (for 2D BN-PAGE) of protein before solubilization which was assessed by Bradford assay. For BN-PAGE and CN-PAGE, linear 3 to 13% gradient gels overlaid with a 3% stacking gel were used in a Hoefer SE 600 system (18 by 16 by 0.15 cm3) (40, 69, 72). CN-PAGE was performed according to the methods of Schägger et al. (72), omitting Coomassie blue dye in the cathode buffer and without the addition of detergent in the gel (46, 61). The apparent molecular masses of the OXPHOS complexes and their supercomplexes were calibrated to digitonin-solubilized bovine heart mitochondria (67) applied to the same first-dimension BN gel as earlier described (45, 47, 64). The supercomplexes were assigned according to their subunit compositions and apparent molecular masses. Lanes from the first-dimension BN-PAGE or CN-PAGE were then excised and used for a second-dimension 13% SDS-PAGE (64) or, alternatively, for a second-dimension 3 to 20% BN-PAGE with the addition of 0.02% (wt/vol) DDM to the cathode buffer (67). Strips used for second-dimension SDS-PAGE were incubated in a solution containing 1% SDS and 1% mercaptoethanol for 2 h at room temperature. The first-dimension gels and 2D BN gels were Coomassie blue R-250 stained, whereas the 2D SDS gels were stained with silver nitrate (64) or SYPRO Ruby (Bio-Rad), respectively.
In-gel activity assays were performed as described elsewhere (45, 47, 48) using the addition of potassium cyanide as specific inhibitor to detect stained bands specific for complex IV, as well as comparative deamino-NADH dehydrogenase in-gel activity specific for complex I in parallel BN gels.
In-gel digestion, MALDI-TOF-MS, and bioinformatics.
Proteins were excised from second-dimension SDS gels and subjected to in-gel digestion by overnight trypsin (Promega) incubation at 37°C. Desalination was achieved using μ-C18 Zip-Tips (Millipore) by peptide elution from the tips onto a matrix-assisted laser desorption ionization (MALDI) sample target using 5 mg/ml 4-hydroxycinnamic acid in 50% acetonitrile-0.1% trifluoroacetic acid. Peptide samples were analyzed by MALDI-time of flight mass spectrometry (MALDI-TOF-MS) peptide mass fingerprint analysis (Applied Biosystems Voyager-DE PRO) as described elsewhere (64, 65).
The peptide mass lists were matched against the fungi database subset from the NCBInr database using the MASCOT search engine (www.matrixscience.com). The search included one possible missing cleavage site as well as possible methionine oxidation and cysteine derivatization by acrylamide (Cys-S-β-propionamide) with a maximal MS error tolerance of 30 to 90 ppm. The probability score calculated by the software was set to be higher than 58 (P < 0.05) as the criterion for correct identification. Furthermore, the spot position and its apparent mass in the second-dimension gel had to be in line with the calculated physical data of the protein found in the database and literature search. Prediction of transmembrane domains was according to the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM-2.0/), and the molar mass, pI, and the grand average of hydropathicity (GRAVY) were calculated using the Protparam tool (us.expasy.org/tools/protparam.html) as described elsewhere (64).
Western blotting technique.
After 2D BN-SDS-PAGE, proteins were electrotransferred onto a polyvinylidene difluoride membrane, purchased from Bio-Rad, using the semidry method. The Whatman paper was soaked in blotting buffer (anode, 300 mM Tris, 100 mM Tricine, pH ∼8.7; cathode, 300 mM 6-aminocaproic acid, 30 mM Tris, 0.05% SDS, pH ∼9.2). The membranes were preincubated in methanol and then in anode blotting buffer, and the gels were washed in cathode blotting buffer. 2D SDS gels were blotted for 2 h at maximal settings of 400 mA and 25 V. After transfer, membranes were treated according to the methods of Towbin et al. (83) and incubated overnight with polyclonal antibodies against the 30.4-, 20.8-, 14-, 11.5-, and 9.8-kDa subunits of N. crassa complex I. Proteins were detected with alkaline phosphatase-conjugated secondary antibodies.
RESULTS
Supramolecular organization of N. crassa wild-type respiratory chain complexes.
To analyze the role of complex I in the supramolecular organization of the OXPHOS system in N. crassa, proteins from crude wild-type mitochondria were solubilized with 3.5 g digitonin/g protein and further separated using 2D BN-SDS-PAGE and BN/BN-PAGE. These analyses revealed characteristic subunit patterns of protein complexes and the individual complexes that form the supercomplexes, respectively (Fig. 1A) in line with in-gel activity staining of BN gels like that of cytochrome c oxidase (Fig. 1A; see also below). In addition, 31 subunits of all five OXPHOS complexes in 2D BN-SDS gels were identified by MALDI-TOF-MS (Fig. 2; Table 1), corroborating the assignment of OXPHOS complexes and supercomplexes as described below.
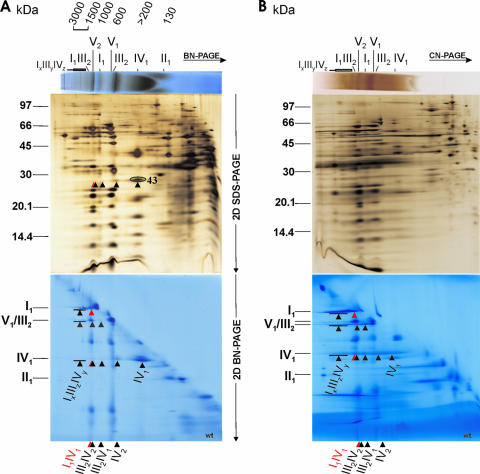
FIG. 1.
Respiratory supercomplexes and ATP synthase dimers in N. crassa wild-type mitochondria. (A and B) Digitonin-solubilized crude mitochondria were analyzed by BN-PAGE (A) and CN-PAGE (B) in the first dimension. In both panels, results are shown for in-gel activity of COX (upper panels) and subsequent 2D SDS-PAGE (silver stained) to resolve the subunits of all OXPHOS complexes and their supercomplexes (middle panels) and 2D BN-PAGE (Coomassie stained) with 0.02% DDM in the cathode buffer to dissociate the OXPHOS supercomplexes into their individual complexes (lower panels). For mass calibration, digitonin-solubilized bovine heart mitochondria were used: individual complexes I to V (130 to 1,000 kDa) and supercomplexes a to e (I1III2IV0-4; 1,500 to 2,300 kDa). Additionally, 31 subunits of all five OXPHOS complexes separated in 2D BN-SDS-PAGE (A) were verified by MALDI-MS, which are marked in detail in Fig. 2 for the sake of clarity. The OXPHOS supercomplexes were assigned according to their subunit compositions and apparent molecular masses. Besides ATP synthase monomers and dimers (V1 and V2), the individual respiratory complexes I to IV as well as the respiratory supercomplexes IxIIIyIVz, I1III2, I1IV1, III2IV2, III2IV1, and IV2 are indicated. Additionally, a subunit of complex IV (spot 43; Table 1) in the 2D SDS-PAGE (A) as well as the separated complex IV monomers, complex III dimers, and complex I monomers, which constitute the supercomplexes IV2, III2IV1, III2IV2, I1IV1, and IxIIIyIVz, in the 2D BN-PAGE (A and B) are marked by arrowheads. Note that supercomplexes I1IV1 and III2IV2 have very similar apparent masses. Similarly, the mobility of dimeric complex IV (IV2) is only slightly higher than that of complex III.
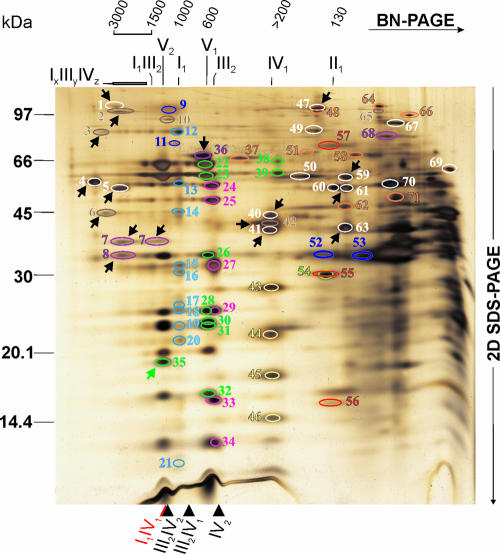
FIG. 2.
Identification of proteins in a representative 2D BN-SDS gel of digitonin-solubilized N. crassa wild-type crude mitochondria. The same gel as displayed in Fig. 1A is shown. The proteins identified by MALDI-TOF-MS peptide mass fingerprint are labeled by circles and numbers in different colors indicating their function, as follows: OXPHOS, complex I (light blue); complex II (red); complex III (pink); complex IV (yellow); complex V (green); tricarboxylic acid cycle and glycolysis (white); lipid metabolism (light green); amino acid metabolism and urea cycle (orange); chaperones (violet); transport and carrier proteins (dark blue); other proteins (brown). The identified proteins are listed by their numbers in Table 1. All proteins described in the text are highlighted by arrows. The other markings correspond to Fig. 1A and were done as described in the legend of Fig. 1.
TABLE 1.
