Abstract
Sporulation of the fission yeast Schizosaccharomyces pombe is a developmental process that generates gametes and that includes the formation of spore envelope precursors called the forespore membranes. Assembly and development of forespore membranes require vesicular trafficking from other intracellular membrane compartments. We have shown that phosphatidylinositol 3-kinase (PtdIns 3-kinase) is required for efficient and proper development of forespore membranes. The role of a FYVE domain protein, Sst4p, a homolog of Vps27p/Hrs, as a downstream factor for PtdIns 3-kinase in sporulation was investigated. sst4Δ asci formed spores with oval-shaped morphology and with reduced viability compared to that of the wild-type spores. The extension of forespore membranes was inefficient, and bubble-like structures emerged from the leading edges of the forespore membranes. Sst4p localization was examined using fluorescent protein fusions and was found to be adjacent to the forespore membranes during sporulation. The localization and function of Sst4p were dependent on its FYVE domain and on PtdIns 3-kinase. Sst4p colocalized and interacted with Hse1p, a homolog of Saccharomyces cerevisiae Hse1p and of mammalian STAM. Mutations in all three UIM domains of the Sst4p/Hse1p complex resulted in formation of spores with abnormal morphology. These results suggest that Sst4p is a downstream factor of PtdIns 3-kinase and functions in forespore membrane formation.
Spores in the fission yeast Schizosaccharomyces pombe are formed through a series of events, including premeiotic DNA synthesis, first and second meiotic nuclear divisions, formation of a spore envelope, and maturation of ascospores (55). During the second meiotic division, precursors of the spore envelope, the forespore membranes, are assembled by fusion of vesicles on the cytoplasmic surfaces of the spindle pole bodies. The forespore membranes then extend and close to form prespores, each harboring a haploid nucleus. These eventually become spores after the accumulation of the spore wall materials in the lumen between the inner and outer layers of the forespore membranes. The vesicles that fuse to assemble and extend the forespore membranes can derive directly or indirectly from various intracellular membrane compartments, such as the endoplasmic reticulum, the Golgi apparatus, or vacuoles, as well as the plasma membrane. Spo14p (homolog of Saccharomyces cerevisiae Sec12p), a GDP/GTP exchange factor for the Sar1 GTPase that is essential for budding of vesicles from the endoplasmic reticulum (38), and Spo20p (homolog of S. cerevisiae Sec14p), a phosphatidylinositol/phosphatidylcholine transfer protein that is essential for the formation of Golgi apparatus-derived vesicles, are required for the proper development of the forespore membrane (39). Ypt7p, a Rab GTPase homolog, localizes to vacuolar membranes and is required for forespore membrane development (25). A t-SNARE protein, Psy1p (homolog of S. cerevisiae Sso1p and Sso2p), translocates from the plasma membrane to the forespore membrane during meiosis and is essential for the assembly of the forespore membranes (37). Taken together, assembly and extension of the forespore membranes are likely to involve membrane trafficking and/or remodeling of the membranous organelles.
We have shown that the fission yeast class III phosphatidylinositol 3-kinase (PtdIns 3-kinase), Pik3p, plays an important role in proper formation of the forespore membranes (41). pik3Δ cells exhibit a pleiotropic phenotype in forespore membrane formation, such as disoriented and inefficient extension, failure of closure of the leading edges, and formation of bubbles, resulting in formation of nonviable spores.
Class III PtdIns 3-kinase is a key enzyme that specifically phosphorylates phosphatidylinositol to produce phosphatidylinositol 3-phosphate [PtdIns(3)P] (50, 60). For the budding yeast Saccharomyces cerevisiae, it has been shown that PtdIns(3)P produced by Vps34p PtdIns 3-kinase is a key regulator of transport of newly synthesized vacuolar proteins, such as carboxypeptidase Y (CPY), from the trans-Golgi network to the vacuoles via the endosomes (15). Recruitment of proteins that interact with PtdIns(3)P is essential for proper transport of CPY. We searched for factors that function downstream of Pik3p in sporulation and identified two phox homology (PX) domain proteins, Vps5p and Vps17p, as good candidates (28). The homologs of these proteins in S. cerevisiae have been well characterized as PtdIns(3)P-dependent mediators of retrograde trafficking from the endosome to the Golgi apparatus. This finding suggested the importance of PtdIns 3-kinase-mediated retrograde trafficking from the endosome or the forespore membrane to the Golgi apparatus in development of the forespore membrane (21, 44, 53). Interestingly, overproduction of Vps5p and/or Vps17p partly restored the viability of pik3Δ spores; nevertheless, the morphology of the forespore membranes was not completely normal, i.e., although the forespore membranes succeeded in encapsulating the nuclei, they frequently formed bubbles (28). These results imply another unidentified downstream factor(s) of Pik3p during sporulation that may contain a PtdIns(3)P-binding motif, such as PX, FYVE (Fab1/YGL023/Vps27/EEA1), or pleckstrin homology domains (9).
Vps27p/Hrs (hepatocyte growth factor 1-regulated tyrosine kinase substrate) is a FYVE domain protein that was originally identified as one of the class E VPS (vacuolar protein sorting) genes required for proper sorting of CPY to the vacuoles in S. cerevisiae (45, 47). In class E vps mutants, instead of being transported to the vacuole, CPY accumulates in exaggerated perivacuolar membrane compartments called class E compartments. Electron microscopic analysis revealed the class E compartment to be stacks of curved membrane cisternae (5, 48). Vps27p/Hrs homologs are conserved among eukaryotes; they are composed of a VHS (Vps27p/Hrs/Stam) domain, a FYVE domain, and two ubiquitin-interacting motifs (UIMs) (46).
