Abstract
The Aspergillus nidulans ambient pH signaling pathway involves two transmembrane domain (TMD)-containing proteins, PalH and PalI. We provide in silico and mutational evidence suggesting that PalI is a three TMD (3-TMD) protein with an N-terminal signal peptide, and we show that PalI localizes to the plasma membrane. PalI is not essential for the proteolytic conversion of the PacC translation product into the processed 27-kDa form, but its absence markedly reduces the accumulation of the 53-kDa intermediate after cells are shifted to an alkaline pH. PalI and its homologues contain a predicted luminal, conserved Gly-Cys-containing motif that distantly resembles a Gly-rich dimerization domain. The Gly44Arg and Gly47Asp substitutions within this motif lead to loss of function. The Gly47Asp substitution prevents plasma membrane localization of PalI-green fluorescent protein (GFP) and leads to its missorting into the multivesicular body pathway. Overexpression of the likely ambient alkaline pH receptor, the 7-TMD protein PalH, partially suppresses the null palI32 mutation. Although some PalH-GFP localizes to the plasma membrane, it predominates in internal membranes. However, the coexpression of PalI to stoichiometrically similar levels results in the strong predominance of PalH-GFP in the plasma membrane. Thus, one role for PalI, but possibly not the only role, is to assist with plasma membrane localization of PalH. These data, considered along with previous reports for both Saccharomyces cerevisiae and A. nidulans, strongly support the prevailing model of pH signaling involving two spatially segregated complexes: a plasma membrane complex containing PalH, PalI, and the arrestin-like protein PalF and an endosomal membrane complex containing PalA and PalB, to which PacC is recruited for its proteolytic activation.
In filamentous fungi and yeasts, regulation of gene expression by the ambient pH involves six dedicated components of a pH signaling pathway that mediates the proteolytic activation of the zinc-finger transcription factor PacC/Rim101p in response to ambient alkaline pH (2, 36, 37). pH regulation has been studied most extensively in Aspergillus nidulans and Saccharomyces cerevisiae genetic models. In A. nidulans, the pal/PacC pathway tailors the synthesis of plasma membrane transporters, extracellular hydrolytic enzymes, and exported metabolites to the needs imposed by the ambient pH. The S. cerevisiae RIM/Rim101p pathway regulates the ambient alkaline pH response, the ion tolerance, and the cell differentiation and additionally contributes to cell wall assembly (8, 24, 25, 44).
Apart from the finding that PalB/Rim13p, one of the six pal/RIM components, is a calpain-like cysteine protease that almost certainly mediates the single proteolytic activating step of yeast Rim101p (15, 27, 53) and the first of the two proteolytic steps involved in PacC activation (11, 12, 33, 38), the characterization of the genes encoding the six pH signaling proteins gave few clues as to their precise molecular function. Recent work with both A. nidulans and S. cerevisiae has dramatically changed this situation and has revealed an unexpected additional role in pH signal transduction for most, but not all, of the components of the multivesicular body pathway cargo-sorting protein complexes (6, 16, 18, 19, 42, 52-54). Two A. nidulans pH signaling proteins are predicted to be membrane residents. The seven-transmembrane domain (7-TMD) protein PalH (32), which has two S. cerevisiae homologues, Rim21p and Dfg16p (5), is almost certainly a component of a pH signaling receptor as its cytosolic tail interacts strongly with the PalF arrestin-like protein, which is ubiquitinated and phosphorylated in an alkaline ambient pH- and PalH-dependent manner (19). PalI, a second pH signaling, dedicated TMD-containing protein, acts upstream of or in concert with PalH (19). Because PalF is a positive-acting arrestin-like protein, multiubiquitination is an endocytic signal, and positive-acting mammalian arrestins promote signaling of their cognate-activated receptors from endosomes (26), PalH, PalF, and PalI hypothetically would form a pH signaling complex at the plasma membrane whose role seemingly also involves endocytic trafficking.
Evidence that a second protein complex on endosomal membranes plays a key role in pH signaling is compelling. With the sole exceptions of Vps24p and Vps2p, whose deletion results in a certain degree of constitutivity (18), all components of the endosomal-sorting-complex-required-for-transport (ESCRT) complexes I, II, and III are required for Rim101p processing (54). PalA/Rim20p interacts with Vps32, a key component of ESCRT-III, through its Bro1 domain (52, 53). Vps32 binds membranes by itself and through its interacting partners ESCRT-II (49) and ESCRT-III Vps20 (3, 28). As Rim20p-containing endosomes segregate spatially from multivesicular body pathway endosomes (6), the prevailing model assumes that the pH signaling pathway hijacks multivesicular body pathway sorting components for its PacC/Rim101p processing purposes (6, 16). Although subcellular localization of PalB/Rim13p, the likely signaling protease for PacC and the sole protease for Rim101p activation, has not yet been reported, this calpain-like signaling protease is included with the endosomal membrane module, as yeast Rim13p is a two-hybrid interactor of Vps32 (23), and PalB can potentially be recruited to ESCRT-III through its MIT interacting domain (43). The transcription factors and signaling protease substrates PacC and Rim101p would be recruited to endosomes through PalA/Rim20p, which bind their respective transcription factor substrates PacC72 and Rim101p (52, 53). As the PalA binding motifs in PacC72 flank the signaling protease cleavage site (52), the suggestion that Rim20p helps to determine the cleavage site specificity of Rim13p on Rim101p (53) is a highly attractive but as-yet-untested possibility which would also agree with the relatively low target sequence specificity of the A. nidulans signaling protease (38).
The existence of two sequentially acting pH signaling protein complexes is strongly buttressed by epistasis analyses in yeast (18) and by data showing that PalA, PalB, and PalC are not required for PalF ubiquitination, arguably the most direct indication of the activation of the ambient pH sensing mechanism (19). PalF ubiquitination and yeast epistasis analyses have also determined that PalI and Rim9p act upstream of the endosomal pH signaling complex, but the finding that PalF ubiquitination is not fully prevented by a null palI allele additionally provided a molecular explanation as to why the A. nidulans null palI mutant shows a leaky pH regulatory phenotype (19).
As PalC (whose likely S. cerevisiae homologue is YGR122w) (5, 16, 42) is a Vps32 interactor, this protein can potentially localize to endosomes. However, PalC is recruited to plasma membrane-associated punctate structures in an alkaline pH- and a PalH-dependent manner but in a PalA-independent manner, which led to the suggestion that PalC can liaise with both the PalA/PalB/PacC endosomal membrane pH signaling complex and the putative pH-sensing PalH/PalI/PalF plasma membrane signaling complex (16). However, while the endosomal association of Rim20p has been unequivocally demonstrated for yeast, the localization of green fluorescent protein (GFP) fusions involving A. nidulans PalH or PalI has not been analyzed, and the localization of their S. cerevisiae homologues Rim9p, Rim21p, and Dfg16p has not been determined (http://yeastgfp.ucsf.edu/).
We report here that PalI and PalH indeed localize to the plasma membrane, but PalH requires the coexpression of PalI at similar levels to predominate in the plasma membrane, strongly suggesting that one (but possibly not the only) role of PalI is to promote the plasma membrane localization of PalH. Notably, when these findings are considered in conjunction with the data on the localization of Rim20p to endosomes reported by Boysen and Mitchell (6), this report represents strong evidence for the existence of two spatially separated complexes in the fungal pH signaling pathway.
MATERIALS AND METHODS
A. nidulans techniques.
A. nidulans strains carried markers in standard use (9). Strains carrying mutant argB2 and pyroA4 alleles were used as recipients of pALCargB and pALCpyroA constructs, respectively. Transformation (51) and phenotypic testing of pH regulatory phenotypes in strains expressing proteins under the control of alcAp have been described previously (13). Strains which have been constructed for this work are shown in Table 1. Sequence changes for palF15, palI30, and palI32 have been published previously (10, 19). palH72 carries a G-to-A transition in nucleotide 39, resulting in the conversion of codon 13 to a stop codon.
