Abstract
Autophagy is the major cellular pathway for bulk degradation of cytosolic material and is required to maintain viability under starvation conditions. To determine the contribution of autophagy to starvation stress responses in the filamentous fungus Aspergillus fumigatus, we disrupted the A. fumigatus atg1 gene, encoding a serine/threonine kinase required for autophagy. The ΔAfatg1 mutant showed abnormal conidiophore development and reduced conidiation, but the defect could be bypassed by increasing the nitrogen content of the medium. When transferred to starvation medium, wild-type hyphae were able to undergo a limited amount of growth, resulting in radial expansion of the colony. In contrast, the ΔAfatg1 mutant was unable to grow under these conditions. However, supplementation of the medium with metal ions rescued the ability of the ΔAfatg1 mutant to grow in the absence of a carbon or nitrogen source. Depleting the medium of cations by using EDTA was sufficient to induce autophagy in wild-type A. fumigatus, even in the presence of abundant carbon and nitrogen, and the ΔAfatg1 mutant was severely growth impaired under these conditions. These findings establish a role for autophagy in the recycling of internal nitrogen sources to support conidiophore development and suggest that autophagy also contributes to the recycling of essential metal ions to sustain hyphal growth when exogenous nutrients are scarce.
Nutrient limitation is one of the most significant stresses encountered by microorganisms in nature. Autophagy is a catabolic membrane trafficking system that counters such nutrient stress by initiating a process of limited intracellular digestion to support the organism during periods of reduced nutrient availability (26, 32, 37, 60). The process begins with the formation of isolation membranes within the cytoplasm, whose origin is not entirely clear. These membranes progressively expand, nonselectively encapsulating cytosolic material into a double-membrane vesicle called the autophagosome. The autophagosome fuses its outer membrane with a vacuole, releasing the autophagic body and its contents into the vacuolar lumen for degradation by resident hydrolases. Autophagy is upregulated in response to starvation stress, resulting in the generation of a pool of recycled molecules that provide the building blocks for continued synthesis of essential components until nutrient conditions improve (26, 37, 39, 60). However, some autophagy remains constitutively active at low levels, even under nutrient-replete conditions, where it serves as a mechanism to remove old or damaged proteins and organelles (17, 25, 28, 46, 54). This provides an important form of quality control that combats the toxic accumulation of abnormal cytoplasmic components.
The degradative functions of autophagy contribute to several important aspects of cell physiology, including autophagy-dependent programmed cell death (2, 14), cellular remodeling during development and differentiation (21, 36, 40, 49, 57), maintenance of endoplasmic reticulum homeostasis (4, 30, 61), removal of damaged or excess mitochondria (25, 46, 54), and defense against invading bacteria (8, 9, 16, 24). These functions are essential to maintain cell and tissue homeostasis, and defects in autophagy function have been implicated in human disease (50). In addition to its prominent role in degradation, the autophagy machinery is also involved in a yeast biosynthetic process that targets at least two resident vacuolar hydrolases, aminopeptidase I and α-mannosidase, to the vacuole (18, 38). This cytoplasm-to-vacuole (Cvt) targeting pathway uses much of the same molecular machinery as autophagy but differs from starvation-induced autophagy in that it operates constitutively, is highly selective for its cargo, and involves Cvt vesicles that are typically smaller than autophagosomes (18, 38).
Although model systems for studying autophagy have been well established for Saccharomyces cerevisiae and higher eukaryotes, little is known about the contribution of autophagy to the biology of filamentous fungi. The mycelium of a filamentous fungus is comprised of a network of interconnected hyphae that forms a colony with a circular margin. At the periphery of the colony, the hyphae are at a low density and exhibit negative autotropism, an avoidance mechanism that promotes the foraging of individual hyphae into uncolonized areas (47). In contrast, hyphae in the central region are nutrient deprived, stop growing, and branch towards each other, forming numerous hyphal fusions (anastomoses). The interconnected hyphae are divided by perforated septa into multinucleated compartments that permit bulk flow of cytoplasm and organelles throughout the mycelium. These interconnections are thought to allow the organism to survive periods of nutrient deprivation by sharing resources throughout the mycelium (13, 47). Unlike colonies of yeast, which stop growing when nutrients are exhausted, nutrient-deprived hyphae are able to grow beyond an area of nutrient depletion by concentrating resources at the hyphal tips (47). This foraging-like response is fundamental to the filamentous lifestyle, but the cellular and molecular mechanisms involved are largely unknown.
In this study, we examined the contribution of autophagy to starvation responses in the filamentous fungus Aspergillus fumigatus, focusing on sporulation and the radial growth of hyphal tips. An autophagy-deficient strain of A. fumigatus was constructed by disrupting the Afatg1 gene, encoding the A. fumigatus homolog of the Atg1 serine/threonine kinase required for autophagy in other species (22, 35, 44, 49, 56). The ΔAfatg1 mutant failed to conidiate normally unless the nitrogen content of the medium was increased, suggesting that starvation-associated conidiation relies upon autophagy to provide sufficient nitrogen to support conidiophore development. Unlike wild-type (wt) A. fumigatus, the ΔAfatg1 mutant was unable to grow radially under starvation conditions, indicating that starvation foraging of hyphal tips is autophagy dependent. Surprisingly, growth under starvation conditions could be rescued by incorporating certain metal ions into the medium. Depleting rich medium of divalent cations by treatment with EDTA was sufficient to induce autophagy in wt A. fumigatus, and the ΔAfatg1 mutant was severely growth impaired under these conditions. These data establish a role for autophagy in starvation-associated foraging and provide an unanticipated link between autophagy and metal ion homeostasis.
MATERIALS AND METHODS
Aspergillus fumigatus strains, media, and culture conditions.
The strains used in this study are listed in Table 1. All strains were maintained and harvested from inhibitory mold agar plates because this medium supports conidiation of the ΔAfatg1 mutant. For analysis of conidiophore development, conidia were inoculated around a plug of potato dextrose agar (PDA; 1% glucose, 2% potato flakes) and a coverslip was placed on top. Conidiophores were examined after 2 to 3 days of incubation at 37°C and photographed using differential interference contrast (DIC) microscopy. The number of conidia produced by each colony was estimated by flooding the plate with 10 ml of sterile water and scraping off the conidia. This was performed four times for each plate to maximize conidial recovery. The average number of conidia obtained from triplicate plates was then determined with a hemacytometer.
TABLE 1.