Vps27p forms a stable protein complex with Hse1p (STAM in mammals), a protein with VHS, Src homology 3, and UIM domains (3, 11). Recent studies identified this complex as the priming machinery essential for sorting the cargo proteins into endosomal structures called multivesicular bodies (MVBs) (12, 26). MVBs are a part of a eukaryotic endosomal system that is conserved from yeast to humans, formed by the invagination and budding of vesicles from the limiting membrane into the lumen of the endosome. During this process, endosomal transmembrane proteins with monoubiquitin tags destined for the vacuolar lumen are recognized and retained by three UIM domains of the Vps27p/Hse1p complex, which subsequently recruits three distinct protein complexes consisting mainly of class E Vps proteins, called ESCRT-I, ESCRT-II, and ESCRT-III (endosomal sorting complex required for transport). The ESCRT complexes function in a sequential manner to sort the cargo proteins into invaginating vesicles (4). Because of its role in initiating the ESCRT-mediated process, the Vps27p/Hse1p complex is also called ESCRT-0 (13). Binding of the FYVE domain of Vps27p to PtdIns(3)P at the limiting membrane of the endosomes is essential for the localization and function of the Vps27p/Hse1p complex (26).
We previously identified sst4, a VPS27 homolog in S. pombe, as a suppressor for the sterility of the ste12Δ mutant, which is defective in vacuolar protein sorting and in cell conjugation (42). In a subsequent study, we showed that Sst4p is required for efficient transport of the fluorescent dye FM4-64 and ubiquitin-green fluorescent protein-carboxypeptidase S (Ub-GFP-CPS) fusion protein to the vacuole (23). This observation suggested the existence of Sst4p-mediated endocytic membrane trafficking and anterograde sorting of biosynthesized proteins into MVBs in S. pombe. Thus, Sst4p is likely to be a functional counterpart of Vps27p in S. cerevisiae. During the course of this study, we noticed that sst4Δ cells formed spores with slightly abnormal morphology.
Here, we show that Sst4p contributes to the development of the forespore membrane and to the maturation of spores in a PtdIns(3)P-dependent manner and that Sst4p interacts with Hse1p to form a complex whose UIM domains are important for its function. These phenotypic and biochemical data suggest that the Sst4p/Hse1p complex contributes to sporulation in S. pombe to enable the formation of the forespore membrane with normal morphology.
MATERIALS AND METHODS
Strains, media, and plasmids.
The S. pombe strains used in this study are listed in Table 1. The yeast strains were grown in YPD (1% yeast extract, 2% polypeptone, 2% dextrose), YE (0.5% yeast extract, 3% dextrose), or EMM medium (32); SSA medium (19) was used to induce sporulation. To induce sporulation, cells were grown for 12 to 18 h on an EMM plate, transferred to an SSA plate, and incubated for 10 h before examination by fluorescence microscopy, unless otherwise indicated.
TABLE 1.
Yeast strains used in this study
| Strain name | Description | Reference or source |
|---|---|---|
| THP17 | h90 ade6-M210 ura4-D18 leu1-32 | 28 |
| MI160 | h90 ade6-M210 ura4-D18 leu1-32 sst4Δ::ura4+ | See text |
| MI368 | h90 ade6-M210 ura4-D18 leu1-32 sst4Δ::kanMX6 | See text |
| MI597 | h90 ade6-M210 ura4-D18 leu1-32 sst4R193A | See text |
| MI578 | h90 ade6-M210 ura4-D18 leu1-32 sst4S271D | See text |
| MI550 | h90 ade6-M216 ura4-D18 leu1-32 sst4S316D | See text |
| MI577 | h90 ade6-M210 ura4-D18 leu1-32 sst4S271,316D | See text |
| MI545 | h90 ade6-M210 ura4-D18 leu1-32 hse1S176D | See text |
| MI596 | h90 ade6-M210 ura4-D18 leu1-32 sst4S271,316Dhse1S176D | See text |
| MI392 | h90 ade6-M210 ura4-D18 leu1-32 sst4Δ::kanMX6 hse1Δ::ura4+ | This studya |
| MI512 | h90 ade6-M210 ura4-D18 leu1-32 sst4+-mRFP5::kanMX6 | See text |
| MI446 | h90 ade6-M210 ura4-D18 leu1-32 sst4+-HA-FLAG::kanMX6 | See text |
| MI301 | h90 ade6-M210 ura4-D18 leu1-32 hse1Δ::ura4+ | See text |
| MI452 | h90 ade6-M210 ura4-D18 leu1-32 sst2Δ::ura4+ | See text |
| MI297 | h90 ade6-M210 ura4-D18 leu1-32 sst6Δ::ura4+ | See text |
| MI532 | h90 ade6-M210 ura4-D18 leu1-32 vps28Δ::ura4+ | See text |
| MI533 | h90 ade6-M210 ura4-D18 leu1-32 vps20Δ::ura4+ | See text |
| MI298 | h90 ade6-M210 ura4-D18 leu1-32 vps32Δ::ura4+ | See text |
| MI534 | h90 ade6-M210 ura4-D18 leu1-32 vps4Δ::ura4+ | See text |
| TW747 | h+/h−ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | 41 |
| MI619 | h90/h90 ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 sst4Δ::ura4+/sst4Δ::ura4+ | This studyb |
| TP3 | h+/h−ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 pik3Δ::ura4+/pik3Δ::ura4+ | 28 |
| YS2 | h+/h−ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 ste12Δ::ura4+/ste12Δ::ura4+ | 42 |
This strain was constructed by crossing MI306 and MI368.
This strain was constructed by mating MI160 to an ade6-M216 segregant from a cross of MI160 to THP18 (h90 ade6-M210 ura4-D18 leu1-32).
