Abstract
The Ccr4-Not complex is a multifunctional regulatory platform composed of nine subunits that controls diverse cellular events including mRNA degradation, protein ubiquitination, and transcription. In this study, we identified the yeast Saccharomyces cerevisiae osmotic and oxidative stress transcription factor Skn7 as a new target for regulation by the Ccr4-Not complex. Skn7 interacts with Not1 in a two-hybrid assay and coimmunoprecipitates with Not5 in a Not4-dependent manner. Skn7-dependent expression of OCH1 and Skn7 binding to the OCH1 promoter are increased in not4Δ or not5Δ mutants. Skn7 purified from wild-type cells but not from not4Δ cells is associated with the Srb10 kinase. This kinase plays a central role in the regulation of Skn7 by Not4, since increased OCH1 expression in not4Δ cells requires Srb10. These results reveal a critical role for the Ccr4-Not complex in the mechanism of activation of Skn7 that is dependent upon the Srb10 kinase.
Eukaryotic cells must adapt to continually changing environments. Therefore, they have adapted molecular mechanisms for very rapid responses that involve in most cases transcriptional activation of stress genes. Responses to stress are particularly critical for unicellular organisms such as the yeast Saccharomyces cerevisiae that must adapt to rapid and extreme changes, for instance, changes in nutrient availability, osmolarity, and temperature. Stresses are sensed at the cell membrane and through specific signaling pathways induce transcriptional activation of stress-responsive genes. Expression of these genes then protects the cells against damage. In yeast, it has been well documented that exposure of cells to one type of stress protects them from exposure to other stresses. This stems from the induction of a similar subset of genes (about 10% of the genome) upon exposure of yeast to any type of stress (12). For the most part, these genes are controlled by the STRE (stress response element) promoter element, which is recognized by two partially redundant transcription factors, Msn2 and Msn4 (8, 22, 38, 46). This transcriptional response is called the environmental stress response.
We recently determined that the S. cerevisiae environmental stress response is activated in cells growing in high glucose that lack an intact Ccr4-Not complex (31). This complex consists of at least nine subunits (Not1 to Not5, Ccr4, Caf1, Caf40, and Caf130) that exist in complexes of 1.2 and 2 MDa from yeast to human. Two of its subunits are enzymes, namely, Not4, which is an E3 ubiquitin ligase, and Ccr4, which is the major yeast deadenylase. Genetic and molecular analyses have suggested that the complex may be composed of different functional modules. Indeed, it has been proposed that the Caf1 and Ccr4 subunits are physically and functionally separated from the Not subunits (1). Nevertheless, the role of the different subunits of the Ccr4-Not complex still remains to be clarified, since it is presently known that multiple cellular functions are under the control of the Ccr4-Not complex. For instance, in addition to its role in protein ubiquitination (44), this complex regulates mRNA metabolism, both at the level of mRNA degradation, via the Ccr4 deadenylase subunit, and at the level of transcription initiation, via a control of the distribution of TFIID across the genome (for reviews, see references 14, 17, and 19). In the case of the environmental stress response, the Ccr4-Not complex controls transcription initiation indirectly through the Msn2 transcription factor. Indeed, Msn2-dependent transcriptional activation is increased and the Msn2 protein is posttranslationally modified in mutants of the Ccr4-Not complex (31). Both of these effects require the Glc7 type I protein phosphatase and its regulatory subunit Bud14 (30).
In this work, we determine that in addition to controlling the environmental stress response in unstressed cells, the Ccr4-Not complex regulates the response to specific stress signals. Indeed, we show that the Ccr4-Not complex controls the Skn7 transcription factor, which is one of the transcription factors that yeast cells have evolved to respond to specific stress responses in addition to the general response mediated by Msn2/4 (for a review, see reference 25). Skn7 contains a receiver domain typical of two-component signal transduction systems (26, 32), which, though commonly used for environmental sensing in eubacteria, are restricted in S. cerevisiae to the response to osmotic and oxidative stresses. Indeed, in response to hypo-osmolarity, Sln1, a membrane-associated histidine kinase, is autophosphorylated, and through its intermediary phosphorelay protein Ypd1, it transfers its phosphate residue to Skn7 on a specific aspartate residue, D427, within the Skn7 receiver domain. Skn7 then activates hypotonic target genes such as OCH1, which encodes a mannosyltransferase involved in N-linked glycoprotein maturation, through binding to SSRE (Sln1*-responsive element) sites in the promoter (33). It has been shown that Skn7 in this pathway is involved in cell wall integrity and the cell cycle (7, 9, 10, 40). However, Skn7 also plays a role in response to oxidative stress (28). Its activation in response to oxidative stress is independent of aspartate 427 (32, 39). Furthermore, different sites in the promoters are recognized by Skn7 in this response, such as the OSRE (oxidative stress response element), which shares a GCC repeat with the SSRE (47).
In this study, we show that Skn7-dependent transcriptional activation is increased in exponentially growing cells lacking subunits of the Ccr4-Not complex and that this increase correlates with increased binding of Skn7 to specific target promoters. We find that Skn7 purified from wild-type cells is associated with the Srb10 kinase, which mediates the regulation of Skn7-dependent OCH1 transcription by Not4. The Srb10 kinase is part of a module of the mediator that includes three other proteins, Srb8, Srb9, and the Srb11 cyclin (5), and which has both positive and negative effects on transcription regulation (2; for a review see also reference 11). Our present results demonstrate that the Ccr4-Not complex plays a widespread role in the regulation of the cellular response to stress, not only through the general Msn2-dependent stress response as previously published but also via the more specific stress response mediated by Skn7.
MATERIALS AND METHODS
Media and strains.
All media were standard. The strains used in this work derive from MY1 (16) (Table 1). Single-step deletions and/or tagging of genes was performed by PCR according to the method in reference 35, and many strains were created by mating of haploid strains and sporulation of the diploids. MY2048 and MY2052 result from crosses with null mutant strains described in reference 43. To create strains expressing Skn7 fused to a tandem affinity purification (TAP) tag at its C terminus, we amplified the cassette with the TAP tag and the marker genes from pFA6a-taptag.8.8.6 (a kind gift from D. Bartford) using oligonucleotides with Skn7 sequences just prior to and beyond the stop codon, leading to strains MY5037 and MY5041. Strain MY4858, derived from BY4741 (see http://web.uni-frankfurt.de/fb15/mikro/euroscarf/data/by.html) and expressing a TAP-tagged Caf40, was a kind gift from Klaas Mulder and Marc Timmers, and MY4910 was created from MY4858 by PCR according to the method in reference 35.
TABLE 1.