TABLE 1.
A. nidulans strains used in this worka
| Strain | Full genotype | GFP reporter |
|---|---|---|
| 1014 | palI30 areAr49 argB2::[pALC::palI-GFP]argB*; pantoB100; fwA1 | PalI |
| 1015 | biA1; wA3; palA1 argB2::[pALC::palH-GFP]argB* areAr49 | PalH |
| 1039 | yA2; argB2::[pALC::palH-GFP]argB*; palF15 | PalH |
| 1113 | yA2 adE20; palI32 argB2::[pALC::palH-GFP]argB* | PalH |
| 1114 | yA2 pabaA1 palH72; palI32 argB2::[pALC::palH-GFP]argB*; inoB2 | PalH |
| 1137 | yA2 adE20 palH72; palI30 areAr49 argB2::[pALC::palI-GFP]argB*; fwA1 | PalI |
| 1139 | areAr49 palI30 argB2::[pALC::palI30-GFP]argB*; pantoB100; fwA1 | PalIG47D |
| 1263 | palH72; argB2::[pALC::palH-GFP]argB*; pyroA4::[pALC::PalI]pyroA*; pantoB100 | PalH |
| 1288 | palH72; argB2::[pALC::palH-GFP]argB*; pyroA4; pantoB100 | PalH |
| 1293 | palH72; argB2::[pALC::palI-GFP]argB*; inoB2; fwA1 | PalI |
| 1421 | palH72; argB2::[pALC::palH-GFP]argB*; pyroA4::[pALC::PalC]pyroA*; pantoB100 | PalH |
| 1500 | palH72; argB2::[pALC::palH-GFP]argB*; pyroA4::[pALC::PalI::HA3]pyroA*; pantoB100 | PalH |
| 1984 | biA1 yA2 pabaA1; palI32; meaA8 pyroA4::[pALC::PalI::HA3]pyroA* | None |
An asterisk indicates a truncated nonfunctional allele.
Selection of new palI alleles.
The palI alleles were selected in palI30 heterozygous diploid strains by using the γ-aminobutyric acid (GABA) selection technique as described previously (50). All mutants were induced by UV and selected at 37°C on minimal medium containing 5 mM GABA as the nitrogen source and 1% glucose as the carbon source (50) and characterized following benlate haploidization and subsequent purification. The palI300 through palI307 alleles were selected in diploid strain no. 1 (pabaA1 yA2 areAr5 palI30 pantoC3 lysD20/areAr5 inoB2 glrA1 fwA1). The palI308 through palI324 alleles were selected in diploid strain no. 2 (pabaA1 yA2 puA2 areAr5 palI30 spsA1 lysD20/areAr5 inoB2 glrA1 fwA1). lysD is located on chromosome VII. lysD20 is a chromosome III-VII translocation. Thus, haploidization was done using complete medium supplemented with 15 mM l-arginine to select against the palI30-containing chromosome III homologue (and unavoidably also against the chromosome VII homologue involved in the lysD20 translocation). To avoid mitotic recombinants homozygous for palI30, only pantoC+ (for diploid no. 1) and spsA+ (for diploid no. 2) haploids were analyzed further. pantoC3 and spsA1 are recessive. Both are linked and are centromere distal to palI, but spsA1 is preferable to pantoC3 because it is closer to palI (1).
Plasmids.
All plasmids used for targeted integration into argB2 were derived from pALC1 (31). The palH and palI coding sequences were PCR amplified and subcloned into the single BamHI site of pALCargB::GFP (30) to yield pALCargB::PalH-GFP and pALCargB::PalI-GFP, respectively. In the PalH- and PalI-GFP fusion proteins, GFP is separated from the Pal moieties by a Gly-Ser-Pro linker. pALCargB::PalIG47D-GFP was constructed by site-directed mutagenesis using a Stratagene QuickChange site-directed mutagenesis kit. Single-copy integration of the plasmids in transformants selected for further analyses were confirmed in all cases by Southern blotting using an argB-specific probe.
Plasmids for targeted integration into pyroA4 were all derived from pAPN1, a derivative of pPAP(M1) (7). This vector is a pUC19-based 7.4-kb expression construct carrying, downstream of the alcAp promoter (ending at the 3′ KpnI site), the coding region of a truncated FLAG-NIMA NimA protein kinase (with a HindIII site 3′ to the FLAG tag), followed by a synthetic linker (StuI-NheI-AgeI) and 0.75 kb of the phoA gene 3′ untranscribed region. Using KpnI-AgeI to digest this construct allows substitution of the NimA coding region by any other gene of interest, obtained by PCR using KpnI- and AgeI-anchored oligonucleotides. The pyroA4 loss-of-function mutation results in the replacement of the tetranucleotide sequence 1328GGAC1331 by T (nucleotide numbering as presented in the GenBank entry no. AF133101 [34]). This mutation results in a Gly89 deletion and a His90Tyr substitution in the mutant protein. The transformation marker in pAPN1 is a 2,271-bp genomic fragment that contains a mutant pyroA gene carrying a 113-bp EagI-BssHII deletion. This results in the truncation of PyroA after Ala222. The integration of pAPN1 into pyroA4 is selected by plating transformants on synthetic medium lacking pyridoxine, as a functional pyroA gene can be reconstructed only if the integration of the transforming covalently closed plasmid into the genome involves a crossover within the 460-bp region separating the chromosomal mutation from the deletion/frameshift mutation in the plasmid. As an additional crossover upstream of pyroA4 results in the replacement of this mutant allele with that of the wild type (without integration of plasmid sequences), transformants were checked by Southern blotting to confirm plasmid (single-copy) integration. pAPN1 derivatives expressing pH regulatory proteins were named pALCpyroA::X, where X indicates a Pal protein.
Constructs pALCpyroA::PalI and pALCpyroA::PalC drive the expression of PalI and PalC, respectively, under the control of the alcAp promoter. The palI and palC coding sequences were amplified from genomic DNA and cloned into pAPN1 as KpnI-AgeI (for untagged versions) or as KpnI-StuI (for subsequent C-terminal tagging of PalI either with three copies of the hemagglutinin epitope [(HA)3] or with GFP) fragments. The GFP- and (HA)3-encoding regions that were used for constructing epitope-tagged derivatives of pALCpyroA::PalI were derived from pAM6 and pAM21. pAM6 and pAM21 are TOPO 2.1-based plasmids containing DNA fragments that encode three copies of the hemagglutinin epitope and a single copy of GFP, respectively, flanked by StuI and AgeI sites. Plasmids pALCpyroA::PalI::(HA)3 and pALCpyroA::PalIG47D::GFP were made by inserting wild-type and Gly47Asp PalI mutant-encoding KpnI-StuI fragments in frame with C-terminal StuI-AgeI (HA)3- and GFP-encoding regions, respectively, between the KpnI and AgeI sites of pAPN1.
Microscopy.
Germlings of strains expressing untagged and epitope-tagged proteins under the control of the alcAp promoter were cultured on glass coverslips submerged in 2.5 ml of appropriately supplemented “watch” minimal medium (WMM) adjusted to an acidic pH with 25 mM NaH2PO4 (35). Coverslips were incubated overnight at 25°C with 0.02% glucose as the sole C source and transferred to a medium containing 1% (vol/vol) ethanol as the sole carbon source for an additional 3 h before microscopy was performed. Microscopy was carried out using a Nikon E-600 upright epifluorescence microscope equipped with a 100× 1.40 numerical aperture Plan apochromat objective and a Nikon B-2A GFP filter. Images were recorded with an Orca-ER camera (Hamamatsu) driven by Metamorph (Universal Image Co.) software. The contrast of 12-bit images was improved using either Metamorph or Wasabi 1.5 (Hamamatsu Photonics GmbH, Germany) software before images were converted to an 8-bit format.
Total membrane protein extraction and Western blotting analysis.