The sst, hse1, and vps deletion strains were created as follows. The coding regions of the genes were amplified by PCR from the S. pombe genomic DNA and cloned into a vector, pT7Blue (Novagen, San Diego, CA). Plasmids containing disrupted genes were then constructed by replacement of portions of the coding regions with a ura4+ marker cassette obtained as a HindIII fragment from a KS-ura4 plasmid (8). The primer sets and restriction enzymes used in this process and the positions replaced by the ura4+ marker are listed in Table 2. The fragments containing the disrupted genes were released from the plasmids by digestion with appropriate restriction enzymes and transformed into a wild-type strain, THP17. For strains MI368, MI512, and MI446, the complete sst4 coding region was deleted or tagged at its C terminus with mRFP1 (16) or hemagglutinin (HA)-FLAG sequences by use of a PCR method with pFA6a-kanMX6 (8), pFA6a-mRFP-kanMX6, or pFA6a-HA-FLAG-kanMX6 (a gift from H. Ishikawa, Tokyo University of Agriculture and Technology, Japan) as the template. pFA6a-mRFP-kanMX6 was constructed by insertion of the BamHI-BglII mRFP1-TADH1 fragment from pFA6a-mRFP5-His5 (a gift from T. Yoko-o, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan) into BamHI/BglII-digested pFA6a-kanMX6. The successfully targeted recombination was verified by either PCR or Southern blotting. Plasmids with genes encoding point mutant alleles of sst4 or hse1 for construction of strains MI597 (sst4R193A), MI578 (sst4S271D), MI550 (sst4S316D), and MI545 (hse1S176D) were generated by site-directed mutagenesis (49) by use of the plasmid pT7(sst4+) or pT7(hse1+) as a template with the primers listed in Table 2. The plasmid for strain MI577 (sst4S271,316D) was generated by the same method by use of pT7(sst4S271D) as a template with the primer for sst4S316D. The coding regions were released from the plasmids by use of appropriate restriction enzymes and transformed into sst4Δ::ura4+ or hse1Δ::ura4+ mutants. Ura− transformants were selected on plates containing 5-fluoroorotic acid and checked by amplification of the gene loci by PCR followed by DNA sequencing to ensure successful integrations of the fragments so that correct mutant alleles were produced. The triple point mutant strain was constructed by crossing MI577 and MI545.
TABLE 2.
Primers used for plasmid construction and primers and restriction enzymes used for gene disruption in this study
| Plasmid or gene | Primer sequence | Site(s) (position[s])a |
|---|---|---|
| Plasmids | ||
| pT7(sst4R193A) | 5′-ATTCACTACCCGGAAACACCATTGTGCAAATTGCGGTGGCG-3′ | |
| pT7(sst4S271D) | 5′-TATTGAGCTTGATCTCAAAGAAA-3′ | |
| pT7(sst4S316D) | 5′-TATTGCTATCGATCTTGAGGAAG-3′ | |
| pT7(hse1S176D) | 5′-TTTAGCATTAGATCTTTCGGAAT-3′ | |
| pAL(Sst4p-GFP) | 5′-CGGATCCTGAGGTATACTAAACTCGTC-3′ | |
| 5′-AGCGGCCGCCTCCACCCGGGAAAAGCTCGATTAAAGATGCTTC-3′ | ||
| pAL(Hse1p-GFP) | 5′-CGGATCCATTTTGCTGAGTAAATGTC-3′ | |
| 5′-GTCTGAGATGAGGAATTTTTTCCCGGGTGGAGGCGGCCGCT-3′ | ||
| pGEX(GST-FYVE) | 5′-CGCGGATCCCGTAGTAAGTCTGTTGATCC-3′ | |
| 5′-AAACTCGAGGCCTCTGAAGGTCTTTCATAAG-3′ | ||
| Genes | ||
| sst4+ | 5′-AAGGATCCGCATGTCTCGGTGGTGGAATTCC-3′ | HincII (+215), HindIII (+1292) |
| 5′-AAGTCGACCCGGGCCTAAAGCTCGATTAAAGATGC-3′ | ||
| hse1+ | 5′-CGGATCCCATGTTTCGAGGAAAACCCAAC-3′ | EcoRV (+701) |
| 5′-GTCTGAGATGAGGAATTTTTTCCCGGGTGGAGGCGGCCGCT-3′ | ||
| sst6+ | 5′-CAGATCTCATGTCAGATCATGCAATCAATG-3′ | HincII (+1059) |
| 5′-TCTCGAGTTAAGAGTATTGAAGCATTTCGTC-3′ | ||
| vps28+ | 5′-AAGGATCCTATGACTGAATACTACGATCTGAACC-3′ | HindIII (+137), EcoRV (+320) |
| 5′-TTCCCGGGTCATAACAAGCTATAGCACTCTGCG-3′ | ||
| vps20+ | 5′-AGATCTTATGGGGGTTAACAGTAGTAAAATTAATGATAAGGACAG-3′ | NdeI (+216), HindIII (+594) |
| 5′-GTCGACCCGGGCCAGATCCATAGTTATCCCAGGACTGG-3′ | ||
| vps32+ | 5′-AAGGATCCAATGTCTGGATTTTTAAGATGGTTTGGGGGC-3′ | MboI (+220), SspI (+370) |
| 5′-TTCCCGGGGCAAGCGTACTGGTCACTTTAAAGAGAGAATTCAGCC-3′ | ||
| sst2+ | 5′-CGGATCCCATGAACATTCTCCAAGGATCTG-3′ | BglII (+726) |
| 5′-GTTGATTTGAGGGTTAAATTCCCGGGTGGAGGCGGCCGCT-3′ | ||
| vps4+ | 5′-AAGGATCCATGTCCAATCCAGATTGTTTAAG-3′ | XbaI (+365) |
| 5′-TTGTCGACTATCCCTCTGCTCCAAAGTCCTTTG-3′ |
A ura4+ marker cassette was inserted as a HindIII fragment into the indicated position. A HindIII linker was inserted at the indicated restriction sites if necessary.
The plasmids expressing Sst4p or Hse1p fused to GFP at their C termini, pAL(Sst4p-GFP) and pAL(Hse1p-GFP), were constructed by insertion of the fragments obtained by amplification of the genomic sst4+ or hse1+ locus, including both promoters and coding regions between the BamHI and NotI sites of pAL(GFP). The primers used are listed in Table 2. Both Sst4p-GFP and Hse1p-GFP were functional, as they suppressed the sporulation defect of the respective deletion mutants. pAL(GFP) was constructed by insertion of a NotI-SacI fragment containing GFP and nmt1 terminator sequences from pGFT1 (61) into NotI/SacI-digested pALSK+ (58). The plasmid pREP41(GFP-Sst4p) for expression of Sst4p tagged with N-terminal GFP was constructed by insertion of the coding sequence of sst4+ into pGFT41, an nmt41 promoter derivative of pGFT1.
