Abstract
Species of the mycoparasitic fungal genus Hypocrea/Trichoderma are prominent producers of peptaibols, a class of small linear peptides of fungal origin. Some of these peptaibols have been shown to act synergistically with cell-wall-degrading enzymes in the inhibition of the growth of other fungi in vitro and in vivo. Here we present the structure of the Hypocrea atroviridis peptaibol synthetase gene (pbs1), deduced from the genome sequence of H. atroviridis. It consists of 19 typical peptide synthetase modules with the required additional modifying domains at the N and C termini. Phylogenetic and similarity analyses of the individual amino acid-activating modules is consistent with its ability to synthesize atroviridins. Matrix-assisted laser desorption ionization-time of flight mass spectrometry of surface-grown cultures of H. atroviridis showed that no peptaibols were formed during vegetative growth, but a microheterogenous mixture of atroviridins accumulated when the colonies started to sporulate. This correlation between sporulation and atroviridin formation was shown to be independent of the pathway inducing sporulation (i.e., light, mechanical injury and carbon starvation, respectively). Atroviridin formation was dependent on the function of the two blue light regulators, BLR1 and BLR2, under some but not all conditions of sporulation and was repressed in a pkr1 (regulatory subunit of protein kinase A) antisense strain with constitutively active protein kinase A. Conversely, however, loss of function of the Gα-protein GNA3, which is a negative regulator of sporulation and leads to a hypersporulating phenotype, fully impairs atroviridin formation. Our data show that formation of atroviridin by H. atroviridis occurs in a sporulation-associated manner but is uncoupled from it at the stage of GNA3.
Peptaibols, a class of linear peptides of fungal origin with 7 to 20 residues, have three structural characteristics: (i) a high proportion of dialkylated amino acids with an abundance of α-aminoisobutyric acid (Aib); (ii) an N-acyl (usually acetyl) terminus; and (iii) a C-terminal amino alcohol, such as phenylalaninol or leucinol. Peptaibols naturally occur as mixtures of isoforms, and more than 300 sequences are now known (http://www.crvstbbk.ac.uk/peptaibol/home.shtml). They are divided into three subclasses, which are (i) the long-sequence peptaibols such as alamethicins or trichorzianins, which contain 18 to 20 amino acid residues; (ii) the short-sequence peptaibols such as harzianins or zervamicins containing 11 to 16 residues; and (iii) the lipopeptaibols with 7 or 11 residues and whose N terminus is acylated by a short fatty acid chain (e.g., trichogin A [11, 43]. Peptaibols generally exhibit antimicrobial activity against gram-positive bacteria and fungi but have also been implicated in the interaction with mammalian cells (39). Their biological activities are believed to be due to their membrane-modifying properties and ability to form transmembrane voltage-dependent channels (23).
Fungi of the anamorphic fungal genus Trichoderma (Hypocreale, Ascomycota), which contains cosmopolitan soilborne fungi with economic importance as biocontrol agents (13, 27), are well known as producers of peptaibols. It has been shown in vitro that peptaibols act synergistically with the cell-wall-degrading enzymes secreted by Trichoderma to inhibit the growth of fungal pathogens (22, 32). This antagonism is thought to be due to the action of the peptaibols on the membrane of the target fungus, thereby inhibiting membrane-associated enzymes involved in cell wall synthesis.
Despite their attractive potential as antimycotica and their potential importance in the physiology of the fungus, little information is available on the physiological and molecular regulation of peptaibol formation. Such a knowledge would in turn be a prerequisite to understanding the role of these compounds for the fungus. Like other fungal peptides that are products of secondary metabolism, peptaibols are synthesized by nonribosomal peptide synthetases, and genes encoding the respective peptaibol synthetases are known from Trichoderma virens (44, 45) and T. harzianum (42). Due to the large molecular mass of the transcript, which makes its intact isolation difficult, almost no studies have been carried out on the regulation of transcript formation. Recently, Vizcaino et al. (42) reported that the peptaibol synthetase transcript of T. harzianum can only be detected under nitrogen starvation.
In the present study, we used a recently developed method for studying peptaibol formation from low amounts of fungal biomass using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) (25) to investigate the formation of H. atroviridis peptaibols. This fungus has been chosen because it is a strong biocontrol agent, and its peptaibols may play a role in its antagonistic and mycoparasitic abilities. In order to correctly align the observed masses with the peptaibols, we have cloned in silico the H. atroviridis pbs1 gene and deduced its protein structure predicting that it produces atroviridin. Here we show that the formation of atroviridin by H. atroviridis occurs by a sporulation-coupled pathway, whose physiological implications are discussed. In addition, we show that atroviridins belong to the group of secondary metabolites which require positive regulation by G proteins.
MATERIALS AND METHODS
Nomenclature.
The nomenclature for the genes and proteins described here has been partially revised in order to follow the recommendations of the consortium annotating the Trichoderma genomes to follow the Neurospora/pyrenomycete nomenclature, and—if already known—to use the name of the respective Neurospora orthologue. Where this led to a change in the name of an already-cited gene or protein is indicated at the first use.
Reagents.
2,5-Dihydroxibenzoic acid from Sigma Chemicals (St. Louis, MO) was used as matrix for MALDI-TOF experiments. Trifluoroacetic acid, ethanol, acetonitrile, and methanol from Merck were used as solvents.
Strains and culture conditions.
The wild-type strains of H. atroviridis used throughout the present study were IMI 206040 and ATCC 74058 (= P1). The following mutants, prepared from these strains, were used: the blue-light regulator (BLR1 and -2) Δblr-1, and Δblr-2 delta strains (5), the G-α protein GNA-delta Δgna3 strain (= tga3) (48) and strain AS-pkr1, in which the protein kinase A (PKA) regulatory subunit (PKR1) gene is inactivated by expression of an antisense copy (6). With the exception of the gna3 strain, which was derived from H. atroviridis P1, all other mutants were from strain IMI 206040. Cultures were routinely grown at 25°C on malt agar plates. To test the influence of exogenous cyclic AMP (cAMP; Sigma), cAMP was added to potato dextrose agar cooled to 48°C after autoclaving.
Colonies were induced to conidiate as previously described (5) by exposure to white light (25 μmol photons m−2 s−1; 1,800 lx). In case of dark-grown cultures, harvesting was done under red safety light, which has been shown to have no effect on the fungus for the short period applied (1 to 2 min).
To induce conidiation by carbon starvation, fungal colonies were grown in darkness for 48 h in MM (1.66 mM MgSO4, 5.16 mM K2HPO4, 2.68 mM KCl, 12.5 mM NH4NO3, 7.19 μM FeSO4, 6.95 μM ZnSO4, 10.1 μM MnCl2), 111 mM glucose (6), and a piece of agar with mycelium and then transferred to glucose-free MM medium and allowed to grow for a further 24 h in either complete darkness or under illumination. For injury-induced conidiation (6), fungal colonies were grown in total darkness at 25°C for 72 h, cut in strips with a scalpel, and incubated for an additional 24 h in the dark at 25°C.
