Abstract
Protein O-mannosyltransferases initiate O mannosylation of secretory proteins, which are of fundamental importance in eukaryotes. In this study, the PMT gene family of the human fungal pathogen Aspergillus fumigatus was identified and characterized. Unlike the case in Saccharomyces cerevisiae, where the PMT family is highly redundant, only one member of each PMT subfamily, namely, Afpmt1, Afpmt2, and Afpmt4, is present in A. fumigatus. Mutants with a deletion of Afpmt1 are viable. In vitro and in vivo activity assays confirmed that the protein encoded by Afpmt1 acts as an O-mannosyltransferase (AfPmt1p). Characterization of the ΔAfpmt1 mutant showed that a lack of AfPmt1p results in sensitivity to elevated temperature and defects in growth and cell wall integrity, thereby affecting cell morphology, conidium formation, and germination. In a mouse model, Afpmt1 was not required for the virulence of A. fumigatus under the experimental conditions used.
O glycosylation is a frequent modification of proteins entering the secretory pathway. In fungi, proteins are O mannosylated at serine or threonine residues during import into the endoplasmic reticulum. Protein O mannosylation is initiated by a family of protein O-mannosyltransferases (PMTs) that are evolutionarily conserved from Saccharomyces cerevisiae to humans (33, 40). The O-glycans in yeast are short, typically containing one or two mannosyl residues. In mammalian cells, the inner O-linked mannose is elongated with the first addition of an N-acetylglucosamine and then various sugars (10). Mammalian homologues of PMTs have been described previously (17, 39), and recently, PMT activity of human POMT proteins was verified (23).
The genetics of fungal O glycosylation has been studied in the yeast species Saccharomyces cerevisiae, Candida albicans, Schizosaccharomyces pombe, and Cryptococcus neoformans (11, 27, 33, 42). In S. cerevisiae, a total of seven PMT family members (ScPmt1-7p) are present (13, 32), which fall into the following three major groups of homology: (i) Pmt1/5/7, (ii) Pmt2/3/6, and (iii) Pmt4. Specific protein substrates that are O glycosylated by ScPmt1p, ScPmt2p, or ScPmt4p have been described previously (8, 14). PMT activity has been demonstrated in vivo for ScPmt1-4p and ScPmt6p (33). Single pmt1 mutants fail to grow on some media under anaerobic conditions (3). pmt1,2,3 triple mutants grow only in osmotically stabilized medium, whereas pmt1,2,4 and pmt2,3,4 triple mutants are not viable under any conditions, indicating that PMT protein activity is essential in S. cerevisiae, although individual genes are dispensable (13).
The genome of the human fungal pathogen C. albicans contains five PMT genes. pmt1 mutants are viable, but they are defective in undergoing cellular differentiation from yeast to a true hyphal growth form under some conditions (34). The cellular differentiation defect of C. albicans pmt mutants is reminiscent of Drosophila melanogaster mutants lacking the PMT homologue rotated abdomen, which show an aberrant morphology (24). pmt1 mutants are supersensitive to aminoglycoside antibiotics and to other agents affecting fungal cell wall synthesis. The virulence of pmt1 mutants is significantly attenuated (34). pmt1,4 double mutants are not viable. Comparisons of mutant phenotypes with the wild type revealed that Pmt isoforms are relevant not only for general growth but also for growth in the presence of antifungal drugs. The pmt phenotypes were closely related to alterations in cell wall components, including cell wall mannoproteins and polysaccharides (29).
In S. pombe, only one member of each PMT subfamily, namely, oma1+, oma2+, and oma4+, is present. They all act as PMTs in vivo. Deletion of oma2+ as well as simultaneous deletion of oma1+ and oma4+ is lethal. Characterization of the viable S. pombe oma1Δ and oma4Δ single mutants showed that a lack of O mannosylation results in abnormal cell wall and septum formation, therefore severely affecting cell morphology and cell-cell separation (42). More recently, Pmt4 in C. neoformans was shown to be essential for morphogenesis and virulence (27).
Among multicellular eukaryotes, human, mouse, and Drosophila genes with significant homology to PMTs have been cloned (17, 24, 39). In comparison with S. cerevisiae, the PMT family is less redundant in higher eukaryotes. In Drosophila, only two PMT family members are present (rotated abdomen and twisted) (24, 39). The same is true for mice and humans (POMT1 and POMT2) (17, 39). Mutations in human POMT1 cause Walker-Warburg syndrome, which is characterized by severe congenital muscular dystrophy, neuronal migration defects, and structural abnormalities of the eye (2). Targeted deletion of the Walker-Warburg syndrome gene Pomt1 in the mouse results in embryonic lethality due to defects in the formation of Reichert's membrane, the first basement membrane to form in the embryo (41). Mutations of the Drosophila PMT homologues alter muscle structures and the alignment of the adult cuticle (16, 24). Taken together, pmt mutants from different species reveal that protein O mannosylation is of fundamental importance in uni- and multicellular eukaryotes.
Aspergillus fumigatus causes fatal invasive aspergillosis among immunocompromised patients (18). The main reason for patient death is the low efficiency of the drug therapies available to treat aspergillosis and the lack of an assay that can detect the fungus early during infection. Although the structure of O-linked oligosaccharides present in the cell wall peptidogalactomannan of A. fumigatus has been characterized (20), little is known about the genes responsible for the initiation of O-mannosylation assembly. As a matter of fact, only the pmtA gene, which encodes a member of the PMT2 subfamily, has been studied so far, in Aspergillus nidulans and Aspergillus awamori (25, 26). Here we report the functional analysis of Afpmt1 of A. fumigatus. We demonstrate that protein O mannosylation catalyzed by AfPmt1p, an O-mannosyltransferase belonging to the PMT1 subfamily, is a vital protein modification which is crucial for growth and cell wall integrity of A. fumigatus at an elevated temperature.
MATERIALS AND METHODS
Strains and growth conditions.
A. fumigatus wild-type strain YJ-407 (CGMCC 0386; China General Microbiological Culture Collection Center) was maintained on potato glucose (2%) agar slants (43). A. fumigatus strain CEA17 (pyrG−) (37), a kind gift from C. d'Enfert, Institut Pasteur, France, was propagated at 37°C on YGA (0.5% yeast extract, 2% glucose, 1.5% Bacto agar), complete medium (5), or minimal medium with 0.5 mM sodium glutamate as a nitrogen source (5). Uridine and uracil were added at a concentration of 5 mM when required. Liquid cultures used for DNA preparation were grown at 30°C for 24 h on a shaker (300 rpm) with 5-mm-diameter glass beads in YG. Vectors and plasmids were propagated in Escherichia coli DH5α (BRL).
Computer analysis.
Sequence analysis of cDNA clones and multiple sequence alignments were performed using Omiga (2.0). Blast searches were performed using the program BLAST (1).
Construction of ΔAfpmt1 mutant and revertant stains.