The plasmids for bacterial expression of GST-FYVE and GST-FYVER193A were constructed by insertion of the DNA fragments corresponding to amino acid residues 40 to 291 of Sst4p, obtained by the PCR method by use of primers listed in Table 2 with pT7(sst4+) or pT7(sst4R193A) as the template, into the BamHI and XhoI sites of pGEX4T3 (GE Healthcare Biosciences, Piscataway, NJ). The resulting plasmids express GST fusion proteins containing a portion of the VHS domain, the entire FYVE domain, and one UIM domain, corresponding to the region of S. cerevisiae Vps27p that has been shown to bind to PtdIns(3)P in vitro (14).
Light microscopy.
Differential interference contrast (DIC) microscopy was done with an Olympus IX71 microscope equipped with a UPlanApo 100× objective.
For fluorescence microscopy, cells were fixed with 3.3% formaldehyde (final concentration) for 30 min at 30°C in EMM or MM-N (EMM lacking NH4Cl) medium at ∼107 cells/ml. The cells were collected by centrifugation and washed three times with PEM buffer {100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.9, containing 1 mM EGTA and 1 mM MgSO4}. For staining of the nuclear chromatin region, cells were treated with PEMS (PEM containing 1.2 M sorbitol) plus 1% Triton X-100 for 1 min, washed three times with PEM, and then stained with 4′,6′-diamidino-2-phenylindole (DAPI) at 1 μg/ml in PEM. For immunostaining of Spo3p-HA, fixed cells were digested with 1 mg/ml Zymolyase 20T in PEMS for 10 min at 37°C prior to treatment with 1% Triton X-100. Spo3-HA was visualized by use of a high-affinity rat anti-HA monoclonal antibody, 3F10 (used at 1:200; Roche, Mannheim, Germany), and Alexa595-conjugated goat anti-rat immunoglobulin G (IgG) (used at 1:10,000; Molecular Probes, Eugene, OR) in PEM containing 1% bovine serum albumin and 1% lysine-HCl.
Labeling of the vacuolar membrane with FM4-64 was carried out without fixation of the cells, as described previously (41). Cells were observed under a Zeiss Axiovert 100 fluorescence microscope with a 100× PlanApochromat objective. To obtain three-dimensional reconstructed images, we used IPLab 3.6 software (Scanalytics, Rockville, MD).
Time-lapse imaging was done as follows. MM-N medium containing 3% agarose-S (Takara Bio, Shiga, Japan) was dropped onto a glass-bottomed dish. The solidified medium was removed from the dish, cells were spotted on the flat surface, and the medium was again applied to the dish so that the cells were sandwiched between the glass and the medium. Images were captured at 1-min intervals under the control of IPLab 3.6.
Electron microscopy.
Cells were mounted on copper grids to form a thin layer and plunged into liquid propane cooled with liquid N2. The frozen cells were transferred to 2% OsO4 in anhydrous acetone, kept at −80°C for 48 h in a solid CO2-acetone bath, and then transferred to −35°C for 2 h, 4°C for 2 h, and room temperature for 2 h. After being washed with anhydrous acetone three times, samples were infiltrated with increasing concentrations of Spurr's resin in anhydrous acetone and finally with 100% Spurr's resin. These samples were then polymerized in capsules at 50°C for 5 h and at 60°C for 50 h. Thin sections were cut on a Reichest Ultracut S and then stained with uranyl acetate and lead citrate. Sections were viewed on a JEOL 100CX electron microscope at 80 kV and JEOL 2010 at 100 kV.
Ethanol treatment.
Cells incubated on SSA plates for 5 days were collected and suspended in 500 μl of 1 mM citrate phosphate buffer (pH 5.6) at an approximate optical density at 600 nm of 0.5. After the numbers of spores were counted, ethanol was added to a final concentration of 30%, and the cells were incubated for 40 min at 30°C with periodic vortexing. The cells were plated on YE plates at serial dilutions, and the number of viable spores was estimated by the numbers of resulting colonies after incubation for 5 days at 30°C.
Protein-lipid overlay assay.
Escherichia coli strain BL21 (Novagen, Madison, WI) harboring plasmids encoding GST fusion proteins was cultured at 20°C for 16 days. The cells were harvested and incubated with 100 μg/ml lysozyme in TNE buffer (100 mM Tris [pH 7.5], 150 mM NaCl, and 0.2 mM EGTA) plus 1 mM phenylmethylsulfonyl fluoride for 30 min on ice, followed by three freeze-thaw cycles. The extract was prepared by centrifugation at 16,000 × g for 10 min. Lipid solution (2.5 μl) containing 0, 100, or 500 pmol of diC16 PtdIns (catalog no. P1368; Sigma) or diC16 PtdIns(3)P (catalog no. 549-01881; Wako Chemicals, Tokyo, Japan) dissolved in chloroform-methanol-H2O (1:2:0.8) was spotted onto a Hybond-C Extra membrane (GE Healthcare Biosciences, Piscataway, NJ) and dried at room temperature. The membrane was incubated at room temperature with 0.1% egg albumin (catalog no. 012-09885; Wako Chemicals, Tokyo, Japan) in TBS-T (10 mM Tris-HCl, pH 8.0, containing 150 mM NaCl and 0.1% Tween 20) for 30 min and then with TBS-T with E. coli extract containing 0.5 μg/ml of the indicated protein for 1 h. After the membrane was washed three times for 10 min with TBS-T at room temperature, proteins interacting with the phosphoinositides were detected with an anti-GST monoclonal antiserum and a horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody (part no. AMI3704; BioSource International, Camarillo, CA).