Extraction and preparation of mycelia for MALDI-TOF analysis.
A few micrograms of fungal mycelia, corresponding to 5 mm2 of the edge of the colonies, were suspended in acetonitrile-methanol-water (1:1:1), and 1 μl of the suspension was directly spotted onto target wells of a 100-position sample plate and immediately mixed with 1 μl of matrix solution (10 mg of 2,5-dihydroxybenzoic acid/ml in acetonitrile-methanol-water [1:1:1] and 0.3% trifluoroacetic acid). The sample matrix mixture was allowed to air dry prior to analysis. Alternatively, freeze-dried mycelium obtained from shaken cultures or fungi grown on plates was homogenized in 60% ethanol and centrifuged. Then, 1 μl of the protein solution was spotted onto a MALDI target plate and mixed with matrix.
MS analysis of low-molecular-mass peptides.
Measurements were performed in the delayed extraction mode, allowing the determination of monoisotopic mass values. A low mass gate of 800 Da improved the measurement by filtering out the most intensive matrix ions. The mass spectrometer was used in the positive ion detection and reflector mode.
Semiquantitative analysis of peptaibol formation.
Because the biomass weight of the part of the fungal colony that was prepared for analysis could not be measured, the values obtained cannot be related to the biomass amount, and the method therefore does not allow a quantitative analysis. Consequently, only very strong differences (present versus not present or present versus almost not present) were considered relevant and used for the interpretation of results.
Bioinformatic methods.
A full-length copy of H. atroviridis pbs1 was identified in its genome sequence by TBLASTN search with the T. virens pbs1 gene as a probe. The amino acids activated by individual ATP-binding (amino acid activating)-domains were identified by neighbor joining (NJ) analysis (MEGA 3.1) (20) of individual A domains of H. atroviridis pbs1 and those from T. virens (44), T. harzianum (42), and H. jecorina (T. reesei [unpublished data]) (Table 1) . For this purpose, the amino acid sequences were aligned with CLUSTAL X; the alignment was manually improved in GENEDOC and then exported to MEGA 3.1. The NJ tree was constructed with 1,000 bootstrap replicas.
TABLE 1.
Comparison of H. atroviridis PBS1 to other peptaibol synthases from Trichoderma/Hypocrea
| Organism | Gene/protein | Product name | Product length | Source or reference |
|---|---|---|---|---|
| H. atroviridis | pbs1/PBS1 | Atroviridin | 19-mer, 20-mer | This study |
| H. virens | tex1/TEX1 | Trichorzin | 18-mer | 46 |
| H. jecorina | par1/PAR1 | Paracelsin | 20-mer | Unpublished |
| har1/HAR1 | Harzianin | 11-14-mer | Unpublished | |
| H. lixii | salps2/SALPS2 | Trichorzin (?) | 18-mer (?) | 42a |
Only three terminal modules were sequenced; however, because of the presence of a C-terminal alcohol dehydrogenase domain and the amino acid activated by the last three A domains, its identity as a large peptaibol synthase was concluded (42).
RESULTS
In silico characterization of the H. atroviridis 19-residue peptaibol synthetase and its putative product.
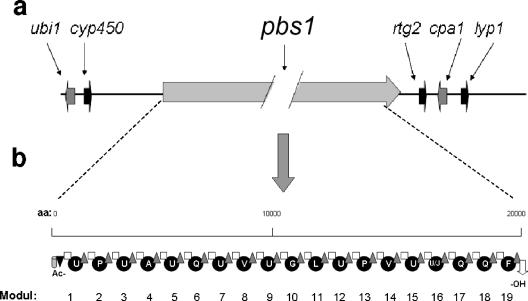
Using the T. virens tex1 sequence as a query in TBLASTN, we identified a continuous open reading frame in the genome sequence of H. atroviridis IMI 206040, which, when translated, would give rise to a 21,879-amino-acid (aa) protein. This deduced polypeptide consists of 19 complete peptide synthetase modules and the respective additional acetyltransferase and alcohol dehydrogenase domains at the N and C termini of the predicted protein (Fig. 1a), thus clearly identifying it as a peptaibol synthetase. It is the only large peptaibol synthase gene in the genome of H. atroviridis. The genes flanking pbs1 in H. atroviridis are shown in Fig. 1b: the immediate downstream region contains sequences putatively encoding proteins similar to S. cerevisiae Rtg2p and an N. crassa calcium/proton exchanger in the same order and orientation as immediately downstream of T. virens tex1 and T. harzianum salps2 (42, 46) (Fig. 1a). The upstream region contains genes encoding proteins of the cytochrome P450 subfamilies and a prenyltransferase (Fig. 1). Both genes are also found in the same orientation upstream of the harzianin synthetase gene of H. jecorina (unpublished data) but are absent from the 5′ flanking area of T. virens tex1. In fact, these two genes occur at two different scaffolds of the T. virens genome database (scaffolds 6 and 17), which are both different from that containing tex1 (scaffold 12 [unpublished data]). These comparisons show that the locus encoding the large peptaibol synthetases in Trichoderma apparently underwent major reorganization during evolution.
FIG. 1.
pbs1 locus of H. atroviridis and modular organization of the predicted PBS1 protein. (a) Chromosomal arrangement of the pbsl locus: ubi1, UbiA-like prenyltransferase; cyp450, cytochrome P450 subfamily protein; pbs1l, 19-mer peptaibol synthetase; rgt2, Rtg2-like protein; cpa1, Ca2+/H+ antiporter; lyp1, lysine permease. With the exception of pbs1, gene lengths are drawn to scale. (b) Modules within the PBS1 protein, numbered consecutively from 1 to 19. The domains within the modules are indicated as follows: gray cylinder, ketoacyl synthetase; black triangle, acyl transferase; white square, condensation domain; black circles: adenylation domains; gray triangles, thiolation domain; white arrow, alcohol dehydrogenase. The amino acids activated by the individual adenylation domains are indicated in the one-letter code (U, α-aminoisobutyric acid; J, isovaline).