Strain list
| Strain | Description | Reference |
|---|---|---|
| EGY48 | MATahis3 trp1 ura3 LEU2::pLexOp6-LEU2 | 48 |
| MY1 | MATaura3-52 trp1 leu2::PET56 gcn4 gal2 | 30 |
| MY2 | MY1 MATα | This work |
| MY3 | MY1 his3::TRP1 | This work |
| MY4 | MY2 his3::TRP1 | This work |
| MY2048 | MY4 not5::LEU2 | This work |
| MY2052 | MY1 not4-3::LEU2 | 30 |
| MY3246 | MY1 srb10::kanMX4 ura3::STRE-lacZ-URA3 | This work |
| MY3247 | MY2 srb10::kanMX4 ura3::STRE-lacZ-URA3 not4-3::LEU2 | This work |
| MY3496 | MY2 msn2::TRP1 | 30 |
| MY3498 | MY3 not4-3::LEU2 msn2::TRP1 | 30 |
| MY3595 | MY2 not4::kanMX6 | This work |
| MY3683 | MY3 skn7::SKN7-Myc13-HIS3 | This work |
| MY3688 | MY3 not4::kanMX6 skn7::SKN7-Myc13-HIS3 | This work |
| MY3725 | MY1 skn7::TRP1 | This work |
| MY3726 | MY1 not4::kanMX6 skn7::TRP1 | This work |
| MY3812 | MY3 not5::LEU2 skn7::TRP1 | This work |
| MY3819 | MY3 not3::URA3 skn7::SKN7-Myc13-HIS3 | This work |
| MY3820 | MY3 not5::LEU2 skn7::SKN7-Myc13-HIS3 | This work |
| MY3824 | MY3 bud14::kanMX6 skn7::SKN7-Myc13-HIS3 | This work |
| MY3825 | MY4 not4::kanMX6 bud14::kanMX6 skn7::SKN7-Myc13-HIS3 | This work |
| MY3826 | MY3 not5::LEU2 bud14::kanMX6 skn7::SKN7-Myc13-HIS3 | This work |
| MY4829 | MY3725 pHA-SKN7-LEU2 | This work |
| MY4844 | MY3726 pHA-SKN7-LEU2 | This work |
| MY4858 | MATα leu2Δ20 ura3Δ met15Δ his3Δ1 caf40::CAF40-TAP tag-URA3 | 41 |
| MY4910 | MY4858 not4::His3MX4 | This work |
| MY5037 | MY3 skn7::SKN7-TAP tag-kanMX4 | This work |
| MY5041 | MY3 skn7::SKN7-TAP tag-kanMX4 not4-3::LEU2 | This work |
| MY5187 | MY3726 pHA-skn7-D427N-LEU2 | This work |
| MY5188 | MY3726 pHA-skn7-D427E-LEU2 | This work |
| MY5189 | MY3725 pHA-skn7-D427N-LEU2 | This work |
| MY5190 | MY3725 pHA-skn7-D427E-LEU2 | This work |
Plasmids.
The plasmid expressing B42-Skn7cl was obtained by subcloning the SmaI-XhoI fragment of a partial SKN7 clone in pACT2 obtained from a large two-hybrid selection into pJG4-5 digested with EcoRI, treated with Klenow fragment, and digested with XhoI. We used gap repair to create a prey plasmid expressing the entire Skn7. Using an oligonucleotide with sequences homologous to the pJG4-5 vector (48) at the end of the B42 activation domain and hemagglutinin (HA) epitope, followed by SKN7 sequences starting at the start codon on one hand, and an oligonucleotide with sequences homologous to the pJG4-5 vector in the ADH1 3′ untranslated region followed by SKN7 sequences covering the stop codon, we amplified SKN7 using genomic DNA. We cotransformed the linearized pJG4-5 vector together with the PCR fragment into the reporter two-hybrid strain EGY48 (48). Transformants that could grow in the absence of tryptophan were selected for, and the expression of the Skn7 fusion protein was verified from transformants growing in glucose or galactose by Western blot analysis with an anti-HA antibody. The plasmid was recovered from transformants expressing a fusion protein, and the EcoRI-SalI fragment covering the entire SKN7 open reading frame (ORF) could be subcloned into pRS316, pRS306, or pBSKS for further use. Site-directed mutagenesis was performed with the SKN7 sequences cloned in pRS306, leading to pMAC561, pMAC562 (D427N) and pMAC566 (D427E), and the mutations were verified by sequencing. The mutant SKN7 sequences were subcloned back into pJG4-5, leading to pMAC653 (D427N) and pMAC654 (D427E). Then we amplified by PCR the wild-type and mutant SKN7 sequences from pJG4-5 including the fused N-terminal HA tag by using a 5′ oligonucleotide containing the end of NCB2 promoter sequences and the sequence for the HA tag and the beginning of SKN7 sequences and a 3′ oligonucleotide containing ADH1 3′ sequences that then allowed us to do gap repair in yeast, by cotransforming a plasmid with the NCB2 promoter and ADH1 3′ sequences (pMAC399) digested with PstI and SphI and the PCR fragments. This allowed the creation of plasmids expressing wild-type (pMAC582) or D427-mutated (pMAC656 for D427N and pMAC661 for D427E) Skn7 with an N-terminal HA tag from the NCB2 promoter. Oligonucleotide sequences are available upon request.
Two-hybrid selection and assay.
For the large-scale two-hybrid selection, full-length and N-terminally truncated Not1 was expressed as a fusion to the Gal4 DNA binding domain and tested with the library described in reference 21. Transformants (6.2 × 106) were obtained and selected for 50 mM or 100 mM 3-aminotriazole resistance. Of the positive clones initially obtained, 78 were still positive after plasmid rescue and retransformation and corresponded to 53 different genes. Most genes were represented only once, and this was the case for SKN7 as well as NOT4, suggesting that the selection was by far not saturated. At most four clones covering the same gene were obtained in two cases. For subsequent two-hybrid analyses, strain EGY48 was transformed with derivatives of plasmid pLex202 expressing full-length or partial Not1 fused to LexA and plasmid pJG4-5 expressing full-length or partial Skn7 fused to the B42 activation domain. Transformants were spotted on glucose or galactose plates lacking or containing leucine and grown for 3 to 10 days.
Coimmunoprecipitations.
To perform coimmunoprecipitation experiments with Skn7, Not1, or Not5, wild-type or not4Δ strains expressing genomically Myc-tagged Skn7 were grown exponentially to an optical density at 600 nm of 0.8 to 1.1. Cells were spun and resuspended in FA lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8, 1% Triton, 0.1% sodium deoxycholate, and phenylmethylsulfonyl fluoride at 1 mM). Following addition of an equal volume of glass beads, cells were broken with a cell disruptor for 15 min at 4°C. The extracts were clarified for 15 min in a microcentrifuge at 14,000 rpm, and the protein concentration of the supernatant was measured by the Bradford assay. One milligram of total lysate was incubated with antibodies against Not1, Not5 (1 μl), or Myc (Covance) overnight at 4°C. Fifteen microliters of protein G-Sepharose slurry was then added for at least 2 h. After two washes with 1 ml FA lysis buffer, the protein G-Sepharose beads were resuspended in 40 μl of 2× sample buffer (62.5 mM Tris [pH 6.8], 25% glycerol, 2% sodium dodecyl sulfate [SDS], 0.01% bromophenol blue, 5% β-mercaptoethanol), boiled for 5 min, and loaded on a 9% polyacrylamide-SDS gel. Twenty-five micrograms of the total lysate was also loaded on the gel. After electrophoresis, proteins were transferred to nitrocellulose for Western blot analysis.