Summary of proteins from 2D BN-SDS gels identified by MALDI-TOF-MS peptide mass fingerprinta
| Protein function (color), complex (color), and protein no. | Protein name or BLAST homology | Mass theor. (kDa) | pI theor. | TM pred. | GRAVY | gI protein | Accession no. | Gene locusb | Score | Mass error tolerance (ppm) |
|---|---|---|---|---|---|---|---|---|---|---|
| OXPHOS, complex I (light blue) | ||||||||||
| 12 | NADH-ubiquinone oxidoreductase 78-kDa subunit precursor | 81.6 | 6.05 | 0 | −0.330 | 85083792 | NCBI XP_957188 | NCU01765.2 | 198 | 30 |
| 13 | NADH-ubiquinone oxidoreductase 49-kDa subunit precursor | 54.0 | 6.26 | 0 | −0.265 | 85119577 | NCBI XP_965665 | NCU02534.2 | 128 | 30 |
| 14 | NADH-ubiquinone oxidoreductase 40-kDa subunit precursor | 42.9 | 6.62 | 0 | −0.445 | 85093948 | NCBI XP_959794 | NCU02373.2 | 202 | 30 |
| 15 | NADH-ubiquinone oxidoreductase 29.9-kDa subunit precursor | 30.9 | 5.12 | 0 | −0.558 | 85105954 | NCBI XP_962070 | NCU05299.2 | 45 | 60 |
| 16 | NADH-ubiquinone oxidoreductase 30.4-kDa subunit precursor | 32.2 | 8.78 | 0 | −0.534 | 85086366 | NCBI XP_957689 | NCU04074.2 | 111 | 30 |
| 17 | NADH-ubiquinone oxidoreductase 21.3-kDa subunit | 21.3 | 7.77 | 0 | −0.322 | 85099504 | NCBI XP_960797 | NCU08930.2 | 90 | 60 |
| 18 | Hypothetical protein, BLAST search: NADH dehydrogenase (ubiquinone) 21.3b | 21.3 | 9.69 | 3 | −0.213 | 85094628 | NCBI XP_959924 | NCU02280.2 | 86 | 30 |
| 19 | 19.3-kDa iron-sulfur subunit | 25.0 | 10.02 | 0 | −0.182 | 2764632 | EMBL CAA04802 | NCU03953.2 | 42 | 60 |
| 20 | NADH dehydrogenase, 20.9-kDa subunit | 21.0 | 9.40 | 1 | −0.280 | 3030 | EMBL CAA43221 | NCU01859.2 | 57 | 60 |
| 34 | NADH-ubiquinone oxidoreductase 10.5-kDa subunit | 10.5 | 9.46 | 0 | −0.509 | 2833211 | Swiss-prot Q07842 | NCU03156.2 | 51 | 90 |
| OXPHOS, complex II (red) | ||||||||||
| 55 | Hypothetical protein; BLAST search, succinate dehydrogenase iron-sulfur protein (Aspergillus fumigatus Af293) | 31.7 | 9.12 | 0 | −0.450 | 85116983 | NCBI XP_965152 | NCU00959.2 | 42 | 30 |
| 56 | Hypothetical protein; BLAST search, succinate dehydrogenase cytochrome b560 subunit (A. fumigatus Af293) | 19.3 | 10.38 | 3 | 0.464 | 85090713 | NCBI XP_958549 | NCU07756.2 | 58 | 60 |
| 57 | Hypothetical protein; BLAST search, succinate dehydrogenase flavoprotein subunit (A. fumigatus Af293) | 65.3c | 5.72 | 0 | −0.397 | 85117365 | NCBI XP_965239 | NCU08336.2 | 77 | 30 |
| OXPHOS, complex III (pink) | ||||||||||
| 24 | Ubiquinol-cytochrome c reductase complex core protein I | 52.5 | 5.63 | 0 | −0.309 | 127289 | Swiss-prot P11913 | NCU02549.2 | 70 | 30 |
| 25 | Ubiquinol-cytochrome c reductase complex core protein 2 precursor | 47.0 | 8.95 | 0 | 0.053 | 18376040 | EMBL CAD21046 | NCU03559.2 | 63 | 30 |
| 27 | Hypothetical protein; BLAST search, cytochrome c1, heme protein, mitochondrial precursor | 35.4 | 7.66 | 0 | −0.305 | 85111062 | NCBI XP_963756 | NCU09816.2 | 106 | 30 |
| 29 | Hypothetical protein; BLAST search, ubiquinol-cytochrome c reductase iron-sulfur subunit percursor (A. fumigatus Af293) | 24.8 | 8.84 | 0 | −0.065 | 85107305 | NCBI XP_962348 | NCU06606.2 | 64 | 60 |
| 33 | Hypothetical protein; BLAST search, probable ubiquinol-cytochrome c reductase complex 14-kDa protein (subunit VII) | 14.0 | 6.59 | 0 | −0.472 | 85099543 | NCBI XP_960807 | NCU08940.2 | 54 | 60 |
| 34 | Hypothetical protein; BLAST search, ubiquinol-cytochrome c reductase chain VIII | 11.8 | 9.89 | 0 | −0.466 | 85099572 | NCBI XP_960814 | NCU08947.2 | 50 | 30 |
| OXPHOS, complex IV (yellow) | ||||||||||
| 43 | cytochrome oxidase subunit 2 prepeptide | 28.7 | 4.44 | 2 | 0.317 | 7145096 | NCBI AAA31959 | 23 | 30 | |
| 44 | Hypothetical protein; BLAST search, cytochrome c subunit Vb (A. fumigatus Af293) | 20.6 | 5.90 | 0 | −0.551 | 85109713 | NCBI XP_963051 | NCU05689.2 | 59 | 30 |
| 45 | Cytochrome c oxidase polypeptide V precursor | 18.8 | 9.88 | 1 | −0.308 | 85110415 | NCBI XP_963448 | NCU05457.2 | 32 | 30 |
| 46 | Hypothetical protein; BLAST search, cytochrome c oxidase subunit Va (A. fumigatus Af293) | 16.8 | 5.79 | 0 | −0.404 | 85100681 | NCBI XP_961010 | NCU06695.2 | 86 | 60 |
| OXPHOS, complex V (green) | ||||||||||
| 22, 38 | ATP synthase alpha chain, mitochondrial precursor | 59.5 | 9.07 | 0 | −0.110 | 85119497 | NCBI XP_965645 | NCU02514.2 | 191 | 30 |
| 23, 39 | ATP synthase beta chain, mitochondrial precursor | 55.6 | 5.10 | 0 | −0.045 | 85074641 | NCBI XP_963253 | NCU05430.2 | 145 | 30 |
| 26 | Hypothetical protein; BLAST search, ATP synthase gamma chain, mitochondrial precursor, putative (A. fumigatus Af293) | 32.3 | 8.27 | 0 | −0.116 | 85091417 | NCBI XP_958891 | NCU09119.2 | 111 | 30 |
| 28 | Hypothetical protein; BLAST search, ATP synthase subunit 4, mitochondrial precursor (Saccharomyces cerevisiae) | 26.3 | 9.35 | 0 | −0.162 | 85112239 | NCBI XP_964306 | NCU00502.2 | 66 | 30 |
| 30 | Probable oligomycin sensitivity-conferring protein | 23.0 | 9.72 | 0 | 0.136 | 85080302 | NCBI XP_956517 | NCU01606.2 | 59 | 60 |
| 31 | Hypothetical protein; BLAST search, ATP synthase D chain, mitochondrial (A. fumigatus Af293) | 19.4 | 9.02 | 0 | −0.503 | 85074817 | NCBI XP_965776 | NCU00636.2 | 97 | 60 |
| 32 | Hypothetical protein; BLAST search, subunit h of the Fo sector (S. cerevisiae) | 13.6 | 4.79 | 0 | −0.456 | 85117672 | NCBI XP_965299 | NCU03199.2 | 40 | 30 |
| 35 | Hypothetical protein; BLAST search, F1Fo ATP synthase g subunit, putative (A. fumigatus Af293) | 22.8 | 10.58 | 0 | 0.037 | 85074833 | NCBI XP_965784 | NCU00644.2 | 80 | 30 |
| Glycolysis and TCA cycle (white) | ||||||||||
| 1, 47 | Oxoglutarate dehydrogenase precursor | 134.1 | 8.19 | 0 | −0.517 | 85074631 | NCBI XP_963248 | NCU05425.2 | 136 | 30 |
| 4 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (PDC-E2) (MRP3) | 48.6 | 6.23 | 0 | −0.196 | 85109166 | NCBI XP_962786 | NCU07659.2 | 78 | 60 |
| 5 | Hypothetical protein; BLAST search, 2-oxoglutarate dehydrogenase, E2 component, dihydrolipoamide succinyltransferase (A. fumigatus Af293) | 46.0 | 8.24 | 0 | −0.332 | 85092528 | NCBI XP_959443 | NCU02438.2 | 91 | 60 |
| 40 | Hypothetical protein; BLAST search, probable isocitrate dehydrogenase [NAD] subunit 1, mitochondrial precursor | 43.8 | 8.76 | 0 | −0.076 | 85115775 | NCBI XP_964931 | NCU00775.2 | 152 | 30 |
| 41 | Hypothetical protein; BLAST search, isocitrate dehydrogenase, NAD dependent (A. fumigatus Af293) | 41.0 | 6.97 | 0 | −0.046 | 85106968 | NCBI XP_962283 | NCU07697.2 | 117 | 30 |
| 49 | Glycerol-3-phosphate dehydrogenase precursor | 76.9 | 7.60 | 0 | −0.309 | 85110409 | NCBI XP_963445 | NCU05454.2 | 79 | 60 |
| 50 | Hypothetical protein; BLAST search, fumarate hydratase, putative (A. fumigatus Af293) | 57.2 | 8.83 | 0 | −0.153 | 85092553 | NCBI XP_959454 | NCU10008.2 | 111 | 30 |
| 59 | Hypothetical protein; BLAST search, dihydrolipoamide dehydrogenase (A. fumigatus Af293) | 56.5 | 7.64 | 0 | −0.078 | 85092766 | NCBI XP_959535 | NCU02407.2 | 107 | 30 |
| 60, 70 | Citrate synthase, mitochondrial | 52.0 | 8.10 | 0 | −0.244 | 85082342 | NCBI XP_956898 | NCU01692.2 | 147 | 30 |
| 61 | Hypothetical protein; BLAST search, pyruvate dehydrogenase complex alpha subunit (E1) putative (A. fumigatus Af293) | 46.1 | 8.49 | 0 | −0.362 | 85083464 | NCBI XP_957122 | NCU06482.2 | 132 | 30 |
| 63 | Probable pyruvate dehydrogenase beta chain precursor (E1) (PDB1) | 40.9 | 6.04 | 0 | 0.018 | 85118132 | NCBI XP_965390 | NCU03004.2 | 64 | 30 |
| 67 | Hypothetical protein; BLAST search, aconitate hydratase, mitochondrial (A. fumigatus Af293) | 85.0 | 6.22 | 0 | −0.335 | 85093919 | NCBI XP_959787 | NCU02366.2 | 152 | 30 |
| 69 | Hypothetical protein; BLAST search, d-lactate dehydrogenase, Dld2p (S. cerevisiae) | 60.8 | 6.12 | 0 | −0.333 | 85084552 | NCBI XP_957332 | NCU06441.2 | 79 | 60 |
| Lipid metabolism (light green) | ||||||||||
| 54 | Hypothetical protein; BLAST search, enoyl-coenzyme A hydratase/isomerase family protein (A. fumigatus Af293) | 32.2 | 9.24 | 0 | −0.134 | 85084582 | NCBI XP_957339 | NCU06448.2 | 96 | 30 |
| Amino acid metabolism and urea cycle (orange) | ||||||||||
| 37 | Hypothetical protein; BLAST search, δ-1-pyrroline-5-carboxylate dehydrogenase (A. fumigatus Af293) | 66.0 | 8.91 | 0 | −0.232 | 85111922 | NCBI XP_964169 | NCU03076.2 | 152 | 60 |
| 48 | Hypothetical protein; BLAST search, glycine dehydrogenase (A. fumigatus Af293) | 120.5 | 6.77 | 0 | −0.228 | 85116528 | NCBI XP_965069 | NCU02475.2 | 66 | 30 |
| 51 | Hypothetical protein; BLAST search, serine hydroxymethyltransferase (A. fumigatus Af293) | 57.5 | 9.08 | 0 | −0.375 | 85095341 | NCBI XP_960065 | NCU05805.2 | 86 | 30 |
| 58 | Hypothetical protein; BLAST search, mitochondrial dihydroxy acid dehydratase (A. fumigatus Af293) | 64.0 | 6.97 | 0 | −0.208 | 85090149 | NCBI XP_958280 | NCU04579.2 | 60 | 30 |