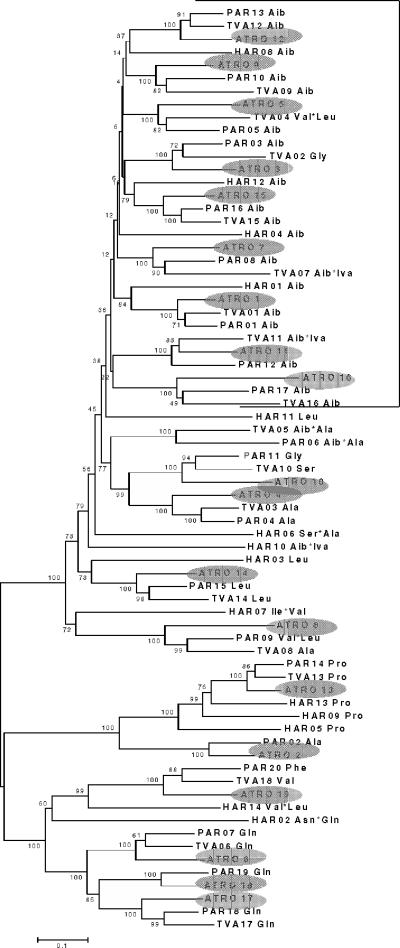
Peptide synthetases consist of a conserved iteration of modules, each consisting in 5→3 order of an ATP-binding domain, an amino acid thioesterification domain, and a condensation domain (37). Therein, the ATP-binding domains specify the substrate specificity and thus the sequence of the formed peptide. In order to predict the latter, we first performed a phylogenetic analysis of all 19-aa activating domains and corresponding domains identified in T. virens TEX1 and H. jecorina paracelsin and harzianin synthetases (PAR1 and HAR1, respectively; H. von Döhren et al., unpublished data) for which the amino acid sequences of the respective peptaibol products are known (Table 1). The rationale for this was the hypothesis that domains activating the same amino acid may be more similar to each other within different Trichoderma spp. than to other domains of the same protein. The resulting tree (Fig. 2) verifies this hypothesis in part: there are consistent and well-supported clades for the P-and Q-activating domains, and—albeit poorly resolved at the central nodes—several terminal clades for Aib and Ala. However, a number of clades were mixed. We also noted that domains which activate the same amino acids but are close to either the N or the C terminus of the resulting peptaibol consistently formed different clusters. This analysis allowed some identification but left several domains unidentified.
FIG. 2.
Phylogenetic relationships of the different adenylation domains of PBS1 to the adenylation domains found in the H. jecorina paracelsin synthetase (PAR [unpublished data]), the H. jecorina harzianin synthetase (HAR [unpublished data]), and T. virens TEX1 (45). Modules in PBS1 (named “ATRO” in this figure) are followed by the respective module number and are shaded in gray. TEX1 is named “TVA” in this figure. In PAR, HAR, and TVA the numbers indicate the positions of the respective modules in the protein, but in addition the amino acid (indicated in the three-letter code) specifies the acti-vated amino acid. When more than a single amino acid is given (linked by an asterisk [*]), this indicates that the module accepts more than a single amino acid. The tree was constructed by NJ analysis, using 1,000 bootstrap replications, whose statistics (percentage of occurrence of the branch in 1,000 trees) are given at the respective nodes. The bracket summarizes all branches leading to modules involved in adenylation of α-aminoisobutyric acid.
Therefore, the domains were further analyzed with respect to the presence of the signature sequences proposed by Stachelhaus et al. (38) and Challis et al. (7). For this purpose, we compared them to those present in H. jecorina PAR1 because the respective paracelsin is structurally most similar to atroviridin (Table 2) (25). Modules putatively acting on the same aa indeed showed conserved signature sequences, differences often being only conserved changes (e.g., L→V). Based on these combined analyses in Fig. 2 and Table 2, the peptaibols “encoded” by H. atroviridis would be compatible with that of atroviridins from a bona fide strain of H. atroviridis (26), Ac-UPUAQUVUGLUPVU(U/J)QQF-OH, with the exception that the U between A4 and Q5 is missing.
TABLE 2.
Signature sequences of the A domains of H. atroviridis PBS1 compared to H. jecorina PAR1a
| Domain | Activated aa | Signature aa at position:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | ||
| A1 | Aib | D | L | G | Y | L | A | G | V | F |
| A2 | Pro | D | I | L | I | C | A | L | I | C |
| v | f | g | ||||||||
| A3 | Aib | D | V | G | F | L | A | G | V | F |
| l | ||||||||||
| A4 | Ala | D | L | G | F | L | A | G | V | F |
| v | v | l | ||||||||
| A4 | Aib | D | M | G | F | I | A | G | V | V |
| l | l | f | ||||||||
| A5 | Gln | D | G | G | M | V | G | G | N | Y |
| A6 | Aib | D | L | G | Y | V | A | G | V | F |
| A7 | Val | D | A | F | L | L | G | I | V | A |
| i | g | l | ||||||||
| A8 | Aib | D | L | G | Y | L | A | G | C | F |
| v | ||||||||||
| A9 | Gly | D | V | G | Y | L | M | A | V | L |
| i | f | |||||||||
| A10 | Leu | D | F | S | Y | L | G | A | V | M |
| l | f | g | v | |||||||
| A11 | Aib | D | L | G | F | L | A | G | V | F |
| A12 | Pro | D | V | L | F | C | G | L | I | C |
| A13 | Val | D | A | A | L | V | V | G | V | F |
| g | m | i | i | |||||||
| A14 | Aib | D | L | G | F | L | A | G | V | F |
| A15 | Aib/Iva | D | M | G | W | M | G | G | V | I |
| f | a | v | ||||||||
| A16 | Gln | D | G | G | M | V | G | G | N | Y |
| A17 | Gln | D | G | G | M | I | G | G | N | Y |
| v | ||||||||||
| A18 | Phe | D | A | A | I | L | V | G | V | G |
| f | l | m | ||||||||
Challis et al. (7). Identical signature amino acids are underlined. If the corresponding signature amino acid is different in PAR1, the respective amino acid is indicated by a lowercase letter below that of PBS1. Since no activation domain for leucine (A10) is present in PAR1, its signature was taken from TEX1, and identical amino acids are indicated in boldface to illustrate this difference. aa, amino acids.
Identification of atroviridins formed by H. atroviridis.
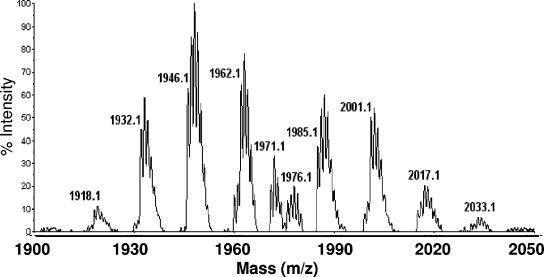
In order to compare the structure of the peptaibols putatively formed by PBS1 to those actually formed by H. atroviridis IMI 206040 and P1, we used MALDI-TOF analysis of surface-grown cultures of H. atroviridis. The rationale for this was that a review of the literature about peptaibol formation by Trichoderma revealed that most researchers have used either surface cultures or submerged cultures, to which solid components had been added (19). The results from such an experiment, which yielded consistent results for both strains, are shown in Fig. 3: as long as the culture of T. atroviride was in the state of vegetative hyphal growth, no peptaibol formation was detected (data not shown). However, as soon as the fungus started to sporulate, peptaibol formation was evident by the observation of a characteristic island of several mass peaks in the range of 1,935 to 2,010 Da. Table 3 compares the masses of these peaks with those of atroviridins and neoatroviridins of H. atroviridis: the main peak at m/z 1,963 corresponds to protonated atroviridin A. Masses at 1,920 and 1,934 correspond to atroviridins A and B without the Aib in position 6 and for which the respective module is missing in PBS1. Masses at 1,949, 2,003, and 2,017 may represent new atroviridin variants because they can be explained by the loss or gain of one or two methylene groups from atroviridin A, respectively. While the identification of all of the components of the peptaibol mixture was beyond the scope of the present study, these findings confirm that this “mass island” corresponds to the atroviridins and that both the 20-residue and the 19-residue peptaibols (which would correlate with the module structure) are detectable. We have thus taken the abundance of this peak island, which did not change in its shape (i.e., ratio of individual peaks to each other) throughout this investigation as a semiqualitative measure (see above) for the formation of peptaibols.