The flanking regions of the Afpmt1 gene were amplified from A. fumigatus wild-type strain YJ-407 genomic DNA by PCR. The forward primer AFPMT1-5-N and the reverse primer AFPMT1-5-C (Table 1) were used to generate a 2.0-kb upstream flanking region, and AFPMT1-3-N and AFPMT1-3-C were used for the 2.0-kb downstream flanking region. The upstream and downstream flanking regions of the Afpmt1 gene were digested with NotI/XbaI and XbaI/EcoRI, respectively. By insertion of the flanking regions into the NcoI/EcoRI sites of pBluescript II SK(+) (Stratagene), p5′3′SK carrying the flanking regions of the Afpmt1 gene was obtained.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Restriction enzyme(s) |
|---|---|---|
| AFPMT1-5-N | GGTGGTGCGGCCGCCTTGCTTCCTCCTCAGGATCGA | NotI |
| AFPMT1-5-C | GGTGGTTCTAGACGACCGCAATCAATTAGGACCAAGT | XbaI |
| AFPMT1-3-N | GGTTCTAGATGAACGCCCGACGGCTGACG | XbaI |
| AFPMT1-3-C | GTGGTGAATTCGCGGCCGCCATTGGCACCCTTGCGCGCTC | EcoRI and NotI |
| AFPMT1-flank-5 | AAATATGCGGCCGCCTTGCTTCCTCCTCAGGATCG | |
| AFPMT1-flank-3 | GTCCGGAATTCGCATTGGCACCCTTGCGCGCTC | |
| N-flanking-5 | CCTGGTTGGCGGCTATCAGA | |
| C-flanking-3 | GCGTTGATGATGCAATTGGG |
Restriction enzyme sites are underlined.
pAFPMT1-pyrG, in which pyrG was flanked by 2.0-kb upstream and 2.0-kb downstream noncoding regions of the Afpmt1 gene, was constructed by subcloning the 4.0-kb XbaI fragment of pCDA14 (kindly provided by C. d'Enfert, Institut Pasteur, France) (6) into XbaI-digested p5′3′SK.
A. fumigatus protoplasts were prepared from germinating conidiospores grown for 5 to 5.5 h at 37°C in YG medium containing uridine and uracil, using 3 mg/ml of Novozyme 234 as described for A. nidulans (28). Transformations were carried out as described previously, and transformation mixtures were mixed with 3.5 ml of minimal medium containing 1% agar, 0.4 M (NH4)2SO4, and 1 M sucrose as osmotic stabilizers and then poured onto minimal medium agar plates containing 0.4 M (NH4)2SO4 and 1 M sucrose. For each transformation, 108 Novozyme-treated conidiospores were mixed with 4 μg of NotI-digested pAFPMT1-pyrG. Transformants became visible after 5 to 7 days of incubation at 30°C.
The revertant strain was constructed by replacement of pyrG in the ΔAfpmt1 mutant with the wild-type copy of Afpmt1. The forward primer AFPMT1-flank-5 and the reverse primer AFPMT1-flank-3 (Table 1) were used to generate a 7.0-kb Afpmt1 region by two-step PCR (94°C for 1 min, 98°C for 20 s, and 68°C for 12 min for 30 cycles, followed by 10 min at 72°C). Transformants with uracil auxotrophy were selected and confirmed first by PCR and then by Southern blotting, using the upstream flanking region as a probe.
Genomic DNA analysis.
PCR analysis of the A. fumigatus ΔAfpmt1 mutant was performed by 35 reaction cycles (94°C for 0.5 min, 52°C for 0.5 min, and 72°C for 4.5 min). N-flanking-5 and C-flanking-3 (Table 1) were used as primers to generate the wild-type 3.5-kb and mutant 4.5-kb fragments, which included Afpmt1 and pyrG-blaster flanked by 0.25-kb upstream and 0.25-kb downstream noncoding regions of Afpmt1, respectively.
Genomic DNAs of A. fumigatus strains were digested with SalI or EcoRI. DNA restriction fragments transferred to nylon membranes (Zeta-probe+; Bio-Rad) were hybridized with the probe in 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% sodium dodecyl sulfate (SDS), 5× Denhardt solution, and 10% dextran sulfate at 65°C for 12 to 16 h. Final washes were performed at 65°C in 2× SSC-0.5% SDS. The 2.0-kb NcoI/XbaI fragment of p5′3′SK was used to probe the Afpmt1 gene in the wild-type strain and the pyrG gene in the mutant strain. Labeling and visualization were performed using a DIG DNA labeling and detection kit (Roche Applied Science) according to the manufacturer's instructions.
In vitro analysis of PMT activity.
The membranes of A. fumigatus cells were prepared from 24-h-old mycelial cultures grown on a shaker (200 rpm) at 30°C in YG liquid medium (1% yeast extract and 2% glucose, pH 8.0). The fungal material was filtered, washed extensively with water, and homogenized in liquid nitrogen. The homogenates were resuspended in 10 ml of 50 mM Tris-HCl (pH 7.5) containing 0.3 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10 mM mercaptoethanol. Cell debris was removed by centrifugation at 12,000 × g at 4°C for 10 min. The membranes in the supernatant were collected by centrifugation at 70,000 × g for 1 h and solubilized in 50 mM Tris-HCl (pH 7.5) containing 0.3 mM MgCl2, 1 mM PMSF, and 33% glycerol. The protein concentration was adjusted to 10 mg/ml for activity assay.
The in vitro peptide assay for PMT1 activity was performed as described by Weston et al. (38). The assay was optimized using 0.02 μCi of Dol-P-[3H]mannose (40 Ci/mmol; American Radiolabeled Chemicals) and the acceptor peptide Ac-YAYAV-NH2 (final concentration, 3.5 mM). Five microliters of 20% Triton X-100, 10 μl of 0.2 M HEPES (pH 7.5), and 25 μg of membrane proteins were added. The assay mixture, in a total volume of 50 μl, was incubated at 25°C for 30 min. The reaction was stopped by the addition of 1 ml of chloroform-methanol (3:2) followed by 200 μl of water. For each sample, the assay was repeated at least three times.
O-glycosylation analysis by HPAEC-PAD.
The chitinase AfCHI44 used for O-glycan analysis was induced and purified from the wild-type, mutant, and revertant strains of A. fumigatus as previously described (43). For comparison of O-glycans from different strains, 200 μg of purified AfCHI44 of the wild-type, mutant, or revertant strain was β-eliminated with 100 μl of 0.1 M NaOH at 25°C overnight to release O-glycans. The proteins were removed by centrifugation, and 20 μl of supernatant was analyzed with a Carbo-PA1 column on a high-performance anion-exchange chromatograph-pulsed ampere detector (HPAEC-PAD; Dionex). The glycans were eluted with 15 mM NaOH at a flow rate of 1.0 ml/min. Mannose was treated by the same method as a standard because the mannose released from protein by β-elimination with NaBH4 is converted into anhydrous mannitol, which shows a different retention time from that of mannose.
Phenotypic analysis of mutants.