| 62 | Hypothetical protein; BLAST search, aspartate aminotransferase, putative (A. fumigatus Af293) | 47.2 | 9.01 | 0 | −0.201 | 85110141 | NCBI XP_963283 | NCU08411.2 | 66 | 30 |
| 64 | Hypothetical protein; BLAST search, carbamoyl-phosphate synthase large subunit (A. fumigatus Af293) | 128.3 | 5.65 | 0 | −0.168 | 85118469 | NCBI XP_965450 | NCU02677.2 | 125 | 30 |
| 66 | Hypothetical protein; BLAST search, aminopeptidase (peptide metabolism) (A. fumigatus Af293) | 101.4 | 5.16 | 0 | −0.311 | 85091989 | NCBI XP_959172 | NCU09228.2 | 49 | 60 |
| 71 | Hypothetical protein; BLAST search, glycine cleavage system T protein (A. fumigatus Af293) | 48.4 | 9.25 | 0 | −0.289 | 85116758 | NCBI XP_965112 | NCU02727.2 | 69 | 30 |
| Chaperones (violet) | ||||||||||
| 7 | Hypothetical protein; BLAST search, subunit Phb2p of the prohibitin complex (S. cerevisiae) | 34.1 | 9.73 | 0 | −0.309 | 85113233 | NCBI XP_964487 | NCU03310.2 | 60 | 30 |
| 8 | Hypothetical protein; BLAST search, probable prohibitin PHB1 (A. fumigatus Af293) | 30.3 | 9.07 | 0 | −0.107 | 85099568 | NCBI XP_960813 | NCU08946.2 | 149 | 30 |
| 36 | Probable heat shock protein Hsp60 | 60.5 | 5.60 | 0 | −0.062 | 85080225 | NCBI XP_956500 | NCU01589.2 | 130 | 30 |
| 68 | Hypothetical protein (AF401236) heat shock protein 70 kDa (Coccidioides immitis) | 56.2 | 5.09 | 0 | −0.389 | 85104522 | NCBI XP_961753 | NCU08693.1 | 49 | 30 |
| Transport and carrier proteins (dark blue) | ||||||||||
| 9 | Plasma membrane ATPase | 99.9 | 5.03 | 9 | 0.148 | 85082294 | NCBI XP_956886 | NCU01680.2 | 143 | 30 |
| 11 | H+-transporting ATPase, vacuolar, 67K chain | 67.1 | 5.32 | 0 | −0.282 | 18376302 | EMBL CAD21414 | NCU01207.2 | 97 | 30 |
| 52 | Outer mitochondrial membrane protein porin (VDAC) | 30.0 | 9.02 | 0 | −0.166 | 85100389 | NCBI XP_960950 | NCU04304.2 | 80 | 30 |
| 53 | ADP, ATP carrier protein (adenine nucleotide translocator) | 33.9 | 9.84 | 3 | 0.049 | 85110027 | NCBI XP_963201 | NCU09477.2 | 94 | 30 |
| Other proteins (brown) | ||||||||||
| 2 | Hypothetical protein (AF323912) matrix AAA protease MAP-1 | 103.0 | 7.21 | 1 | −0.538 | 85081618 | NCBI XP_956756 | NCU01479.2 | 77 | 30 |
| 3 | Hypothetical protein (AF323913) intermembrane space AAA protease IAP-1 | 80.1 | 8.98 | 1 | −0.242 | 85080016 | NCBI XP_956468 | NCU03359.2 | 60 | 90 |
| 6 | h Hypothetical protein; BLAST search, Homo sapiens stomatin (EPB72)-like 2 | 45.8 | 9.20 | 0 | −0.171 | 85095578 | NCBI XP_960112 | NCU05633.2 | 107 | 30 |
| 10 | Hypothetical protein; BLAST search, RNA12 protein (A. fumigatus Af293] | 99.9 | 9.17 | 0 | −0.292 | 85109974 | NCBI XP_963176 | NCU09598.2 | 80 | 60 |
| 42 | Mitochondrial import receptor subunit TOM40 | 38.1 | 5.65 | 0 | −0.201 | 85103565 | NCBI XP_961545 | NCU01179.2 | 95 | 30 |
| 65 | Hypothetical protein; BLAST search, pitrilysin family metalloprotease Cym1 (A. fumigatus Af293) | 112.8 | 5.84 | 0 | −0.398 | 85099301 | NCBI XP_960750 | NCU01272.2 | 108 | 30 |
The proteins are listed according to their functions and the numbers (colors) marked in Fig. 2. The theoretical molecular masses (Mr), pI, and GRAVY values were calculated from the amino acid sequences found in the NCBInr database without including possible processing, posttranslational modifications, and cofactors. The number of transmembrane helices (TM) was predicted by the TMHMM algorithm (http://www.cbs.dtu.dk/services/TMHMM/).
Mass calculated according to the N. crassa genome annotation.
In line with results from mammalian, plant, and P. anserina mitochondria (45-48, 67, 68), large amounts of large respiratory supercomplexes comprising complexes I, III, and IV (I1IV1, I1III2, and IxIIIyIVz) as well as the smaller ones (III2IV1 and III2IV2) were found (Fig. 1). The supercomplex I1IV1 has essentially the same apparent mass as dimeric ATP synthase (V2, ∼1,250 kDa), whereas the supercomplex III2IV2 runs slightly faster. The small supercomplex I1IV1, indicating a direct complex I-IV interaction, has only been previously found in bovine heart mitochondria by native electrophoresis (46, 48, 67). The two largest I-III-IV supercomplexes had mobilities similar to that of the ketoglutarate dehydrogenase complex (∼2,800 kDa) (Fig. 2; Table 1), suggesting compositions like I1III2IV5-6. Comparable supercomplexes with molecular masses up to 3,300 kDa were also observed in 2D BN-SDS gels of digitonin-solubilized mitochondria isolated from fresh bovine heart (40, 46, 48). About half of the total ATP synthase was recovered as dimers (V2), as for digitonin-solubilized Podospora wild-type mitochondria (45). Another observation was the reduced mobility of individual dimeric complex III (III2) migrating nearly at the same position as monomeric ATP synthase with an apparent molecular mass of ∼600 kDa, confirming the results described for Podospora (45). This is probably due to boundary lipids (45), known to be important for the stability and function of complex III (51). Similarly, the mobilities of individual monomeric complex IV (IV1) and its dimers (IV2) were slightly reduced compared to those of bovine heart, likewise due to tightly bound lipids (73), apparently removed by solubilization with Triton X-100 and DDM (see below).
Although at a slightly reduced resolution, the proportion of preserved supercomplexes could be enhanced with the gentler CN-PAGE method (Fig. 1B) as seen in other organisms (45, 46, 48).
Identification of protein complexes from wild-type mitochondria by MALDI-TOF-MS.
Overall, 2D BN-SDS-PAGE of mitochondrial detergent extracts followed by MALDI-TOF-MS enables a mitoproteome analysis of protein complexes (11, 40, 46, 64). Importantly, the simultaneous separation of various protein complexes (11, 40, 46, 64), particularly if most of the complexes are already known, mutually supports their physiological significance, arguing against artificial aggregation (64).
Through MALDI-TOF-MS analysis, 67 different proteins were identified, among which 31 subunits are components of the five OXPHOS complexes, in either individual complexes or supercomplexes. Interestingly, a protein spot that could only be detected as a subunit of the ATP synthase dimer but not of the monomer was identified as subunit g (Fig. 2, spot 35), which is also known to be dimer specific in yeast (4). This suggests a close structural relationship between yeast and filamentous fungal ATP synthase dimerization.
The 36 non-OXPHOS proteins identified represent a variety of functions, such as metabolic pathways, chaperones, and transporters, of which most are constituents of known complexes. Two such examples are the pyruvate dehydrogenase complex (∼8,000 to 10,000 kDa) and the ketoglutarate dehydrogenase complex (∼2,800 kDa), both mitochondrial multienzyme assemblies, as well as their subcomplexes, which have also been identified in mammalian mitochondria (40, 64, 69). The mitochondrial import receptor Tom40 (Fig. 2, spot 42) was also identified as a subunit of the TOM holo or core complex (2, 63, 75, 76, 82) and found to comigrate with monomeric complex IV and heterooligomeric NAD+-dependent isocitrate dehydrogenase (Fig. 2, spots 40 and 41). Tom40 is the main component of the TOM complex and essential for viability of N. crassa (2, 82).
In the chaperone category we found the heat shock protein 60 (Hsp60) (Fig. 2, spot 36), which is required for the assembly of proteins into oligomeric complexes and for the stabilization of preexisting proteins under stress conditions (14, 41). The apparent mass in the first dimension was ∼650 kDa, suggesting a tetradecameric assembly built by two stacked rings of seven monomers each, in contrast to the mammalian Hsp60 complex, which displays an apparent size of ∼420 kDa in BN gels (64). Another group of chaperones are the highly conserved prohibitins, which stabilize newly synthesized subunits of mitochondrial respiratory chain complexes (9, 57). The prohibitin complex (apparent mass of 1,200 kDa), comprising the two homologues Phb1 (Fig. 2, spot 8) and Phb2 (Fig. 2, spot 7), was detected as it has been for yeast (57), plant (38), nematode (5), and mammalian (57, 64) mitochondria. Additionally, we also observed that ≥50% of the prohibitins migrated as an even heavier complex with a molecular mass greater than 2,500 kDa. This large complex was visualized in BN gels of digitonin-solubilized yeast mitochondria and assigned as a supercomplex of the prohibitin complex with the m-AAA protease (81), which was previously detected by coimmunoprecipitation and gel filtration analysis (78). In fact, the only Neurospora homologue of the two yeast m-AAA protease subunits, MAP-1 (44), was found comigrating with this heavier prohibitin complex (Fig. 2, spot 2) and displayed the same band shape, which strongly suggests that this low-mobility species is a prohibitin/MAP-1 supercomplex.