Immunoprecipitation and Western blotting.
Approximately 2 × 108 cells grown in EMM medium were collected and suspended in 500 μl of TNET buffer (10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 150 mM NaCl, 1% Triton X-100) plus 2 mM phenylmethylsulfonyl fluoride. The cells were vortexed vigorously with acid-washed glass beads three times for 1 min each and centrifuged at 16,000 × g for 10 min. The supernatant was subjected to immunoprecipitation by use of anti-HA antibody (12CA5) prebound to protein A beads (GE Healthcare Biosciences, Piscataway, NJ) at 4°C for 1 h. The proteins in the precipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting with an anti-GFP antibody (catalog no. sc-8334; Santa Cruz Biotechnology, CA) or with the anti-HA antibody. The bands were visualized by an HRP-conjugated goat anti-rabbit IgG antibody (catalog no. 4050-05; Southern Biotech, Birmingham, CA) or an HRP-conjugated goat anti-mouse IgG antibody.
RESULTS
sst4Δ cells form spores with abnormal morphology.
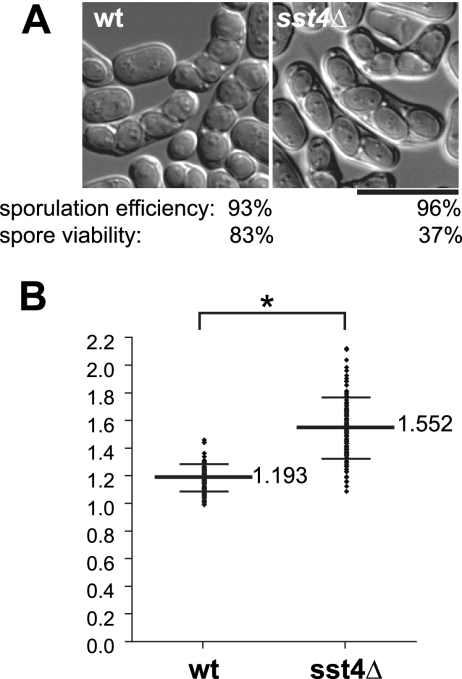
The morphology of spores in sst4Δ zygotic asci was examined. Homothallic wild-type and sst4Δ cells were incubated on sporulation plates for 48 h and observed by DIC microscopy (Fig. 1A). Under these conditions, wild-type cells formed spores predominantly with a round and sharp appearance, and some of them had already been released from asci. Though sst4Δ cells form spores with an efficiency comparable to that of wild-type cells, the majority of spores were oval shaped with fuzzy rims, exhibiting a low-contrast outline by DIC microscopy (Fig. 1A and Table 3). The viability of the sst4Δ spores was considerably lower than that of the wild-type spores (Fig. 1A). Quantification of the ratio of a long axis and a short axis of individual spores showed a significant difference between the wild-type and sst4Δ spores (Fig. 1B). Because newly formed immature prespores in wild-type cells have elongated oval shapes which eventually become spheres (55), it is possible that the shapes of the spores in sst4Δ cells are due to some defect in maturation.
FIG. 1.
Formation of oval-shaped spores in sst4Δ cells. (A) Strains THP17 (wild type [wt]) and MI160 (sst4Δ) were incubated on a sporulation plate for 48 h and examined for morphology of the spores in the zygotic asci. Overall efficiency of spore formation and viability of spores from each strain after ethanol treatment (see Materials and Methods) are indicated. Bar, 10 μm. (B) Quantitative analysis of the shapes of the spores shown in panel A. The ratios of a long axis to a short axis of individual spores are plotted. The long bars and short bars indicate means and standard deviations, respectively (n = 100). *, P < 0.0001.
TABLE 3.
Percentages of mutant asci with different types of sporesa
| Cell type | % Sporulation efficiencyb | % Asci with wild-type-like sporesc | % Asci with sst4Δ-like sporesd | % Asci with aberrant numberse | Description (reference[s]) |
|---|---|---|---|---|---|
| Wild type | 93 | 79 | 16 | 4 | |
| sst4Δ | 96 | 10 | 85 | 5 | VPS27/Hrs homolog, ESCRT-0 |
| hse1Δ | 95 | 9 | 79 | 12 | STAM homolog, ESCRT-0 |
| sst4R193A | 94 | 19 | 75 | 6 | Mutation in the FYVE domain |
| sst4S271D | 95 | 85 | 9 | 6 | Mutation in one UIM domain |
| sst4S316D | 94 | 80 | 11 | 9 | Mutation in one UIM domain |
| sst4S271,316D | 94 | 74 | 21 | 5 | Mutations in two UIM domains |
| hse1S176D | 94 | 78 | 12 | 10 | Mutation in one UIM domain |
| sst4S271,316Dhse1S176D | 97 | 21 | 76 | 3 | Mutations in three UIM domains |
| sst6Δ | 75 | 20 | 60 | 20 | VPS23 homolog, ESCRT-I |
| vps28Δ | 74 | 22 | 54 | 24 | ESCRT-I |
| vps20Δ | 84 | 23 | 54 | 23 | ESCRT-III |
| sst2Δ | 78 | 12 | 55 | 33 | Homolog of mammalian AMSH which binds to the Vps27/STAM complex and ESCRT-III (1, 31) |
| vps32Δ | 64 | 13 | 14 | 73 | ESCRT-III |
| vps4Δ | 63 | 11 | 17 | 72 | AAA-type ATPase which binds to ESCRT-III (6) |
Mutant cells with the indicated genotypes were incubated on SSA plates for 48 h and then observed by DIC microscopy. More than 200 asci were counted for each mutant and classified. For columns 3, 4, and 5, percentages above 70 are indicated by boldface type.
Asci with visible spores, regardless of morphology and number.
Asci with round spores with sharp rims.
Asci with oval-shaped spores with fuzzy rims.
Asci with less than four spores, ignoring whether the spores were round or oval shaped.
sst4Δ cells form forespore membranes with bubbles.