FIG. 3.
Detailed view of a MALDI-TOF mass spectrum from a peptaibol producing sporulating culture of the wild-type strain H. atroviridis IMI 206040, grown in the presence of light. The m/z values of the individual peaks are given.
TABLE 3.
Interpretation of the H. atroviridis atroviridin by MALDI-TOF analysis
| m/z value(s) | Compound identity | Explanation |
|---|---|---|
| 1,920 | Atroviridin A | —Ala + H = 1919; position 6 deletion |
| 1,934 | Atroviridin B | -Ala + H = 1933, position 6 deletion |
| 1,948, 1,949 | New atroviridin | -CH2 (1961 minus 14) |
| 1,963 | Atroviridin A | (1961) + H = 1962 |
| 1,987, 1,988 | ? | |
| 2,001 | ? | |
| 2,003, 2,004 | New atroviridin | +CH2, 1989 + 14 |
| 2,017, 2,018 | New atroviridin | +2CH2, 1989 + 28 |
Peptaibol formation depends on the BLRl and BLR2 proteins and is stimulated by light.
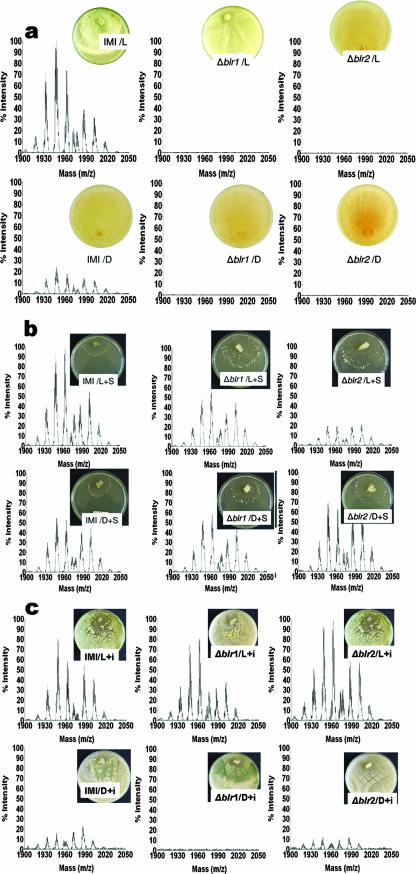
Since atroviridin formation was only detected under conditions of conidiation, we investigated this in more detail. The process of conidiation by fungi is subject to several regulatory influences (47), and the correlation observed above may therefore be simply coincidence, i.e., independent regulation by the same physiological event, or due to a common signal upstream of the conidiation event. We therefore investigated whether different methods for promoting conidiation would always correlate with peptaibol formation or whether the latter specifically occurs only under one of these conditions. As a first condition, we chose white light, which is known to trigger photoconidiation in H. atroviridis via the function of the blue light regulator proteins BLR1 and BLR2 (5). Figure 4a shows that the triggering of conidiation by light was indeed paralleled by a strong accumulation of peptaibols, and only traces were seen in dark cultivated cultures. The latter may have been due to some sporulation, since they were completely absent in a parallel set of experiments (data not shown). This accumulation was completely blocked in the blr1 and blr2 delta mutants. These data indicate that peptaibol formation under conditions of light-induced sporulation depends on the BLR1/2 proteins.
FIG. 4.
BLR1- and BLR2-dependent formation of atroviridin in H. atroviridis by different sporulation-inducing pathways. Graphs show the MALDI-TOF spectra of cultures of the wild-type strain (IMI), the delta-blr1 mutant strain (blr1), and the delta blr2 mutant strain (blr2) under illumination (L) and in darkness (D) on ME agar (a), under conditions of starvation (S) (b), and under conditions of mechanical injury (i) (c). See Materials and Methods for details. The ranges of the intensity of the MALDI-TOF spectra were 0 to 28,000 (a), 0 to 23,000 (b), and 0 to 25,000 (c). The insets show photographs of the respective cultures immediately before peptaibol extraction. Data consistent with the claims drawn were obtained in at least one additional, separate experiment.
Starvation-induced sporulation triggers peptaibol formation independently of light and the BLR proteins.
Another universal inducer of conidiation in fungi is nutrient starvation, and we therefore tested whether the accumulation of peptaibols would also be stimulated under these conditions. Figure 4b shows that carbon starvation indeed led to the accumulation of peptaibols and that under these conditions the accumulation was independent of the blrl and blr2 genes and, consistent with this, was also independent of light. This would in theory be in contrast to previous reports that carbon deprivation-induced sporulation depends on the BLRl/BLR2 proteins (6). However, more recent data have shown that the BLR1/2-dependent response is only observed when cultures are subjected to a starvation for not more than 12 h and absent during incubation under starvation conditions for a longer time (A. Herrera-Estrella, unpublished data). The present data therefore show that the response to light observed above (i.e., peptaibol accumulation and dependence on the BLR proteins) is due to its sporulation-associated nature and not because of a direct regulatory role of the light triggering signal cascade on peptaibol synthetase.
Induction of conidiation-associated peptaibol formation by mycelial injury is light dependent but BLR independent.
As a third, independent approach to study the correlation of peptaibol formation with conidiation, we used mechanical stress (injury with a scalpel (see reference 5) of the mycelia. This conidiation pathway has also been shown to be independent of the BLR proteins (5), and consequently there is also sporulation in the darkness. Interestingly, however, peptaibol formation under these conditions in H. atroviridis was completely dependent on the presence of light, and virtually no peptaibols were detectable in its absence in spite of sporulation; essentially the same findings were also observed in the blrl and blr2 mutants (data not shown). While this is consistent with the above findings that injury-promoted conidiation is BLR independent, it documents that there is also a BLR-independent pathway of stimulation of peptaibol formation by light under stress.
The hypersporulating H. atroviridis delta-gna3 mutant is defective in peptaibol formation.