In the inner mitochondrial membrane there are two AAA proteases, which form independent oligomeric complexes involved in quality control by degrading misfolded membrane proteins. The m-AAA protease (MAP-1) has its catalytic site facing the matrix, whereas that of the i-AAA protease is exposed to the mitochondrial intermembrane space. The activity of the m-AAA protease is modulated by the prohibitin complex, as suggested by their physical interaction (78, 81). The N. crassa homologue of the i-AAA protease, IAP-1 (Fig. 2, spot 3), was identified for the first time in a putative supercomplex with an apparent molecular mass of ∼4,000 to 6,000 kDa, much higher than that reported based on gel filtration (44). Supporting its high-molecular-weight organization, we identified a comigrating protein homologous to the human stomatin-like 2 protein (Fig. 2, spot 6), a mitochondrial inner membrane polypeptide (16) with an apparent mass of ∼1,800 kDa (64) which is widely expressed in different mammalian tissues (59, 92). Recently, an interaction of stomatin-like 2 protein with mitofusin 2 in HeLa cells was found by coimmunoprecipitation (36). As such, and based on their mobility in BN-PAGE, we suggest that IAP-1 and stomatin-like 2 protein form a large supercomplex in the inner membrane of N. crassa mitochondria.
Other interesting protein complexes (Fig. 2; Table 1), of which some are putative novel ones, will be described elsewhere.
Putative complex I dimers in N. crassa wild-type mitochondria.
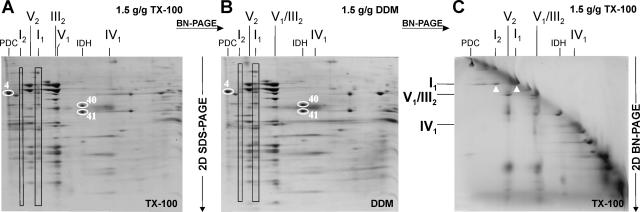
To complement the analysis of OXPHOS supercomplexes, we utilized the less-mild detergents Triton X-100 and DDM for solubilization of wild-type mitochondria; these are known to disrupt OXPHOS supercomplexes and other mitochondrial protein complexes more easily than digitonin (43, 46, 67). Interestingly, using either Triton X-100 or DDM at a detergent/protein ratio of 1.5 g/g, minor amounts of low-mobility species of complex I were observed, presumably containing no additional proteins (Fig. 3A and B), which was further supported by 2D BN/BN-PAGE (Fig. 3C). Its apparent molecular mass (∼1,900 kDa) was consistent with that of the dimeric complex I previously detected in COX-deficient strains of P. anserina (45). An increase of the detergent/protein ratio to 3 g/g resulted in the dissociation of the putative complex I dimers and the ATP synthase dimers into the respective monomers (not shown). In addition, ATP synthase monomers (complex V) migrate a little faster (Fig. 3A) or at the same position as complex III (Fig. 3B) when solubilized with Triton X-100 or DDM, respectively. Likewise, complex IV monomers migrated with higher mobility than those obtained upon digitonin solubilization (Fig. 1A, see above).
FIG. 3.
Putative complex I dimers in wild-type mitochondria after solubilization with Triton X-100 or DDM. 2D BN-SDS-PAGE of crude mitochondria solubilized with Triton X-100 (A) or DDM (B) at a detergent/protein ratio of 1.5 g/g. The subunits of monomeric complex I (I1) and putative complex I dimers (I2) are marked by boxes. One subunit of the pyruvate dehydrogenase complex (PDC; spot 4 of Table 1) and two subunits of the NAD+-dependent isocitrate dehydrogenase (IDH; spots 41 and 42 of Table 1) are indicated as in Fig. 2. (C) 2D BN/BN-PAGE of crude mitochondria solubilized with Triton X-100 at a detergent/protein ratio of 1.5 g/g. The separated complex I monomers of individual complex I (I1) and dimeric complex I (I2) are marked by arrowheads.
Comparative analysis of the OXPHOS system from eight complex I mutants by in-gel activity assays after BN-PAGE.
The availability of a nearly complete set of N. crassa complex I mutants (55, 89) offers a unique opportunity to investigate not only the assembly process of complex I but also its role in the formation of OXPHOS supercomplexes. We selected eight deletion mutants (nuo9.8, nuo11.5, nuo14, nuo20.8, nuo21, nuo29.9, nuo30.4, and nuo51), representing the whole range of the various reported defects in complex I biogenesis. Their mitochondrial detergent extracts were analyzed in the same manner as those of the wild type through native gel electrophoresis. The mutants nuo21, nuo29.9, and nuo51 are able to assemble an intact complex I lacking the respective peripheral arm subunit, although the nuo29.9 mutant assembles reduced amounts (approximately 20%) of complex I (23, 28, 29, 87). The fourth peripheral arm mutant used in this study, nuo30.4, cannot assemble a peripheral arm and accumulates membrane arm intermediates (22). The other four mutants investigated (nuo9.8, nuo11.5, nuo14, and nuo20.8) each lack a membrane arm subunit of complex I and are able to assemble the peripheral arm and fragments of the membrane arm (17, 54, 55).
For direct comparison of the OXPHOS system in the wild type and the eight mutants, their digitonin-solubilized mitochondrial proteins were separated by BN-PAGE and probed for in-gel activity of NADH dehydrogenase (complex I), cytochrome c oxidase (complex IV), ATP hydrolase (complex V), and succinate dehydrogenase (complex II), giving a qualitative indication of these four OXPHOS complexes (Fig. 4). The NADH dehydrogenase (NADH-DH) bands attributed to complex I were assigned by comparative gels tested for deamino-NADH dehydrogenase activity specific for complex I (not shown). The specificity of cytochrome c oxidase activity of complex IV was confirmed by inhibition with cyanide in parallel experiments (not shown).
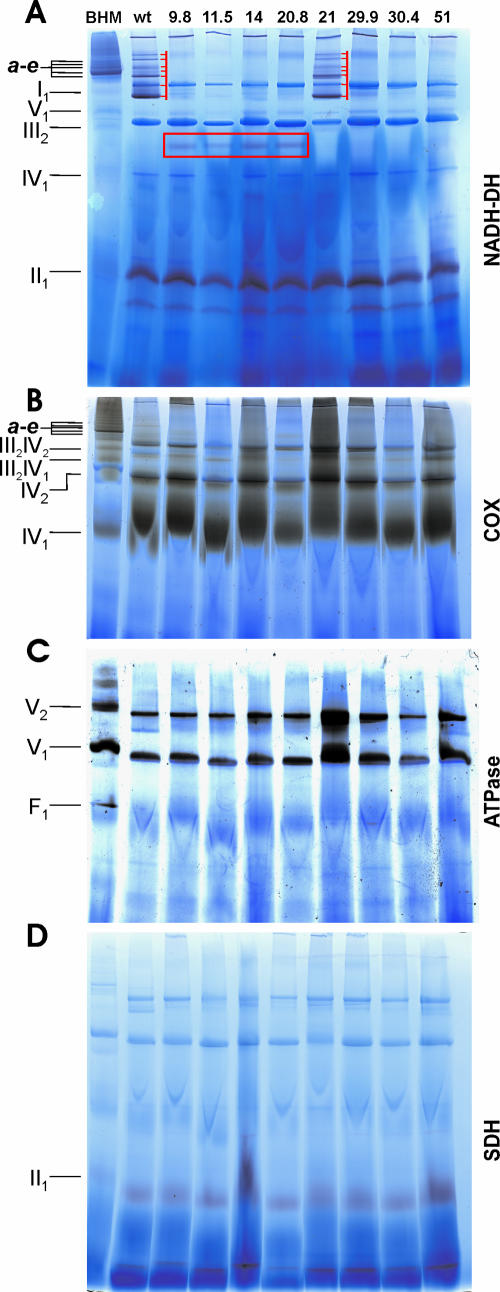
FIG. 4.
In-gel activity assays of NADH dehydrogenase, cytochrome c oxidase, ATP hydrolase, and succinate dehydrogenase. BN-PAGE of digitonin-solubilized mitochondria from bovine heart (BHM) as a con-trol, N. crassa wild type (wt), and eight complex I deletion mutants (nuo9.8, nuo11.5, nuo14, nuo20.8, nuo21, nuo29.9, nuo30.4, and nuo51). (A) NADH dehydrogenase (purple bands), reactive bands of wt and nuo21 corresponding to complex I are marked by red bars, and the red box indicates the active band of the peripheral arm of complex I from the four membrane arm mutants. (B to D) Cytochrome c oxidase (brown-yellowish bands) (B), ATP hydrolase (black-white bands) (C), and succinate dehydrogenase (purple band) (C). In all panels, some OXPHOS complexes and supercomplexes, like a to e (I1III2IV0-4) of bovine heart mitochondria, are marked on the left side.
All nine strains contained similar amounts of complex V (ATP synthase) in about an equal proportion (∼50%) of monomers and dimers (Fig. 4C), as well as of active complex IV monomers (Fig. 4B) and complex II (Fig. 4D). Furthermore, in all strains two prominent low-molecular-weight bands with NADH-DH activity were visualized which were not detected in bovine heart mitochondria. Additionally, these two bands were almost undetectable when deamino-NADH was used instead of NADH, suggesting that they may correspond either to alternative NADH dehydrogenases or to non-OXPHOS enzymes (Fig. 4A). The differences in the staining intensities are partially due to deviations in the loaded amounts.
In the wild type, the in-gel NADH-DH activity staining revealed bands corresponding to individual complex I as well as to its supercomplexes with complex III and IV, like I1III2, I1IV1, and I1III2IVX (Fig. 4A). In addition, monomeric complex IV, its dimer (IV2), and the smaller supercomplexes III2IV1 and III2IV2, as well as complex I supercomplexes containing one or more copies of complex IV, displayed cyanide-sensitive COX activity in the wild type (Fig. 4B). The same pattern of active supercomplexes containing complexes I and/or IV found in the wild type was also detected in the mutant nuo21 (Fig. 4A and B), indicating that the loss of the 21-kDa subunit does not impair the formation and stability of III-IV and I-III-IV supercomplexes.