Stains used in this study
| Strain | Genotype |
|---|---|
| H237 | Wild type |
| ΔAfatg1 | Afatg1::hph |
| C′ | ΔAfatg1 (Afatg1/ble) |
| GFP-Afatg8 | wt (GFP-Afatg8/ble) |
| ΔAfatg1(GFP-atg8) | ΔAfatg1 (GFP-Afatg8/ble) |
For analysis of the starvation-induced foraging response, 100 conidia were spread for isolation onto the surface of a YG plate (2% glucose, 0.5% yeast extract) and incubated for 24 h at 37°C. Hyphal plugs from individual overnight colonies were then obtained using the narrow end of a 5 3/4-inch Pasteur pipette and transferred to the center of a water-agarose (WA) plate either containing no additional nutrients (1% agarose in Milli-Q water) or supplemented with the indicated concentrations of ZnSO4, CuSO4, MnSO4, MgSO4, or FeSO4 (Sigma). The plates were incubated at 37°C, and colony diameters were measured daily.
Sensitivity to the chelating agent EDTA was determined by inoculating 2 × 105 conidia into liquid cultures of YG containing 0.5 mM EDTA and incubating them at 37°C for 3 to 6 days. To quantitate EDTA sensitivity, 1 × 106 conidia/ml of the wt, ΔAfatg1 mutant, and reconstituted strains were incubated at 37°C for 3 to 6 days in YG medium in the presence or absence of 0.75 mM EDTA. The percent germination was then calculated for 100 conidia analyzed microscopically. A conidium was scored as germinated if it had extended a germ tube with a length that was equal to or greater than the length of the conidium.
Disruption and reconstitution of the A. fumigatus Afatg1 gene.
Oligonucleotides used in this study are shown in Table 2. The A. fumigatus Afatg1 gene (GenBank accession no. Q4WPF2) was disrupted using the split-marker strategy (7). The left arm of Afatg1 was PCR amplified from genomic DNA by using Pfu Turbo polymerase (Stratagene) with primers 481 and 482, creating PCR product 1. The first two-thirds of the hygromycin resistance cassette was amplified from plasmid pAN7-1 by using primers 395 and 398, making PCR product 3. PCR products 1 and 3 were then combined in an overlap PCR with primers 395 and 481 to create PCR product 5. PCR product 5 was then cloned into pCR-Blunt II-TOPO (Invitrogen) to create p513.
TABLE 2.
PCR primers used in this study
| Primer | Gene | Sequence (5′-3′)a |
|---|---|---|
| 395 | hph | CTCCATACAAGCCAACCACGG |
| 396 | hph | CGTTGCAAGACCTGCCTGAA |
| 398 | hph | CGCCAGGGTTTTCCCAGTCACGACAAGTGGAAAGGCTGGTGTGC |
| 399 | hph | AGCGGATAACAATTTCACACAGGATCGCGTGGAGCCAAGAGCGG |
| 481 | Afatg1 | TAGGTATCGCAGCTTTGAAG |
| 482 | Afatg1 | GTCGTGACTGGGAAAACCCTGGCGCTGGAGTGAATAGGCAGAAGA |
| 483 | Afatg1 | TCCTGTGTGAAATTGTTATCCGCTTTCGAAGGCGCTCAATATGG |
| 484 | Afatg1 | CGCTTACTTCGGCGGCGTTG |
| 536 | Afatg1 | CTTAAGCGGTTTAGCCGCTTCTTGT |
| 540 | Afatg1 | AATAAGTCGGAGTGGACCGAAACAC |
| 570 | Afatg8 | GGAATTCCATATGCGGTCGAAGTTCAAGGA |
| 571 | Afatg8 | ATAAGAATGCGGCCGCCCATTTGCTAAGAAGGACCC |
M13-derived sequences used for overlap PCR are underlined.
The right arm of the ΔAfatg1 gene was PCR amplified from genomic DNA by using primers 483 and 484 to generate PCR product 2. The second two-thirds of the hygromycin resistance cassette was amplified from pAN7-1 with primers 396 and 399 to make PCR product 4, and PCR products 2 and 4 were combined in an overlap PCR with primers 396 and 484 to create PCR product 6. PCR product 6 was then cloned into pCR-Blunt II-TOPO to create p514. The inserts from p513 and p514 were gel purified following digestion with KpnI, XbaI, and SacI, and 5 μg of each was used to transform wt protoplasts as previously described (6). Hygromycin-resistant colonies were screened by PCR, and confirmation of disruption was performed on monoconidial isolates by genomic Southern blot analysis as described in Results. Complementation of the ΔAfatg1 mutant was accomplished by introducing the Afatg1 gene into the ΔAfatg1 mutant as an ectopic transgene. The Afatg1 gene, containing 1,046 bp upstream of the predicted translation start site, was PCR amplified from genomic DNA by using primers 536 and 540 and cloned into pCR-Blunt II-TOPO to create p531. A phleomycin resistance cassette containing the ble gene flanked by the Aspergillus nidulans gpdA promoter and the yeast CYC1 terminator was then excised from p402, using XbaI and SpeI, and inserted into the XbaI site of plasmid p531 to create the complementation plasmid p532. Ten micrograms of p532 was linearized with XbaI and introduced into ΔAfatg1 mutant protoplasts as previously described (6). Phleomycin-resistant transformants were screened by PCR and Southern blot analysis to confirm the presence of Afatg1 (data not shown).
Analysis of autophagy.
For microscopic analysis of autophagosome accumulation in vacuoles, overnight cultures were washed in sterile distilled water, and the medium was replaced either with sterile distilled water containing 2 mM phenylmethylsulfonyl fluoride (PMSF) or with YG containing 2 mM PMSF and 0.5 mM EDTA. After 4 h of incubation at 37°C, the presence of autophagic bodies within vacuoles was visualized by DIC microscopy.
To construct a green fluorescent protein (GFP)-tagged Atg8 protein, soluble GFP was first PCR amplified from pMCB32 (12), using a 5′ primer containing an NcoI cloning site together with a 3′ primer that replaced the stop codon with a six-His linker and an NdeI cloning site. The PCR product was then cloned into pCR-TOPO2.1 (Invitrogen) to make psGFP. The A. nidulans gpdA promoter was then excised from pAN7-1 as a BamHI-NcoI fragment and inserted into the corresponding sites of psGFP to create plasmid pPgpdA-GFP6His. Next, a phleomycin resistance cassette containing the A. nidulans gpdA promoter, the Streptoalloteichus hindustanus ble gene encoding phleomycin resistance, and the S. cerevisiae CYC1 terminator was excised from pBCphleo as a 3-kb SalI-HindIII fragment and inserted into the XhoI-HindIII sites of the cloning vector pSL1180 to make pSL1180-phleo. A 0.8-kb segment of the A. nidulans trpC terminator was then PCR amplified from pAN7-1 and cloned upstream of the phleomycin resistance cassette in pSL1180-phleo to make pSL1180-TtrpC-phleo. The PgpdA-GFP6His gene was then excised from pPgpdA-GFP6His as a BamHI-NotI fragment and cloned into the BglII-NotI sites of pSL1180-TtrpC-phleo to create p331. This plasmid allows a gene to be cloned as an N-terminal fusion to GFP, using the available NdeI and NotI cloning sites located between GFP and the trpC terminator.