Membrane protein extracts were prepared by following a previously reported procedure for S. cerevisiae (45), which was adapted to A. nidulans strains expressing epitope-tagged PalH or PalI proteins under the control of the alcAp promoter, as follows. An appropriately supplemented minimal medium solution containing 0.05% glucose (wt/vol) and 5 mM ammonium tartrate as the carbon and nitrogen sources, respectively, was inoculated with a conidiospore suspension and incubated for 14 h at 28°C and shaking at 200 rpm. Mycelia were harvested by filtration, transferred to the same medium containing 1% ethanol (vol/vol) (inducing conditions) or 1% glucose (wt/vol) (repressing conditions) as the sole carbon source, and incubated for an additional 3 h before ∼200-mg (wet weight) samples were harvested into FastPrep 2-ml plastic tubes, which were stored at −70°C. Samples were removed from −70°C storage and thawed in the presence of 0.75 ml of MPE buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, and 25 mM N-ethylmaleimide), using one tablet of complete protease inhibitor cocktail (EDTA-free; Roche) per 10 ml of buffer. Cells were ground in a FastPrep, using 0.5 ml of glass beads and four 45-s pulses under cold conditions. Lysates were centrifuged at 800 × g for 3 min at 4°C. The resulting pellet was resuspended in an additional 0.75 ml of MPE buffer and centrifuged at the above settings. The supernatants from these centrifugation steps were combined, and a crude membrane fraction was collected by centrifugation for 45 min at 12,000 × g and 4°C. Membrane-enriched pellets were resuspended in MPE buffer containing 5 M urea for 30 min at 4°C, using a rotating wheel. Membrane-enriched material was collected by centrifugation for 45 min at 12,000 × g and 4°C and resuspended in 0.2 ml of MPE buffer without urea. Total membrane protein was determined using a bicinchoninic acid protein assay kit (Sigma). GFP- and (HA)3-tagged protein extracts (15-μg and 0.5-μg samples, respectively) were incubated with Laemmli loading buffer (6 M urea, 2% sodium dodecyl sulfate [SDS], 5% mercaptoethanol) at 37°C for 10 min. Samples were loaded onto 7.5% SDS-polyacrylamide gels (Bio-Rad) adjacent to Rainbow molecular mass markers (10,000 to 250,000 Da; Amersham Biosciences). Proteins were transferred to nitrocellulose membranes that were reacted with mouse monoclonal anti-GFP cocktail (clones 7.1 and 13.1; Roche) or rat anti-HA (catalog no. 3F10; Roche) primary antibodies. The secondary antibodies were peroxidase-coupled goat anti-mouse immunoglobulin G (Jackson) and peroxidase-coupled goat anti-rat immunoglobulin G (Southern Biotechnology), respectively. Peroxidase activity was revealed using an ECL Western blotting detection system (Amersham Pharmacia).
RESULTS
In silico analyses in combination with mutational evidence suggest that PalI is a TMD protein and contains a signal peptide.
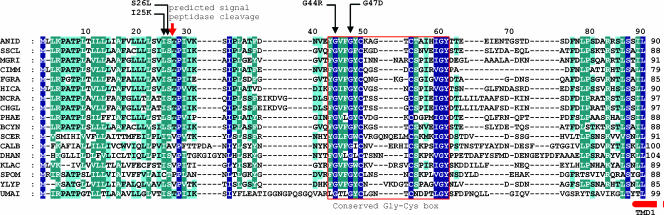
PalI and its S. cerevisiae homologue Rim9p contain four hydrophobic segments which might represent TMD regions. However, Li and Mitchell (27) noted that the most N-terminal of these regions is located at or very close to the N terminus of Rim9p and suggested the possibility that it represents a signal peptide. Indeed SignalP 3.0 (Technical University, Denmark) predicted, with a probability of 0.96, that the N-terminal hydrophobic segment in PalI is a signal peptide, with a high probability (0.84) that the cleavage site corresponds to the peptide bond located between Ser26 and Thr27. Multiple alignments using amino acid sequences of PalI/Rim9p homologues from ascomycetous fungi and yeasts revealed that Ser26 and Thr27 are within a notably conserved motif comprising PalI residues 25 through 33 within the clearly conserved hydrophobic N-terminal 35 residues (Fig. 1). The functional role of this motif is strongly supported by its marked conservation in the less closely related basidiomycete Ustilago maydis. We isolated loss-of-function mutations in palI (Table 2). TMDs dictate both the folding and the topology of a membrane protein. In agreement, most of these mutations resulted in premature truncation, removing one or more of the TMDs. However, among these new alleles are four missense mutations. Two, Ile25Lys and Ser26Leu, result in nonconservative substitutions of residues within this motif, including Ser26, involved in the predicted scissile bond. These data support the contention that residues 1 to 26 in PalI represent a signal peptide and thus that this protein (and, by extension, its homologues) is a 3-TMD protein with the basic C terminus facing the cytosol.
FIG. 1.
The N-terminal region of PalI and Rim9p family members. Multiple sequence alignment of the ∼100 N-terminal residues of PalI/Rim9p homologues in filamentous and yeast ascomycetes and the dimorphic basidiomycete Ustilago maydis. Single-residue substitutions of residues within this region that led to complete or partial loss of function are indicated (Table 2). The position of the signal peptide cleavage site predicted by SignalPeptide 3.0 (http://www.cbs.dtu.dk/services/SignalP/) with a 0.84 probability is indicated by a red arrow. The red box area is the conserved Gly-Cys motif. The start of the first TMD, corresponding to the second hydrophobic region from the N termini of the preproteins (10), is indicated by a broken bar. Conserved residues (according to the Blosum62 matrix) were shaded with navy blue, blue-green, and blue, indicating full, more than 80% but less than full conservation, and more than 60% but less than 80% conservation, respectively. ANID, A. nidulans; SSCL, Sclerotinia sclerotiorum; MGRI, Magnaporthe grisea; CIMM, Coccidioides immitis; FGRA, Fusarium graminearum; HICA, Histoplasma capsulatum; NCRA, Neurospora crassa; CHGL, Chaetomium globosum; PHAE, Phaeosphaeria nodorum; BCYN, Botrytis cinerea; SCER, S. cerevisiae RIM9p; CALB, Candida albicans Rim9; DHAN, Debaryomyces hansenii; KLAC, Kluyveromyces lactis; SPOM, Schizosaccharomyces pombe; YLYP, Yarrowia lipolytica; UMAI, Ustilago maydis.
TABLE 2.
New mutations characterized in palIa
| Allele | Truncating mutations
|
Missense mutations
|
||
|---|---|---|---|---|
| DNA change | Protein | DNA change | Substitution | |
| palI320 | ΔC64-T71 | L22fs | ||
| palI304 | T67A + ΔC69-C70 | S23fs | ||
| palI315 | ΔC83-G93 | P28fs | ||
| palI309 | DT84-T90 | P28fs | ||
| palI303 | 85 insA | I29fs | ||
| palI312 | A91T | K31stop | ||
| palI321 | T96C + ΔA97 | S32fs | ||
| palI317 | T137C + G139T + ΔG140 | G47fs(F46S) | ||
| palI314 | ΔT274 | F76fs | ||
| palI310 | ΔT342-C343 | F98fs | ||
| palI300 | ΔA414 | L122fs | ||
| palI311 | T413G | L122stop | ||
| palI308 | ΔT470-T477 | L141fs | ||
| palI313 | ΔT498 | H150fs | ||
| palI316 | G516insA | I157fs | ||
| palI307 | T74A | I25K | ||
| palI322 | C77T | S26L | ||
| palI306 | G130A | G44R | ||
| palI319 | A326C | H93P | ||
Nucleotide numbering starts from the A in the initiation codon of the genomic palI sequence (GenBank entry AJ007629). The abbreviation fs indicates a frameshift, while stop denotes a nonsense codon. In contrast to the null phenotype of the palI30 (G47D) mutation, all four missense mutations listed here retain some residual activity.
PalI is a plasma membrane protein.