S1 analysis.
Total cellular RNA was extracted by the hot acid phenol method, and 50 μg was hybridized to DED1 or OCH1 probes (sequences available upon request) and then digested with S1 nuclease and analyzed on a sequencing gel as described previously (15).
ChIP experiments.
Chromatin immunoprecipitation (ChIP) experiments and quantitative real-time PCR were performed as described previously (18). Monoclonal anti-Myc and polyclonal anti-TATA binding protein (anti-TBP) antibodies were used for the immunoprecipitations of cross-linked extracts. Oligonucleotide sequences used for quantitative real-time PCR are available upon request; they amplify sequences between −350 and −200 relative to the ORF for AHP1, from −247 to −147 for CCP1, and from −207 to −57 for TSA1. The oligonucleotides of the intergenic region amplify 150 nucleotides starting at 1516197 on chromosome IV.
TAP.
For TAP, 20 liters of cells expressing TAP-tagged Skn7 or TAP-tagged Caf40 was grown to an optical density at 600 nm of 2 to 3. Cells were spun for 10 min at 5,000 rpm at 4°C, and then the pellets were washed with 1 liter of cold E buffer (20 mM HEPES, pH 8, 350 mM NaCl, 0.1% Tween 20, 10% glycerol) and spun for 10 min at 5,000 rpm at 4°C. The final pellets were resuspended in 50 ml of cold E buffer completed with protease inhibitors. Fifty milliliters of cell suspension was broken eight times during 30 seconds using a Bead Beater filled up to one-third of the volume with glass beads. Cell lysates were spun for 10 min at 5,000 rpm at 4°C. The extracts were clarified for 45 min in a microcentrifuge at 43,000 rpm at 4°C, and the protein concentration of the supernatants was measured by the Bradford assay. Two grams of proteins was used for purification over 400 μl of an immunoglobulin G-Sepharose column (IgG Sepharose Fast Flow; Pharmacia). After 2 h of protein binding with rotation at 4°C, the column was washed with 10 ml of E buffer and 10 ml of tobacco etch virus (TEV) protease cleavage buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.5 mM EDTA, 0.1% Tween). TEV protease (100 U) cleavage was performed in 1 ml buffer at 18°C for 2 h. The TEV eluate was bound to 100 μl of calmodulin affinity resin (Stratagene) in 3.6 ml of calmodulin binding buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 10 mM CaCl2, 0.1% Tween, 10% glycerol) by rotating for 1 h at room temperature. The column was washed with 10 ml of calmodulin binding buffer, and bound proteins were recovered in three elutions using calmodulin elution buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 10 mM EGTA, 0.1% Tween, 10% glycerol).
RESULTS
The Ccr4-Not complex interacts with the Skn7 transcription factor.
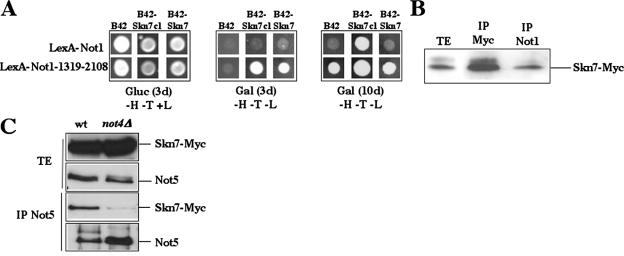
We performed a large screen for proteins that interact with Not1 in the two-hybrid assay (see Materials and Methods). We chose Not1 as a bait because it is the scaffold of the complex and might lead to the isolation of proteins interacting with any of the subunits of the complex. In turn, this might give clues to the mechanism of function of the Ccr4-Not complex. In this screen, we identified a clone expressing a portion of Skn7 that could interact with a clone expressing full-length Not1 and with a clone expressing an N-terminal truncation of Not1, namely, amino acids 1319 to 2108. Sequencing of the isolated SKN7 clone revealed that it expressed amino acids 191 to 300 of Skn7, containing a predicted coiled-coil domain of the protein, fused to the Gal4 activation domain. This result was confirmed using a different two-hybrid system in which we fused Not1 to the LexA DNA binding domain (vector pLex202) and the interacting Skn7 amino acids to the B42 activation domain (vector pJG4-5) (48) (Fig. 1A). We also created a fusion of the B42 activation domain to full-length Skn7 (622 amino acids) and observed that full-length Skn7 also gave a two-hybrid interaction with the C-terminal region of Not1. In contrast, we could not detect a two-hybrid interaction between the two full-length fusions (Fig. 1A), suggesting that in the two long fusion proteins, the interaction may not be favorable.
FIG. 1.
Skn7 interacts with two subunits of the Ccr4-Not complex. (A) Strain EGY48 was transformed with plasmids expressing the LexA-Not1 or LexA-Not1-1319-2108 fusions together with pJG4-5 expressing B42, pJG4-5 expressing a fusion of the B42 activation domain to a portion of Skn7 (amino acids 191 to 300, B42-Skn7cl), or pJG4-5 expressing a fusion of B42 to full-length Skn7 (B42-Skn7). The transformants were spotted on glucose minimal medium lacking histidine and tryptophan (−H−T+L) or on galactose minimal medium lacking histidine, tryptophan, and leucine (−H−T−L). After 3 or 10 days of growth, pictures were taken. (B) Wild-type cells expressing genomically tagged Skn7-Myc (MY3683) were grown exponentially in rich medium. Total protein extracts (TE) were incubated with the indicated antibodies (IP) followed by the addition of protein G-Sepharose. The immunoprecipitates (IP) were washed and then loaded on 9% SDS-polyacrylamide gels to be analyzed by Western blotting with anti-Myc antibodies. (C) Wild-type (MY3683) and not4Δ cells (MY3688) expressing genomically tagged Skn7-Myc were analyzed as for panel B, except that immunoprecipitation was with anti-Not5 antibodies (IP Not5), and visualization of the blots was with Myc or Not5 antibodies, as indicated.
We then created a strain expressing Myc-tagged Skn7 from its own promoter and its own locus (according to reference 35; see Materials and Methods). We prepared total protein extracts from this strain growing in high glucose and immunoprecipitated Not1 or Skn7 from the total extract. We found that tagged Skn7 was in the immunoprecipitate (Fig. 1B), demonstrating that in vivo Not1 can interact with Skn7. We then investigated whether another subunit of the Ccr4-Not complex, Not5, was able to coimmunoprecipitate with Skn7. We chose to look at Not5 because, like Skn7, it interacts in the two-hybrid assay with the C-terminal domain of Not1 (37). We found that indeed Skn7-Myc coimmunoprecipitated with Not5 (Fig. 1C).