To construct a GFP-AfAtg8 fusion protein, the A. fumigatus Afatg8 gene (GenBank accession no. Q4WJ27) was PCR amplified from genomic DNA by using primers 570 and 571, incorporating an NdeI site in the 5′ primer and a NotI site in the 3′ primer. The PCR product was then cloned into the corresponding sites of p331 to make pGFP-Afatg8. Ten micrograms of the pGFP-atg8 expression construct was linearized with HindIII and introduced into the wt and ΔAfatg1 strains by protoplast transformation as previously described (6). The intracellular localization of the expressed fusion protein was visualized by live-cell imaging with a 100× oil objective on a Leica TCS SP2 laser scanning confocal microscope set for GFP detection.
Mouse model of invasive aspergillosis.
C57BL/6 mice were rendered neutropenic with a single dose of the monoclonal antibody RB6-8C5 (25 μg injected intraperitoneally on day −1) and with cyclophosphamide (150 mg/kg of body weight injected intraperitoneally on day −1 and readministered on day +3) (52). Mice were inoculated intratracheally on day 0 with 5 × 105 conidia from the wt, ΔAfatg1 mutant, or complemented strain (n = 11, 11, and 13 mice per group, respectively), and mortality was monitored for 14 days. For the sham control, six immunosuppressed mice were inoculated intratracheally with phosphate-buffered saline containing 0.05% Tween 20.
RESULTS
Construction of an autophagy-deficient mutant of A. fumigatus.
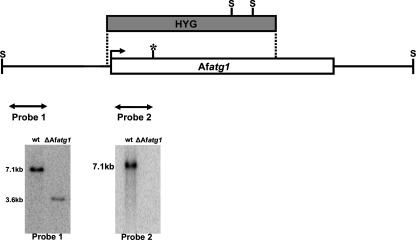
To determine the contribution of autophagy to the biology of A. fumigatus, we disrupted the A. fumigatus Afatg1 gene, encoding the predicted ortholog of S. cerevisiae Atg1p, a serine/threonine kinase that is essential for autophagy in S. cerevisiae, Podospora anserina, and Dictyostelium discoideum (22, 35, 44, 49, 56). Disruption of Afatg1 was accomplished by gene replacement with a hygromycin resistance cassette, and confirmation of homologous insertion was obtained by genomic Southern blot analysis of SacII-digested genomic DNA, using the two probes shown in Fig. 1.
FIG. 1.
Disruption strategy for Afatg1. The split-marker approach was used to create the ΔAfatg1 mutant. Homologous recombination between the disruption cassette and the Afatg1 gene inserted the hygromycin resistance gene (HYG) at the indicate sites (dotted lines). The predicted active site in the kinase domain is shown by an asterisk. Southern blot analysis of SacII (S)-digested genomic DNA by using probe 1 (flanking the disruption cassette) identified the expected 7.1-kb wt band, which was truncated to 3.6 kb in the ΔAfatg1 mutant. A second probe derived from the deleted region (probe 2) confirmed that no duplication had occurred.
Disruption of Afatg1 inhibits autophagy in A. fumigatus.
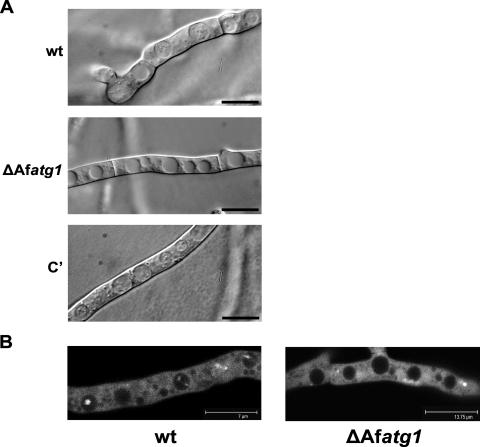
To determine if autophagy was disrupted in the ΔAfatg1 mutant, overnight cultures of the wt and the ΔAfatg1 mutant were washed and placed under starvation conditions in the presence of the vacuolar protease inhibitor PMSF. These conditions prevent the degradation of autophagic bodies within vacuoles, and their accumulation can be used as a marker of active autophagy (35, 45, 53). In this assay, the wt, ΔAfatg1 mutant, and complemented strains all demonstrated extensive vacuolization, but only the wt and complemented strains accumulated autophagic bodies within their vacuoles (Fig. 2A).
FIG. 2.
ΔAfatg1 mutant is defective in autophagy. (A) Conidia from the wt, ΔAfatg1 mutant, and complemented (C′) strains were incubated in YG for 16 h at 37°C. The hyphae were washed, the medium was replaced with sterile distilled water containing 2 mM PMSF, and the hyphae were incubated at 37°C for 4 h. Bar, 10 μm. (B) Conidia from wt and ΔAfatg1 strains expressing GFP-AfAtg8 were incubated in Aspergillus minimal medium for 16 h at 37°C. The hyphae were washed, the medium was replaced with sterile distilled water containing 2 mM PMSF, and the hyphae were incubated at 37°C for 4 h. The strains were examined by laser scanning confocal microscopy, and images shown are single optical sections through the hyphae. A series of optical sections were taken throughout the hyphae and compiled into a movie (see the supplemental material).