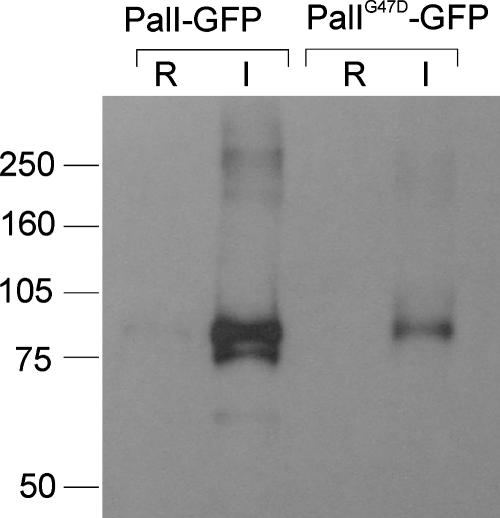
In view of the above-described conclusion, we tagged PalI with GFP at the C terminus, using alcAp, the promoter of the A. nidulans alcohol dehydrogenase gene, to express the fluorescent protein fusion (31). This PalI-GFP fusion protein is functional, as determined by its ability to complement the phenotypically null palI30 mutation (data not shown). A strain carrying an alcAp::palI-GFP transgene integrated into a single copy at the argB locus was constructed by transformation. This strain expressed high levels of the fluorescent fusion protein under inducing conditions (with ethanol as the carbon source), as determined by Western blotting analyses of membrane-enriched fractions, whereas growth under repressing conditions (1% glucose as the carbon source) largely prevented transgene expression (Fig. 2).
FIG. 2.
Western blotting analysis of cells expressing PalI-GFP and PalIG47D-GFP fusion proteins. Strains expressing wild-type and Gly47Asp PalI-GFP fusion proteins under the control of the alcAp promoter were cultured overnight in minimal medium with 0.05% (wt/vol) glucose and shifted for 3 h to either 1% ethanol-containing medium (I, inducing conditions for alcAp) or 1% glucose-containing medium (R, repressing conditions for alcAp) before proceeding to membrane protein extraction. Proteins were analyzed by Western blotting, which was reacted using a cocktail of monoclonal anti-GFP antibodies (Roche). Approximately equal loading of the different lanes was confirmed after protein staining of a duplicate gel. Standards (at left) are in kDa.
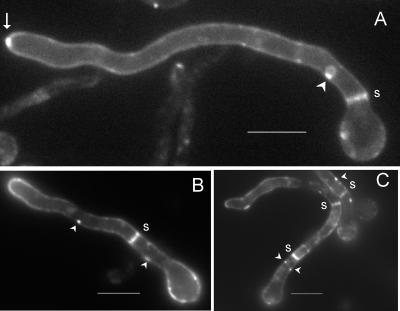
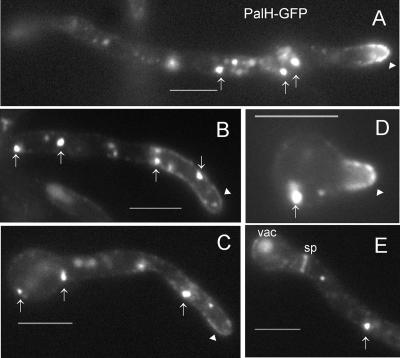
PalI-GFP robustly localized to the plasma membrane at the cell periphery and septae in germlings cultured for 14 to 16 h under inducing conditions (Fig. 3A). Such localization is characteristic of resident plasma membrane proteins. In addition, we observed a marked spot of strong fluorescence at the hyphal apices, at the position corresponding to the Spitzenkörper (Fig. 3A). This is in agreement with the current view that secretion is highly polarized in filamentous fungi (17, 47) and that the Spitzenkörper acts as a vesicle supply center (4). It is also in agreement with the strong labeling of the A. nidulans Spitzenkörper with FM4-64, presumably after exocytic recycling of endocytosed membranes (14), and with the clustering of vesicles, of which most if not all are believed to be secretory vesicles, at the apical region of A. nidulans hyphae (22). Prominent localization at the plasma membrane was also noticeable for germlings subjected to a 3-h induction period (Fig. 3B and C). In this case, the marked apical spot was not visible, although the fusion protein was clearly polarized, as indicated by the brighter fluorescence seen at the hyphal tips (Fig. 3B). It is notable that PalI-GFP distribution in the plasma membrane was patchy, and we frequently observed labeling of punctate structures adjacent to the plasma membrane (Fig. 3B and C). Thus, the fact that PalI plays a definite but nonessential role upstream of the PalF arrestin (19) and localizes to the plasma membrane is consistent with this protein playing an accessory role upstream or in concert with a plasma membrane ambient pH sensor.
FIG. 3.
PalI localizes to the plasma membrane. Epifluorescence microscopy using a filter set specific for GFP of germlings cultured in acidic WMM. (A) With ethanol as the sole carbon source. The arrow indicates strong labeling in the position corresponding to the Spitzenkörper. (B and C) Germlings germinated in 0.02% glucose and shifted to WMM with 1% ethanol for 3 h. Membrane-associated punctate structures are indicated by arrowheads, whereas septae are indicated by s. Bars, 5 μm.
A single-residue substitution involving a conserved Gly leads to PalI mislocalization.
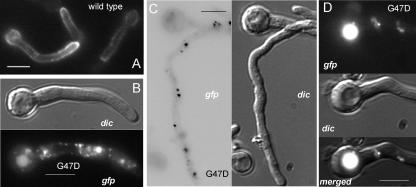
The most N-terminal TMD (formerly the second TMD) predicted in PalI extends from Ile89 through Ala111. The His93Pro substitution leading to two consecutive Pro residues within this predicted α-helical region (Table 2) leads to partial loss of function. Within the region delimited by the predicted signal peptidase cleavage site and this N-terminal TMD, amino acid sequence alignments revealed a strongly conserved motif with consensus sequence Y/FGVFGψC(X)4-9CSXφXφGY (where ψ indicates an aromatic residue, φ indicates an aliphatic residue, X indicates any amino acid, and underlining indicates conservation in PalI homologues from every ascomycetous filamentous fungus and yeast examined and in the basidiomycete U. maydis) (Fig. 1). We named this motif the Gly-Cys box, corresponding to residues 43 through 61 in PalI and containing three invariant Gly residues and two invariant Cys residues. The phenotypically null palI30 mutation (10) results in the replacement of Gly47 by Asp, whereas the phenotypically leaky palI306 mutation (Table 2) results in the replacement of Gly44 by Arg, indicating functional and/or structural roles for the invariants Gly44 and Gly47. We constructed an alcAp transgene driving the expression of PalIG47D-GFP. Western blotting demonstrated that Gly47Asp leads to a conspicuous reduction in the levels of the fusion protein detected in membrane-enriched fractions (Fig. 2). In agreement, epifluorescence microscopy demonstrated that, in marked contrast with the robust plasma membrane localization of the wild type, the Gly47Asp mutant PalI-GFP fusion protein localizes to cytosolic structures possibly representing mature endosomes and to the lumens of vacuoles, indicating that the fusion protein is sorted into the multivesicular body pathway (Fig. 4). We conclude that Gly47 within the Gly-Cys box is required for the plasma membrane localization of PalI-GFP.
FIG. 4.
PalIG47D-GFP mislocalization to the endosomal system and the vacuole. (A) GFP fluorescence microscopy of a strain expressing wild-type PalI-GFP. (B, C, and D) Fluorescence microscopy of germlings expressing PalIG47D-GFP, including Nomarski differential interference contrast (dic) and GFP images, as indicated. (C) Fluorescent cytosolic dots are shown, using reversed contrast for clarity. (D) Vacuole labeled with GFP fluorescence, as seen in mutant but not in wild-type germlings. Germlings were cultured overnight at 25°C in acidic WMM with 0.02% glucose and shifted for 3 h to the same medium with ethanol as the sole carbon source. Bars, 5 μm.