The same numbers of wild-type, mutant, and revertant conidiospores were collected and grown in complete medium containing uridine and uracil or containing 1 M sucrose at 37°C, 42°C, or 50°C for 10 to 16 h. For mycelia growth or conidial germination in liquid complete medium, the strains were incubated at 37°C or 42°C for 6 to 8 h and examined with a microscope (magnification, ×250).
For antifungal sensitivity testing, A. fumigatus was grown in complete medium containing 0 to 100 μg/ml of calcofluor white (Sigma) or 25 to 100 μg/ml of hygromycin B (Roche). Growth end points were determined visually on plates after 24 to 48 h of growth at 37°C or 42°C.
Electron microscopy.
Conidia produced in solid or liquid medium and fixed with 2.5% glutaraldehyde and 1% osmium tetroxide were examined with a Quanta 200 scanning electron microscope (FEI).
Conidia were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.0, for 4 h or overnight at 4°C. Cells were fixed in 2.5% glutaraldehyde in 0.1 M phosphate, washed three times with 0.1 M phosphate, postfixed in 1% osmium tetroxide, incubated for 2 to 4 h in 0.1 M phosphate and then for 15 to 20 min in 30%, 50%, 70%, 85%, 95%, and 100% methanol, and postfixed in 2% uranyl acetate-30% methanol. Cells were rinsed, dehydrated, and embedded in Epon 812 by the floating sheet method. The sections were examined with an H-600 electron microscope (Hitachi).
Chemical analysis of cell walls.
Conidia were inoculated into 100 ml of yeast extract-peptone-dextrose liquid medium at a concentration of 106 conidia/ml and incubated at 37°C with shaking (300 rpm) for 48 h. The mycelium was then harvested by filtering the culture through two layers of Miracloth, washed twice with distilled water, and lyophilized. Three aliquots of 5 mg dry mycelium were used as independent samples for cell wall analysis. The same experiment was repeated twice. To remove unbound cell wall proteins and water-soluble sugar, each sample was boiled in 2 ml of 2% SDS in 50 mM Tris-HCl buffer supplemented with 100 mM Na-EDTA, 40 mM β-mercaptoethanol, and 1 mM PMSF for 5 min (9, 15, 31). Mannoprotein was extracted with 3% NaOH at 75°C for 1 h and quantitatively determined by Lowry protein assay (21). Glucan and chitin were digested in 96% formic acid at 100°C for 4 h. Formic acid was evaporated by lyophilization, and the residues were dissolved in 10 ml of distilled water. Glucan and chitin contents were estimated by determining the released glucose and N-acetylglucosamine after digestion. Glucose was measured by the phenol-sulfuric acid method (7). N-Acetylglucosamine was measured by the method described by Lee et al. (19).
Analysis of mutant virulence.
The mutant, revertant, and wild-type strains were used for experimental infections of white male BALB/c mice (20 to 22 g). Conidia were suspended in 0.01% Tween 20 in saline to give a challenge inoculum of 3 × 105 CFU/g of mouse body weight in a 30-μl volume. Mice were immunosuppressed by intraperitoneal injection of cyclophosphamide (150 mg/kg of body weight) on days −3 and −1 and by one subcutaneous injection of hydrocortisone acetate (40 mg/kg of body weight) on day −1. On day 0, mice were anesthetized by inhalation of diethyl ether and infected intranasally with spores in a 30-μl volume containing 6 × 106 conidia. A concurrent control group consisted of mice that were immunosuppressed and inoculated with 30 μl 0.01% Tween 20 in saline. Immunosuppression was prolonged by cyclophosphamide injections (150 mg/kg) on days 3, 6, and 9. Mice were kept in sterile cages with filter tops and received sterile food and bedding. Tetracycline (1 mg/ml) and uridine (100 mM) were added to the drinking water, which was changed twice daily. Four groups containing 15 mice each were inoculated and monitored twice each day for 30 days after inoculation, and mortality was recorded. Mice surviving the course of the experiment were terminated humanely on day 30. The survival rate was analyzed statistically by the methods of Kaplan-Meier, using SPSS11 software. P values of <0.01 were considered significant in this analysis.
RESULTS
PMT gene family in A. fumigatus.
To identify the genes responsible for the initiation of O mannosylation in A. fumigatus, we performed a tBLASTn search (1) of the A. fumigatus genome database at the TIGR (The Institute for Genomic Research) website (http://tigrblast.tigr.org/er-blast/index.cgi?project=afu1), using protein sequences of S. cerevisiae PMT1-7. The database search revealed the presence of three genes encoding proteins with significant homology to PMT1, PMT2, and PMT4: AfPmt1p (Afu3g06450 on chromosome 3, encoding a protein of 947 amino acids [aa]) has an overall protein sequence homology of 62% and identity of 45% to PMT1; AfPmt2p (Afu1g07690 on chromosome 1, encoding a protein of 760 aa) has 68% homology and 51% identity to PMT2; and AfPmt4p (Afu8g04500 on chromosome 8, encoding a protein of 781 aa) has 65% homology and 49% identity to PMT4. Phylogenetic analyses of known PMT proteins placed AfPmt1p within the PMT1 subfamily, AfPmt2p within the PMT2 subfamily, and AfPmt4p within the PMT4 subfamily.
Deletion of the Afpmt1 gene in A. fumigatus.
We tried to express the Afpmt1 gene in E. coli and Pichia yeast, respectively. However, we did not detect any recombinant protein. To analyze the physiological function of the PMT1 (AfPmt1p) in A. fumigatus, we created the null mutant of Afpmt1 by replacement of the Afpmt1 gene with pyrG (see Materials and Methods). A fragment of NcoI-digested pAFPMT1-pyrG which contains 2.0-kb upstream and 2.0-kb downstream noncoding regions of Afpmt1 separated by the pyrG gene was used to transform protoplasts of A. fumigatus strain CEA17 (pyrG−) to uridine/uracil prototrophy. Only one viable ΔAfpmt1 mutant was obtained. In order to ensure that all phenotypes noted for the ΔAfpmt1 strain were due to specific deletion of Afpmt1, the revertant strain was constructed by reintroduction of a wild-type copy of Afpmt1 directly into the mutated locus under the control of its own promoter.
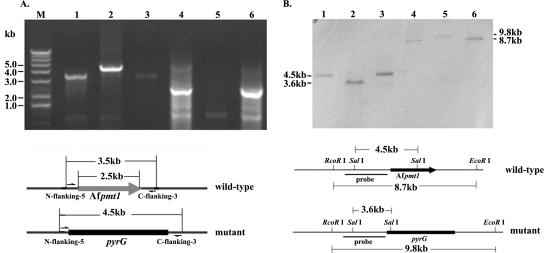
Using PCR analysis, a 4.5-kb fragment containing the pyrG gene was amplified from the mutant genome (lane 2 in Fig. 1A), instead of a 3.5-kb fragment containing the Afpmt1 gene from the wild-type or revertant genome (lanes 1 and 3 in Fig. 1A). A 2.5-kb fragment containing the Afpmt1 open reading frame was unable to be amplified from the ΔAfpmt1 mutant (lane 5 in Fig. 1A). Southern analysis of the SalI-digested genomic DNA of this mutant strain demonstrated that the 4.5-kb SalI fragment in the wild-type or revertant strain was converted into a 3.6-kb SalI fragment and the 8.7-kb EcoRI fragment of the wild-type or revertant strain was changed into a 9.8-kb EcoRI fragment (Fig. 1B). These results clearly demonstrated that the Afpmt1 gene in A. fumigatus was replaced by a pyrG gene and that the revertant strain was successfully constructed.