In contrast, the nuo51 mutant displayed no complex I NADH-DH activity (Fig. 4A) but rather a wild-type-like pattern of high-molecular-weight species with COX activity (Fig. 4B), suggesting that these active bands correspond to the III-IV and I-III-IV supercomplexes observed in the wild-type and mutant nuo21 strains. In fact, this mutant strain does not possess any rotenone-sensitive NADH:ubiquinone oxidoreductase activity of complex I, due to the absence of the highly conserved 51-kDa subunit that binds NADH (28).
The mutant nuo29.9 showed no NADH-DH activity related to complex I, possibly due to assay sensitivity (Fig. 4A). Moreso, hardly detectable high-molecular-weight COX bands at the positions of the wild-type I-III-IV supercomplexes were noticed, whereas the bands of the small supercomplexes III2IV1 and III2IV2 were clearly identified after COX activity staining (Fig. 4B).
Strikingly, the four membrane arm mutant strains, nuo9.8, nuo11.5, nuo14, and nuo20.8, displayed a specific single band with both NADH-DH and deamino-NADH-DH activities with an apparent molecular mass of ∼450 kDa that seemingly corresponded to the peripheral arm domain (Fig. 4A). Accordingly, the NADH-DH activity 450-kDa band could not be detected in either the nuo30.4 mutant or the wild-type and nuo21 strains (Fig. 4A). The four membrane arm mutants displayed both monomeric complex IV and high-molecular-weight complexes with cyanide-sensitive COX activity which corresponded to IV2, III2IV1, and III2IV2 as found in the other strains and, in addition, some weakly stained bands with an apparent mass comparable to those of the wild-type complex I supercomplexes (Fig. 4D). Overall, the absence of an assembled complex I does not influence the assembly of the other four OXPHOS complexes.
Respiratory supercomplexes are assembled even with an inactive complex I.
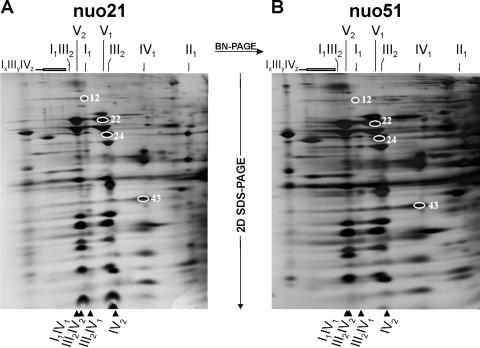
We further investigated the supramolecular organization of the OXPHOS complexes in the mutants nuo21, nuo29.9, and nuo51 by 2D BN-SDS-PAGE. Supporting the results obtained by the in-gel activity assays (Fig. 4), the second-dimension gels of nuo21 (Fig. 5A) and nuo 51 (Fig. 5B) mitochondria revealed a wild-type-like distribution of individual OXPHOS complexes and supercomplexes, in particular III2IV1, III2IV2, and I-III-IV supercomplexes.
FIG. 5.
Wild-type-like respiratory supercomplexes in peripheral arm mutants nuo21 and nuo51. The nuo51 mutant lacking the 51-kDa subunit forms stable respirasomes containing complexes I, III, and IV without NADH oxidase activity. Results shown are from 2D BN-SDS-PAGE of digitonin-solubilized crude mitochondria of nuo21 (A) and nuo51 (B). The markings are according to the legend of Fig. 1. Additionally, each one subunit of complex I (spot 12 in Table 1), complex III (spot 24 in Table 1), complex IV (spot 43 in Table 1), and complex V (spot 22 in Table 1) are indicated as described for Fig. 2.
The wild-type-like I-III-IV supercomplexes in the nuo51 mutant are the first examples of respirasomes without overall enzymatic activity (NADH oxidase, I-III-IV) only able to oxidize ubiquinol by molecular oxygen (ubiquinol oxidase, III-IV). This finding is the first direct demonstration that the assembly of the complexes I, III, and IV into a supercomplex is independent of complex I activity and does not represent a transient-like interaction required for efficient electron transfer from complex I to III. In fact, the biogenesis of supercomplexes is an elaborate and energy-consuming process, and complexes III and IV of these “inactive respirasomes” rely on alternative NADH dehydrogenases or FADH2-linked enzymes to deliver electrons to ubiquinol to be operable. Taken together, these results suggest that it is conceivable that III-IV supercomplexes play a role in the assembly/stability of complex I.
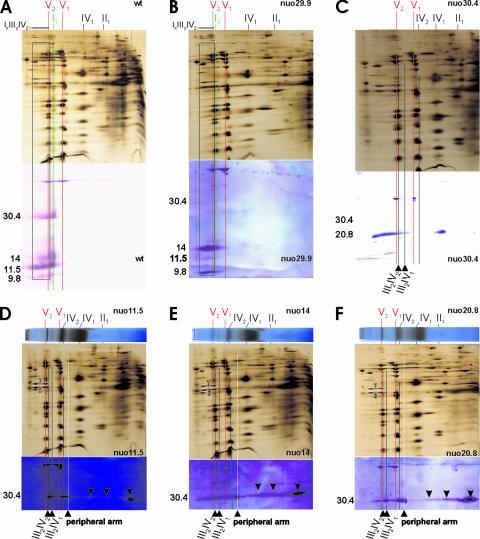
High-molecular-mass species of the 30.4-kDa subunit comigrating with III-IV supercomplexes and the prohibitin complex in the membrane arm mutants.
To achieve a sensitive detection of any complex I species in mutants nuo9.8, nuo11.5, nuo14, nuo20.8, nuo30.4, and nuo29.9, we used immunoblots of 2D BN-SDS gels from digitonin-solubilized mitochondria probed with a set of polyclonal antibodies each raised against a particular subunit of complex I from N. crassa. We used antibodies against the 30.4-kDa subunit of the peripheral arm and either the 9.8-kDa, the 11.5-kDa, the 14-kDa, or the 20.8-kDa subunits of the membrane arm. In particular, the screening for the 30.4-kDa subunit was performed due to its crucial role in the assembly of the peripheral arm and thus of intact complex I (22, 86, 90).
In wild-type mitochondria, the corresponding subunits of individual complex I as well as its supercomplexes were decorated with the four employed antibodies, indicating the presence of fully assembled complex I (Fig. 6A, lower panel). Note that the 30.4-kDa subunit of individual complex I was also identified by MALDI-TOF-MS (Fig. 2 and Table 1). The only other significant signals arose from a cross-reaction with the β-subunit of both monomeric and dimeric ATP synthase (complex V), which could actually be used as a helpful marker to align the blots with parallel silver-stained 2D SDS gels (Fig. 6A to F) and 1D BN gels probed for in-gel COX activity (Fig. 6D to F, upper panels). The corresponding immunoblot of nuo29.9 mitochondria (Fig. 6B, lower panel) displayed a result similar to that of the wild type (Fig. 6A, lower panel). This corroborates the presence of low amounts of assembled complex I separated as individual complex I and its supercomplexes also detected in silver-stained 2D gels (Fig. 6B, lower panel). Notably, the signal of the 30.4-kDa subunit was rather weak, since this protein, located in close vicinity to the 29.9-kDa subunit, is strongly reduced in the mutant nuo29.9 (87). No subcomplexes of complex I could be immunodetected, indicating that in the nuo29.9 mutant essentially all of the assembled peripheral arm joins with the membrane arm, occurring in surplus quantity, to form “complete” complex I. Additionally, it is likely that the excess hydrophobic membrane arm forms low-mobility membrane aggregates that contribute to the immunosignals in Fig. 6B (see below).
FIG. 6.
High-molecular-mass complexes of the 30.4-kDa subunit in complex I-deficient mutants each lacking a membrane arm subunit. (A to F) Alignment between silver-stained 2D BN-SDS gels and corresponding immunoblots probed with antibodies against complex I subunits, as well as corresponding 1D BN gel strips tested for COX in-gel activity from digitonin-solubilized crude mitochondria of (A) wild type (wt) (A), nuo29.9 (B), nuo30.4 (C), nuo11.5 (D), nuo14 (E), and nuo20.8 (F). The employed antibodies against the 30.4-kDa subunit of the peripheral arm and the 9.8-, 11.5-, 14-, and 20.8-kDa subunits of the membrane arm are indicated on the left side of the immunoblots. The subunits of ATP synthase monomers (V1) and dimers (V2), complex IV dimers, and supercomplexes III2IV1 and III2IV2, as well as of complex I and its peripheral arm are indicated by red, black, green, and white continuous vertical lines. (A and B) The subunits of supercomplexes of complex I (IxIIIyIVz, inclusively I1III2 and I1IV1) are marked by boxes. (D to F) The membrane arm mutants display distinct high-molecular-mass species of the 30.4-kDa subunit which contain none of the tested membrane subunits and comigrate with supercomplexes IV2, III2IV1, and III2IV2 as well as the prohibitin complex, whose subunits (spots 7 and 8 of Table 1) are encircled. Furthermore, the 30.4-kDa subunit of the peripheral arm and at least three distinct subcomplexes containing the 30.4-kDa subunit marked by arrows were immunodetected.
In the nuo30.4 mutant, a smear-like panel of high-molecular-mass species of the 20.8-kDa subunit was immunodetected (Fig. 6C, lower panel), where the most intense ones were near the top of the first-dimension gel. Such high-molecular-weight species of membrane subunits were previously observed after density gradient centrifugation of Triton X-100 extracts from mitochondria of peripheral arm mutants like nuo30.4 (22), but also from nuo29.9 (87), which generates a surplus of the membrane assembly intermediate (84) as mentioned above. Most probably, the largest 20.8-kDa species represented unspecific aggregates of partially assembled hydrophobic membrane subunits, as previously suggested (22, 23). However, it cannot be ruled out that there might be assembly intermediates of the membrane arm bound to “assembly factors” among those high-molecular-weight species.
The 2D BN-SDS gels and the corresponding representative immunoblots of the investigated membrane arm mutants were remarkably consistent. In particular, the membrane arm mutants nuo11.5 (Fig. 6D), nuo14 (Fig. 6E), nuo20.8 (Fig. 6F), and nuo9.8 (not shown) revealed essentially the same pattern for the 30.4-kDa subunit, as parts of complexes with apparent masses from ∼50 kDa to at least 1,500 kDa. The 450-kDa subcomplex detected by in-gel NADH-DH staining (Fig. 4A) represented the peripheral arm, whose constituents, like the 78-kDa, 49-kDa, and 40-kDa subunits, were discernible as weakly stained spots in the 2D BN-SDS gels (Fig. 6D to F). The peripheral arm could also be separated by BN-PAGE of membrane arm mutants after solubilization with DDM (53). Due to tiny amounts of assembled peripheral arm in the mutant nuo11.5, no peripheral arm could be detected in the first report, in which we used less-sensitive approaches (55). It is unlikely that the subcomplexes smaller than the peripheral arm originate from the breakdown of the assembled peripheral arm induced by the experimental conditions. In fact, their mobilities correspond to assembly intermediates of the 30-kDa subunit (the mammalian homologue of the 30.4-kDa subunit) and/or the free 30-kDa subunit found in human mitochondria, which appears to represent an early indispensable assembly step of the peripheral arm (86, 90).