Evidence for concerted actions of PalI and PalH.
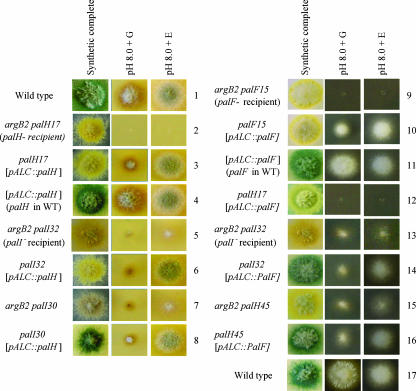
PalH and PalI are 7-TMD and 3-TMD proteins, respectively. They act upstream of the ambient pH-dependent ubiquitination of the arrestin-like protein PalF. However, null palI mutants, in contrast to null palH mutants, impair but do not prevent PalF ubiquitination (19). In agreement with these molecular phenotypes and in stark contrast with null mutations in palA, -B, -C, -F, and -H, which prevent growth completely at an alkaline pH, null palI mutations allow some growth on alkaline pH plates (10) (Fig. 5, compare rows 2 and 5). As growth on alkaline pH plates is the most sensitive test of pH regulation (see below), this suggests that PalI plays an important role but not an essential one.
FIG. 5.
Overexpression of PalH and, to a lesser extent, PalF suppresses the reduced ability of palI loss-of-function mutant strains to grow at an alkaline pH. Forced expression of PalH and PalF was driven by single-copy transgenes integrated at the argB locus, using the alcAp promoter. Thus, recipient strains carry the argB2 allele in addition to the relevant pH regulatory mutations (as shown also in Fig. 7), and transgene expression mirrors that of the alcA gene, which is induced by ethanol (E) and repressed by glucose (G). Under conditions of arginine supplementation, the argB2 mutation does not affect growth on pH media. The synthetic complete (pH 6.5) medium is permissive for the acidity-mimicking pal mutants. However, in contrast to the wild type (strains 1, 4, and 11), the pal mutants grow poorly at pH 8.0, unless this phenotype is complemented (control strains 3 and 10) or suppressed (see relevant strains) by expression of the transgene. Growth on alkaline pH plates is the most sensitive test for ambient pH signaling, as shown by the residual growth under repressing conditions of strains 3 and 10, due to very low levels of expression of alcAp under repressing conditions. palH17 and palF15 carry phenotypically null mutations. palH45 is a leaky palH mutation allowing some growth at alkaline pH. As noted in the text, palI mutations characteristically allow some growth at alkaline pH (strains 5, 7, and 13). Note that strains carry different spore color markers (wild-type green, mutant yellow, or mutant chartreuse).
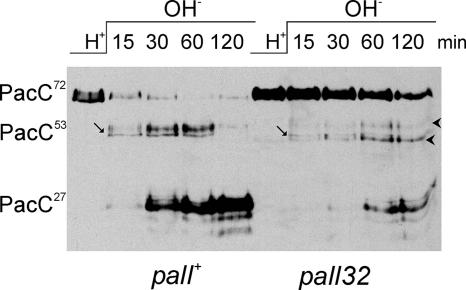
Transferring cells grown under acidic conditions to alkaline ambient pH conditions results in the two-step conversion of PacC72 into PacC53 and then into PacC27. To analyze the effects of the null palI32 mutation in this short-term response to ambient alkalinization, we followed the two-step processing of PacC in a pH shift experiment (12, 38). palI32, unlike the complete loss-of-function mutations in palA, palB, and palC (12, 16, 38), does not fully prevent processing under these conditions and allows slow yet detectable formation of PacC27, which is most conspicuous at the 60- and 120-min time points (Fig. 6). Slow processing to PacC27 in these mutant cells occurs with a markedly reduced accumulation of PacC53 compared to that of the wild type (Fig. 6, PacC53 is indicated with an arrow; PacC53 heterogeneity results from multiple phosphorylation [A. Hervás-Aguilar, J. M. Rodríguez, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva, unpublished results]). As signaling proteolysis is largely, but not fully, prevented by palI32, one possible interpretation is that the reduced rate of PacC72 conversion to PacC53 in the mutant makes processing of PacC53 to PacC27 nonlimiting. The palI32 mutation also results in the slow accumulation of two bands showing slightly reduced and increased electrophoretic motility, respectively, relative to that of PacC53 (Fig. 6).
FIG. 6.
The complete loss-of-function mutation palI32 does not fully prevent the formation of PacC27 in pH shift experiments. Shown is Western blotting analysis of Myc-tagged PacC in extracts from wild-type or palI32 cells cultured under acidic conditions (H+) and transferred to alkaline conditions (OH−) for the indicated time points before proceeding with protein extraction. Arrows indicate (nonphosphorylated) PacC53 (see text), whereas arrowheads indicate the two abnormal bands seen in the mutant but not in the wild type.
As molecular data and growth tests strongly indicated that PalI contributes to, but is not completely essential for, pH signaling, we tested whether the overexpression of every other Pal protein, expressed under the control of the strong alcAp promoter, bypasses the need for PalI. Growth on pH 8 plates is our most sensitive test to detect any function of pH signal transduction, as even minimal expression from the alcAp promoter with glucose as the sole carbon source is sufficient to allow a detectable degree of complementation of the palH and palF phenotypically null mutations by their respective transgenes (Fig. 5, rows 2 to 3 and 9 to 10). Notably, the overexpression (with ethanol induction) of PalH resulted in a wild-type level of growth of a null palI32 strain and of a phenotypically null palI30G47D missense mutant strain on alkaline pH plates (Fig. 5, rows 5 to 8), whereas the overexpression of PalF weakly improved growth of the null palI32 mutant (palI30 was not tested) (Fig. 5, rows 13 and 14). Weak growth improvement on alkaline pH plates after PalF overexpression was also observed for the partial loss-of-function palH45 mutant (Fig. 5, rows 15 and 16), which came as no surprise as the truncated palH45 product is missing only one of the two PalF binding sites (19). In contrast, the overexpression of PalA, PalB, or PalC had no effect on the inability of palH or palI mutants to grow on alkaline pH plates (data not shown).
Although growth on alkaline plates is the most sensitive test for pH regulation, loss-of-function mutations in pal genes additionally result in hypersensitivity to molybdate and increased acid phosphatase expression. Because the overexpression of PalH did not detectably improve the tolerance to molybdate of the palI32 mutant, nor did it reduce its elevated acid phosphatase levels (data not shown), we conclude that increased levels of PalH and, to a lesser extent, of PalF can bypass the requirement for PalI only partially. As the overexpression of PalI did not suppress null mutations in any other pal gene, including palH (data not shown), and since PalF acts downstream of PalH and PalI (19), these data are consistent with a model in which PalI plays an accessory role, determining the functional levels of PalH, and strongly support the contention that PalI, PalH, and PalF act in concert and upstream of PalC, PalA, and PalB. The finding that overexpressed PalI cannot functionally replace PalH indicates that PalI by itself does not constitute a plasma membrane pH sensor.
Overexpressed PalH-GFP localizes to the plasma membrane but predominates in cytosolic punctate structures.
To confirm that PalH is a plasma membrane protein, we constructed a PalH-GFP chimera and expressed it under the control of the alcAp promoter, as described above for PalI. This PalH-GFP fusion protein was functional, as determined by its ability to complement the palH72 null mutation (data not shown). The subcellular localization of PalH-GFP was examined in this null palH72 background under acidic growth conditions, in which the pH signaling pathway receptor would be expected to be expressed at the cell surface. PalH-GFP is indeed seen in the plasma membrane (Fig. 7). However, in marked contrast with PalI-GFP, PalH-GFP does not label the peripheral plasma membrane uniformly, and instead, it predominates at the hyphal tips (Fig. 7A through D). Such predominance at the hyphal tips is most noticeable in very young germlings pictured shortly after polarity establishment, when the hyphal tip region involves the entire length of the emerging germ tube (Fig. 7D). In addition, we occasionally observed the labeling of septae and vacuolar fluorescence (Fig. 7E), indicating that the fusion protein may be sorted into the multivesicular body pathway. (The GFP moiety is known to be recalcitrant to degradation by vacuolar proteases [48)].) However, the most conspicuous localization of PalH-GFP is not that in the plasma membrane, as the fusion protein predominates in cytosolic, highly fluorescent specks of various sizes (Fig. 7), likely representing endosomal or Golgi compartments. Localization of PalH-GFP to these specks was not prevented by null palA, palF, and palI mutations (data not shown). Thus, we conclude that while overexpressed PalH-GFP can localize to the plasma membrane, it accumulates in an internal compartment under acidic conditions.