FIG. 1.
Confirmation of ΔAfpmt1 mutant and revertant by PCR (A) and Southern blotting (B). (A) PCR analysis was carried out as described in Materials and Methods. Lanes 1 to 3, N-flanking-5 and C-flanking-3 were used as primers to generate a wild-type 3.5-kb or mutant 4.5-kb fragment that includes Afpmt1 or pyrG-blaster flanked by 0.25-kb upstream and 0.25-kb downstream noncoding regions of Afpmt1, respectively; lanes 4 to 6, primers were used to amplify the 2.5-kb coding region of Afpmt1. (B) Genomic DNA digested with SalI (lanes 1 to 3) or EcoRI (lanes 4 to 6) was probed with the NcoI/XbaI fragment of p5′3′SK, which contains a 2.0-kb upstream noncoding region of Afpmt1. The electrophoretic positions and sizes of DNA are indicated in both panels. Lanes for both panels: 1 and 4, wild type; 2 and 5, ΔAfpmt1 mutant; 3 and 6, revertant.
O-mannosyltransferase activity in the ΔAfpmt1 mutant.
In vitro O-mannosyltransferase activity of the ΔAfpmt1 mutant strain was determined by measuring the transfer of [3H]mannose from Dol-P-[3H]Man to the synthetic acceptor peptide Ac-YATAV-NH2 (see Materials and Methods). As summarized in Table 2, in vitro O-mannosyltransferase activity of the ΔAfpmt1 mutant dropped to 40%, indicating that AfPmt1p acts as one of the O-mannosyltransferases in the PMT family.
TABLE 2.
In vitro O-mannosyltransferase activity of ΔAfpmt1 mutanta
| Strain | Activity (dpm/mg/h) | Relative activity (%) |
|---|---|---|
| Wild type | 4,907 ± 33 | 100 |
| ΔAfpmt1 | 1,969 ± 221 | 40 |
| Revertant | 5,257 ± 40 | 107 |
| CEA17 | 5,512 ± 532 | 112 |
Twenty-five micrograms of membrane protein fraction from the wild-type (YJ-407), ΔAfpmt1, revertant, or CEA17 strain was incubated for in vitro mannosyltransferase assay following the transfer of [3H]mannose from Dol-P-[3H]Man to the peptide Ac-YATAV-NH2 (see Materials and Methods). The assay was repeated at least three times. Average values (± standard deviations) from a typical experiment are shown.
Characterization of O glycosylation in the ΔAfpmt1 mutant.
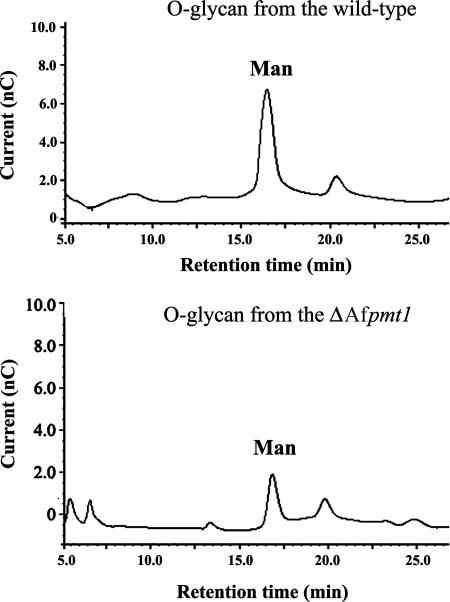
Previously, we purified a secretory glycoprotein, the chitinase AfCHI44, from A. fumigatus (43). The cDNA encoding AfCHI44 has been cloned (35). The C-terminal end of AfCHI44 is rich in serine and threonine, which are potential O-glycosylation sites. Using the purified AfCHI44 protein as an indicator molecule, we were able to evaluate the in vivo O-mannosylation status of the ΔAfpmt1 mutant by analyzing the O-glycans on AfCHI44 produced by the mutant. As shown in Fig. 2, the O-linked mannose attached to AfCHI44 from the ΔAfpmt1 mutant dropped to about 40% of that in the wild type. It can be concluded that the decrease of O mannosylation on AfCHI44 resulted from the absence of the Afpmt1 gene. In S. cerevisiae, PMT1 and PMT2 act on the same set of protein substrates, but mannosylation may occur at different serine and threonine residues (13). Therefore, the retained residual O-mannosylation activity observed in the ΔAfpmt1 mutant might be contributed by AfPmt2p.
FIG. 2.
In vivo O mannosylation in the ΔAfpmt1 mutant. Two hundred micrograms of AfCHI44 purified from the wild-type or ΔAfpmt1 mutant strain was used in each assay as described in Materials and Methods. The O-glycans were analyzed with a Carbo-PA1 column on an HPAEC-PAD. The glycans were eluted with 15 mM NaOH at a flow rate of 1.0 ml/min. The mannose standard used for HPAEC-PAD analysis was treated by β-elimination before injection, and thus the peaks marked “Man” indicate the retention times of anhydrous mannitol, which is derived from mannose treated by β-elimination reaction with NaBH4. This experiment was repeated at least three times, and representative data are shown.
Growth phenotypes of the ΔAfpmt1 mutant.
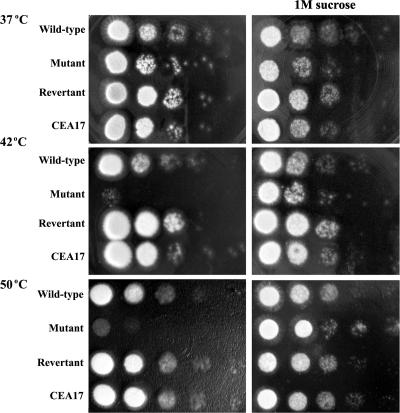
It has been shown that S. cerevisiae pmt1,4, pmt2,3, and pmt2,4 double mutants, as well as pmt1,2,3 triple and pmt1,3,4 triple mutants, are temperature sensitive. In addition, the pmt2,3 and pmt2,4 double mutants, as well as pmt1,2,3 triple mutants, are osmolabile (13). Since wild-type A. fumigatus is highly thermotolerant and grows comfortably at 30 to 50°C, we tested the growth of the mutant at various temperatures. As shown in Fig. 3, when the ΔAfpmt1 mutant was grown on solid complete medium at 37°C, no difference was found between the mutant and the wild-type or revertant strain. Strongly retarded growth, however, was observed when the ΔAfpmt1 mutant was grown on agar at 42°C and 50°C. Thus, we concluded that the ΔAfpmt1 mutant displayed a temperature-sensitive growth defect at higher temperatures. Interestingly, this temperature-sensitive phenotype could be complemented by the addition of 1 M sucrose to the medium, suggesting a defective cell wall of the ΔAfpmt1 mutant at elevated temperatures (Fig. 3).