To determine whether this interaction required an integral Ccr4-Not complex, we created a not4Δ strain expressing tagged Skn7. We chose this strain because Not4, like Not5, is part of the Not module of the Ccr4-Not complex. The deletion of NOT4 led to the loss of coimmunoprecipitation between Not5 and Skn7 (Fig. 1C) and to the loss of coimmunoprecipitation between Not1 and Skn7 but did not abolish the interaction between Not5 and Not1 (data not shown). We also immunoprecipitated Myc-tagged Skn7 from wild-type and not4Δ cell extracts and detected low levels of Not5 in the immunoprecipitate from wild-type cell extracts but none in the immunoprecipitate from not4Δ cell extracts, despite efficient immunoprecipitation of Skn7 itself from both cell extracts (data not shown).
Taken together these results show that Skn7 can interact, directly or indirectly, with at least two subunits of the Ccr4-Not complex in vivo and that the association of Skn7 with Not5 requires Not4.
Skn7-dependent expression of OCH1 is increased in mutants of the Ccr4-Not complex.
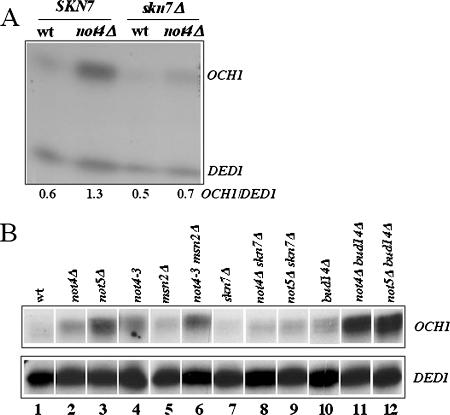
Skn7 is important for the response to osmotic stress, and it activates genes such as OCH1 in response to hypotonic stress. We tested the expression of OCH1 in wild-type and not4Δ cells, by S1 analysis of total cellular RNA, and for the same amount of total cellular RNA we compared OCH1 mRNA levels in the different strains. The levels of DED1 mRNA were measured as an internal control. We observed that OCH1 mRNA levels were increased in cells lacking NOT4. This increased expression of OCH1 required Skn7, as could be determined by measuring expression of OCH1 in not4Δ cells lacking SKN7 (Fig. 2A). This result was confirmed in multiple experiments (data not shown).
FIG. 2.
Skn7-dependent and Msn2-independent increase of OCH1 mRNA in not4Δ or not5Δ cells. (A) Equal amounts of total cellular RNA from wild-type (MY1), not4Δ (MY3595), skn7Δ (MY3725), and not4Δ skn7Δ (MY3726) strains were analyzed by S1 digestion for the levels of OCH1 mRNA and DED1 as an internal control. The results were quantified using a phosphorimager, and the OCH1 signal relative to the DED1 signal is indicated below each lane. Similar results were obtained in several experiments. (B) Equal amounts of total cellular RNA from wild-type (MY3683), not4Δ (MY3688), not5Δ (MY3820), bud14Δ (MY3824), not4Δ bud14Δ (MY3825), and not5Δ bud14Δ (MY3826) strains expressing genomically tagged Skn7-Myc and not4-3 (MY2052), msn2Δ (MY3496), not4-3 msn2Δ (MY3498), skn7Δ (MY3725), not4Δ skn7Δ (MY3726), and not5Δ skn7Δ (MY3812) strains, grown exponentially in rich medium, were analyzed for the levels of OCH1 or DED1 mRNA by S1 analysis. not4-3 is an allele of NOT4 in which the 5′ half of the NOT4 ORF is disrupted by insertion of the LEU2 gene (strain YOU584 [43]), which phenotypically is similar to a total disruption of NOT4 and was already used in our studies of Msn2.
We then investigated whether Msn2 contributed to increased OCH1 expression in not4Δ cells by measuring OCH1 mRNA levels in wild-type or not4Δ cells, expressing or lacking MSN2. The deletion of MSN2 had no effect on OCH1 mRNA levels in wild-type or not4Δ cells (Fig. 2B, compare lanes 1 and 5 and lanes 4 and 6), whereas, as before, we saw that increased OCH1 mRNA levels in not4Δ cells required Skn7 (Fig. 2B, compare lanes 1 and 2 to lanes 7 and 8). As in not4Δ cells, OCH1 mRNA levels were increased in not5Δ cells through Skn7 (Fig. 2B, compare lanes 1 and 3 to lanes 7 and 9). Deletion of the Glc7 type I protein phosphatase regulatory subunit Bud14, which mediates increased activity of Msn2 in mutants of the Ccr4-Not complex, did not reduce OCH1 expression in either not4Δ or not5Δ cells (Fig. 2B, compare lanes 1 to 3 to lanes 10 to 12). On the contrary, in the absence of Bud14, the activation of OCH1 in not4Δ and not5Δ cells was remarkably higher.
Taken together, these results show that Skn7-dependent expression of OCH1 is increased under normal growth conditions upon mutation of the Ccr4-Not complex. Msn2 plays no role in this increase. Furthermore, Bud14, which mediates Msn2 activation upon mutation of the Ccr4-Not complex (30), is in contrast inhibitory to Skn7-dependent expression of OCH1 in ccr4-not mutants.
Residue 427 of Skn7 and Not4 independently set thresholds of Skn7-dependent OCH1 expression.
Previous studies have demonstrated that a reporter construct driven by the OCH1 promoter responds to activated alleles of SLN1 (called sln1*) in an Skn7-D427-residue-dependent manner (33). Indeed, D427N Skn7 led to reduced activity and D427E Skn7 resulted in increased activity in this reporter system. To test if the Ccr4-Not complex might impact on D427-dependent transcriptional activation by Skn7, we created plasmids expressing HA-tagged wild-type or D427N or D427E mutant forms of Skn7 (see Materials and Methods). These plasmids were transformed into wild-type or not4Δ cells lacking the chromosomal copy of SKN7. We then grew the transformants in high glucose to exponential phase and extracted total cellular RNA for S1 analysis of OCH1 mRNA levels. We also measured the levels of rRNA in these reactions as a loading control. We observed that in wild-type cells the expression of OCH1 was higher in cells expressing D427E Skn7 and lower in cells expressing D427N Skn7 than in cells expressing wild-type Skn7 (Fig. 3, lanes 1 to 3). Similarly, in not4Δ cells, expression of OCH1 was higher in cells expressing D427E Skn7 and lower in cells expressing D427N Skn7 than in not4Δ cells expressing wild-type Skn7 (Fig. 3, lanes 4 to 6). Thus, not4Δ cells retain the response to residue D427 modification observed in wild-type cells. These results suggest that residue D427 and Not4 independently set thresholds of Skn7-dependent expression of OCH1.
FIG. 3.