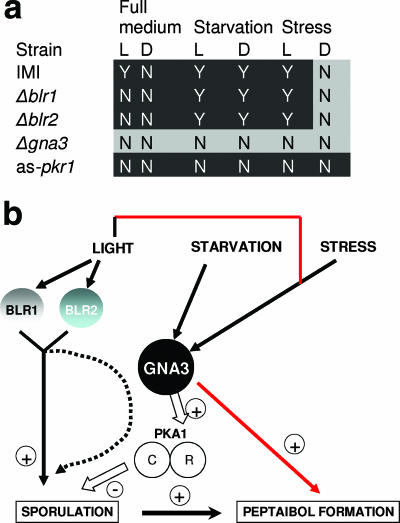
Conidiation in Neurospora crassa is negatively regulated by the Gα-protein Gna3 (17), and consistent data have also been reported also for H. atroviridis (48). Consequently, a gna3-delta mutant leads to hypersporulation. We wondered whether this mutant would consequently overproduce peptaibols. However, in contrast to these expectations, the results shown in Fig. 5 document that this mutant is almost completely impaired in peptaibol formation and that this impairment is apparent under all conditions tested above, i.e., light, carbon starvation, and mechanical injury, while the hypersporulating phenotype indeed formed under all of these (with the exception of starvation) conditions. As has been reported previously (48), the addition of exogenous cAMP (1 and 5 mM) did not alter the slowly growing, hypersporulating phenotype of the mutants. We therefore tested whether the addition of cAMP would rescue peptaibol formation in these mutants, but we found that this also was not the case (data not shown). Consequently, these data imply that peptaibol formation is positively controlled by GNA3 via a pathway that must be different from that repressing conidiation and that this control overrides the correlation with sporulation.
FIG. 5.
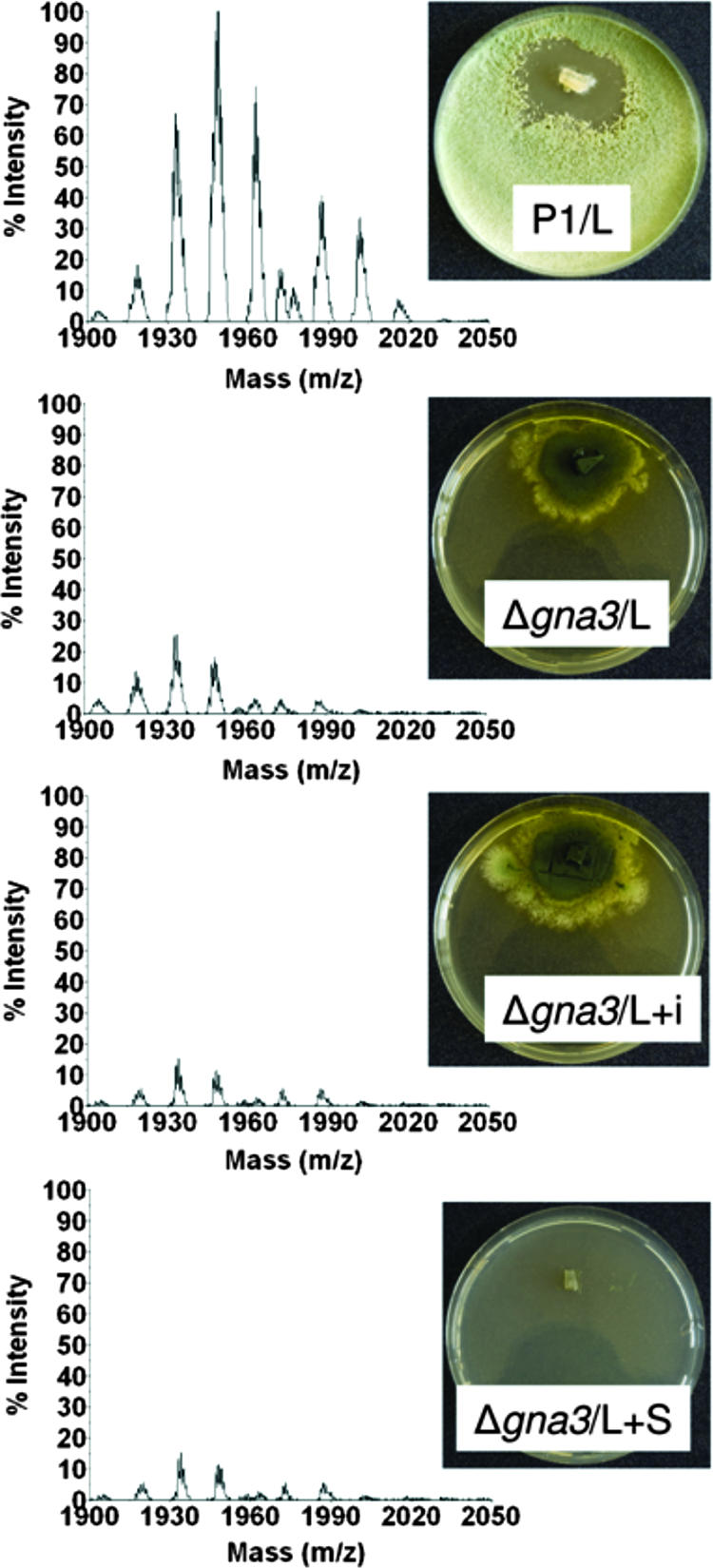
GNA3 is required for peptaibol production by H. atroviridis. H. atroviridis P1, and its Δgna3 mutant were used. Other symbols are used as explained in Fig. 4. Only experiments performed with illuminated cultures are shown since none of the nonilluminated cultures of the delta-gna3 mutant showed any peptaibol production. The range of intensity is 0 to 18,000 to make the traces of peptaibols which are still formed more visible. The experiment shown is typical for three independent experiments, whose results were consistent with the data shown here.
Constitutive activation of PKA leads to impairment of peptaibol formation.
Despite of the lack of influence of addition of exogenous cAMP, the disparity of the effect of loss of function of GNA3 on conidiation and peptaibol formation prompted us to investigate whether the effect on peptaibol formation would involve PKA (PKA1). G-protein-mediated inhibition of sporulation is known to involve the downstream action of PKA (35). To this end, we used a strain overexpressing the antisense version of the regulatory subunit of PKA1 (6) and whose PKA1 activity is thus constitutively activated. Consistent with what is known in other fungi, sporulation in this strain was essentially absent (see also reference 6). Also consistent with the findings that the addition of cAMP had no effect, peptaibol formation was not observed in this strain, irrespective of the conditions used (light/darkness, mechanical injury, starvation, etc. [data not shown]). Therefore, the positive action of GNA3 on peptaibol formation does not involve PKA1.
DISCUSSION
Peptaibols are among the largest peptide-like components formed by fungi and also have a number of interesting applications. However, apart of their isolation and chemical characterization, little information has thus far become available about the reasons for their biodiversity and regulation of formation. Here we used in silico analysis to predict the structure of the peptaibol synthetase of H. atroviridis, and we report that the formation of the respective peptaibols—atroviridins—partially correlates with sporulation and is GNA3 dependent. To facilitate the subsequent discussion, these findings and the interaction of components are summarized in Fig. 6.
FIG. 6.