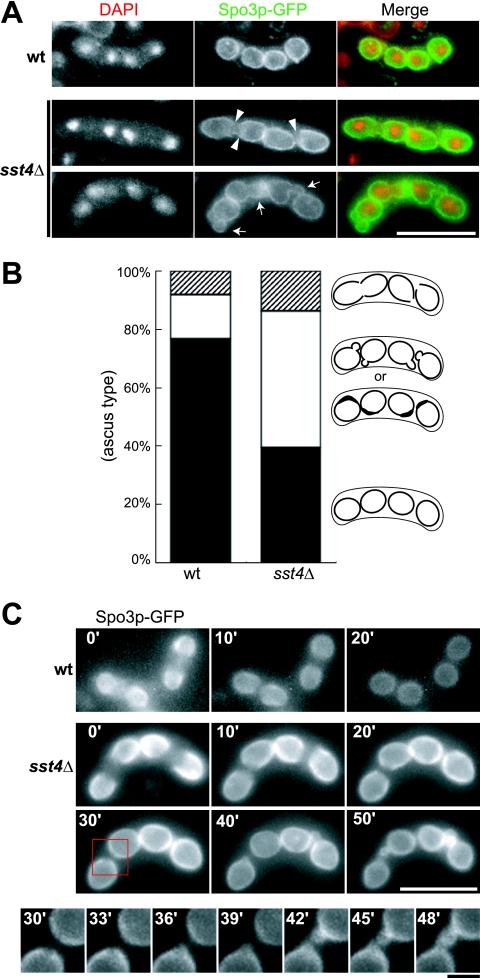
The morphology of forespore membranes in sst4Δ cells was analyzed by fluorescence microscopy with use of the forespore membrane marker Spo3p-GFP (37). After incubation for 10 h on sporulation plates, many of the wild-type cells displayed Spo3p-GFP fluorescence as circles with uniform brightness, representing the forespore membranes that had already closed with each haploid nucleus inside (Fig. 2A). In contrast, although most forespore membranes extended around the nuclei in sst4Δ cells, some of them remained open (Fig. 2A). In addition, many forespore membranes contained bubbles or bright regions, which were rarely seen in the wild-type cells (Fig. 2A). After further incubation in the sporulation medium for 6 h, the number of forespore membranes with bubbles increased, whereas those with open leading edges decreased, suggesting that the bubble formation might be linked to the defect in closure of the leading edges (Fig. 2B). Consistent with this interpretation, the time-lapse experiment revealed that the bubbles emerged from the gaps of unclosed leading edges in sst4Δ cells (Fig. 2C).
FIG. 2.
Fluorescence microscopic analysis of the forespore membrane in sst4Δ zygotic asci. (A) Morphology of the forespore membrane in the wild-type (wt) and the sst4Δ zygotes. Sporulation of wild-type and sst4Δ cells carrying the plasmid pAL(Spo3-GFP) was induced for 10 h on a sporulation plate. The arrowheads and the arrows indicate unclosed leading edges and bubbles, respectively, observed in sst4Δ cells. Bar, 10 μm. (B) Cells shown in panel A were incubated for a further 6 h, and asci were observed to fall into three categories according to the shapes of their forespore membranes: normal (black), bubbles or thick regions (white), and unclosed leading edges (stripes). The percentage in each category is shown. Illustrations for each category are shown on the right. (C) Time-lapse observation of Spo3p-GFP in a wild-type ascus and an sst4Δ ascus. In the wild-type ascus, the forespore membranes closed to form circles. In the sst4Δ ascus, the leading edges of the forespore membranes remained unclosed even after observation for 30 min (30′), and bubbles emerged from the gaps. High-magnification images of the region framed by the red square are shown at the bottom. Strains used were THP17 (wt) and MI160 (sst4Δ). Long bar, 10 μm; short bar, 2 μm.
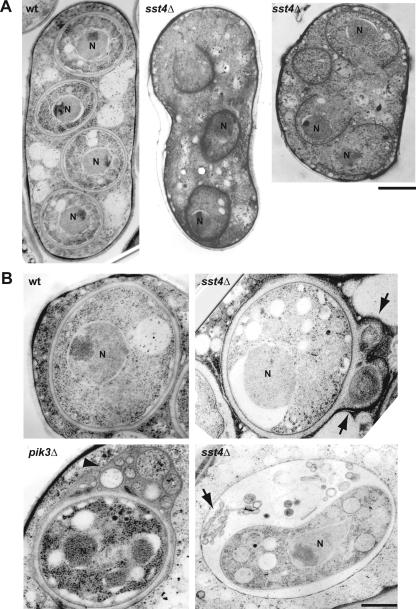
The spore formation defect of sst4Δ cells was also analyzed by electron microscopy. Whereas most spores displayed mature spore walls appearing as white, thick layers in the wild-type cells, the forespore membranes in sst4Δ cells were frequently open (Fig. 3A). Even if the forespore membranes appeared to be closed, they were accompanied by bubbles in the lumen between the inner and outer leaflets of the forespore membranes (Fig. 3B). These results were consistent with the fluorescence microscopic analysis. Bubble formation is one of the defects of pik3Δ cells (Fig. 3B) (41), implying that Sst4p may contribute to the PtdIns 3-kinase pathway during sporulation.
FIG. 3.
Electron microscopic analysis. (A) Extension of the forespore membrane was retarded in sst4Δ cells. Shown are representative images of a wild-type (wt) ascus and two sst4Δ azygotic asci after incubation on sporulation plates for 8 h. Many forespore membranes in sst4Δ asci remained unclosed. Bar, 1 μm. N, nucleus. (B) High-magnification images of spores in a wild-type ascus, two sst4Δ asci, and a pik3Δ azygotic ascus. Bubbles and membranous structures were found in sst4Δ asci (arrows), which were also observed in pik3Δ asci (arrowheads) (41). Strains used were TW747 (wt), MI619 (sst4Δ), and TP3 (pik3Δ). Bar, 0.5 μm.