FIG. 3.
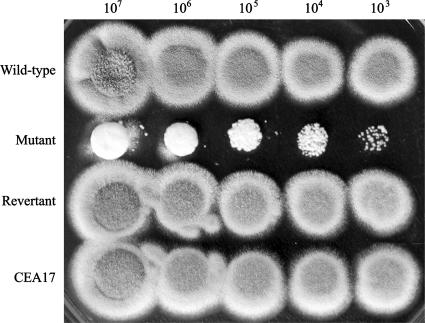
Temperature-sensitive and osmotically stabilized phenotypes of ΔAfpmt1 mutant. A series of 10-fold dilutions (107 to 103 cells) of YJ-407, ΔAfpmt1, revertant, and CEA17 strains of A. fumigatus were cultivated on solid complete medium at 37°C, 42°C, or 50°C for 16 to 18 h. The wild-type strain YJ-407 (Afpmt+ pyrG+) and the parental strain of the mutant, CEA17 (Afpmt+ pyrG−), were set as controls for the ΔAfpmt1 mutant (Afpmt− pyrG+) and the revertant (Afpmt+ pyrG−), respectively.
Cell wall defects of the ΔAfpmt1 mutant.
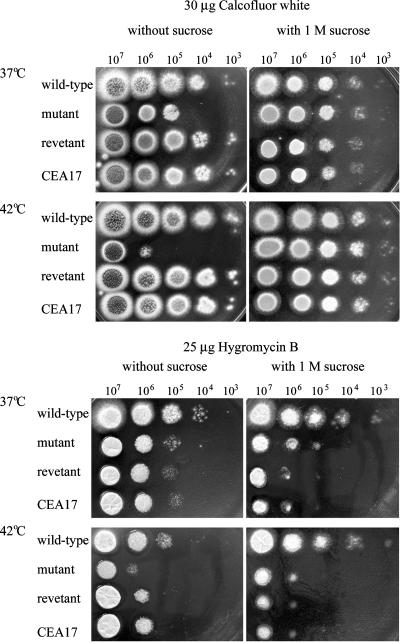
It has been shown that S. cerevisiae strains defective in cell wall biosynthesis are supersensitive to calcofluor white, a reagent known to bind to β-1,4-linked polysaccharides, specifically chitin and cellulose (30). To verify the cell wall defect of the ΔAfpmt1 mutant, the strains were grown on solid medium containing 30 μg/ml of calcofluor white at 37°C and 42°C. As shown in Fig. 4 (top), the ΔAfpmt1 mutant showed an increased sensitivity to calcofluor white at 37°C, but the mutant did not display any increased sensitivity at 42°C in comparison with its growth on complete medium without calcofluor white at 42°C (Fig. 3), suggesting a predominant temperature-sensitive phenotype. It is interesting that the increased sensitivity to calcofluor white at 37°C could be supplemented by the addition of an osmotic stabilizer (1 M sucrose). These observations suggested that deletion of Afpmt1 caused a defect in cell wall integrity.
FIG. 4.
Effects of calcofluor white and hygromycin B on growth of the ΔAfpmt1 mutant. (Top) A series of 10-fold dilutions (107 to 103 cells) of A. fumigatus strains were spotted on solid complete medium containing 25 μg/ml of calcofluor white. Cells were incubated for 24 h at 37°C or 42°C. (Bottom) A series of 10-fold dilutions (107 to 103 cells) of A. fumigatus strains were inoculated on solid medium containing 25 μg/ml of hygromycin B and incubated at 37°C for 36 h. The wild-type strain YJ-407 (Afpmt+ pyrG+) and the parental strain of the mutant, CEA17 (Afpmt+ pyrG−), were set as controls for the ΔAfpmt1 mutant (Afpmt− pyrG+) and the revertant (Afpmt+ pyrG−), respectively.
Hygromycin B, an aminoglycoside antibiotic originally isolated from Streptomyces hygroscopicus, is known to inhibit protein synthesis by acting on bacterial and eukaryotic ribosomes (4, 22). When we cultivated the ΔAfpmt1 mutant on solid medium containing 25 μg/ml of hygromycin B, the growth of the mutant was inhibited at both 37°C and 42°C (Fig. 4, bottom panels). In this case, the retarded growth of the mutant was caused by the inhibition of protein synthesis, not a defective cell wall, since it could not be complemented by the addition of the osmotic stabilizer. Interestingly, we found the mutant grown at 42°C without hygromycin B exhibited a growth phenotype similar to that shown in Fig. 3. Taking the predominant temperature-sensitive phenotype shown in Fig. 4, top panel, into consideration, it is likely that deletion of the Afpmt1 gene would affect the function of certain unknown O-mannosylated proteins that could be essential for A. fumigatus to grow comfortably at elevated temperatures.
To further investigate the cell wall defect at various temperatures, the mycelial cell wall contents, including mannoproteins, glucans, and chitin, were analyzed. As summarized in Table 3, the mannoproteins, α-glucans, and chitin in the mycelial cell wall of the mutant grown at 37°C were induced 1.5-, 2-, and 1.3-fold compared to those in the wild type, respectively, while β-glucans decreased by 8%. When the temperature was elevated to 42°C, the α-glucans were induced 1.5-fold relative to the wild type, the β-glucans decreased by 16%, and the mannoprotein and chitin contents remained at the same levels as those of the wild type.
TABLE 3.
Chemical analysis of mutant cell walla
| Temperature (°C) | Strain | Amt of cell wall component (μg)
|
|||
|---|---|---|---|---|---|
| Mannoprotein | α-Glucan | Chitin | β-Glucan | ||
| 37 | Wild type | 101 ± 3 | 537 ± 9 | 312 ± 11 | 1,700 ± 60 |
| ΔAfpmt1 | 156 ± 3 | 1,005 ± 7 | 400 ± 13 | 1,560 ± 30 | |
| 42 | Wild type | 117 ± 5 | 377 ± 6 | 299 ± 8 | 1,150 ± 40 |
| ΔAfpmt1 | 122 ± 4 | 590 ± 17 | 301 ± 6 | 960 ± 10 | |
Conidia were inoculated into 100 ml of yeast extract-peptone-dextrose liquid medium at a concentration of 106 conidia/ml and incubated at 37°C or 42°C with shaking (300 rpm) for 48 h. The mycelium was then harvested and lyophilized. Three aliquots of 5 mg dry mycelium were used as independent samples for analyses of unbound cell wall proteins and water-soluble sugar as described in Materials and Methods. The same experiment was repeated twice. The numbers shown are micrograms of cell wall component per 10 mg dry mycelium. Data are means ± standard deviations.