Schematic summary of the regulation of peptaibol biosynthesis, based on results from the present study. (a) Summary of peptaibol formation under the various conditions in the mutants tested. L, light; D, dark. Strains are abbreviated as indicated in Materials and Methods. “Y” (yes) indicates peptaibol formation; “N” (no) indicates no peptaibol formation. Conditions in which peptaibol formation correlates with sporulation are in white on a black background. (b) Model for the putative interaction of the factors, as studied here, on sporulation and peptaibol formation. Red arrows indicate signaling mechanisms in which peptaibol formation is uncoupled from sporulation. +, activation; −, inhibition. The dotted arrow indicates the possibility of BLR1 and BLR2 action via GNA3 (unpublished data), for which no evidence is presented here.
Although we have not produced a delta mutant of H. atroviridis pbs1, in silico analysis predicts that the respective gene encodes all of the domains necessary for synthesizing at least 19-residue peptaibols. Amino acid residues 232 to 430 of H. atroviridis PBS1 showed 52% identity over 80% of the length of the ketoacyl synthetase domain of H. jecorina PAR1, and residues 431 to 815 were 74% identical over 100% of the length of the PAR1 acyltransferase domains and were also highly similar to T. virens TEX1. These domains therefore encode the proteins acetylating of the peptaibol N terminus. Amino acid residues 22694 to 23110, on the other hand, were 73% identical over 100% of the length of the alcohol dehydrogenase domain (pfam00106) of PAR1, which is necessary for the reductive cleavage of the final amino acid to generate the C-terminal alcohol. The same domain has been found at the same place in T. virens TEX1 and T. harzianum SALPS2 (42, 46). Therefore, together with the 19-aa activating, transferring, and condensation domains, the deduced PBS1 protein theoretically contains all of the enzymatic activities necessary and typical to produce peptaibols.
Peptaibols are notoriously microheterogeneous. Wiest et al. (46) emphasized that this multiplicity of products is likely a result of the ability of the activating modules to bind multiple substrates. In addition, this ability may be reflected in different Km values for different amino acids, because it is known that the microheterogeneity can be influenced by supplementing cultures with a specific amino acid, thereby likely increasing its intracellular concentration (see reference 19). The possibility that multiple peptaibol synthetases would be responsible for generation of this microheterogeneous mixture has been ruled out by Wiest et al. (46), who showed that a tex1 knockout in T. virens eliminated the formation of all 18-residue peptaibols. In addition, Wei et al. (45) proved that the small 11-14-mer peptaibols are products of a second, smaller peptaibol synthase. Our genome sequence data support such a claim: in fact, pbs1 is the only gene in the H. atroviridis genome that encodes a peptaibol synthetase consisting of the required number of modules for synthesis of peptaibols with more than 16 residues (C. P. Kubicek, unpublished data). Similar findings have also been made for H. jecorina (H. von Doehren, unpublished data). However, our data contribute a new aspect on the multiplicity of peptaibol production: the atroviridins formed contain the sequence Aib5-Aib6-Gln7. while the structure of PBS1 does not contain a second Aib domain preceding the Gln domain. The only explanation that can be offered for this finding is that the U5 module acts twice, forming a hexapeptide intermediate. Such cases of the iterative use of peptide synthetase modules are known in fungal nonribosomal peptide synthetases (43) but have thus far not been reported for peptaibol synthetases. Our data suggest that this could be a further mechanism contributing to peptaibol heterogeneity.
The phylogenetic analysis, which was performed to identify the individual activating domains, revealed a very high diversity and, with the exception of the domains specific for Pro, Gln, Phe, and in some cases Aib, phylogeny was unable to predict the amino acids bound by these modules. Similar difficulties were observed with the signature sequences proposed by Challis et al. (7) based on a limited set of domains. This high amino acid diversity contrasts with that of the otherwise rather highly conserved transfer and condensation domains (>80% identity). This and the fact that some of the modules activating the same amino acid (e.g., Aib or Ala) occur in different clades of the NJ tree suggests a history of gene duplication and a high mutation rate that ultimately leads to an alteration of the substrate specificity. In addition, the fact that the pbs1 locus has only partially maintained synteny in Trichoderma suggests the possibility of recombination as an additional origin of the diversity of peptaibols in this fungal genus.
The pbs1 mRNA, in a stretched form, must be about 10 μm long, and it was already emphasized (46) that the mechanism for the transcription of such large mRNAs in eukaryotes is unknown, and the correlation between transcript abundance and expression level is questionable. Therefore, rather than quantifying the pbs1 transcript, we used MALDI-TOF MS identification of the peptaibols as a means to learn about their regulation of formation. Also, the MALDI-TOF procedure has the advantage that it allows the analysis of a very small part of the growing colony, thereby ensuring the analysis of a homogenous fungal tissue. Unfortunately, it also has the drawback that absolute quantification is impossible since the weight of the fungal material that was extracted could not be determined. Consequently, only extreme changes (present or not present) are used for interpretation. However, using this semiquantitative approach, we could show that the formation of “long” peptaibols by the wild-type of H. atroviridis is associated with conidiation. Such an association is not without precedent: as an example, sporulation-deficient mutants of Aspergillus spp. (2), Claviceps purpurea (28), or Fusarium verticillioides (34) have also been shown to be defective in the production of their respective secondary metabolites. Calvo et al. (4) grouped secondary metabolites associated with sporulation into three broad categories: (i) metabolites that are required for the sporulation process (for example, the sporulation-activating linoleic acid-derived compounds produced by A. nidulans) (8-10), (ii) metabolites that are required for sporulation structures (for example, pigments such as melanins, which are required for the integrity of sexual and asexual spores) (1, 15), and (iii) metabolites that are toxic and whose formation coincides with the approximate time of sporulation (for example, the biosynthesis of mycotoxins) (14, 41). Based on their fungicidal action, peptaibols would fit into the last group. However, an important difference is that they are not secreted but remain bound to the spores of Trichoderma. So could they be important for sporulation in the sense of groups i and ii listed above? Peptaibol synthetase-delta mutants of T. virens (46) and of T. harzianum (D. Keszenman-Pereyra et al., unpublished data) still sporulate, so the answer would be no. However, both the T. harzianum and the T. virens delta mutants still form the 11-14 residue peptaibols (46; Keszenman-Pereyra et al., unpublished), and they could compensate for the loss of formation of the 18- to 20-residue peptaibols. In addition, the peptaibols might have subtle effects on sporulation that escape inspection of the morphology only. For example, they are known to function as voltage-gated membrane channels, primarily transporting K+ (11, 12, 18, 33), which could be important for ion transport and/or homeostasis during sporulation. In fact, gramicidin, another ion channel-forming peptide antibiotic, induces sporulation in its producer Bacillus subtilis (24, 29). The necessity for the presence of specific membrane channels in fungal spores has recently been stressed by Sidoux-Walter et al. (36), who showed that sporulation in S. cerevisiae is accompanied by the expression of a specific aquaporin, which is responsible for water outflow from spores. Thus, in analogy, one could speculate that the peptaibols are important for K+ and other monovalent cation homeostasis during the conidiation of Trichoderma. We have been unable to find a report describing the effect of peptaibols on the producer organisms, which in the light of the present findings clearly warrants investigation.