To confirm that the observed vacuolar structures were autophagic bodies, a second assay was employed to monitor the fluorescence of a GFP-tagged Atg8 protein. Atg8 is conjugated to the autophagosome during autophagy, where it remains associated until it is finally degraded in the vacuole (27). Thus, vacuolar accumulation of fluorescent GFP-Atg8 can be used as a marker of active autophagy. The A. fumigatus homolog of Atg8 was tagged at the N terminus with GFP, as described in Materials and Methods, and then expressed in the wt and ΔAfatg1 strains. As expected, the GFP-AfAtg8 protein was predominantly cytoplasmic under nutrient-rich conditions, with some localization to punctate perivacuolar structures in both the wt and ΔAfatg1 strains (Fig. 2B; see the supplemental material). Similar findings have been described for Atg8 localization in Podospora anserina and Aspergillus oryzae (23, 44). Under starvation conditions, fluorescently labeled Atg8 accumulated within the vacuoles of wt A. fumigatus (Fig. 2B; see the supplemental material). In contrast, fluorescently labeled Atg8 failed to accumulate in the vacuoles of the ΔAfatg1 mutant under starvation conditions (Fig. 2B; see the supplemental material). Taken together, these observations confirm that autophagy is defective in the ΔAfatg1 mutant.
A. fumigatus Atg1 facilitates conidiophore development and conidiation.
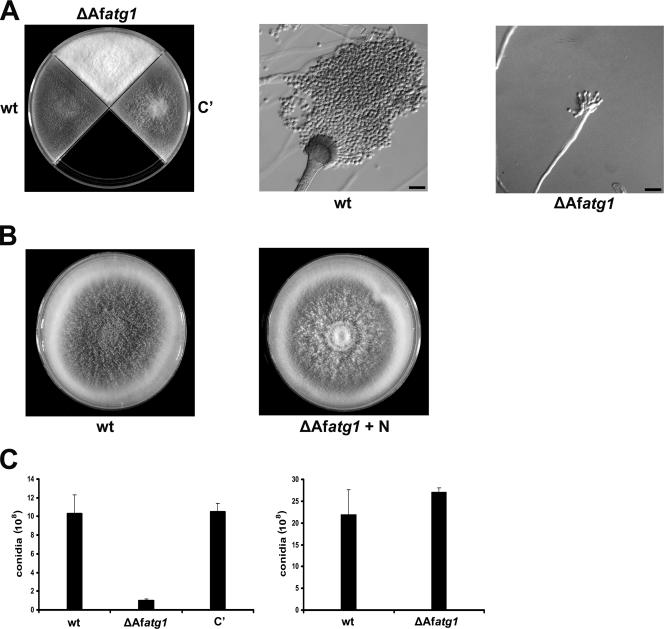
The ΔAfatg1 mutant generated white colonies on PDA medium because of reduced conidiation, a phenotype that was corrected by complementing the ΔAfatg1 mutation (Fig. 3A). Microscopic examination revealed that the reduced conidiation was due to abnormal conidiophore development: the wt strain produced numerous conidiophores and conidia on PDA medium, but the ΔAfatg1 mutant produced only attenuated conidiophores with abnormal phialides under the same conditions (Fig. 3A). Surprisingly, the white appearance of ΔAfatg1 colonies could be rescued by supplementing the medium with additional nitrogen (Fig. 3B), and the number of conidia produced was restored to normal levels (Fig. 3C). Supplementation of PDA medium with 40 mM ammonium tartrate, ammonium chloride, ammonium sulfate, or sodium nitrate was equally effective at restoring conidiation to wt levels (data not shown), suggesting that conidiophore development requires an abundant source of nitrogen and that autophagy provides this nitrogen by recycling internal proteins when external supplies are low. The ΔAfatg1 mutant also showed a conidiation defect on Aspergillus minimal medium that could be reversed by supplementation with the same nitrogen sources listed above (data not shown).
FIG. 3.
ΔAfatg1 mutant is defective in conidiophore development and conidiation. (A) Colony and microscopic morphologies of the wt, ΔAfatg1 mutant, and complemented (C′) strains after 3 days of growth on PDA medium at 37°C. Bar, 10 μm. (B) Colony morphology of the wt and ΔAfatg1 strains grown for 3 days at 37°C on PDA medium supplemented with 40 mM ammonium tartrate. (C) Numbers of conidia recovered from the wt, ΔAfatg1 mutant, and complemented strains grown on PDA for 4 days at 37°C in the absence or presence (+ N) of 40 mM ammonium tartrate. The experiment was performed in triplicate, and the values represent the means ± standard errors of the means (SEM).
Afatg1 is required for starvation-induced foraging of hyphal tips.
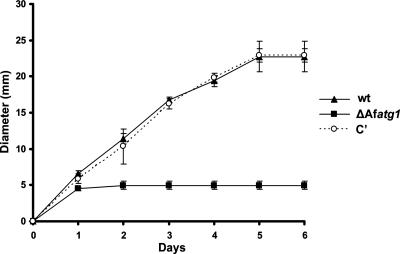
The ability to direct growth to the hyphal tips when exogenous nutrients are exhausted is an important aspect of filamentous growth that allows the mycelium to expand into areas of unexplored substrate (47). To test the hypothesis that autophagy is involved in this process, the ΔAfatg1 mutant was compared with the wt for the ability to grow radially on starvation medium. Hyphal plugs were transferred from rich medium (YG) to WA starvation plates, and colony diameters were measured daily for 6 days at 37°C. Unlike growth on a plate of nutrient-rich medium, where the hyphal tips are able to acquire nutrients from both endogenous and exogenous sources, this assay forces the hyphal tips to rely on nutrients from subapical hyphae to support radial outgrowth. In contrast to the wt and complemented strains, which were able to expand radially from the initial hyphal plug for 6 days, the ΔAfatg1 mutant stopped growing after 1 day (Fig. 4). The limited amount of growth observed for the ΔAfatg1 mutant under these conditions was most likely due to some nutrient carryover from the original plug of YG. However, since the ΔAfatg1 mutant grew no further, even after 14 days of incubation (data not shown), we concluded that radial growth of hyphal tips under starvation conditions is Atg1 dependent in this organism.
FIG. 4.
ΔAfatg1 mutant is unable to grow under starvation conditions. Hyphal plugs from the wt, ΔAfatg1 mutant, and complemented (C′) strains were placed onto WA plates and incubated at 37°C for 6 days. Colony diameters were measured daily. The experiment was performed in triplicate, and the values represent the means ± SEM.
In yeast, autophagy mutants rapidly lose viability under starvation conditions (35, 57), raising the possibility that the inability of the ΔAfatg1 mutant to grow on starvation medium was due to death of the colony. However, the ΔAfatg1 mutant retained the ability to grow normally on rich medium after 2 weeks of incubation on WA starvation plates at 37°C (data not shown), indicating that the failure of the ΔAfatg1 mutant to undergo starvation-associated growth was not related to complete death of the colony.
Metal ions restore the ability of the ΔAfatg1 mutant to grow under starvation conditions.