Sst4p localizes as dots adjacent to the vacuoles and the forespore membranes.
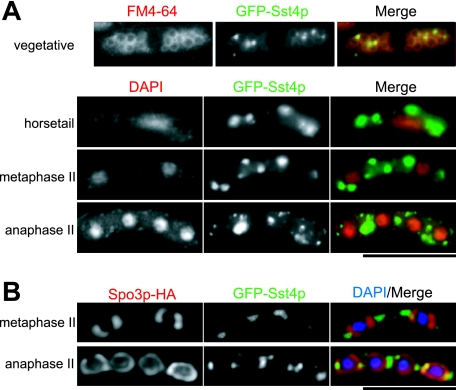
To analyze the localization of Sst4p, we adopted several expression strategies for Sst4p fused with GFP or red fluorescent protein (RFP) either at the N terminus or at the C terminus (see Materials and Methods). All of the plasmids for expression of GFP-tagged Sst4 proteins complemented the defect of sst4Δ, and an sst4+-mRFP5 strain exhibited no defect, indicating that the tagged Sst4 proteins were functional. In vegetative cells, GFP-Sst4p localized as intracellular dots adjacent to the vacuoles visualized by the endocytic membrane dye FM4-64 (Fig. 4A). Although the distribution of the endosomes in S. pombe has not been analyzed precisely, the perivacuolar dot localization of Sst4p was similar to that of Vps27p in S. cerevisiae or Hrs in mammalian cells (7, 26, 45). It is likely that Sst4p localizes at the endosomal membranes, as is the case with its homologs. The dot localization of Sst4p was preserved throughout the process of meiosis and sporulation, although the dots became enlarged in size and reduced in number, as if they were integrated into larger compartments or fused with each other (Fig. 4A). To test whether the enlarged Sst4p-containing dots were integrated into the forespore membrane, we immunostained wild-type cells expressing GFP-Sst4p and Spo3-HA with anti-HA antibody. To exclude false colocalization because of vertical overlapping, single-plane images were reconstituted by the deconvolution method. As shown in Fig. 4B, the GFP-Sst4p signals were localized adjacent to but remained mostly separated from the forespore membranes. All of the other tagged Sst4 proteins behaved very similarly, confirming the results of the experiments with GFP-Sst4p (M. Iida, unpublished). Thus, it is likely that Sst4p does not function directly on the forespore membranes but acts on a membrane compartment adjacent to the forespore membrane.
FIG. 4.
Sst4p localizes as intracellular dots adjacent to the vacuoles and to the forespore membranes. (A) Strain MI160 (sst4Δ) carrying plasmid pREP41(GFP-Sst4p) was grown in EMM medium without thiamine for 12 h, stained with FM4-64, and observed by fluorescence microscopy (vegetative). For sporulation, cells were pregrown on an EMM plate without thiamine for 12 h, moved onto an SSA plate, and further incubated for 8 h. The cells were fixed and stained with DAPI to specify the stages of meiosis. (B) Strain MI392 (sst4Δ) was transformed with plasmids pREP41(GFP-Sst4p) and pAU(Spo3-HA), incubated as described above, and immunostained for the Spo3p-HA protein. Single-plane images obtained by the deconvolution method are shown. Bars, 10 μm.
Localization and function of Sst4p are dependent on PtdIns(3)P.
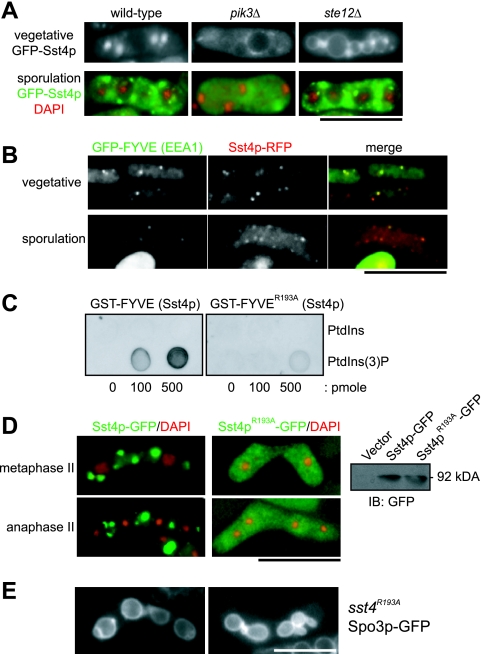
In S. pombe, pik3+ and ste12+ encode PtdIns 3-kinase and PtdIns(3)P 5-kinase, respectively. pik3Δ cells lack both PtdIns(3)P and phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2], whereas ste12Δ cells lack PtdIns(3,5)P2 but preserve PtdIns(3)P (27, 34, 57). The localization of Sst4p in pik3Δ and ste12Δ mutants was analyzed. As shown in Fig. 5A, GFP-Sst4p in pik3Δ cells lost its localization as punctate dots throughout vegetative growth and sporulation, except for some residual signal. In ste12Δ cells, GFP-Sst4p localization looked more diffuse than in wild-type cells perhaps because of a slight signal at the limiting membrane of the enlarged vacuoles; however, the dot localization was preserved, and the dots were located adjacent to the large vacuoles (Fig. 5A). During sporulation, the behavior of GFP-Sst4p in ste12Δ cells as dots was very similar to that in the wild-type cells (Fig. 5A). Therefore, the dot localization of Sst4p depends on PtdIns(3)P but not on PtdIns(3,5)P2. Consistently, Sst4p-RFP colocalized with the FYVE domain of EEA1, a well-characterized PtdIns(3)P-binding domain (20), in vegetative and sporulating cells (Fig. 5B).
FIG. 5.