Due to the fact that the 30.4-kDa subunit was separated to a large extent as part of high-mobility species, it is unlikely that the distinct low-mobility species of the rather hydrophilic 30.4-kDa subunit arose from any aberrant aggregation triggered by the experimental conditions or in vivo. Indeed, these immunosignals represent distinct high-molecular-weight complexes which comigrated with the supercomplexes IV2, III2IV1, and III2IV2 and the prohibitin complex with apparent masses of ∼550 up to ∼1,250 kDa, as indicated in Fig. 6D to F. In nuo14 and nuo20.8 mutants, even larger complexes up to about 2,000 kDa of the 30.4-kDa subunit were found comigrating with III-IV supercomplexes which display COX in-gel activity (Fig. 6E and F). Taken together, these high-molecular-weight complexes seem to be assembly intermediates of the peripheral arm which likely function in assisting the biogenesis of the peripheral arm and/or the whole complex I. Nonetheless, the comigration of proteins in the first-dimension BN-PAGE with consistent band shapes in the second-dimension SDS gel is a necessary precondition to propose an interaction between them (46, 72) and, thus, III-IV supercomplexes and/or the prohibitin complex, respectively, are conceivable constituents of the various large 30.4-kDa species. If subcomplexes of complex I containing the 30.4-kDa subunit were actually bound to the supercomplexes IV2, III2IV1, and III2IV2, it would follow that these subcomplexes are indeed rather small, well below the size of the peripheral arm, since no major migration shift of the supercomplexes III2IV1 and III2IV2 was recognized in the first-dimension BN gel.
DISCUSSION
Mammalian-like respiratory I-III-IV supercomplexes and complex I dimers in N. crassa mitochondria.
Herein we report the first extensive survey of the OXPHOS system in the N. crassa wild type and a representative set of complex I deletion mutants displaying a variety of complex I assembly defects. In particular, we detected complexes I, III, and IV associated in mammalian-like supercomplexes in both the wild type and nuo21, nuo29.9, and nuo51 mutants, as previously reported in wild-type mitochondria of the fungus P. anserina (45). In addition, we could detect the digitonin-stable supercomplex I1IV1 comigrating with ATP synthase dimers (Fig. 1A), which was not previously identified in Podospora mitochondria (45). This indicates a direct complex I-IV interaction, demonstrated to occur in bovine heart mitochondria by native electrophoresis (48, 67) and in particular by a 2D (66) and 3D projection map of a single particle structure of the bovine heart supercomplex I1III2IV1 (66a). Furthermore, the I-III-IV supercomplexes (Fig. 1A) with apparent masses higher than 2,300 kDa and up to ≥3,000 kDa seem to be larger fragments of a respirasome network of complexes I, III, and IV, as proposed for bovine heart (67, 68, 70, 95), and possibly composed of I1III2IV5-6. Such stoichiometries could arise from the breakdown of two copies of supercomplex I1III2IV4 or else from one copy each of supercomplexes I1III2IV4 and III2IV4, which could be connected by a direct interaction between adjacent complex IV dimers (70, 95). Indeed, similar supercomplexes with masses up to about 3,300 kDa have been observed in 2D BN-SDS gels of digitonin-solubilized mitochondria isolated from fresh bovine heart (40, 46, 48). Taken together, these results indicate a very similar respirasome organization in fungi and mammals. Moreover, when disrupting all I-III-IV supercomplexes upon solubilization with either Triton X-100 or DDM, we found evidence for the existence of complex I dimers (I2) in the respiratory chain of N. crassa wild type (Fig. 3) as well as in the nuo21, nuo29.9, and nuo51 mutants (not shown). It is likely that small amounts of complex I dimers are also stable upon digitonin solubilization, although their identification may be hampered due to comigration with I-III-IV supercomplexes during BN-PAGE. In fact, complex I dimers as well as the larger supercomplex I2III2 were found in BN gels of digitonin-solubilized mitochondria of COX-deficient Podospora strains (45). It is possible that the presence of complex I dimers occurs only when alternative respiratory enzymes are present in the organism, hence the reason why they have not been detected so far in mammalian mitochondria. Whereas AOX as well as the alternative NADH dehydrogenases were found to be constitutively expressed and active in wild-type P. anserina (34), there is no evidence for any AOX respiration in the wild type of N. crassa under normal growth conditions (20, 80). Therefore, we propose that the occurrence of dimeric complex I is a general feature of the respiratory chain in N. crassa.
III-IV supercomplexes are formed without the presence of assembled complex I.
Our results demonstrate that complex I in N. crassa is not essential for assembly/stabilization of the other OXPHOS complexes or for assembly/stabilization of III-IV supercomplexes and ATP synthase dimers (Fig. 6). On the other hand, analysis of the three mutants that assemble complex I indicates that the biogenesis of complex I is obligatorily linked with its assembly into supercomplexes. Furthermore, we demonstrated that neither the 21-kDa, 29.9-kDa, nor 51-kDa peripheral arm subunit plays a pivotal role in the physical interaction of complex I with complexes III and IV, suggesting that the binding interfaces are predominantly provided by the membrane arm, in accordance with structural data of the bovine heart supercomplex I1III2IV1 (66, 66a), and/or by other peripheral arm subunits. Mitochondria of the nuo21 and nuo51 mutants have wild-type-like amounts of assembled complex I as part of supercomplexes, which results in a similar distribution of individual complex I and its supercomplexes (I-III-IV and I2) after solubilization with digitonin (Fig. 5) or either Triton X-100 or DDM, respectively. Strikingly, while complex I in nuo21 is enzymatically active, the complex I of nuo51 lacks the highly conserved 51-kDa subunit hosting the NADH-binding site and has therefore no respiratory activity. This is the first direct demonstration that the incorporation of complex I into supercomplexes as part of a proposed supramolecular network has major nonrespiratory significance and cannot be solely related to conceivable enzymatic advantages, like substrate channeling. Furthermore, the nuo29.9 mutant can assemble only low amounts of complex I, about 20% of that of the wild type (23, 87), which is nonetheless incorporated into supercomplexes as in wild-type, nuo21, and nuo51 strains (Fig. 6B). Altogether, the results from nuo21, nuo29.9, and nuo51 imply that III-IV supercomplexes “trap” essentially all complex I, even when complex I is inactive. This may occur either after initial formation or at the early stages of complex I biogenesis, suggesting that III-IV supercomplexes could be involved in assembly/stabilization of complex I. The situation in COX-deficient mutants of P. anserina possessing I-III supercomplexes is different from that of the nuo51 mutant (45). Indeed, this suggests the assembly/stabilization of complex I as a key function of complex III in Podospora, but complex III is active, although it does not contribute to respiration (45). Evidence for a role of either complex III or IV in the assembly of complex I has also been described in human patients and mammals (1, 7, 18, 21, 56, 71) as well as in bacteria (79).
III-IV supercomplexes and/or the prohibitin complex might serve as assembly factors for complex I.
Herein we have provided direct evidence that high-molecular-weight complexes bind subcomplexes of the peripheral arm containing at least the 30.4-kDa subunit (Fig. 6), as shown in Western blots of 2D BN-SDS gels of digitonin-solubilized mitochondria from the four membrane arm mutants nuo9.8, nuo11.5, nuo14, and nuo20.8. These subcomplexes were found to be comigrating with III-IV supercomplexes and/or the prohibitin complex, thus pointing to them as putative candidates for interaction partners. This is in line with the above discussion on mutants forming complex I. In fact, an interaction of the prohibitin complex with a subcomplex of complex I, containing at least the 23-, 30-, and 49-kDa subunits, has been demonstrated by coimmunoprecipitation in human cells missing mitochondria-encoded membrane subunits, similar to what we observed in the four membrane arm mutants (9). The prohibitin complex (Fig. 2; Table 1) can interact with subunits of respiratory complexes; thus, it probably has a chaperone-like function (9, 57). In addition, the interaction of both the human 30-kDa (homologous to the 30.4-kDa subunit) and 49-kDa subunits has been proposed as the first assembly step towards the peripheral arm (86). These results were corroborated in a very recent study which traced complex I assembly in human cell culture by using a green fluorescent protein-tagged 30-kDa subunit (90). The poorly understood biogenesis of complex I, the largest and most complicated respiratory enzyme, is probably assisted by many proteins and/or protein complexes, of which only a few are known to date (25, 50, 58, 91). In particular, there are two proteins that transient interact with two membrane arm intermediates which are essential for assembly of the membrane arm in N. crassa (50). Of these, a human homologue appears to have a corresponding role in assembly/stabilization of complex I (25, 91).
In summary, the present survey demonstrates the importance of III-IV supercomplexes in assembly/stabilization of complex I, highlighting the unique potential of the comprehensive set of Neurospora complex I deletion mutants to decipher the biogenesis of complex I.
Acknowledgments
I.M. was awarded an EMBO short-term fellowship (ASTF 39.00-06). This work was supported by Fundação para a Ciência e a Tecnologia and POCTI program of QCA III (coparticipated by FEDER) to A.V. as well as by EC FP6 contract number LSHM-CT-2004-512020 and Deutsche Forschungsgemeinschaft grant SFB 472 to N.A.D.
This article is part of the Ph.D. thesis of I.M.