FIG. 7.
PalH-GFP expressed from the alcAp localizes to the plasma membrane but predominates in cytosolic specks. (A) A germling where PalH-GFP localization at the apical plasma membrane of the shorter germ tube is clearly visible (arrowhead), although PalH-GFP predominates in cytosolic specks (arrows). (B and C) Plasma membrane localization of PalH-GFP (arrowheads) in longer germlings is less prominent than that seen in internal specks (arrows). (D) Very young germling showing clear polarization of PalH-GFP at the apical plasma membrane. (E) PalH-GFP also labels septae (sp) and eventually reaches the lumen of the vacuole (vac). Bars, 5 μm.
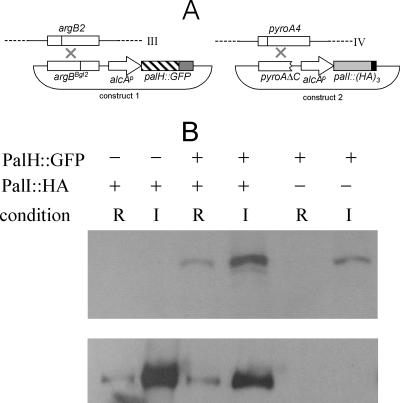
A system for cooverexpression of two proteins from single-copy transgenes targeted to argB and pyroA.
As inactivation of other pH signaling pathway components did not prevent PalH-GFP localization to internal specks, we reasoned that the overexpression of the fusion protein might elicit a deficiency of another pal product required for the localization of PalH at the plasma membrane. PalI is one obvious candidate for such a role, as it seemingly plays a less prominent role in signaling, and this role can be partially bypassed by PalH overexpression. Thus, we set out to design a system permitting the simultaneous cooverexpression of PalI and PalH, using the alcAp promoter (Fig. 8A). The expression of PalH-GFP is driven by a transgene integrated in a single copy at argB, which is located on the right arm of chromosome III. Integration of the alcAp-driven transgene is targeted to argB by using as the recipient a mutant strain carrying the argB2 truncating loss-of-function mutation in combination with a different frameshift mutation in the argB selective marker in the transforming construct (31, 39). This is added to competent protoplasts as a covalently closed form, such that a functional argB gene is reconstructed when a single crossover takes place between the two mutations. We used a similar strategy to target constructs to the pyroA locus located on chromosome IV. Thus, we combined a truncated pyroA allele in the alcAp transforming construct with the pyroA4 loss-of-function mutation in the recipient strain (Fig. 8A and Materials and Methods). We next constructed strains expressing PalH-GFP and either PalI or PalI-(HA)3 (PalI tagged with three copies of the hemagglutinin epitope in the C terminus and shown to be functional by complementation of the null palI32 mutation) from argB- and pyroA-targeted single-copy transgenes driven by alcAp. Western blotting analysis demonstrated that these strains expressed full-length versions of PalH-GFP and PalI-(HA)3 and that the expression of transgenic proteins was largely dependent on ethanol induction (Fig. 8B).
FIG. 8.
A system for the simultaneous expression of two proteins from single-copy transgenes targeted to argB and pyroA. (A) Targeted integration of the transforming plasmids to the argB and pyroA genes, located at chromosomes III and IV, respectively. Use of the argBBgl2 allele for site-directed integration has been reported previously (31, 39). (B) Western blotting analysis of PalH-GFP and PalI-(HA)3 expression driven by alcAp. Strains carrying (+) or lacking (−) the transgenes are indicated. The PalH-GFP transgene was integrated at argB, whereas that driving expression of PalI-(HA)3 was integrated at pyroA. The top and bottom panels were revealed with anti-GFP and anti-HA antibodies, respectively.
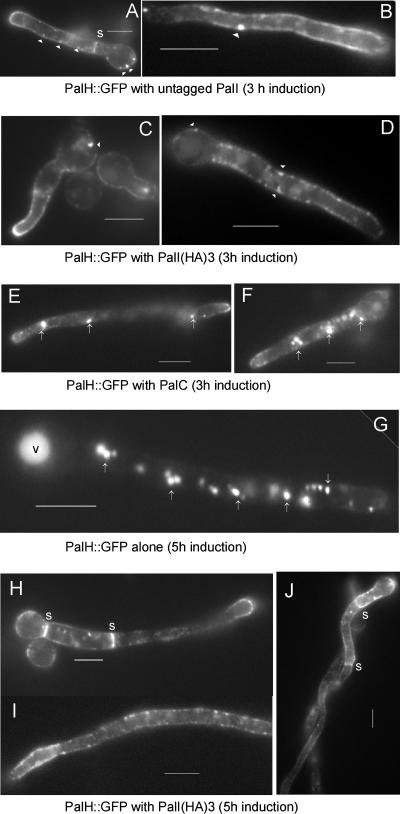
Cooverexpression of PalI but not of PalC promotes the plasma membrane expression of PalH-GFP.
We used this system to determine the subcellular localization of PalH-GFP, with and without PalI coexpression under the control of the same promoter. In stark contrast to the strain expressing PalH-GFP alone (see Fig. 7), strongly fluorescent internal specks were notably absent in both the PalI and the PalI-(HA)3 coexpressing strains, which showed a strong predominance of PalH-GFP at the plasma membrane [Fig. 9A and B, untagged PalI, and C and D, PalI-(HA)3]. Interestingly, the PalH-GFP distribution was not uniform, which suggests segregation of the protein in plasma membrane subdomains. PalH-GFP was additionally seen in peripheral punctate structures that might represent sites of endocytosis (Fig. 9A to D). Because the predominance of PalH-GFP at the plasma membrane was not seen when PalC was coexpressed instead of PalI (Fig. 9E and F), we conclude that PalI specifically assists the localization of PalH-GFP at the plasma membrane. This conclusion was strongly reinforced by observations after a longer (5.5-h as opposed to the normal 3-h) promoter induction period. Under such conditions, PalH-GFP expressed alone predominated in highly fluorescent relatively immotile compartments probably corresponding to mature endosomes and to vacuoles (35) (Fig. 9G). This localization is remarkably different from that seen in the PalI-(HA)3 coexpressing strain, where the “patchy” plasma membrane predominance of PalH-GFP was evident over a cytosolic haze that reflects the longer period of expression of the reporter (Fig. 9H through J). Thus, we conclude that PalI assists the plasma membrane localization of PalH.
FIG. 9.
Coexpression of PalI from the alcAp promoter results in the plasma membrane localization of PalH-GFP. Germlings of strains expressing the indicated proteins under the control of the alcAp gene were cultured as described in the legend to Fig. 3, with a 3-h (A to F) or a 5-h (G to J) induction period, as indicated. (A and B) Coexpression of PalI with PalH-GFP promotes the plasma membrane localization of the latter. (C and D) As described in the legend to panels A and B above, using PalI-(HA)3 rather than untagged PalI. (E and F) Coexpression of PalC does not promote plasma membrane localization of PalH-GFP. Note that the distribution of PalH-GFP in this strain cannot be distinguished from that shown in Fig. 7 (in the absence of PalC coexpression). (G) After a relatively long period of PalH-GFP transgene induction, the reporter almost exclusively localizes to strongly fluorescent cytosolic specks (arrows) and to the vacuole (v). (H, I, and J) In marked contrast, PalH-GFP predominates at the plasma membrane if PalI-(HA)3 is coexpressed using the same induction regimen. Note the clearly patchy appearance of PalH-GFP at the plasma membrane, the strong labeling of septae (s), and the peripheral punctate structures. Bars, 5 μm.