Sporulation of H. atroviridis is dependent on light (4, 6), and it was therefore not unexpected that the formation of a conidiation-associated secondary metabolite would consequently respond to light as well. However, the present data show that the relationship between light and peptaibol formation is more complex: the fact that mechanical injury can override control by BLR1 and -2 but is still affected by light (i.e., no peptaibol formation was observed in the dark, despite sporulation) suggests that there is a second, light-dependent mechanism that is essential for peptaibol formation. The occurrence of light stimulation of gene expression by an BLR1 and two independent pathways in H. atroviridis has been recently shown (31), and it is possible that this pathway may contribute to peptaibol formation under conditions of mechanical injury-induced sporulation.
The G-protein signaling pathway has been shown to be involved in the control of secondary metabolism and conidiation in Aspergillus species, particularly in A. nidulans (4), but there is also evidence indicating the regulation of trichothecene production in Fusarium (40) and γ-pentylpyrone formation in H. atroviridis (30) by a similar pathway. Interestingly, not all effects of G proteins (as would be expected from a sporulation-associated process) are negative: while in A. nidulans the dominant-activating fadAG42R allele represses conidiation and sterigmatocystin biosynthesis, it stimulates penicillin biosynthesis at the same time (4). Introduction of the fadAG42R allele into F. sporotrichioides results in reduced conidiation but increased trichothecene production (40). It is also worth noting that all of the reports on a role of G proteins in fungal secondary metabolism were done with the Aspergillus G-protein FadA or its Trichoderma orthologue TGA1 (GNA1). In contrast, the present study was undertaken with another G protein, GNA3. Although both GNA1 and GNA3 (previously termed TGA3 [48]) are negative regulators of conidiation in H. atroviridis (30, 48), N. crassa GNA1 is regulated by the transmembrane receptor protein GPR-4, which is responsible for carbon source signaling (21). The receptor to which GNA3 binds is not known yet. Thus, our data show that not only FadA/GNA1 mediate secondary metabolism, but at least GNA3 also does so. It is tempting to speculate that all G proteins might be able to regulate secondary metabolism, because null mutants in all of them affect sporulation (16). This is understandable in view of the multiple signals (pH, sugar, nitrogen content, light, and many others) that determine whether a fungal cell can maintain a vegetative mode of growth or whether it is advisable to conidiate.
The subsequent steps required for triggering of peptaibol formation by GNA3 are unclear. If this occurred via activation of adenylate cyclase and thus the formation of cAMP, one would assume that the H. atroviridis strain harboring the antisense gene for the regulatory subunit of PKA and thus bearing a constitutively active PKA would overproduce peptaibols. This was shown not to be the case, and indeed, at the level of PKA, peptaibol formation again strictly correlated with sporulation and was therefore absent in the PKA-overproducing mutant. The finding of loss of sporulation is consistent with the hypersporulating phenotype of the delta-gna3 strain and suggests that the pathway triggering sporulation in H. atroviridis is subject to a negative control by a G-protein/PKA pathway similar to that in A. nidulans (35). Hence, the positive effect of GNA3 on peptaibol biosynthesis must involve another signaling pathway for which, thus far, only the requirement for GNA3 is known.
Acknowledgments
This study was supported by the Fifth (EC) Framework program (Quality of Life and Management of Living Resources; project EUROFUNG 2 [QLK3-1999-00729]) and by a grant from the Austrian Science Foundation (P 17325-B17) to C.P.K. The genomic sequence for H. atroviridis was provided by the DOE Joint Genome Institute, Walnut Creek, CA. This study was performed under the auspices of the U.S. Department of Energy's Office of Science, Biological, and Environmental Research Program and the by the University of California, Lawrence Livermore National Laboratory, under contract W-7405-Eng-48; the Lawrence Berkeley National Laboratory under contract DE-AC03-76SF00098; and the Los Alamos National Laboratory under contract W-7405-ENG-36.
We thank S. Zeilinger for the tga3-delta strain of H. atroviridis and S. Baker for help with the H. atroviridis genome sequence.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, J. W., and K. E. Papa. 1988. The aflatoxigenic Aspergillus, p. 264-280. In D. S. Ingram and P. A. Williams (ed.), Genetics of plant pathology, vol. 6. Academic Press, London, United Kingdom. [Google Scholar]
- 3.Reference deleted.
- 4.Calvo, A. M., R. A. Wilson, J. W. Bok, and N. P. Keller. 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66:447-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casas-Flores, S., M. Rios-Momberg, M. Bibbins, P. Ponce-Noyola, and A. Herrera-Estrella. 2004. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 150:3561-3569. [DOI] [PubMed] [Google Scholar]
- 6.Casas-Flores, S., M. Rios-Momberg, T. Rosales-Saavedra, P. Martinez-Hernandez, V. Olmedo-Monfil, and A. Herrera-Estrella. 2006. Cross talk between a fungal blue-light perception system and the cyclic AMP signaling pathway. Eukaryot. Cell 5:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 8.Champe, S. P., and A. A. E. El-Zayat. 1989. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J. Bacteriol. 171:3982-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champe, S. P., P. Rao, and A. Chang. 1987. An endogenous inducer of sexual development in Aspergillus nidulans. J. Gen. Microbiol. 133:1383-1388. [DOI] [PubMed] [Google Scholar]
- 10.Chang, P.-K., J. W. Cary, D. Bhatnagar, T. E. Cleveland, J. W. Bennett, J. E. Linz, C. P. Woloshuk, and G. A. Payne. 1993. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chugh, J. K., and B. A. Wallace. 2001. Peptaibols: models for ion channels. Biochem. Soc. Trans. 29:565-570. [DOI] [PubMed] [Google Scholar]
- 12.Duclohier, H., G. M. Alder, C. L. Bashford, H. Bruckner, J. K. Chugh, and B. A. Wallace. 2004. Conductance studies on trichotoxin_A50E and implications for channel structure. Biophys. J. 87:1705-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harman, G. E., and C. P. Kubicek. 1998. Trichoderma and Gliocladium, vol. 2. Enzymes, biocontrol, and commercial applications. Taylor & Francis, London, United Kingdom.