Based on our results, we concluded that deletion of the Afpmt1 gene led to a cell wall defect, which activated the cell wall integrity pathway and, in turn, compensated for the cell wall defect by increasing the contents of mannoproteins, α-glucans, and chitin. Although the mechanism of the cell wall integrity pathway in A. fumigatus is not clear, it seems that compensatory increases of the mannoprotein and chitin contents are temperature sensitive in the ΔAfpmt1 mutant. Thus, the deletion of the Afpmt1 gene resulted in a more severe cell wall defect at an elevated temperature.
Morphological changes of conidia of the ΔAfpmt1 mutant.
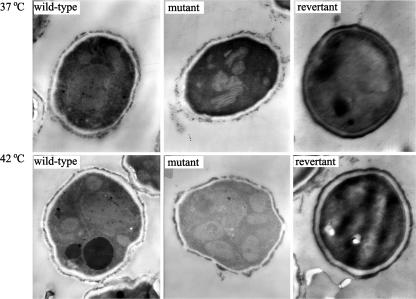
When strains were grown on solid medium at 37°C or 42°C for 48 h, the hyphae differentiated into conidiophore vesicles and phialides. As shown in Table 4, in comparison to the wild type, the number of conidiospores produced by the ΔAfpmt1 mutant was reduced 35% and 77% at 37°C and 42°C, respectively. When the temperature was elevated to 50°C, no conidiospores were counted. This observation was confirmed by the cultivation of A. fumigatus strains at 50°C (Fig. 5). The wild-type, revertant, and CEA17 strains could produce green conidiospores, whereas the mutant formed a white smooth colony, suggesting a totally loss of the ability to produce conidiospores at 50°C. Furthermore, electron microscopy analysis showed that the conidia formed at 42°C had a thinner cell wall (Fig. 6).
TABLE 4.
Number of conidia formed by ΔAfpmt1 mutant at various temperaturesa
| Temperature (°C) | No. of conidia (107)
|
|||
|---|---|---|---|---|
| Wild type | Mutant | Revertant | CEA17 | |
| 37 | 58.7 ± 4.2 | 38.0 ± 7.1 | 25.3 ± 5.4 | 32.0 ± 8.3 |
| 42 | 65.0 ± 13.1 | 15.2 ± 9.4 | 15.7 ± 4.9 | 22.2 ± 12.3 |
| 50 | 9.8 ± 5.8 | 0 | 0.8 ± 0.3 | 1.3 ± 0.5 |
Conidia (105) were spread on CMU plates (solid complete medium containing 5 mM of uridine and uracil) and incubated at 37°C, 42°C, or 50°C for 60 h. The conidia were then harvested and counted by microscopy. The same experiment was repeated twice. Data are means ± standard deviations.
FIG. 5.
Conidium formation of ΔAfpmt1 mutant at 50°C. A series of 10-fold dilutions (107 to 103 cells) of YJ-407, ΔAfpmt1, revertant, and CEA17 strains of A. fumigatus were cultivated on solid complete medium at 50°C for 48 h.
FIG. 6.
Electron microscopy of ΔAfpmt1 mutant conidia. The conidia produced on solid medium were fixed and examined with an H-600 electron microscope (Hitachi) as described in Materials and Methods. Magnification, ×3,000.
Germination of ΔAfpmt1 mutant conidia.
When the conidia of filamentous fungi break dormancy, nuclear division is accompanied by a series of ordered morphological events, including the switch from isotropic to polar growth, the emergence of a second germ tube from the conidium, and septation.
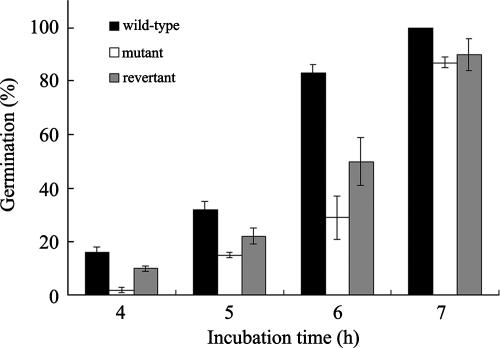
Conidia produced by the ΔAfpmt1 mutant remained viable. These conidia could germinate normally at 37°C (data not shown), but they showed lower germination rates at 42°C. As shown in Fig. 7, the wild-type conidia germinated at rates of 16%, 32%, and 83% after incubation at 42°C for 4 h, 5 h, and 6 h, respectively, while only 2%, 15%, 29%, and 87% of the mutant conidia could germinate at incubation times of 4 h, 5 h, 6 h, and 7 h, respectively. These results demonstrate that the germination of mutant conidia was 1 h later than that of the wild type.
FIG. 7.
Germination of conidia in liquid complete medium at 42°C. The same amounts of wild-type, mutant, and revertant conidiospores were collected and grown in complete medium containing uridine and uracil at 42°C. Conidial germination was examined and counted under a microscope (magnification, ×250). Three independent experiments were carried out.
Effects of Afpmt1 mutation on virulence.
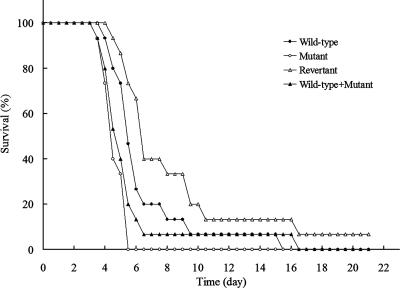
To determine directly whether the ΔAfpmt1 mutant is involved in the pathogenicity of A. fumigatus, we infected immunosuppressed mice (n = 15) with 6 × 106 conidia of the wild-type, ΔAfpmt1, revertant, and mixed wild-type and ΔAfpmt1 conidia. The survival of animals was tested. Mice infected with the ΔAfpmt1 mutant conidia or mixed wild-type and mutant conidia showed a mean survival time of 5 days, while animals treated with the same amount of wild-type or revertant conidia survived this challenge for one more day, displaying a mean survival time of 6 days (Fig. 8). However, the difference between the wild type and the ΔAfpmt1 mutant was not statistically significant (P > 0.01 in all three series). Thus, we concluded that the Afpmt1 gene does not make a significant contribution to the virulence of A. fumigatus, at least under the infection conditions used.
FIG. 8.
Virulence of ΔAfpmt1 mutant. Fifteen BALB/c mice were infected intranasally with 6 × 106 conidia as described in Materials and Methods. The survival of mice was determined. The wild-type strain (•), the ΔAfpmt1 mutant (○), the revertant (▵), and a mixture of the wild-type and mutant strains (▴) were compared.
DISCUSSION
A. fumigatus is one of the most ubiquitous of airborne saprophytic fungi and has been shown to be an opportunistic pathogen, causing pneumonia and other fatal invasive infections in immunocompromised hosts, particularly among patients undergoing cytotoxic chemotherapy or bone marrow transplantation (18). There has been a dramatic increase in severe and usually fatal cases of invasive aspergillosis caused by A. fumigatus. Therefore, the investigation of virulence factors and potential chemotherapeutic targets of A. fumigatus is of clinical interest.