We thank Monika Bloch for technical help in MALDI-MS analysis, Patrícia Carneiro for the English corrections, and Margarida Duarte for helpful discussions.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Acín-Peréz, R., M. P. Bayona-Bafaluy, P. Fernández-Silva, R. Moreno-Loshuertos, A. Pérez-Martos, C. Bruno, C. T. Moraes, and J. A. Enríquez. 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahting, U., C. Thun, R. Hegerl, D. Typke, F. E. Nargang, W. Neupert, and S. Nussberger. 1999. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonicka, H., I. Ogilvie, T. Taivassalo, R. P. Anitori, R. G. Haller, J. Vissing, N. G. Kennaway, and E. A. Shoubridge. 2003. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278:43081-43088. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, I., K. Pfeiffer, W. Neupert, R. A. Stuart, and H. Schägger. 1998. Yeast mitochondrial F1FO-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 17:7170-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artal-Sanz, M., W. Y. Tsang, E. M. Willems, L. A. Grivell, B. D. Lemire, H. van der Spek, and L. G. Nijtmans. 2003. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 278:32091-32099. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi, C., M. L. Genova, G. Parenti Castelli, and G. Lenaz. 2004. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. J. Biol. Chem. 279:36562-36569. [DOI] [PubMed] [Google Scholar]
- 7.Blakely, E. L., A. L. Mitchell, N. Fisher, B. Meunier, L. G. Nijtmans, A. M. Schaefer, M. J. Jackson, D. M. Turnbull, and R. W. Taylor. 2005. A mitochondrial cytochrome b mutation causing severe respiratory chain enzyme deficiency in humans and yeast. FEBS J. 272:3583-3592. [DOI] [PubMed] [Google Scholar]
- 8.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourges, I., C. Ramus, B. Mousson de Camaret, R. Beugnot, C. Remacle, P. Cardol, G. Hofhaus, and J. P. Issartel. 2004. Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem. J. 383:491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandner, K., D. U. Mick, A. E. Frazier, R. D. Taylor, C. Meisinger, and P. Rehling. 2005. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth syndrome. Mol. Biol. Cell 16:5202-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brookes, P. S., A. Pinner, A. Ramachandran, L. Coward, S. Barnes, H. Kim, and V. M. Darley-Usmar. 2002. High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics 2:969-977. [DOI] [PubMed] [Google Scholar]
- 12.Carneiro, P., M. Duarte, and A. Videira. 2007. The external alternative NAD(P)H dehydrogenase NDE3 is localized both in the mitochondria and in the cytoplasm of Neurospora crassa. J. Mol. Biol. 368:1114-1121. [DOI] [PubMed] [Google Scholar]
- 13.Chance, B., and G. R. Williams. 1955. A method for the localization of sites for oxidative phosphorylation. Nature 176:250-254. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, M. Y., F. U. Hartl, J. Martin, R. A. Pollock, F. Kalousek, W. Neupert, E. M. Hallberg, R. L. Hallberg, and A. L. Horwich. 1989. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337:620-625. [DOI] [PubMed] [Google Scholar]
- 15.Cruciat, C. M., S. Brunner, F. Baumann, W. Neupert, and R. A. Stuart. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275:18093-18098. [DOI] [PubMed] [Google Scholar]
- 16.Da Cruz, S., I. Yenarios, J. Langridge, F. Vilbois, P. A. Parone, and J. C. Martinou. 2003. Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 278:41566-41571. [DOI] [PubMed] [Google Scholar]
- 17.da Silva, M. V., P. C. Alves, M. Duarte, N. Mota, A. Lobo-da-Cunha, T. A. Harkness, F. E. Nargang, and A. Videira. 1996. Disruption of the nuclear gene encoding the 20.8-kDa subunit of NADH:ubiquinone reductase of Neurospora mitochondria. Mol. Gen. Genet. 252:177-183. [DOI] [PubMed] [Google Scholar]
- 18.D'Aurelio, M., C. D. Gajewski, G. Lenaz, and G. Manfredi. 2006. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum. Mol. Genet. 15:2157-2169. [DOI] [PubMed] [Google Scholar]
- 19.Davis, R. H., and D. D. Perkins. 2002. Timeline. Neurospora: a model of model microbes. Nat. Rev. Genet. 3:397-403. [DOI] [PubMed] [Google Scholar]
- 20.Descheneau, A. T., I. A. Cleary, and F. E. Nargang. 2005. Genetic evidence for a regulatory pathway controlling alternative oxidase production in Neurospora crassa. Genetics 169:123-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz, F., H. Fukui, S. Garcia, and C. T. Moraes. 2006. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 26:4872-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duarte, M., N. Mota, L. Pinto, and A. Videira. 1998. Inactivation of the gene coding for the 30.4-kDa subunit of respiratory chain NADH dehydrogenase: is the enzyme essential for Neurospora? Mol. Gen. Genet. 257:368-375. [DOI] [PubMed] [Google Scholar]
- 23.Duarte, M., R. Sousa, and A. Videira. 1995. Inactivation of genes encoding subunits of the peripheral and membrane arms of Neurospora mitochondrial complex I and effects on enzyme assembly. Genetics 139:1211-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudkina, N. V., H. Eubel, W. Keegstra, E. J. Boekema, and H. P. Braun. 2005. Structure of a mitochondrial supercomplex formed by respiratory chain complexes I and III. Proc. Natl. Acad. Sci. USA 102:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunning, C. J., M. McKenzie, C. Sugiana, M. Lazarou, J. Silke, A. Connelly, J. M. Fletcher, D. M. Kirby, D. R. Thorburn, and M. T. Ryan. 2007. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26:3227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eubel, H., L. Jänsch, and H. P. Braun. 2003. New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 133:274-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eubel, H., J. Heinemeyer, and H. P. Braun. 2004. Identification and characterization of respirasomes in potato mitochondria. Plant Physiol. 134:1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fecke, W., V. D. Sled, T. Ohnishi, and H. Weiss. 1994. Disruption of the gene encoding the NADH-binding subunit of NADH:ubiquinone oxidoreductase in Neurospora crassa. Formation of a partially assembled enzyme without FMN and the iron-sulphur cluster N-3. Eur. J. Biochem. 220:551-558. [DOI] [PubMed] [Google Scholar]
- 29.Ferreirinha, F., M. Duarte, A. M. Melo, and A. Videira. 1999. Effects of disrupting the 21 kDa subunit of complex I from Neurospora crassa. Biochem. J. 342:551-554. [PMC free article] [PubMed] [Google Scholar]
- 30.Gabaldón, T., D. Rainey, and M. A. Huynen. 2005. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 348:857-870. [DOI] [PubMed] [Google Scholar]
- 31.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 32.Grad, L. I., and D. Lemire. 2004. Mitochondrial complex I mutations in Caenorhabditis elegans produce cytochrome c oxidase deficiency, oxidative stress and vitamin-responsive lactic acidosis. Hum. Mol. Genet. 13:303-314. [DOI] [PubMed] [Google Scholar]
- 33.Grad, L. I., and D. Lemire. 2006. Riboflavin enhances the assembly of mitochondrial cytochrome c oxidase in C. elegans NADH-ubiquinone oxidoreductase mutants. Biochim. Biophys. Acta 1757:115-122. [DOI] [PubMed] [Google Scholar]
- 34.Gredilla, R., J. Grief, and H. D. Osiewacz. 2006. Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Exp. Gerontol. 41:439-447. [DOI] [PubMed] [Google Scholar]
- 35.Hackenbrock, C. R., B. Chazotte, and S. S. Gupte. 1986. The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J. Bioenerg. Biomembr. 18:331-368. [DOI] [PubMed] [Google Scholar]
- 36.Hajek, P., A. Chomyn, and G. Attardi. 2007. Identification of a novel mitochondrial complex containing mitofusin 2 and stomatin-like protein 2. J. Biol. Chem. 282:5670-5681. [DOI] [PubMed] [Google Scholar]
- 37.Hatefi, Y. 1985. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 54:1015-1069. [DOI] [PubMed] [Google Scholar]
- 38.Heazlewood, J. L., K. A. Howell, J. A. Whelan, and H. Millar. 2003. Towards an analysis of the rice mitochondrial proteome. Plant Physiol. 132:230-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinemeyer, J., H. P. Braun, E. B. Boekema, and R. Kouřil. 2007. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 282:12240-12248. [DOI] [PubMed] [Google Scholar]
- 40.Hunzinger, C., W. Wozny, G. P. Schwall, S. Poznanović, W. Stegmann, H. Zengerling, R. Schoepf, K. Groebe, M. A. Cahill, H. D. Osiewacz, N. Jägemann, M. Bloch, N. A. Dencher, F. Krause, and A. Schrattenholz. 2006. Comparative profiling of the mammalian mitochondrial proteome: multiple aconitase-2 isoforms including N-formylkynurenine modifications as part of a protein biomarker signature for reactive oxidative species. J. Proteome Res. 5:625-633. [DOI] [PubMed] [Google Scholar]
- 41.Hutchinson, E. G., W. Tichelaar, G. Hofhaus, H. Weiss, and K. R. Leonard. 1989. Identification and electron microscopic analysis of a chaperonin oligomer from Neurospora crassa mitochondria. EMBO J. 8:1485-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janssen, R. J. R. J., L. G. Nijtmans, L. P. van den Heuvel, and J. A. M. Smeitink. 2006. Mitochondrial complex I: structure, function and pathology. J. Inherit. Metab. Dis. 29:499-515. [DOI] [PubMed] [Google Scholar]
- 43.Kiebler, M., R. Pfaller, T. Söllner, G. Griffiths, H. Horstmann, N. Pfanner, and W. Neupert. 1990. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature 348:610-616. [DOI] [PubMed] [Google Scholar]
- 44.Klanner, C., H. Prokisch, and T. Langer. 2001. MAP-1 and IAP-1, two novel AAA proteases with catalytic sites on opposite membrane surfaces in mitochondrial inner membrane of Neurospora crassa. Mol. Biol. Cell 12:2858-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krause, F., C. Q. Scheckhuber, A. Werner, S. Rexroth, N. H. Reifschneider, N. A. Dencher, and H. D. Osiewacz. 2004. Supramolecular organization of cytochrome c oxidase- and alternative oxidase-dependent respiratory chains in the filamentous fungus Podospora anserina. J. Biol. Chem. 279:26453-26461. [DOI] [PubMed] [Google Scholar]
- 46.Krause, F. 2006. Detection and analysis of protein-protein interactions in organellar and prokaryotic proteomes by native gel electrophoresis: (membrane) protein complexes and supercomplexes. Electrophoresis 27:2759-2781. [DOI] [PubMed] [Google Scholar]