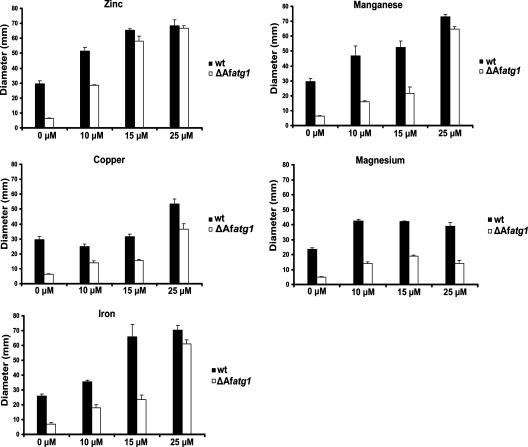
Since the growth rate of the ΔAfatg1 mutant was indistinguishable from that of the wt on defined Aspergillus minimal medium, the components of this medium were tested individually for the ability to restore growth to the ΔAfatg1 mutant on WA starvation plates. Metal ions increased the ability of wt A. fumigatus to expand radially on WA plates (Fig. 5), suggesting that metal ions are limiting for growth in this assay. Surprisingly, the same metal ions were able to restore growth to the ΔAfatg1 mutant, even in the absence of a carbon or nitrogen source (Fig. 5). Zinc was the most effective of the metals tested, followed by manganese and iron. The effects of zinc, manganese, and iron were dose dependent for both the wt and the ΔAfatg1 mutant, and the growth of the two strains became indistinguishable at the higher concentrations of these metals (Fig. 5). In contrast, the same concentrations of copper and magnesium were less effective at rescuing growth. These results suggest that some metal ions are limiting for hyphal extension under starvation conditions and that autophagy contributes to metal ion homeostasis, possibly by recycling intracellular sources.
FIG. 5.
Metal ions are sufficient to restore starvation-associated growth to the ΔAfatg1 mutant. Hyphal plugs from the wt and the ΔAfatg1 mutant were transferred to WA starvation plates, with or without supplementation with the indicated concentrations of ZnSO4, MnSO4, CuSO4, MgSO4, or FeSO4, and colony diameter was measured after 6 days at 37°C. The experiment was performed in triplicate, and the values represent the means ± SEM.
Cation depletion of rich medium is sufficient to induce autophagy in A. fumigatus.
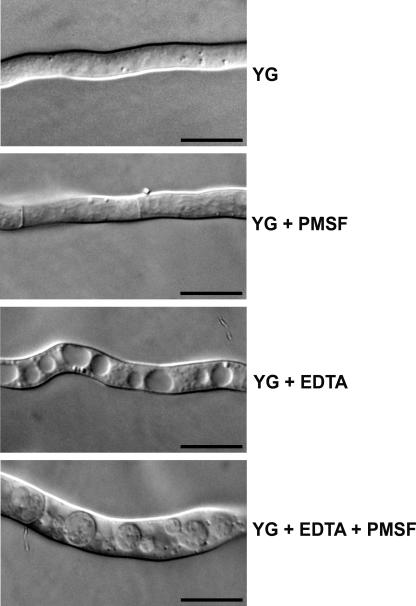
The ability of metal ions to restore normal growth to the ΔAfatg1 mutant under starvation conditions suggested that autophagy is a response to metal ion deficiency. To determine whether metal ion depletion would be sufficient to induce autophagy, the accumulation of autophagic bodies within vacuoles was examined in rich medium supplemented with the chelating agent EDTA. Overnight cultures of wt A. fumigatus were washed and incubated in fresh YG medium containing EDTA and/or PMSF for 4 h at 37°C. As shown in Fig. 6, EDTA induced extensive vacuolization of the hyphae, and the presence of PMSF allowed autophagic bodies to accumulate within the vacuoles, indicating that cation depletion by EDTA is sufficient to induce autophagy. As expected, these autophagic bodies were fluorescent in the wt strain expressing the GFP-Atg8 fusion protein (data not shown).
FIG. 6.
EDTA induces autophagy in wt A. fumigatus. Overnight cultures in YG were washed, and the medium was replaced with fresh YG, YG plus 2 mM PMSF, YG plus 0.5 mM EDTA, or YG plus 2 mM PMSF plus 0.5 mM EDTA and incubated at 37°C for 4 h before visualization by DIC microscopy. Bar, 10 μm.
The ΔAfatg1 mutant is hypersensitive to EDTA.
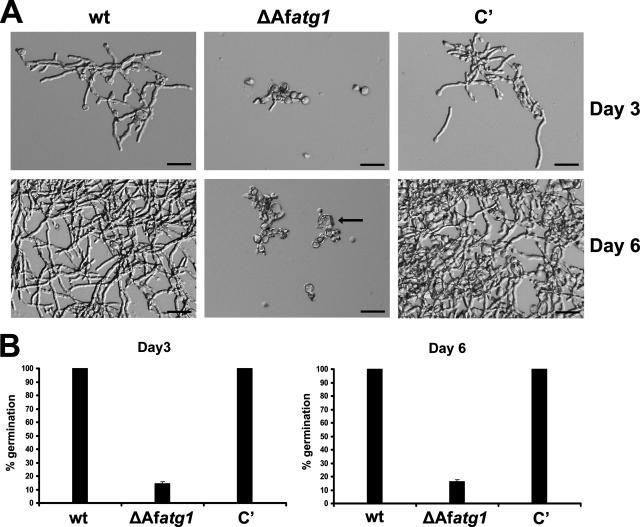
The stimulation of autophagy by EDTA suggested that autophagy has a role in protecting the organism from the deleterious consequences of divalent cation depletion. To test this hypothesis, we compared wt and ΔAfatg1 conidia for the ability to germinate in the presence of EDTA. A total of 2 × 105 wt and ΔAfatg1 conidia were inoculated separately into liquid cultures of YG containing 0.5 mM EDTA and incubated at 37°C for 6 days. Although this concentration of EDTA slowed the germination of wt conidia, the morphology remained normal and the hyphae eventually overgrew the culture, by day 6 (Fig. 7A). In contrast, ΔAfatg1 conidia were unable to switch from isotropic growth to polarized growth under the same conditions, resulting in abnormal swelling and the appearance of reduced cell wall integrity (Fig. 7A). To quantitate this defect, equal numbers of conidia were inoculated into YG-EDTA medium and the percent germination was determined microscopically. As shown in Fig. 7B, all of the wt conidia had germinated within 3 days of incubation, whereas the majority of the ΔAfatg1 mutant conidia failed to germinate after 6 days of incubation. The few ΔAfatg1 conidia that managed to germinate after 6 days were morphologically abnormal, with over half of them showing the abnormal swelling phenotype shown in Fig. 7A. The addition of excess zinc, manganese, or iron to the medium was able to restore wt growth to the ΔAfatg1 mutant under these conditions (data not shown).