Previously, protein O mannosylation in eukaryotes was recognized as an evolutionarily conserved protein modification catalyzed by a family of PMTs. Evidence has shown that the PMT family is crucial for viability, cell wall integrity, and morphogenesis in several fungal species, such as S. cerevisiae, S. pombe, C. albicans, and C. neoformans (11, 27, 33, 42). Recently, pmtA, encoding an O-mannosyltransferase of the PMT2 subfamily, was shown to be required for the formation of a normal cell wall in Aspergillus species (25, 26). It is assumed that the PMT family is also crucial for the growth and, therefore, virulence of A. fumigatus, as seen for C. albicans and C. neoformans.
The sequencing of the A. fumigatus genome now enables us to easily analyze the PMT family in A. fumigatus. Our investigation of O mannosylation in A. fumigatus was initiated before the completion of the genome sequencing of A. fumigatus, and only the Afpmt1 gene was identified. In the last release of the TIGR database (12; http://tigrblast.tigr.org/er-blast/index.cgi?project=afu1), the A. fumigatus PMT family comprises only three genes, lacking homologues of the ScPmt3, ScPmt5, ScPmt6, and ScPmt7 proteins, in contrast to S. cerevisiae, which contains seven PMT genes. Since only one member of each PMT subfamily was found in A. fumigatus, it seems that Afpmt1, Afpmt2, and Afpmt4 provide sufficient genetic redundancy in haploid A. fumigatus, a function that is secured by multiple PMT loci in S. cerevisiae, which has a haploid or a diploid genome. Similarly, the genomes of A. nidulans, Neurospora crassa, S. pombe, and Fusarium gramineum contain only three PMT genes (including PMT1, PMT2, and PMT4) (29, 42).
In S. cerevisiae, no significant growth phenotype is observed for single PMT1 gene mutants, and temperature-sensitive phenotypes are seen only with pmt1,4 double, pmt1,2,3 triple and pmt1,3,4 triple mutants (13). In this study, we have shown that the single deletion mutant of Afpmt1 in A. fumigatus exhibited a defect in cell wall integrity and retarded growth at an elevated temperature. In addition, S. cerevisiae pmt1 mutants do not show increased sensitivity toward hygromycin B and calcofluor white, while the A. fumigatus ΔAfpmt1 mutant was more sensitive to these agents. These significant phenotypes associated with the ΔAfpmt1 mutant could be explained by the fewer members of the PMT family present in A. fumigatus. Similar observations were also obtained for C. albicans and S. pombe. In addition to the temperature-sensitive phenotype, abnormal cell wall and septum formation was observed in S. pombe merely by deleting the PMT1 gene (42). Significant phenotypes, featuring retarded growth and cell aggregation, were observed in pmt1 mutants of C. albicans (29). Although PMT1 fulfills similar basic functions in different fungal species, the resulting phenotypes differ significantly in different fungal species, suggesting a species-specific function of O-mannosyltransferase 1.
To evaluate the activity of Pmt1p, glycoproteins such as chitinase, Als1p, Sec20p, Kre9p, Pir2p, and Wsc1p have been used as in vivo substrates for various fungal species (29, 34, 36, 42). Previously, an extracellular chitinase secreted by A. fumigatus was purified and characterized in our lab (43). Taking advantage of the simple purification procedure for this protein, we were able to precisely evaluate the in vivo O-mannosylation status of the ΔAfpmt1 mutant. Our data show that deletion of Afpmt1 in A. fumigatus caused a 60% loss of O-linked mannose (Fig. 2). We have also shown that deletion of Afpmt1 in A. fumigatus causes temperature-sensitive phenotypes, which include a reduced growth rate, a deficient ability to differentiate into conidia, and a reduced germination rate of conidia at 42°C. These temperature-sensitive phenotypes, together with the cell wall defect observed, might be due to the lack of O-linked mannose modifications on certain proteins that are required for cell wall integrity and thus the normal growth of A. fumigatus at elevated temperatures.
Attenuated virulence has been documented for pmt1 mutants of C. albicans. The reason for the involvement of pmt1 in C. albicans virulence is proposed as follows: the reduced O glycosylation of Als1, a likely adhesin for C. albicans, results in reduced adhesive properties of the pmt1 mutant and thus affects its virulence (34). Our results showed that the Afpmt1 gene was not essential for the virulence of A. fumigatus, at least in the animal model we used. The difference in virulence between the oma1+ mutant of C. albicans and the ΔAfpmt1 mutant of A. fumigatus could be explained as follows: the cell wall defect in the ΔAfpmt1 mutant occurred only at temperatures higher than 37°C, and thus the mutant did not show any defects at 37°C, the body temperature of the mouse.
Acknowledgments
This project was supported by the National Basic Research Program of China (grant 2004CB719606), the National Natural Science Foundation of China (grants 30621005 and 30470023), and the Chinese Academy of Sciences of China (grant KSCX2-3-02-01) (C.J.).
We thank C. d'Enfert for his kind supply of A. fumigatus strain CEA17 and plasmid pCDA14. Genomic sequence data for A. fumigatus were obtained from the Institute for Genomic Research website at http://tigrblast.tigr.org/er-blast/index.cgi?project=afu1.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran-Valero de Bernabe, D., S. Currier, A. Steinbrecher, J. Celli, E. van Beusekom, B. van der Zwaag, H. Kayserili, L. Merlini, D. Chitayat, W. B. Dobyns, B. Cormand, A. E. Lehesjoki, J. Cruces, T. Voit, C. A. Walsh, H. van Bokhoven, and H. G. Brunner. 2002. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am. J. Hum. Genet. 71:1033-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourdineaud, J.-P., J. M. van der Vaart, M. Donzeau, G. de Sampaio, C. T. Verrips, and G. J.-M. Lauquin. 1998. Pmt1 mannosyltransferase is involved in cell wall incorporation of several proteins in Saccharomyces cerevisiae. Mol. Microbiol. 27:85-98. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen, D. E., W. M. J. Clemons, A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 5.Cove, D. J. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51-56. [DOI] [PubMed] [Google Scholar]
- 6.d'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine 5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 7.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and released substances. Anal. Chem. 28:350-356. [Google Scholar]
- 8.Ecker, M., V. Mrsa, I. Hagen, R. Deutzmann, S. Strahl, and W. Tanner. 2003. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep. 4:628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elorza, M. V., A. Murgui, and R. Sentandreu. 1985. Dimorphism in Candida albicans: contribution of mannoproteins to the architecture of yeast and mycelial cell walls. J. Gen. Microbiol. 131:2209-2216. [DOI] [PubMed] [Google Scholar]