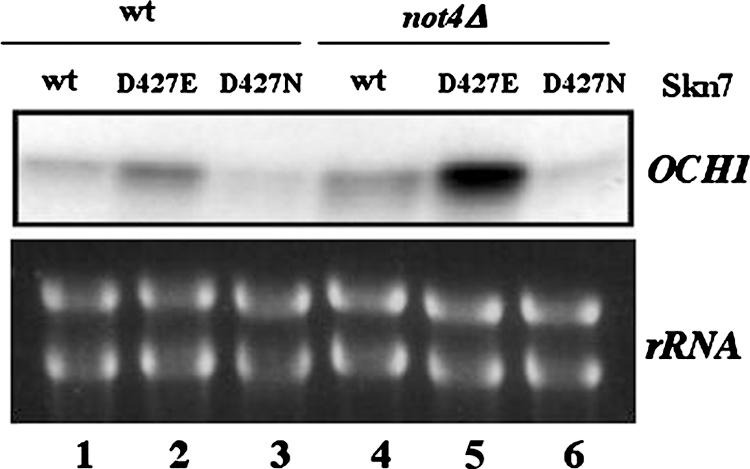
Regulation of Skn7-dependent activation of OCH1 by residue D427 occurs in wild-type and not4Δ cells. skn7Δ strains expressing wild-type or mutant forms of Skn7 from a plasmid (MY5187 to MY5190) as indicated, and additionally a wild-type (MY3725) or not4Δ (MY3726) strain as indicated, were grown exponentially in rich medium. Equal amounts of total cellular RNA were extracted, and OCH1 mRNA levels were revealed by S1 analysis. rRNA is indicated below as a control for RNA loading in the reactions.
The binding of Skn7 to the OCH1 promoter specifically is increased in cells lacking NOT4.
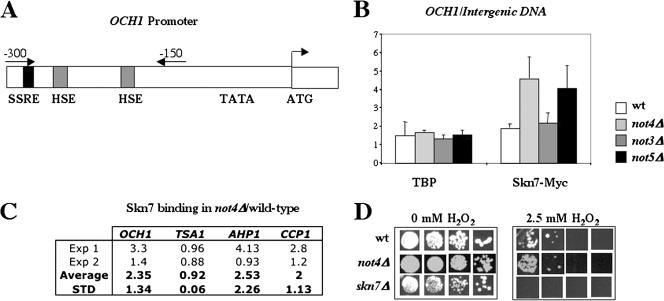
The results presented above suggest that increased Skn7-dependent expression of OCH1 in cells lacking NOT4 is not mediated by activation of residue D427. This would suggest that the Ccr4-Not complex controls Skn7 by a mechanism that is distinct from the control of Skn7 by hypotonic stress. We thus decided to look at the binding of Skn7 to DNA. Indeed, Skn7 has been reported to be exclusively nuclear (36), making it unlikely that the Ccr4-Not complex regulates the localization of Skn7. Moreover, the binding of Skn7 to DNA has been shown to not depend upon residue D427, and thus, an effect of the Ccr4-Not complex on Skn7 binding seemed a reasonable hypothesis. To assay for this, we performed ChIP experiments with wild-type, not3Δ, not4Δ, or not5Δ cells expressing Myc-tagged Skn7. We grew the cells exponentially in high glucose, cross-linked the cells with formaldehyde, prepared total extracts, shared chromatin, and immunoprecipitated Skn7. The presence of several DNA sequences in the immunoprecipitates was investigated by real-time PCR. On one hand we looked at the OCH1 promoter with primers that amplify a region containing one SSRE and two HSEs (heat shock promoter elements), which can all be recognized by Skn7, which has a DNA binding domain with similarity to that of the heat shock factor (10, 40, 45) (Fig. 4A), and on the other hand we amplified an intergenic region that contains no distinguishable HSEs or SSREs. We then expressed the amount of the OCH1 promoter, likely to be bound by Skn7, relative to the intergenic region (unlikely to be associated with Skn7) and observed that there was a significant increase in the association of Skn7 with the OCH1 promoter in cells lacking Not4 compared to wild-type cells (Fig. 4B). A similar increase was observed in cells lacking Not5 but not in cells lacking Not3 (Fig. 4B). No significant change in the association of TBP with OCH1 was found in cells lacking either NOT4 or NOT5 compared to wild-type cells (Fig. 4B). This finding suggests that activation of OCH1 by Skn7 in cells lacking Not4 is likely to act at a step subsequent to TBP binding.
FIG. 4.
The binding of Skn7 is specifically increased on the OCH1 promoter in not4Δ cells compared to wild-type cells. (A to C) The indicated strains, wild-type (MY3683), not3Δ (MY3819), not4Δ (MY3688), and not5Δ (MY3820) expressing genomically tagged Skn7-Myc, were grown exponentially in rich medium, fixed, lysed, and incubated with anti-Myc or anti-TBP antibodies in the presence of protein G-Sepharose beads. The presence of the DNAs of interest in the immunoprecipitates was revealed by real-time PCR after purification of the DNA. (A) The positions of the oligonucleotides used for the amplification of the OCH1 promoter are depicted by arrows, and the positions of the SSRE and HSEs in the OCH1 promoter are indicated. (B) The results presented are the averages of three independent experiments and are expressed as the ratio of the presence of the OCH1 promoter (relative to the input DNA) to that of an intergenic region (relative to input DNA) in the immunoprecipitates. This value is significantly higher in the not4Δ strain than in the wild-type strain (t test, P value of 0.05). Control experiments were done to titrate the amount of Myc antibody required, and cells expressing endogenous untagged Skn7 were used as a control for specific coimmunoprecipitation of DNA by tagged Skn7. (C) The presence of several different promoters in the immunoprecipitates was assessed in two independent experiments and normalized to the intergenic DNA. Ratios of levels in the not4Δ strain to levels in the wild-type strain are presented together with the average and standard deviation (STD). (D) The indicated cells, wild-type (MY1), not4Δ (MY3595), and skn7Δ (MY3725) cells, were grown exponentially in rich medium, and then equal volumes (5 μl) of serial dilutions were spotted on yeast extract-peptone-dextrose plates containing 0 or 2.5 mM H2O2.
These results demonstrate that increased binding of Skn7 to the OCH1 promoter correlates with increased transcription of the OCH1 gene in cells lacking NOT4. We next wanted to determine whether binding of Skn7 to other promoters, in particular to those of oxidative stress-inducible genes that are mediated by different Skn7 binding sites (23, 47), is also increased in cells lacking Not4. Thus, we repeated the ChIP experiments with wild-type and not4Δ cells and evaluated the presence of oxidative stress-inducible promoters, namely, TSA1, AHP1, and CCP1, in addition to that of the OCH1 promoter and intergenic DNA, in the immunoprecipitates by real-time PCR. As before, we found that there was an increase in the association of Skn7 with OCH1 (Fig. 4C). A milder but measurable increase of Skn7 association with CCP1, but not with TSA1, and not reproducibly with AHP1, relative to intergenic DNA, in cells lacking Not4 was also observed (Fig. 4C). In good correlation with the finding of no general effect of Not4 on the association of Skn7 with oxidative stress-responsive promoters, we observed that not4Δ cells had resistance to 2.5 mM hydrogen peroxide similar to that of wild-type cells, resistance that is Skn7 dependent (Fig. 4D). Taken together these findings suggest that Not4 controls preferentially the association of Skn7 with the OCH1 osmotic stress-responsive promoter and that with certain oxidative stress-responsive promoters.
Srb10 mediates OCH1 activation in cells lacking Not4.