FIG. 7.
ΔAfatg1 mutant is hypersensitive to EDTA. (A) A total of 2 × 105 conidia from the wt, ΔAfatg1 mutant, and complemented (C′) strains were inoculated separately into liquid cultures of YG containing 0.5 mM EDTA and incubated at 37°C for 3 to 6 days. The arrow identifies an abnormally swollen conidium. Bar, 30 μm. (B) To quantitate EDTA sensitivity, 1 × 106 conidia/ml from the wt, ΔAfatg1, and reconstituted strains were incubated at 37°C for 3 to 6 days in YG medium in the presence of 0.75 mM EDTA. The percent germination was calculated from 100 conidia analyzed microscopically. The experiment was performed in triplicate, and the values represent the means ± SEM.
Autophagy is dispensable for the virulence of A. fumigatus.
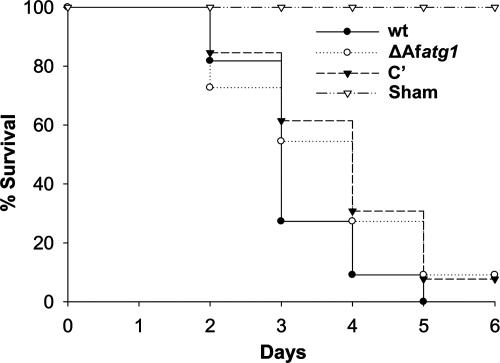
A. fumigatus is a potent opportunistic fungal pathogen that is responsible for life-threatening infections in immunosuppressed patients (3). Current evidence suggests that the host environment is a source of nutritional stress to A. fumigatus, requiring the organism to undergo metabolic adjustments to sustain the infection (19, 20, 29, 43, 48). To determine whether autophagy is necessary for growth in vivo, we tested the virulence of the ΔAfatg1 mutant in a neutropenic mouse model of invasive aspergillosis. Mice were immunosuppressed with a single dose of the monoclonal antibody RB6-8C5 on day −1, in addition to cyclophosphamide on days −1 and +3. As shown in Fig. 8, the ΔAfatg1 mutant retained wt virulence in this model, demonstrating that Afatg1 is not required for virulence in A. fumigatus. Comparable results were also obtained using cortisone acetate as the mode of immunosuppression (data not shown).
FIG. 8.
Virulence comparison. Immunosuppressed mice were infected intratracheally with 5 × 105 conidia of the indicated strains, and mortality was monitored for 14 days. No deaths occurred in the remaining mice after day 5.
DISCUSSION
Autophagy plays a pivotal role in the extensive structural remodeling that accompanies the development and differentiation of multiple species (32, 37). In this report, we demonstrate that A. fumigatus also relies upon autophagy to support asexual sporulation, an important developmental process that is tightly linked to nutrient limitation (1). However, the impaired conidiation of the ΔAfatg1 mutant could be rescued by increasing the nitrogen content of the medium. This finding argues against a direct role for Atg1 in the intracellular remodeling events that are required for conidiophore development but is consistent with a model in which autophagy provides a source of recycled nitrogen to support conidiation when exogenous levels are insufficient. Since developmental defects associated with autophagy mutations have also been reported for other filamentous fungi, such as A. oryzae and P. anserina (23, 44), it will be of interest to determine whether these developmental abnormalities are also influenced by nutrient availability.
In yeast, autophagy is best understood as a mechanism that serves to protect the organism during periods of nutrient stress (37, 60). Yeast cells respond to limiting nutrients by entering a nondividing resting state (15), and autophagy contributes to the maintenance of cell viability under these conditions (35, 57). In contrast, filamentous fungi respond to starvation by targeting growth to hyphal tips, a mechanism that promotes expansion of the colony into areas of new substrate (47). This foraging-like response is facilitated by the fact that fungal hyphae are divided by perforated septa into multinucleated compartments that permit sharing of nutrients throughout the mycelium (13, 47). It has been suggested that vacuole-mediated recycling in the older portions of the mycelium supports the growth of the hyphal tips when exogenous nutrients are scarce (51), but the contribution of autophagy to this process has not been tested directly. Here we demonstrate that Afatg1-dependent autophagy is dispensable for normal radial growth rates on rich or minimal medium but is essential for the hyphal growth that permits colonial expansion under starvation conditions.
As a major component of the biomass in decaying vegetation, A. fumigatus is in direct competition with many other microbes for limited resources and must therefore be able to tolerate periods of fluctuating nutrient availability (31, 55). Although several lines of evidence suggest that A. fumigatus encounters nutrient stress in the host environment (19, 20, 29, 43, 48), we found that the disruption of autophagy caused by the loss of Afatg1 had no major impact on virulence in a neutropenic mouse model, suggesting that the organism does not require Atg1 for growth in the host. Similar findings have been described for an autophagy mutant of Candida albicans, where the loss of Atg9 was associated with a dramatic loss of viability in response to nitrogen starvation but was dispensable for survival within macrophages and for virulence in a mouse infection model (41, 42). These observations contrast with the requirement for autophagy for the virulence of the plant fungal pathogen Magnaporthe grisea. In this organism, autophagy is required for a type of programmed cell death that contributes to the elaboration of an appressorium that is required for virulence (33, 59). The importance of autophagy to virulence has also been demonstrated for the parasite Leishmania major, where autophagy mutants fail to differentiate into the obligate infective form (5). Although A. fumigatus does not produce specialized structures required for infection, it is intriguing to speculate that autophagy may contribute to other aspects of programmed cell death that are relevant to the physiology of this organism, and efforts to examine this are in progress.
Metal ions are essential micronutrients that provide catalytic or structural roles in many enzymes (10). We have shown that zinc is the most effective at restoring growth to the ΔAfatg1 mutant under starvation conditions, a finding that is consistent with previous evidence showing marked susceptibility of pathogenic fungi, including A. fumigatus, to zinc deprivation (34). The majority of intracellular zinc is bound to proteins, serving as both a structural component in domains such as zinc fingers and an essential cofactor for approximately 3% of the yeast proteome (11). Manganese, copper, magnesium, and iron were also able to rescue the growth of the ΔAfatg1 mutant, although higher concentrations were required to achieve the same amount of growth as that provided by zinc. The exact reason for this is presently unclear, but we speculate that it may involve displacement of zinc from cellular components, analogous to the effects of heavy metals on the displacement of zinc from metallothioneins or other cellular components (62).