- 14.Hicks, J., J.-H. Yu, N. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G-alpha protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamura, C., T. Tsujimoto, and T. Tsuge. 1999. Targeted disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol. Plant-Microbe Interact. 12:59-63. [DOI] [PubMed] [Google Scholar]
- 16.Kays, A. M., and K. A. Borkovich. 2004. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric G alpha proteins. Genetics 166:1229-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kropacheva, T. N., and J. Raap. 2002. Ion transport across a phospholipid membrane mediated by the peptide trichogin GA IV. Biochim. Biophys. Acta 1567:193-203. [DOI] [PubMed] [Google Scholar]
- 19.Kubicek, C. P., M. Zelazowska-Komon, E. Sandor, and I. S. Druzhinina. 2007. Facts and challenges in the understanding of the biosynthesis of peptaibols by Trichoderma. Chem. Biodiv. 4:1068-1082. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Li, L., and K. A. Borkovich. 2006. GPR-4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa. Eukaryot. Cell 5:1287-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorito, M., V. Farkas, S. Rebuffat, B. Bodo, and C. P. Kubicek. 1996. Cell wall synthesis is a major target of mycoparasitic antagonism by Trichoderma harzianum. J. Bacteriol. 178:6382-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucaciu, M., S. Rebuffat, C. Goulard, H. Duclohier, G. Molle, and B. Bodo. 1997. Interaction of the 14-residue peptaibols, harzianins HC, with lipid bilayers: permeability modifications and conductance properties. Biochim. Biophys. Acta 1323:85-96. [DOI] [PubMed] [Google Scholar]
- 24.Marahiel, M. A., M. M. Nakano, and P. Zuber. 1993. Regulation of peptide antibiotic production in Bacillus. Mol. Microbiol. 7:631-636. [DOI] [PubMed] [Google Scholar]
- 25.Neuhof, T., R. Dieckmann, I. S. Druzhinina, C. P. Kubicek, and H. von Döhren. 2007. Intact-cell MALDI-TOF mass spectrometry analysis of peptaibol formation by the genus Trichoderma: can molecular phylogenic knowledge predict peptaibol structures? Microbiology 153:3417-3437. [DOI] [PubMed] [Google Scholar]
- 26.Oh, S. U., B. S. Yun, S. J. Lee, J. H. Kim, and I. D. Yoo. 2002. Atroviridins A-C and neoatroviridins A-D, novel peptaibol antibiotics produced by Trichoderma atroviride F80317. I. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. 55:557-564. [DOI] [PubMed] [Google Scholar]
- 27.Papavizas, G. C. 1985. Trichoderma and Gliocladium: biology, ecology, and potential for biocontrol. Annu. Rev. Phytopathol. 23:23-54. [Google Scholar]
- 28.Pazoutova, S., V. Pokorny, and Z. Rehacek. 1977. The relationship between conidiation and alkaloid production in saprophytic strains of Claviceps purpurea. Can. J. Microbiol. 23:1182-1187. [DOI] [PubMed] [Google Scholar]
- 29.Pschorn, W., H. Paulus, J. Hansen, and H. Ristow. 1982. Induction of sporulation in Bacillus brevis. 2. Dependence on the presence of the peptide antibiotics tyrocidine and linear gramicidin. Eur. J. Biochem. 129:403-407. [DOI] [PubMed] [Google Scholar]
- 30.Reithner, B., K. Brunner, R. Schuhmacher, I. Peissl, V. Seidl, R. Krska, and S. Zeilinger. 2005. The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet. Biol. 42:749-760. [DOI] [PubMed] [Google Scholar]
- 31.Rosales-Saavedra, T., E. U. Esquivel-Naranjo, S. Casas-Flores, P. Martinez-Hernandez, E. Ibarra-Laclette, C. Cortes-Penagos, and A. Herrera-Estrella. 2006. Novel light-regulated genes in Trichoderma atroviride: a dissection by cDNA microarrays. Microbiology 152:3305-3317. [DOI] [PubMed] [Google Scholar]
- 32.Schirmböck, M., M. Lorito, Y.-L. Wang, C. K. Hayes, I. Arisan-Atac, F. Scala, G. E. Harman, and C. P. Kubicek. 1994. Molecular mechanisms involved in biocontrol by Trichoderma harzianum: co-induction and synergism of hydrolytic enzymes and pentaibol antibiotics. Appl. Environ. Microbiol. 60:4364-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shenkarev, Z. O., T. A. Balashova, Z. A. Yakimenko, T. V. Ovchinnikova, and A. S. Arseniev. 2004. Peptaibol zervamicin IIb structure and dynamics refinement from transhydrogen bond J couplings. Biophys. J. 86:3687-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shim, W.-B., and C. P. Woloshuk. 2001. Regulation of fumonisin B1 biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl. Environ. Microbiol. 67:1607-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G-protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidoux-Walter, F., N. Pettersson, and S. Hohmann. 2004. The Saccharomyces cerevisiae aquaporin Aqy1 is involved in sporulation. Proc. Natl. Acad. Sci. USA 101:17422-17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stachelhaus, T., and M. A. Marahiel. 1995. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol. Lett. 125:3-14. [DOI] [PubMed] [Google Scholar]
- 38.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 39.Szekeres, A., B. Leitgeb, L. Kredics, Z. Antal, L. Hatvani, L. Manczinger, and C. Vagvolgyi. 2005. Peptaibols and related peptaibiotics of Trichoderma: a review. Acta Microbiol. Immunol. Hung. 52:137-168. [DOI] [PubMed] [Google Scholar]
- 40.Tag, A., J. Hicks, G. Garifullina, C. Ake, T. D. Phillips, M. Beremand, and N. P. Keller. 2000. G-protein signaling mediates differential production of toxic secondary metabolites. Mol. Microbiol. 38:658-665. [DOI] [PubMed] [Google Scholar]
- 41.Trail, F., N. Mahanti, and J. Linz. 1995. Molecular biology of aflatoxin biosynthesis. Microbiology 141:755-765. [DOI] [PubMed] [Google Scholar]
- 42.Vizcaino, J. A., R. E. Cardoza, L. Dubost, B. Bodo, S. Gutierrez, and E. Monte. 2006. Detection of peptaibols and partial cloning of a putative peptaibol synthetase gene from Trichoderma harzianum CECT 2413. Folia Microbiol. 51:114-120. [DOI] [PubMed] [Google Scholar]
- 43.von Döhren, H. 2004. Biochemistry and general genetics of non-ribosomal peptide synthetases in fungi. Adv. Biochem. Eng. Biotechnol. 88:217-264. [DOI] [PubMed] [Google Scholar]
- 44.Wallace, B. A. 2000. Common structural features in gramicidin and other ion channels. Bioessays 22:227-234. [DOI] [PubMed] [Google Scholar]
- 45.Wei, X., F. Yang, and D. C. Straney. 2005. Multiple non-ribosomal peptide synthetase genes determine peptaibol synthesis in Trichoderma virens. Can. J. Microbiol. 51:423-429. [DOI] [PubMed] [Google Scholar]
- 46.Wiest, A., D. Grzegorski, B. W. Xu, C. Goulard, S. Rebuffat, D. J. Ebbole, B. Bodo, and C. Kenerley. 2002. Identification of peptaibols from Trichoderma virens and cloning of a peptaibol synthetase. J. Biol. Chem. 277:20862-20868. [DOI] [PubMed] [Google Scholar]
- 47.Yoder, O. C., and B. G. Turgeon. 2001. Fungal genomics and pathogenicity. Curr. Opin. Plant Biol. 4:315-321. [DOI] [PubMed] [Google Scholar]
- 48.Zeilinger, S., B. Reithner, V. Scala, I. Peissl, M. Lorito, and R. L. Mach. 2005. Signal transduction by Tga3, a novel G protein alpha subunit of Trichoderma atroviride. Appl. Environ. Microbiol. 71:1591-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]