DISCUSSION
PalI, PalH, and PalF are three components of a hypothetical pH signaling complex located at the A. nidulans plasma membrane. This model is strongly supported by our findings that overexpressed GFP-tagged PalI localizes to the plasma membrane on its own and that overexpressed PalH-GFP, which localizes to the plasma membrane but is most abundant in internal membrane compartments, shows plasma membrane predominance when expressed in the presence of stoichiometrically similar amounts of PalI. Together with previous work (6, 18, 19, 53), this report provides strong support for the currently prevailing models of the ambient pH signaling pathway that involve two spatially segregated complexes. The “upstream” plasma membrane complex would comprise PalH/Rim21p (and/or Dfg16p [5]), PalI/Rim9p, and PalF/Rim8p, whereas the “downstream” endosomal membrane complex would involve PalB/Rim13p and PalA/Rim20p, as well as the transcription factor PacC/Rim101p. The sixth pH signaling protein, PalC, has been suggested to play a bridging role between the plasma and the endosomal membrane complexes (16). The recent finding that S. cerevisiae Ygr122w (42) is the almost certain orthologue of PalC completes the molecular basis for a unifying model of pH regulation in filamentous fungi and yeasts (16).
Ubiquitination of arrestin-like PalF is arguably the most reliable indicator of pH signaling (19). The palF orthologues S. cerevisiae RIM8 and Candida albicans PRR1 are acid-expressed genes formally repressed by Rim101p (and Rim13p) (24, 40), strongly supporting the existence, at least in these yeasts, of a negative feedback loop that would downregulate levels of Rim8p in response to an alkaline ambient pH and suggesting that PalF/Rim8p activity would be one limiting factor in the pH signaling pathway (24). PalF binds to the cytosolic tail of 7-TMD PalH, which is required for PalF phosphorylation and ubiquitination. As ubiquitination is a landmark of endocytic internalization in fungi (see references 21 and 46), mammalian arrestin ubiquitination leads to the endocytic internalization of their cognate 7-TMD receptors (26), and PalF/Rim8p promotes rather than attenuates pH signaling, the prevailing view is that pH signaling requires the endocytic internalization of PalH/Rim21p (and/or Dfg16p) (6, 16, 19).
We have determined here that PalH and PalI localize to the plasma membrane under acidic pH conditions, but we have not yet addressed whether either or both are internalized under alkaline pH conditions. In our protocol, microscopic observation of PalH requires overexpression, which makes PalH localization to the plasma membrane dependent on PalI cooverexpression. Because the ambient pH-dependent internalization of PalH would be driven by PalF ubiquitination and because the three proteins in the plasma membrane signaling complex very likely act as a single entity, experiments addressing this possibility should involve coexpression of stoichiometrically equivalent amounts of PalF, PalH, and PalI to provide physiologically meaningful information. Future work will involve the design of genetic systems that allow simultaneous coexpression in A. nidulans of three proteins at stoichiometrically similar levels as well as the identification of resident plasma membrane proteins whose internalization is not promoted by alkaline pH to be used as negative controls.
Mutational and bioinformatic analyses of palI provide evidence that, as suggested by Li and Mitchell (27) for Rim9p, the N-terminal hydrophobic region of the protein is a signal peptide and therefore that PalI and Rim9p are 3-TMD proteins with their N termini located in the lumens of the endoplasmic reticulum (ER). We show that PalI/Rim9p family members contain, within the N-terminal region preceding the first TMD, a diagnostic sequence motif comprising three Gly and one Cys invariant residues. Single-residue substitutions of Gly44 (this work) and Gly47 (10) within this motif lead to loss of function. We show that Gly47Asp results in the missorting of PalI-GFP to endosome-like compartments and into the multivesicular body pathway/vacuole (Fig. 3) and leads to reduced fusion protein levels (Fig. 2). The role of the PalI Gly-Cys-containing motif is currently unknown, but we note that in the glycophorin A dimer, Gly residues in the context of a hydrophobic α-helix facilitate van der Waals interactions involving side chain and backbone atoms in the α-helical element that mediates homodimerization (29). While glycophorin A homodimerization occurs within the lipid membrane, whereas the Gly-Cys motif is predicted to be in the ER lumen, we speculate that the invariant Cys residue within the motif might cooperate in PalI dimerization by forming a disulfide bond within the highly oxidizing environment of the ER. The hypothetical lack of assembly of a PalI homodimer in the Gly47Asp mutant might expose to the lipid bilayer relatively hydrophilic residues that would normally be masked by intramolecular interactions in the quaternary structure. Exposure of polar residues within a TMD leads to transmembrane protein ubiquitination and subsequent biosynthetic sorting into the multivesicular body pathway (20, 41), which would explain the endosomal/vacuolar localization of mutant Gly47Asp PalI-GFP. However, we have not determined if PalIG47D-GFP reaches endosomes following a biosynthetic pathway from the Golgi complex or whether it reaches the plasma membrane and is missorted to these compartments due to abnormally increased endocytosis. To distinguish these possibilities, conditional mutations preventing endocytic internalization would be required, as this process appears to be essential in A. nidulans (our unpublished data).
PalI might stoichiometrically assist the plasma membrane localization of PalH in several ways. One possibility is that PalI assists the hypothetical oligomerization of the 7-TMD receptor that has been suggested for Rim21p/Dfg16 (5), such that overexpressed misfolded/monomeric PalH is inappropriately sorted to endosomal/Golgi compartments. A second possibility is that PalI might escort PalH along the secretory pathway, helping to sort it into vesicle carriers exiting the ER and/or the Golgi complex. A third possibility is that PalH has an intrinsic tendency to segregate into membrane domains where endocytic internalization is strongly favored and PalI acts by preventing its excessive endocytosis. Future research will address these possibilities by colocalization studies of PalH-GFP and monomeric red fluorescent protein-tagged markers of these membrane compartments.
Finally, we note that the overexpression of PalH only partially suppresses the requirement for PalI, as determined with less sensitive diagnostic tests of pH regulation based on molybdate sensitivity or extracellular phosphatase staining (see references 38 and 50). Therefore, assisting the plasma membrane localization of PalH is possibly not the only role that PalI plays in pH signaling.
Acknowledgments
This work was supported by D.G.I.C.Y.T. grant BIO2006-0556 to M.A.P. and Wellcome Trust grant 067878 to H.N.A. and J.T.
We thank Elena Reoyo for technical assistance.