The ability of zinc and other metal ions to rescue the impaired growth of the ΔAfatg1 mutant suggests that autophagy facilitates growth when metals are limiting. This hypothesis predicted that metal ion depletion would be sufficient to induce autophagy in wt A. fumigatus and that the ΔAfatg1 mutant would be hypersensitive to metal depletion. This was confirmed by showing that autophagy is induced in wt A. fumigatus by the incorporation of EDTA into rich medium and that the ΔAfatg1 mutant is severely growth impaired under these conditions. Since the abundant nitrogen in rich medium would normally be expected to suppress autophagy through TOR signaling (22), this implies that metal ion deficiency alone is sufficient to induce autophagy, even in a nitrogen-replete environment.
Taken together, the data presented in this study are consistent with a model in which zinc and other metal ions are limiting for hyphal growth under starvation conditions. Their absence triggers an autophagy response, resulting in the transport of preexisting metal-associated components to the vacuole for degradation, followed by recycling of the metals to support further hyphal tip growth. Failure to induce autophagy, as in the case of the ΔAfatg1 mutant, would result in a critical deficiency in free metal ions, thereby preventing further growth. The interconnected mycelium of A. fumigatus has a considerable amount of biomass from which it can obtain these metals in order to sustain the outward expansion of the colony under starvation conditions, and our data indicate that autophagy is integral to this process. In contrast, unicellular yeasts possess a more limited amount of biomass that is restricted to a single cell. Thus, it is conceivable that filamentous fungi have expanded the role of autophagy to include more extensive intracellular recycling of metal ions and other molecules to meet the growth demands of the hyphal tips when exogenous nutrient supplies are exhausted. Consistent with this, the Zap1 zinc-responsive transcription factor shows no difference in zinc regulation in a Δatg1 mutant of S. cerevisiae, suggesting that autophagy does not have a major role in zinc homeostasis in this organism (David Eide, personal communication). Similarly, the C. albicans atg9Δ autophagy mutant has normal sensitivity to EDTA, and zinc is unable to correct the starvation-associated decrease in viability of this mutant (Glen Palmer, personal communication).
Since carbon and nitrogen were absent from the starvation medium used in this study, it would appear that A. fumigatus has sufficient reserves of carbon and nitrogen to support a limited amount of hyphal growth or, alternatively, that amino acid pools can be generated from another mechanism. The identification of a role for the ubiquitin-proteasome system in the degradation of proteins during acute nutrient stress supports this possibility (58). It will be of interest in future studies to determine whether there are specific signaling pathways that link metal ion availability to the induction of autophagy or perhaps to other proteolytic pathways in A. fumigatus.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants R21AI061495 to D.S.A. and R01AI061497 to J.C.R. and by an NIH F31 predoctoral fellowship (AI064121) to D.L.R.
We thank D. J. Eide, University of Wisconsin-Madison, for critically reviewing the manuscript and Jay Card for photography assistance.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baehrecke, E. H. 2003. Autophagic programmed cell death in Drosophila. Cell Death Differ. 10:940-945. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, P. D., and K. A. Marr. 2006. Aspergillosis: spectrum of disease, diagnosis, and treatment. Infect. Dis. Clin. N. Am. 20:545-561. [DOI] [PubMed] [Google Scholar]
- 4.Bernales, S., K. L. McDonald, and P. Walter. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4:e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besteiro, S., R. A. Williams, L. S. Morrison, G. H. Coombs, and J. C. Mottram. 2006. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 281:11384-11396. [DOI] [PubMed] [Google Scholar]
- 6.Bhabhra, R., M. D. Miley, E. Mylonakis, D. Boettner, J. Fortwendel, J. C. Panepinto, M. Postow, J. C. Rhodes, and D. S. Askew. 2004. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect. Immun. 72:4731-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catlett, N. L., B.-N. Lee, O. C. Yoder, and B. G. Turgeon. 2002. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. News 50:9-11. [Google Scholar]
- 8.Deretic, V. 2006. Autophagy as an immune defense mechanism. Curr. Opin. Immunol. 18:375-382. [DOI] [PubMed] [Google Scholar]
- 9.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 2002. Bacterial interactions with the autophagic pathway. Cell. Microbiol. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 10.Eide, D. J. 2000. Metal ion transport in eukaryotic microorganisms: insights from Saccharomyces cerevisiae. Adv. Microb. Physiol. 43:1-38. [DOI] [PubMed] [Google Scholar]
- 11.Eide, D. J. 1998. The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu. Rev. Nutr. 18:441-469. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Abalos, J. M., H. Fox, C. Pitt, B. Wells, and J. H. Doonan. 1998. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 27:121-130. [DOI] [PubMed] [Google Scholar]
- 13.Glass, N. L., D. J. Jacobson, and P. K. Shiu. 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34:165-186. [DOI] [PubMed] [Google Scholar]
- 14.Gozuacik, D., and A. Kimchi. 2004. Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23:2891-2906. [DOI] [PubMed] [Google Scholar]
- 15.Gray, J. V., G. A. Petsko, G. C. Johnston, D. Ringe, R. A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez, M. G., S. S. Master, S. B. Singh, G. A. Taylor, M. I. Colombo, and V. Deretic. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119:753-766. [DOI] [PubMed] [Google Scholar]
- 17.Hara, T., K. Nakamura, M. Matsui, A. Yamamoto, Y. Nakahara, R. Suzuki-Migishima, M. Yokoyama, K. Mishima, I. Saito, H. Okano, and N. Mizushima. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885-889. [DOI] [PubMed] [Google Scholar]
- 18.Harding, T. M., K. A. Morano, S. V. Scott, and D. J. Klionsky. 1995. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel, M., H. N. Arst, Jr., A. Aufauvre-Brown, and D. W. Holden. 1998. The role of the Aspergillus fumigatus areA gene in invasive pulmonary aspergillosis. Mol. Gen. Genet. 258:553-557. [DOI] [PubMed] [Google Scholar]
- 20.Hissen, A. H., A. N. Wan, M. L. Warwas, L. J. Pinto, and M. M. Moore. 2005. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding l-ornithine N5-oxygenase, is required for virulence. Infect. Immun. 73:5493-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhasz, G., G. Csikos, R. Sinka, M. Erdelyi, and M. Sass. 2003. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 543:154-158. [DOI] [PubMed] [Google Scholar]