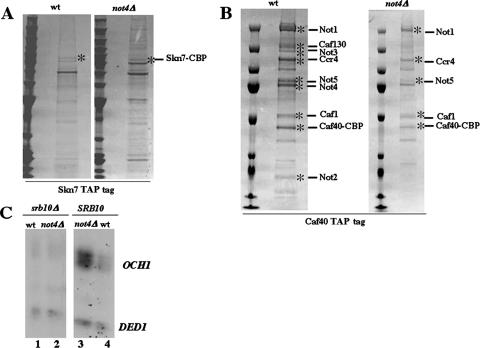
To investigate further the regulation of Skn7 activation by Not4, we purified by tandem affinity chromatography Skn7 TAP tagged at its C terminus from its endogenous promoter and locus. Interestingly, when we visualized the proteins by Coomassie blue staining of the gel, we observed that the profiles of proteins purified with Skn7 from wild-type and not4Δ cells were different (Fig. 5A), and analysis by mass spectrometry of the purified proteins prior to separation by SDS-polyacrylamide gel electrophoresis revealed the presence of a kinase, namely, Srb10, in the preparation of Skn7 from wild-type cells only. We also purified the Ccr4-Not complex from wild-type or not4Δ cells expressing TAP-tagged Caf40 from its endogenous promoter and locus (Fig. 5B). This revealed the absence of Caf130, Not3, and Not2 in Caf40-purified Ccr4-Not complexes when cells lacked NOT4 as determined by both Western blotting and mass spectrometry analysis of the purified material, but most excitingly it revealed the presence of Srb9 in the purification of Ccr4-Not complexes from wild-type but not mutant cells. Indeed, the interest of this finding lies in the fact that Srb9 and Srb10 are part of the same module of the mediator that has been shown to play both negative and positive roles in the regulation of transcription (5). Thus, we considered the possibility that the Srb10 kinase might play a role in the regulation of Skn7 by the Ccr4-Not complex. We created wild-type and not4Δ strains lacking the SRB10 gene and analyzed the expression of OCH1 in these strains. There was no increase of OCH1 expression in not4Δ cells relative to wild-type cells in the absence of Srb10, yet the absence of SRB10 did not seem to have an effect per se on the expression of OCH1 in wild-type cells (Fig. 5C).
FIG. 5.
Purification of TAP-tagged Skn7 and Caf40 in wild-type and not4Δ cells reveals a connection to the Srb10 kinase that mediates activation of OCH1 in not4Δ cells. (A) Wild-type (MY5037) and not4Δ (MY5041) cells expressing TAP-tagged Skn7 were grown to exponential phase, for total extract preparation and TAP. The eluates were loaded on SDS-polyacrylamide gels and stained with Coomassie blue. The indicated proteins (*) were confirmed by mass spectrometry. The most prominent bands in both eluates correspond to heat shock proteins of the Hsp90 and Hsp70 families found in most TAPs. Srb10 cannot be attributed to a particular band as it was identified by mass spectrometry in the purification prior to separation of the proteins by SDS-polyacrylamide gel electrophoresis. (B) Wild-type (MY4858) and not4Δ (MY4910) cells expressing TAP-tagged Caf40 were grown to exponential phase, for total extract preparation and TAP. The eluates were loaded on SDS-polyacrylamide gels and stained with Coomassie blue. The indicated proteins (*) were confirmed by mass spectrometry. The Hsp70 proteins comigrate with Not4; therefore, one does not see Not4 disappear in the not4Δ background. (C) Wild-type (MY1), not4Δ (MY3595), srb10Δ (MY3246), or srb10Δ not4Δ (MY3247) cells were grown exponentially for total RNA preparation. Equal amounts of total cellular RNA were analyzed by S1 digestion for the levels of OCH1 mRNA and DED1 as an internal control.
Taken together, these results show that Srb10 is required for Skn7-dependent increased expression of OCH1 in cells lacking Not4. This correlates with the importance of Not4 for the integrity of the Ccr4-Not complexes that can be purified through Caf40 and for the association of components of the Srb10 kinase module with either the Ccr4-Not complex or Skn7.
DISCUSSION
Skn7, a new target for regulation by the Ccr4-Not complex.
In this work, we have determined that Skn7, a transcription factor involved in the oxidative and osmotic stress responses, is a new component of the transcription machinery that is controlled by the Ccr4-Not complex. We investigated its relationship to the Ccr4-Not complex after it was isolated in a two-hybrid selection with Not1 as bait. We were able to confirm an interaction between Skn7 and two subunits of the Ccr4-Not complex, namely, Not1 and Not5, by coimmunoprecipitation, and we could show that the interaction between Not5 and Skn7 requires Not4. We further found that strains with deletions of NOT4 or NOT5 present an increase in expression of OCH1, a gene that is activated by Skn7 in response to a low osmotic signal, in mutant cells under normal growth conditions. Finally, we confirmed that the induction of OCH1 is mediated by Skn7, because the deletion of SKN7 mostly abolished the induction of OCH1 in cells lacking Not4. The identification of a signal-specific stress transcription factor as a new target for regulation by the Ccr4-Not complex extends the role of this complex, known so far only to regulate the environmental stress response through Msn2, as an integrator of specific stress signals.
Ccr4-Not: an integrator of specific signals for Skn7 activation.
Our results show that activation of OCH1 by Skn7 in cells lacking Not4 correlates with increased binding of Skn7 to OCH1. This gene is activated by Skn7 in response to hypotonic shock through Skn7 binding to the SSRE. Residue D427 of Skn7 is required for activation of Skn7 through SSREs, but it does not affect Skn7 binding to DNA (33). Consistently residue D427 determines the level of OCH1 expression, even in cells lacking Not4. These results show that residue D427 of Skn7 and Not4 independently set thresholds for the expression of OCH1 by Skn7.
Skn7 binds to a number of different sites besides the SSRE, including HSEs (10, 40, 45) or the OSRE, which mediates Skn7-dependent expression of TRX2 and GPX2 in response to oxidative stress (47). Recently yet another Skn7 binding site has been identified in the CCP1 promoter that is responsible for its activation in response to oxidative stress (23). Our work shows that it is mostly the binding of Skn7 to OCH1, and also to some extent to some oxidative stress-responsive promoters such as CCP1, but not to others such as TSA1, that is increased in cells lacking Not4. It could be that Not4 regulates the capacity of Skn7 to associate specifically with SSRE- rather than with OSRE-containing promoters. Alternatively, Not4 might regulate the capacity of Skn7 to associate with other factors present at the OCH1 promoter and other regulated promoters specifically. In any case, it is clear that these findings demonstrate the capacity of Not4 to contribute to the regulation of Skn7 in specific responses, rather than to overall Skn7 activity. Thus, this work provides evidence not only for a new target of the Ccr4-Not complex but also for an additional level of control of the Skn7 transcription factor acting upon its DNA binding to specific target genes.
The Srb10 kinase mediates increased activation of OCH1 by Skn7 in cells lacking Not4.