- 10.Endo, T. 1999. O-mannosyl glycans in mammals. Biochim. Biophys. Acta 1473:237-246. [DOI] [PubMed] [Google Scholar]
- 11.Ernst, J. F., and S. K.-H. Prill. 2001. O-glycosylation. Med. Mycol. 39(Suppl. 1):67-74. [PubMed] [Google Scholar]
- 12.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Baçstürkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Peñalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 13.Gentzsch, M., and W. Tanner. 1996. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. EMBO J. 15:5752-5759. [PMC free article] [PubMed] [Google Scholar]
- 14.Gentzsch, M., and W. Tanner. 1997. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology 7:481-486. [DOI] [PubMed] [Google Scholar]
- 15.Hearn, V. M., and J. H. Sietsma. 1994. Chemical and immunological analysis of the Aspergillus fumigatus cell wall. Microbiology 140:789-795. [DOI] [PubMed] [Google Scholar]
- 16.Ichimiya, T., H. Manya, Y. Ohmae, H. Yoshida, K. Takahashi, R. Ueda, T. Endo, and S. Nishihara. 2004. The twisted abdomen phenotype of Drosophila POMT1 and POMT2 mutants coincides with their heterophilic protein O-mannosyltransferase activity. J. Biol. Chem. 279:42638-42647. [DOI] [PubMed] [Google Scholar]
- 17.Jurado, L. A., A. Coloma, and J. Cruces. 1999. Identification of a human homolog of the Drosophila rotated abdomen gene (POMT1) encoding a putative protein O-mannosyltransferase, and assignment to human chromosome 9q34.1. Genomics 58:171-180. [DOI] [PubMed] [Google Scholar]
- 18.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. I., Y. M. Yu, Y. M. Rho, B. C. Park, J. H. Choi, H. M. Park, and P. J. Maeng. 2005. Differential expression of the chsE gene encoding a chitin synthase of Aspergillus nidulans in response to developmental status and growth conditions. FEMS Microbiol. Lett. 249:121-129. [DOI] [PubMed] [Google Scholar]
- 20.Leitão, E. A., V. C. W. Bittencourt, R. M. T. Haido, A. P. Valenate, J. Peter-Katalinie, M. Letzel, L. M. de Souza, and E. Barreto-Bergter. 2003. β-Galactofuranose-containing O-linked oligosaccharides present in the cell wall peptidogalactomannan of Aspergillus fumigatus contain immunodominant epitopes. Glycobiology 13:681-692. [DOI] [PubMed] [Google Scholar]
- 21.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randal. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 22.Mann, R. L., and W. W. Bromer. 1958. The isolation of a second antibiotic from Streptomyces hygroscopicus. J. Am. Chem. Soc. 80:2714-2716. [Google Scholar]
- 23.Manya, H., A. Chiba, A. Yoshida, X. Wang, Y. Chiba, Y. Jigami, R. U. Margolis, and T. Endo. 2004. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. USA 101:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Blanco, E., and A. Garcia-Bellido. 1996. Mutations in the rotated abdomen locus affect muscle development and reveal an intrinsic asymmetry in Drosophila. Proc. Natl. Acad. Sci. USA 93:6048-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka, T., T. Hamaguchi, Y. Sameshima, M. Goto, and K. Furukawa. 2004. Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans. Microbiology 150:1973-1982. [DOI] [PubMed] [Google Scholar]
- 26.Oka, T., Y. Sameshima, T. Koga, H. Kim, M. Goto, and K. Furukawa. 2005. Protein O-mannosyltransferase A of Aspergillus awamori is involved in O-mannosylation of glucoamylase I. Microbiology 151:3657-3667. [DOI] [PubMed] [Google Scholar]
- 27.Olson, G. M., D. S. Fox, P. Wang, J. A. Alspaugh, and K. L. Buchanan. 2006. Role of protein O-mannosyltransferase Pmt4 in the morphogenesis and virulence of Cryptococcus neoformans. Eukaryot. Cell 6:222-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osmani, S. A., G. S. May, and N. R. Morris. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104:1495-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prill, S. K.-H., B. Klinkert, C. Timpel, C. A. Gale, K. Schröppel, and J. F. Ernst. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546-560. [DOI] [PubMed] [Google Scholar]
- 30.Ram, A. M. J., A. Wolters, R. ten Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 31.Schoffelmeer, E. A., F. M. Klis, J. H. Sietsma, and B. J. Cornelissen. 1999. The cell wall of Fusarium oxysporum. Fungal Genet. Biol. 27:275-282. [DOI] [PubMed] [Google Scholar]
- 32.Strahl-Bolsinger, S., T. Immervoll, R. Deutzmann, and W. Tanner. 1993. PMT1, the gene for a key enzyme of protein O-glycosylation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:8164-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strahl-Bolsinger, S., and A. Scheinost. 1999. Transmembrane topology of pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 274:9068-9075. [DOI] [PubMed] [Google Scholar]
- 34.Timpel, C., S. Strahl-Bolsinger, K. Ziegelbauer, and J. F. Ernst. 1998. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J. Biol. Chem. 273:20837-20846. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Y., J. Wang, H. Hu, X. Yang, S. Yang, X. Yu, and C. Jin. 2004. Cloning and expression of chitinase gene from Aspergillus fumigatus YJ-407. Chin. J. Biotechnol. 20:843-850. [Google Scholar]
- 36.Weber, Y., S. K.-H. Prill, and J. F. Ernst. 2004. Pmt-mediated O mannosylation stabilizes an essential component of the secretory apparatus, Sec20p, in Candida albicans. Eukaryot. Cell 3:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weidner, G., C. d'Enfert, A. Koch, C. P. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]
- 38.Weston, A., P. M. Nassau, C. Henly, and S. Marriott. 1993. Protein O-mannosylation in Candida albicans determination of the amino acid sequences of peptide acceptors for protein O-mannosyltransferase. Eur. J. Biochem. 215:845-849. [DOI] [PubMed] [Google Scholar]
- 39.Willer, T., W. Amselgruber, R. Deutzmann, and S. Strahl. 2002. Characterization of POMT2, a novel member of the PMT protein O-mannosyltransferase family specifically localized to the acrosome of mammalian spermatids. Glycobiology 12:771-783. [DOI] [PubMed] [Google Scholar]
- 40.Willer, T., M. C. Valero, W. Tanner, J. Cruces, and S. Strahl. 2003. O-mannosyl glycans: from yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 13:621-630. [DOI] [PubMed] [Google Scholar]
- 41.Willer, T., B. Prados, J. M. Falcon-Perez, I. Renner-Muller, G. K. Przemeck, M. Lommel, A. Coloma, M. C. Valero, M. H. de Angelis, W. Tanner, E. Wolf, S. Strahl, and J. Cruces. 2004. Targeted disruption of the Walker-Warburg syndrome gene Pomt1 in mouse results in embryonic lethality. Proc. Natl. Acad. Sci. USA 101:14126-14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willer, T., M. Brandl, M. Sipiczki, and S. Strahl. 2005. Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 57:156-170. [DOI] [PubMed] [Google Scholar]
- 43.Xia, G., C. Jin, J. Zhou, S. Yang, S. Zhang, and C. Jin. 2001. A novel chitinase having a unique mode of action from Aspergillus fumigatus YJ-407. Eur. J. Biochem. 268:4079-4085. [DOI] [PubMed] [Google Scholar]