- 22.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuma, T., M. Ohneda, M. Arioka, and K. Kitamoto. 2006. Functional analysis of the ATG8 homologue AoAtg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot. Cell 5:1328-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkegaard, K., M. P. Taylor, and W. T. Jackson. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2:301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kissova, I., B. Salin, J. Schaeffer, S. Bhatia, S. Manon, and N. Camougrand. 2007. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 3:329-336. [DOI] [PubMed] [Google Scholar]
- 26.Klionsky, D. J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klionsky, D. J., A. M. Cuervo, and P. O. Seglen. 2007. Methods for monitoring autophagy from yeast to human. Autophagy 3:181-206. [DOI] [PubMed] [Google Scholar]
- 28.Komatsu, M., S. Waguri, T. Chiba, S. Murata, J. Iwata, I. Tanida, T. Ueno, M. Koike, Y. Uchiyama, E. Kominami, and K. Tanaka. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880-884. [DOI] [PubMed] [Google Scholar]
- 29.Krappmann, S., E. M. Bignell, U. Reichard, T. Rogers, K. Haynes, and G. H. Braus. 2004. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 52:785-799. [DOI] [PubMed] [Google Scholar]
- 30.Kruse, K. B., J. L. Brodsky, and A. A. McCracken. 2006. Autophagy: an ER protein quality control process. Autophagy 2:135-137. [DOI] [PubMed] [Google Scholar]
- 31.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine, B., and D. J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6:463-477. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X. H., J. P. Lu, L. Zhang, B. Dong, H. Min, and F. C. Lin. 2007. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 6:997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lulloff, S. J., B. L. Hahn, and P. G. Sohnle. 2004. Fungal susceptibility to zinc deprivation. J. Lab. Clin. Med. 144:208-214. [DOI] [PubMed] [Google Scholar]
- 35.Matsuura, A., M. Tsukada, Y. Wada, and Y. Ohsumi. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245-250. [DOI] [PubMed] [Google Scholar]
- 36.Melendez, A., Z. Talloczy, M. Seaman, E. L. Eskelinen, D. H. Hall, and B. Levine. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301:1387-1391. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima, N. 2005. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12(Suppl. 2):1535-1541. [DOI] [PubMed] [Google Scholar]
- 38.Nair, U., and D. J. Klionsky. 2005. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 280:41785-41788. [DOI] [PubMed] [Google Scholar]
- 39.Onodera, J., and Y. Ohsumi. 2005. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 280:31582-31586. [DOI] [PubMed] [Google Scholar]
- 40.Otto, G. P., M. Y. Wu, N. Kazgan, O. R. Anderson, and R. H. Kessin. 2003. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 278:17636-17645. [DOI] [PubMed] [Google Scholar]
- 41.Palmer, G. E. 2007. Autophagy in the invading pathogen. Autophagy 3:251-253. [DOI] [PubMed] [Google Scholar]
- 42.Palmer, G. E., M. N. Kelly, and J. E. Sturtevant. 2007. Autophagy in the pathogen Candida albicans. Microbiology 153:51-58. [DOI] [PubMed] [Google Scholar]
- 43.Panepinto, J. C., B. G. Oliver, J. R. Fortwendel, D. L. Smith, D. S. Askew, and J. C. Rhodes. 2003. Deletion of the Aspergillus fumigatus gene encoding the Ras-related protein RhbA reduces virulence in a model of invasive pulmonary aspergillosis. Infect. Immun. 71:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinan-Lucarre, B., A. Balguerie, and C. Clave. 2005. Accelerated cell death in Podospora autophagy mutants. Eukaryot. Cell 4:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinan-Lucarre, B., M. Paoletti, K. Dementhon, B. Coulary-Salin, and C. Clave. 2003. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol. Microbiol. 47:321-333. [DOI] [PubMed] [Google Scholar]
- 46.Priault, M., B. Salin, J. Schaeffer, F. M. Vallette, J. P. di Rago, and J. C. Martinou. 2005. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 12:1613-1621. [DOI] [PubMed] [Google Scholar]
- 47.Robson, G. 1999. Hyphal cell biology, p. 164-184. In R. Oliver and M. Schweizer (ed.), Molecular fungal biology. Cambridge University Press, Cambridge, United Kingdom.
- 48.Schrettl, M., E. Bignell, C. Kragl, C. Joechl, T. Rogers, H. N. Arst, Jr., K. Haynes, and H. Haas. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott, R. C., O. Schuldiner, and T. P. Neufeld. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7:167-178. [DOI] [PubMed] [Google Scholar]
- 50.Shintani, T., and D. J. Klionsky. 2004. Autophagy in health and disease: a double-edged sword. Science 306:990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoji, J. Y., M. Arioka, and K. Kitamoto. 2006. Possible involvement of pleiomorphic vacuolar networks in nutrient recycling in filamentous fungi. Autophagy 2:226-227. [DOI] [PubMed] [Google Scholar]
- 52.Stephens-Romero, S. D., A. J. Mednick, and M. Feldmesser. 2005. The pathogenesis of fatal outcome in murine pulmonary aspergillosis depends on the neutrophil depletion strategy. Infect. Immun. 73:114-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeshige, K., M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tal, R., G. Winter, N. Ecker, D. J. Klionsky, and H. Abeliovich. 2007. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J. Biol. Chem. 282:5617-5624. [DOI] [PubMed] [Google Scholar]
- 55.Tekaia, F., and J. P. Latge. 2005. Aspergillus fumigatus: saprophyte or pathogen? Curr. Opin. Microbiol. 8:385-392. [DOI] [PubMed] [Google Scholar]
- 56.Tekinay, T., M. Y. Wu, G. P. Otto, O. R. Anderson, and R. H. Kessin. 2006. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot. Cell 5:1797-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169-174. [DOI] [PubMed] [Google Scholar]
- 58.Vabulas, R. M., and F. U. Hartl. 2005. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310:1960-1963. [DOI] [PubMed] [Google Scholar]
- 59.Veneault-Fourrey, C., M. Barooah, M. Egan, G. Wakley, and N. J. Talbot. 2006. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312:580-583. [DOI] [PubMed] [Google Scholar]
- 60.Yorimitsu, T., and D. J. Klionsky. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl. 2):1542-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yorimitsu, T., and D. J. Klionsky. 2007. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy 3:160-162. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, B., O. Georgiev, M. Hagmann, C. Gunes, M. Cramer, P. Faller, M. Vasak, and W. Schaffner. 2003. Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol. Cell. Biol. 23:8471-8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.