An exciting finding in this study is the positive role of the Srb10 kinase in the Skn7-dependent activation of OCH1 in cells lacking Not4. Srb10 and its cyclin Srb11 form a stable complex with Srb8 and Srb9 that loosely associates with the 21-subunit core mediator complex (5). This complex is evolutionarily conserved and integrates signals from sequence-specific activators and repressors to the general transcription machinery (3, 4, 27). A recent study has suggested that the Srb10 module may sterically block mediator interactions with RNA polymerase II to inhibit transcription (20). However, Srb10 can phosphorylate the C-terminal domain of the polymerase, but it has many other substrates that may be more relevant for its function in vivo. For instance, negative regulation of transcriptional activators such as Ste12, Gcn4, and Msn2 by Srb10 has been reported (13, 42). Srb10 phosphorylation promotes the rapid degradation of Gcn4. It also promotes Msn2 degradation, but in addition it promotes Msn2 nuclear export, and these different effects are mediated through separable domains (6, 13, 29). In contrast, positive regulation of Gal4 activation by Srb10 has been documented (24). Our current study identifies another positive role for Srb10, namely, promotion of OCH1 activation by Skn7 that correlates with increased DNA binding of Skn7 to this specific promoter.
The copurification of the Srb10 kinase with Skn7 and that of Srb9 with the Ccr4-Not complex, in wild-type but not mutant cells, lead us to propose a model (Fig. 6) in which the interaction of the Srb10 kinase module with the Ccr4-Not complex inhibits the activation of OCH1 by Skn7. Activation might rely on phosphorylation of Skn7 itself or on phosphorylation of other components of the transcription machinery by Srb10 at the OCH1 promoter. In this context it should be mentioned that Skn7 extracted from wild-type cells or not4Δ cells migrates with different apparent sizes on SDS-polyacrylamide gels, suggesting that it exists in different posttranslational states (data not shown).
FIG. 6.
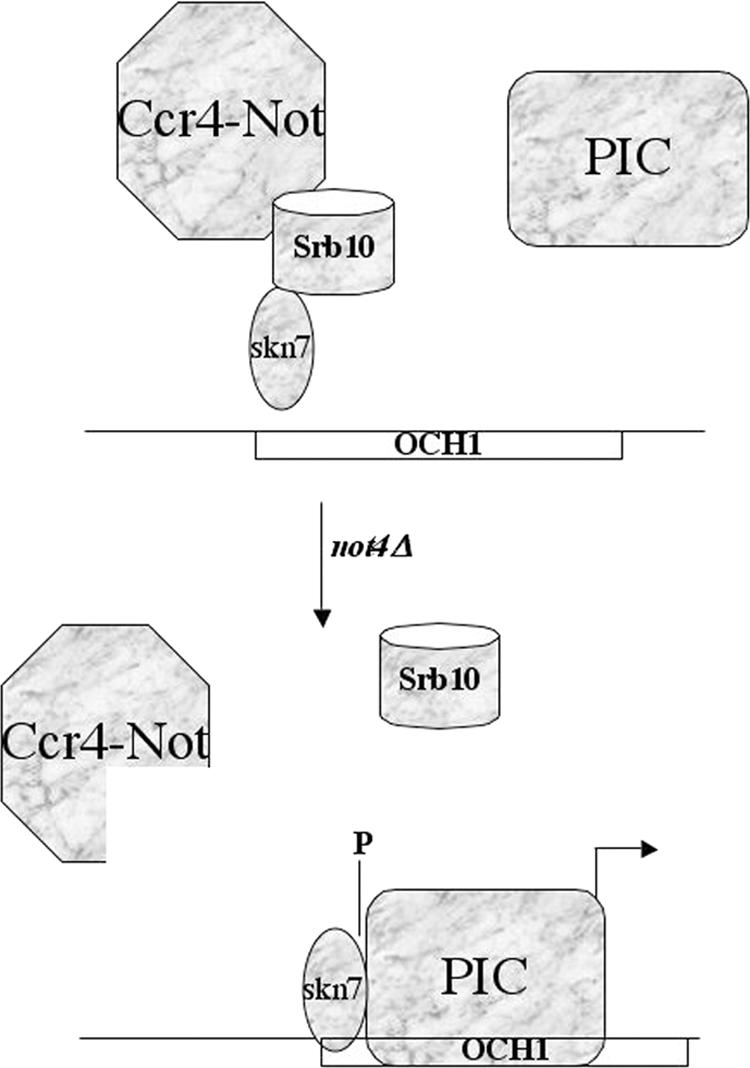
Model for the role of the Ccr4-Not complex in Srb10-dependent regulation of OCH1 activation by Skn7. In wild-type cells, subunits of the Srb10 kinase module (depicted as a cylinder) interact with both Skn7 and the Ccr4-Not complex, and Skn7 interacts with the Ccr4-Not complex, though maybe not directly, or not in a stable way, as this interaction can be detected by two-hybrid assay, but not after purification of the Ccr4-Not complex. This interaction prevents Skn7 from activating OCH1. Upon deletion of the Not4 subunit of the Ccr4-Not complex, interaction between Skn7 and the Ccr4-Not complex is lost. In this mutant, association of Skn7 with the OCH1 promoter increases and the Srb10 kinase module is no longer detectably associated with either Skn7 or the Ccr4-Not complex. Nevertheless, Srb10 is required for increased activation of OCH1 by Skn7 in this mutant, presumably because it has phosphorylated either Skn7 itself or some component of the preinitiation complex (PIC).
Model for regulation by the Ccr4-Not complex.
Our present finding that a component of the Srb10 module is associated with wild-type but not mutant Ccr4-Not complexes raises the question of whether the Ccr4-Not complex regulates specifically the role of Srb10 in Skn7 activation of OCH1 or whether it regulates globally the activity of Srb10. One previous study has already connected the Ccr4-Not complex to the Srb10 module of the holoenzyme by two-hybrid experiments (34), and we have several observations compatible with the idea that the Ccr4-Not complex may have a global rather than specific regulatory impact on the function of the Srb10 kinase. First, GAL1 transcripts accumulate more rapidly in not4Δ cells than in wild-type cells upon transfer from glucose to galactose (data not shown), and Srb10 is known to play a role in GAL1 activation. Second, though we have published that activation of Msn2 in mutants of the Ccr4-Not complex is mediated by the Glc7 type I protein phosphatase and its regulatory subunit Bud14 (30), we have found that Srb10 also contributes to STRE activation in these mutants (data not shown). Thus, we can imagine that the Ccr4-Not complex, which has been reported to be both nuclear and cytoplasmic (for reviews see references 14, 17, and 19), serves as a scaffold for Srb10 and its substrates in response to the appropriate physiological signals, a situation possibly mimicked by mutation of the complex. Alternatively, the complex might function to sequester or inhibit components of the kinase module, releasing or activating them only upon the appropriate signals (Fig. 6). Investigating these different exciting models will most certainly help us understand the mechanism and function of the Ccr4-Not complex.
Acknowledgments
This work was supported by a grant from the National Science Foundation (3100A0-100793) and a grant from the Leenards foundation awarded to M.A.C., and Agnès Michel was funded by the Biomedizin Naturwissenschaft Forschung (BNF).