- 47.Krause, F., N. H. Reifschneider, D. Vocke, H. Seelert, S. Rexroth, and N. A. Dencher. 2004. “Respirasome”-like supercomplexes in green leaf mitochondria of spinach. J. Biol. Chem. 279:48369-48375. [DOI] [PubMed] [Google Scholar]
- 48.Krause, F., N. H. Reifschneider, S. Goto, and N. A. Dencher. 2005. Active oligomeric ATP synthases in mammalian mitochondria. Biochem. Biophys. Res. Commun. 329:583-590. [DOI] [PubMed] [Google Scholar]
- 49.Kronekova, Z., and G. Rödel. 2005. Organization of assembly factors Cbp3p and Cbp4p and their effect on bc1 complex assembly in Saccharomyces cerevisiae. Curr. Genet. 47:203-212. [DOI] [PubMed] [Google Scholar]
- 50.Küffner, R., A. Rohr, A. Schmiede, C. Krüll, and U. Schulte. 1998. Involvement of two novel chaperones in the assembly of mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 283:409-417. [DOI] [PubMed] [Google Scholar]
- 51.Lange, C., J. H. Nett, B. L. Trumpower, and C. Hunte. 2001. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20:6591-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mannhaupt, G., C. Montrone, D. Haase, H. W. Mewes, V. Aign, J. D. Hoheisel, B. Fartmann, G. Nyakatura, F. Kempken, J. Maier, and U. Schulte. 2003. What's in the genome of a filamentous fungus? Analysis of the Neurospora genome sequence. Nucleic Acids Res. 31:1944-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marques, I., A. V. Ushakova, M. Duarte, and A. Videira. 2007. Role of the conserved cysteine residues of the 11.5 kDa subunit in complex I catalytic properties. J. Biochem. 141:489-493. [DOI] [PubMed] [Google Scholar]
- 54.Marques, I., M. Duarte, and A. Videira. 2003. The 9.8 kDa subunit of complex I, related to bacterial Na+-translocating NADH dehydrogenases, is required for enzyme assembly and function in Neurospora crassa. J. Mol. Biol. 329:283-290. [DOI] [PubMed] [Google Scholar]
- 55.Marques, I., M. Duarte, J. Assunção, A. V. Ushakova, and A. Videira. 2005. Composition of complex I from Neurospora crassa and disruption of two “accessory” subunits. Biochim. Biophys. Acta 1707:211-220. [DOI] [PubMed] [Google Scholar]
- 56.McKenzie, M., M. Lazarou, D. R. Thorburn, and M. T. Ryan. 2006. Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J. Mol. Biol. 361:462-469. [DOI] [PubMed] [Google Scholar]
- 57.Nijtmans, L. G., L. de Jong, M. Artal Sanz, P. J. Coates, J. A. Berden, J. W. Back, A. O. Muijsers, H. van der Spek, and L. A. Grivell. 2000. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 19:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogilvie, I., N. G. Kennaway, and E. A. Shoubridge. 2005. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Investig. 115:2784-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Owczarek, C. M., H. R. Treutlein, K. J. Portbury, L. M. Gulluyan, I. Kola, and P. J. Hertzog. 2001. A novel member of the STOMATIN/EPB72/mec-2 family, stomatin-like 2 (STOML2), is ubiquitously expressed and localizes to HSA chromosome 9p13.1. Cytogenet. Cell. Genet. 92:196-203. [DOI] [PubMed] [Google Scholar]
- 60.Perales, M., H. Eubel, J. Heinemeyer, A. Colaneri, E. Zabaleta, and H. P. Braun. 2005. Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I+III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 350:263-277. [DOI] [PubMed] [Google Scholar]
- 61.Pfeiffer, K., V. Gohil, R. A. Stuart, C. Hunte, U. Brandt, M. L. Greenberg, and H. Schägger. 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278:52873-52880. [DOI] [PubMed] [Google Scholar]
- 62.Pineau, B., C. Mathieu, C. Gérard-Hirne, R. De Paepe, and P. Chétrit. 2005. Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild type phenotype in the Nicotiana sylvestris CMS II mutant lacking nad7. J. Biol. Chem. 280:25994-26001. [DOI] [PubMed] [Google Scholar]
- 63.Rapaport, D., and W. Neupert. 1999. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 146:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reifschneider, N. H., S. Goto, H. Nakamoto, R. Takahashi, M. Sugawa, N. A. Dencher, and F. Krause. 2006. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. J. Proteome Res. 5:1217-1232. [DOI] [PubMed] [Google Scholar]
- 65.Rexroth, S., J. M. W. Meyer zu Tittingdorf, H. J. Schwaβmann, F. Krause, H. Seelert, and N. A. Dencher. 2004. Dimeric H+-ATP synthase in the chloroplast of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1658:202-211. [DOI] [PubMed] [Google Scholar]
- 66.Schäfer, E., H. Seelert, N. H. Reifschneider, F. Krause, N. A. Dencher, and J. Vonck. 2006. Architecture of active mammalian respiratory chain supercomplexes. J. Biol. Chem. 281:15370-15375. [DOI] [PubMed] [Google Scholar]
- 66a.Schäfer, E., N. A. Dencher, J. Vonck, and D. Parcej. 2007. Three-dimensional structure of the respiratory chain supercomplex I1 III2 IV1 from bovine heart mitochondria. Biochemistry 46:12579-12585. [DOI] [PubMed] [Google Scholar]
- 67.Schägger, H., and K. Pfeiffer. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schägger, H., and K. Pfeiffer. 2001. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 276:37861-37867. [DOI] [PubMed] [Google Scholar]
- 69.Schägger, H. 2001. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell. Biol. 65:231-244. [DOI] [PubMed] [Google Scholar]
- 70.Schägger, H. 2001. Respiratory chain supercomplexes. IUBMB Life 52:119-128. [DOI] [PubMed] [Google Scholar]
- 71.Schägger, H., R. de Coo, M. F. Bauer, S. Hofmann, C. Godinot, and U. Brandt. 2004. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279:36349-36353. [DOI] [PubMed] [Google Scholar]
- 72.Schägger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 73.Sedlak, E., and N. C. Robinson. 1999. Phospholipase A2 digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38:14966-14972. [DOI] [PubMed] [Google Scholar]
- 74.Selker, E. U. 2002. Repeat-induced gene silencing in fungi. Adv. Genet. 46:439-450. [DOI] [PubMed] [Google Scholar]
- 75.Sherman, E. L., N. E. Go, and F. E. Nargang. 2005. Functions of the small proteins in the TOM complex of Neurospora crassa. Mol. Biol. Cell 16:4172-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sherman, E. L., R. D. Taylor, N. E. Go, and F. E. Nargang. 2006. Effect of mutations in Tom40 on stability of the TOM complex, assembly of Tom40, and import of mitochondrial preproteins. J. Biol. Chem. 281:22554-22565. [DOI] [PubMed] [Google Scholar]
- 77.Spivey, H. O., and J. Ovádi. 1999. Substrate channeling. Methods 19:306-321. [DOI] [PubMed] [Google Scholar]
- 78.Steglich, G., W. Neupert, and T. Langer. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19:3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stroh, A., O. Anderka, K. Pfeiffer, T. Yagi, M. Finel, B. Ludwig, and H. Schägger. 2004. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 279:5000-5007. [DOI] [PubMed] [Google Scholar]
- 80.Tanton, L. L., C. E. Nargang, K. E. Kessler, Q. Li, and F. E. Nargang. 2003. Alternative oxidase expression in Neurospora crassa. Fungal Genet. Biol. 39:176-190. [DOI] [PubMed] [Google Scholar]
- 81.Tatsuta, T., K. Model, and T. Langer. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16:248-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor, R. D., B. J. McHale, and F. E. Nargang. 2003. Characterization of Neurospora crassa Tom40-deficient mutants and effect of specific mutations on Tom40 assembly. J. Biol. Chem. 278:765-775. [DOI] [PubMed] [Google Scholar]
- 83.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tuschen, G., U. Sackmann, U. Nehls, H. Haiker, G. Buse, and H. Weiss. 1990. Assembly of NADH:ubiquinone reductase (complex I) in Neurospora mitochondria. Independent pathways of nuclear-encoded and mitochondrially encoded subunits. J. Mol. Biol. 213:845-857. [DOI] [PubMed] [Google Scholar]
- 85.Ugalde, C., R. J. R. J. Janssen, L. P. van den Heuvel, J. A. M. Smeitink, and L. G. J. Nijtmans. 2004. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 13:659-667. [DOI] [PubMed] [Google Scholar]
- 86.Ugalde, C., R. Vogel, R. Huijbens, B. Van Den Heuvel, J. Smeitink, and L. Nijtmans. 2004. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 13:2461-2472. [DOI] [PubMed] [Google Scholar]
- 87.Ushakova, A. V., M. Duarte, A. D. Vinogradov, and A. Videira. 2005. The 29.9 kDa subunit of mitochondrial complex I is involved in the enzyme active/de-active transitions. J. Mol. Biol. 351:327-333. [DOI] [PubMed] [Google Scholar]
- 88.van der Laan, M., N. Wiedemann, D. U. Mick, B. Guiard, P. Rehling, and N. Pfanner. 2006. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 16:2271-2276. [DOI] [PubMed] [Google Scholar]
- 89.Videira, A., and M. Duarte. 2002. From NADH to ubiquinone in Neurospora mitochondria. Biochim. Biophys. Acta 1555:187-191. [DOI] [PubMed] [Google Scholar]
- 90.Vogel, R. O., C. E. Dieteren, L. P. van den Heuvel, P. H. Willems, J. A. Smeitink, W. J. Koopman, and L. G. J. Nijtmans. 2007. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 282:7582-7590. [DOI] [PubMed] [Google Scholar]
- 91.Vogel, R. O., R. J. Janssen, C. Ugalde, M. Grovenstein, R. J. Huijbens, H. J. Visch, L. P. van den Heuvel, P. H. Willems, M. Zeviani, J. A. Smeitink, and L. G. J. Nijtmans. 2005. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 272:5317-5326. [DOI] [PubMed] [Google Scholar]
- 92.Wang, Y., and J. S. Morrow. 2000. Identification and characterization of human SLP-2, a novel homologue of stomatin (band 7.2b) present in erythrocytes and other tissues. J. Biol. Chem. 275:8062-8071. [DOI] [PubMed] [Google Scholar]
- 93.Werner, S. 1977. Preparation of polypeptide subunits of cytochrome oxidase from Neurospora crassa. Eur. J. Biochem. 79:103-110. [DOI] [PubMed] [Google Scholar]
- 94.Wittig, I., and H. Schägger. 2005. Advantages and limitations of clear-native PAGE. Proteomics 5:4338-4346. [DOI] [PubMed] [Google Scholar]
- 95.Wittig, I., R. Carrozzo, F. M. Santorelli, and H. Schägger. 2006. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta 1757:1066-1072. [DOI] [PubMed] [Google Scholar]
- 96.Yaguzhinsky, L. S., V. I. Yurkov, and I. P. Krasinskaya. 2006. On the localized coupling of respiration and phosphorylation in mitochondria. Biochim. Biophys. Acta 1757:408-414. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, M., E. Mileykovskaya, and W. Dowhan. 2005. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280:29403-29408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang, M., E. Mileykovskaya, and W. Dowhan. 2002. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277:43553-43556. [DOI] [PubMed] [Google Scholar]