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Arst, H. N., Jr., E. Bignell, and J. Tilburn. 1994. Two new genes involved in signaling ambient pH in Aspergillus nidulans. Mol. Gen. Genet. 245:787-790. [DOI] [PubMed] [Google Scholar]
- 2.Arst, H. N., Jr., and M. A. Peñalva. 2003. pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet. 19:224-231. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 4.Bartnicki-García, S., F. Hergert, and G. Gierz. 1989. Computer simulation of fungal morphogenesis and the mathematical basis for hyphal tip growth. Protoplasma 153:46-57. [Google Scholar]
- 5.Barwell, K. J., J. H. Boysen, W. Xu, and A. P. Mitchell. 2005. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 4:890-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boysen, J. H., and A. P. Mitchell. 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bussink, H. J., and S. A. Osmani. 1998. A cyclin-dependent kinase family member (PHOA) is required to link developmental fate to environmental conditions in Aspergillus nidulans. EMBO J. 17:3990-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castrejon, F., A. Gómez, M. Sanz, A. Durán, and C. Roncero. 2006. The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot. Cell 5:507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clutterbuck, A. J. 1993. Aspergillus nidulans, p. 3.71-3.84. In S. J. O'Brien (ed.), Genetic maps. Locus maps of complex genomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Denison, S. H., S. Negrete-Urtasun, J. M. Mingot, J. Tilburn, W. A. Mayer, A. Goel, E. A. Espeso, M. A. Peñalva, and H. N. Arst, Jr. 1998. Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in Saccharomyces are homologous. Mol. Microbiol. 30:259-264. [DOI] [PubMed] [Google Scholar]
- 11.Denison, S. H., M. Orejas, and H. N. Arst, Jr. 1995. Signaling of ambient pH in Aspergillus involves a cysteine protease. J. Biol. Chem. 270:28519-28522. [DOI] [PubMed] [Google Scholar]
- 12.Díez, E., J. Álvaro, E. A. Espeso, L. Rainbow, T. Suárez, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva. 2002. Activation of the Aspergillus PacC zinc-finger transcription factor requires two proteolytic steps. EMBO J. 21:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-Martínez, J., C. V. Brown, E. Díez, J. Tilburn, H. N. Arst, Jr., M. A. Peñalva, and E. A. Espeso. 2003. Overlap of nuclear localisation signal and specific DNA binding residues within the zinc finger domain of PacC. J. Mol. Biol. 334:667-684. [DOI] [PubMed] [Google Scholar]
- 14.Fischer-Parton, S., R. M. Parton, P. C. Hickey, J. Dijksterhuis, H. A. Atkinson, and N. D. Read. 2000. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198:246-259. [DOI] [PubMed] [Google Scholar]
- 15.Futai, E., T. Maeda, H. Sorimachi, K. Kitamoto, S. Ishiura, and K. Suzuki. 1999. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 260:559-568. [DOI] [PubMed] [Google Scholar]
- 16.Galindo, A., A. Hervás-Aguilar, O. Rodríguez-Galán, O. Vincent, H. N. Arst, Jr., J. Tilburn, and M. A. Peñalva. 2007. PalC, one of two Bro1 domain proteins in the fungal pH signaling pathway, localizes to cortical structures and binds Vps32. Traffic 8:1346-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, S. D., N. D. Read, R. W. Roberson, B. Shaw, S. Seiler, M. Plamann, and M. Momany. 2005. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi, M., T. Fukuzawa, H. Sorimachi, and T. Maeda. 2005. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 25:9478-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herranz, S., J. M. Rodríguez, H. J. Bussink, J. C. Sánchez-Ferrero, H. N. Arst, Jr., M. A. Peñalva, and O. Vincent. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. USA 102:12141-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hettema, E. H., J. Valdez-Taubas, and H. R. Pelham. 2004. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277-287. [DOI] [PubMed] [Google Scholar]
- 22.Hohmann-Marriott, M. F., M. Uchida, A. M. van de Meene, M. Garret, B. E. Hjelm, S. Kokoori, and R. W. Roberson. 2006. Application of electron tomography to fungal ultrastructure studies. New Phytol. 172:208-220. [DOI] [PubMed] [Google Scholar]
- 23.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850-1856. [DOI] [PubMed] [Google Scholar]
- 26.Lefkowitz, R. J., and S. K. Shenoy. 2005. Transduction of receptor signals by beta-arrestins. Science 308:512-517. [DOI] [PubMed] [Google Scholar]
- 27.Li, W. S., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, Y., L. A. Kimpler, T. V. Naismith, J. M. Lauer, and P. I. Hanson. 2005. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 280:12799-12809. [DOI] [PubMed] [Google Scholar]
- 29.MacKenzie, K. R., J. H. Prestegard, and D. M. Engelman. 1997. A transmembrane helix dimer: structure and implications. Science 276:131-133. [DOI] [PubMed] [Google Scholar]
- 30.Mingot, J. M., E. A. Espeso, E. Díez, and M. A. Peñalva. 2001. Ambient pH signaling regulates nuclear localization of the Aspergillus nidulans PacC transcription factor. Mol. Cell. Biol. 21:1688-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mingot, J. M., J. Tilburn, E. Díez, E. Bignell, M. Orejas, D. A. Widdick, S. Sarkar, C. V. Brown, M. X. Caddick, E. A. Espeso, H. N. Arst, Jr., and M. A. Peñalva. 1999. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signaling is bypassed. Mol. Cell. Biol. 19:1390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negrete-Urtasun, S., W. Reiter, E. Díez, S. H. Denison, J. Tilburn, E. A. Espeso, M. A. Peñalva, and H. N. Arst, Jr. 1999. Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol. Microbiol. 33:994-1003. [DOI] [PubMed] [Google Scholar]
- 33.Orejas, M., E. A. Espeso, J. Tilburn, S. Sarkar, H. N. Arst, Jr., and M. A. Peñalva. 1995. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 9:1622-1632. [DOI] [PubMed] [Google Scholar]
- 34.Osmani, A. H., G. S. May, and S. A. Osmani. 1999. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J. Biol. Chem. 274:23565-23569. [DOI] [PubMed] [Google Scholar]
- 35.Peñalva, M. A. 2005. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 42:963-975. [DOI] [PubMed] [Google Scholar]
- 36.Peñalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peñalva, M. A., and H. N. Arst, Jr. 2004. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58:425-451. [DOI] [PubMed] [Google Scholar]
- 38.Peñas, M. M., A. Hervás-Aguilar, T. Múnera-Huertas, E. Reoyo, M. A. Peñalva, H. N. Arst, Jr., and J. Tilburn. 2007. Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot. Cell 6:960-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pérez-Esteban, B., M. Orejas, E. Gómez-Pardo, and M. A. Peñalva. 1993. Molecular characterization of a fungal secondary metabolism promoter: transcription of the Aspergillus nidulans isopenicillin N synthetase gene is modulated by upstream negative elements. Mol. Microbiol. 9:881-895. [DOI] [PubMed] [Google Scholar]
- 40.Ramón, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181:7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reggiori, F., and H. R. Pelham. 2002. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 4:117-123. [DOI] [PubMed] [Google Scholar]
- 42.Rothfels, K., J. C. Tanny, E. Molnar, H. Friesen, C. Commisso, and J. Segall. 2005. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 25:6772-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott, A., J. Gaspar, M. D. Stuchell-Brereton, S. L. Alam, J. J. Skalicky, and W. I. Sundquist. 2005. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA 102:13813-13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Ariño. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signaling. Mol. Microbiol. 46:1319-1333. [DOI] [PubMed] [Google Scholar]
- 45.Springael, J. Y., and B. Andre. 1998. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol. Biol. Cell 9:1253-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Springael, J. Y., J. M. Galan, R. Haguenauer-Tsapis, and B. Andre. 1999. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J. Cell Sci. 112:1375-1383. [DOI] [PubMed] [Google Scholar]
- 47.Steinberg, G. 2007. Hyphal growth: a tale of motors, lipids, and the Spitzenkörper. Eukaryot. Cell 5:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stimpson, H. E., M. J. Lewis, and H. R. Pelham. 2006. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 25:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teo, H., D. J. Gill, J. Sun, O. Perisic, D. B. Veprintsev, Y. Vallis, S. D. Emr, and R. L. Williams. 2006. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125:99-111. [DOI] [PubMed] [Google Scholar]
- 50.Tilburn, J., J. C. Sánchez-Ferrero, E. Reoyo, H. N. Arst, Jr., and M. A. Peñalva. 2005. Mutational analysis of the pH signal transduction component PalC of Aspergillus nidulans supports distant similarity to BRO1 domain family members. Genetics 171:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tilburn, J., C. Scazzocchio, G. G. Taylor, J. H. Zabicky-Zissman, R. A. Lockington, and R. W. Davies. 1983. Transformation by integration in Aspergillus nidulans. Gene 26:205-211. [DOI] [PubMed] [Google Scholar]
- 52.Vincent, O., L. Rainbow, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva. 2003. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue AIP1/Alix. Mol. Cell. Biol. 23:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, W., and A. P. Mitchell. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]