We thank Laurent Maillet and Michel Werner for their contribution to this work by the initial two-hybrid screen, Klaas Mulder and Marc Timmers for strains (41) and protocols, David Bartford for plasmids and protocols, and Marc Feuermann for his unpublished microarray results. We thank Laurie Stargell and Patrick Masson for a critical reading of the manuscript.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Bai, Y., C. Salvadore, Y.-C. Chiang, M. A. Collart, H.-Y. Liu, and C. L. Denis. 1999. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 19:6642-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becerra, M., L. J. Lombardía-Ferreira, N. C. Hauser, J. D. Hoheisel, B. Tizon, and M. E. Cerdán. 2002. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 43:545-555. [DOI] [PubMed] [Google Scholar]
- 3.Biddick, R., and E. T. Young. 2005. Yeast mediator and its role in transcriptional regulation. C. R. Biol. 328:773-782. [DOI] [PubMed] [Google Scholar]
- 4.Bjorklund, S., and C. M. Gustafsson. 2005. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30:240-244. [DOI] [PubMed] [Google Scholar]
- 5.Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 2002. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277:44202-44207. [DOI] [PubMed] [Google Scholar]
- 6.Bose, S., J. A. Dutko, and R. S. Zitomer. 2005. Genetic factors that regulate the attenuation of the general stress response of yeast. Genetics 169:1215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouquin, N., A. L. Johnson, B. A. Morgan, and L. H. Johnston. 1999. Association of the cell cycle transcription factor Mbp1 with the Skn7 response regulator in budding yeast. Mol. Biol. Cell 10:3389-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, J. L., H. Bussey, and R. C. Stewart. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, J. L., S. North, and H. Bussey. 1993. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175:6908-6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, M. 1997. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13:1-23. [DOI] [PubMed] [Google Scholar]
- 12.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collart, M. A. 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313:1-16. [DOI] [PubMed] [Google Scholar]
- 15.Collart, M. A., and K. Struhl. 1993. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 12:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collart, M. A., and K. Struhl. 1994. NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8:525-537. [DOI] [PubMed] [Google Scholar]
- 17.Collart, M. A., and M. H. T. Timmers. 2004. The eukaryotic Ccr4-Not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways. Prog. Nucleic Acid Res. Mol. Biol. 77:289-322. [DOI] [PubMed] [Google Scholar]
- 18.Deluen, C., N. James, L. Maillet, M. Molinete, G. Theiler, M. Lemaire, N. Paquet, and M. A. Collart. 2002. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol. Cell. Biol. 22:6735-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denis, C. D., and J. Chen. 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73:221-250. [DOI] [PubMed] [Google Scholar]
- 20.Elmlund, H., V. Baraznenok, M. Lindahl, C. O. Samuelsen, P. J. Koeck, S. Holmberg, H. Hebert, and C. M. Gustafsson. 2006. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. USA 103:15788-15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromont-Racine, M., J. C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277-282. [DOI] [PubMed] [Google Scholar]
- 22.Görner, W., E. Durchschlag, M. T. Martínez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schüller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He, X. J., and J. S. Fassler. 2005. Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 58:1454-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirst, M., M. S. Kobor, N. Kuriakose, J. Greenblatt, and I. Sadowski. 1999. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 3:673-678. [DOI] [PubMed] [Google Scholar]
- 25.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ketela, T., J. L. Brown, R. C. Stewart, and H. Bussey. 1998. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet. 259:372-378. [DOI] [PubMed] [Google Scholar]
- 27.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30:235-239. [DOI] [PubMed] [Google Scholar]
- 28.Krems, B., C. Charizanis, and K. D. Entian. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327-334. [DOI] [PubMed] [Google Scholar]
- 29.Lallet, S., H. Garreau, C. Poisier, E. Boy-Marcotte, and M. Jacquet. 2004. Heat shock-induced degradation of Msn2p, a Saccharomyces cerevisiae transcription factor, occurs in the nucleus. Mol. Genet. Genomics 272:353-362. [DOI] [PubMed] [Google Scholar]
- 30.Lenssen, E., N. James, I. Pedruzzi, F. Dubouloz, E. Cameroni, R. Bisig, L. Maillet, M. Werner, J. Roosen, K. Petrovic, J. Winderickx, M. A. Collart, and C. De Virgilio. 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25:488-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenssen, E., U. Oberholzer, J. Labarre, C. de Virgilio, and M. A. Collart. 2002. Saccharomyces cerevisiae Ccr4-Not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 43:1023-1037. [DOI] [PubMed] [Google Scholar]
- 32.Li, S., A. Ault, C. L. Malone, D. Raitt, S. Dean, L. H. Johnston, R. J. Deschenes, and J. S. Fassler. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17:6952-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, S., S. Dean, Z. Li, J. Horecka, R. J. Deschenes, and J. S. Fassler. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13:412-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, H.-Y., Y.-C. Chiang, J. Pan, J. Chen, C. Salvadore, D. C. Audino, V. Badarinarayana, V. Palaniswamy, B. Anderson, and C. L. Denis. 2001. Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem. 276:7541-7548. [DOI] [PubMed] [Google Scholar]
- 35.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 36.Lu, J. M., R. J. Deschenes, and J. S. Fassler. 2003. Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot. Cell 2:1304-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maillet, L., C. Tu, Y. K. Hong, E. O. Shuster, and M. A. Collart. 2000. The essential function of NOT1 lies within the CCR4-NOT complex. J. Mol. Biol. 303:131-143. [DOI] [PubMed] [Google Scholar]
- 38.Martínez-Pastor, M. T., G. Marchler, C. Schüller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan, B. A., N. Bouquin, G. F. Merrill, and L. H. Johnston. 1995. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 14:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulder, K. W., A. Inagaki, E. Cameroni, F. Mousson, G. S. Winkler, C. De Virgilio, M. A. Collart, and H. T. Timmers. 2007. Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics 176:181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson, C., S. Goto, K. Lund, W. Hung, and I. Sadowski. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421:187-190. [DOI] [PubMed] [Google Scholar]
- 43.Oberholzer, U., and M. A. Collart. 1998. Characterization of NOT5 that encodes a new component of the NOT protein complex. Gene 207:61-69. [DOI] [PubMed] [Google Scholar]
- 44.Panasenko, O., E. Landrieux, M. Feuermann, A. Finka, N. Paquet, and M. A. Collart. 2006. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J. Biol. Chem. 281:31389-31398. [DOI] [PubMed] [Google Scholar]
- 45.Raitt, D. C., A. L. Johnson, A. M. Erkine, K. Makino, B. Morgan, D. S. Gross, and L. H. Johnston. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11:2335-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuzi, D., K. Maeta, Y. Takatsume, S. Izawa, and Y. Inoue. 2004. Distinct regulatory mechanism of yeast GPX2 encoding phospholipid hydroperoxide glutathione peroxidase by oxidative stress and a calcineurin/Crz1-mediated Ca2+ signaling pathway. FEBS Lett. 569:301-306. [DOI] [PubMed] [Google Scholar]
- 48.Zervos, A. S., J. Gyuris, and R. Brent. 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72:223-232. [DOI] [PubMed] [Google Scholar]