Localization and function of Sst4p depend on PtdIns(3)P. (A) Sst4p loses its localization in cells lacking PtdIns(3)P. For this experiment, we used diploid strains because pik3Δ and ste12Δ cells are sterile. Strains TW747 (wild type), TP3 (pik3Δ), and YS2 (ste12Δ) were transformed with the plasmid pREP41(GFP-Sst4p) and incubated as described in the legend for Fig. 4A. (B) Sst4p colocalizes with the FYVE domain of EEA1. Strain MI512 (sst4+-mRFP5) was transformed with plasmid pREP41(GFP-FYVEEEA1) (41) and incubated as described above. The cells were fixed with formaldehyde for 3 min before the pictures were taken. (C) The R193A mutation abolishes the binding of the FYVE domain of Sst4p to PtdIns(3)P. The binding of GST-FYVE and GST-FYVER193A was examined by protein-lipid overlay assay. (D) Sst4pR193A completely loses its specific localization. Strain MI160 (sst4Δ) was transformed with plasmid pAL(Sst4p-GFP) or pAL(Sst4pR193A-GFP). The panel on the right shows proper expression of both proteins confirmed by Western blotting. IB, immunoblot. (E) sst4R193A cells form forespore membranes with bubbles. Sporulation of strain MI597 (sst4R193A) carrying plasmid pAL(Spo3p-GFP) was induced for 14 h. Bars, 10 μm.
We introduced a single amino acid substitution (R193A) into the FYVE domain of Sst4p. This arginine residue in the R(R/K)HHCRXCG motif (the residue corresponding to Arg193 of Sst4p is italicized) is well conserved among FYVE domains and has been suggested to be important for the activity of the domain by interacting directly with the phosphate group at the D3 position of PtdIns(3)P (56). The direct binding of the FYVE domain of Sst4p to PtdIns(3)P and the effect of the mutation were tested by a protein-lipid overlay assay (18) with use of bacterially expressed GST-FYVE domains (Fig. 5C). As expected, wild-type GST-FYVE displayed strong binding to PtdIns(3)P, whereas binding of GST-FYVER193A to PtdIns(3)P was reduced at least fivefold relative to that for the wild-type protein (Fig. 5C). In vivo, Sst4pR193A-GFP was stably expressed; however, punctate localization was lost and the protein was dispersed evenly throughout the cell (Fig. 5D). As expected, Sst4pR193A-GFP did not complement the sporulation defect of sst4Δ cells (data not shown).
We constructed an sst4R193A strain and compared its spore morphology with those of wild-type and sst4Δ cells. The sst4R193A cells formed spores with oval-shaped morphology, similar to those of sst4Δ cells (Table 3). The forespore membranes in these cells were frequently accompanied by bubble-like structures (Fig. 5E). These results suggest that Sst4p is a downstream factor of PtdIns 3-kinase during sporulation which is required for formation of the forespore membrane with proper morphology.
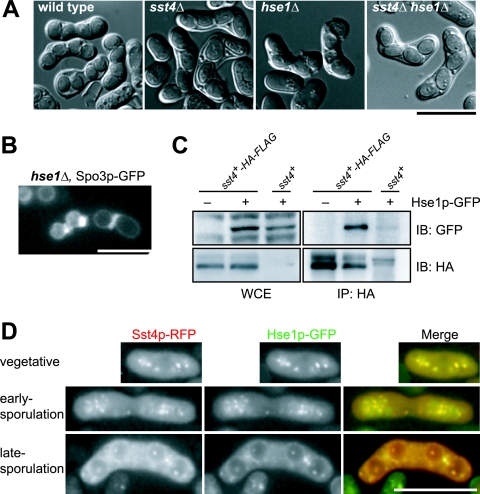
hse1Δ cells show the same defect as that of sst4Δ cells during sporulation.
In a previous screen for the sst genes, we identified genes for another class E vps homolog, sst6/vps23, and a mammalian AMSH homolog, sst2, besides sst4 (42). Both genes were required for proper sorting and processing of the Ub-GFP-CPS protein, classifying them as class E vps genes in S. pombe (23). We analyzed the morphology of spores formed in deletion mutants of these class E vps genes by DIC microscopy (the genes tested are listed in Table 3). All of the mutants exhibited defects in sporulation. However, only the hse1Δ mutant formed predominantly asci with four oval-shaped spores similar to those in sst4Δ cells, suggesting that Hse1p plays a role similar to that of Sst4p (Fig. 6A and Table 3). Other mutants formed asci with less than four spores, though all of the spores were oval shaped when asci harbored four spores (Table 3). These results suggest that many class E genes are required both for maturation of spores and for some additional function, in which Sst4p and Hse1p do not participate, to determine the number of spores formed. Nevertheless, these results suggest that Sst4p and Hse1p share closely related functions during sporulation in S. pombe, as they do in the MVB pathway in S. cerevisiae.
FIG. 6.
Hse1p interacts genetically and physically with Sst4p. (A) hse1Δ and sst4Δ strains exhibit indistinguishable sporulation defects. Sporulation of strains THP17 (wild type), MI160 (sst4Δ), MI301 (hse1Δ), and MI392 (sst4Δ hse1Δ) was induced for 48 h. (B) hse1Δ cells form forespore membranes with bubbles. Sporulation of strain MI301 carrying plasmid pAL(Spo3p-GFP) was induced for 14 h. (C) Hse1p interacts with Sst4p. One milligram of whole-cell extract (WCE) from strain THP17 (sst4+) or MI446 (sst4+-HA-FLAG) carrying plasmid pAL-SK or pAL(Hse1-GFP) was immunoprecipitated (IP) with an anti-HA antibody. Fifty micrograms of the WCE and whole precipitates was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. IB, immunoblot. (D) Hse1p colocalizes with Sst4p. Strain MI512 (sst4+-mRFP5) carrying plasmid pAL(Hse1p-GFP) was analyzed. Bars, 10 μm.
Sst4p and Hse1p form a complex, and their UIM domains are important for sporulation.
Genetic and physical interactions of Sst4p and Hse1p were examined. The sst4Δ hse1Δ double mutant exhibited essentially the same defect in sporulation as the sst4Δ or hse1Δ single mutant. The sst4Δ hse1Δ cells formed oval-shaped spores (Fig. 6A and Table 3), and the forespore membrane was accompanied by bubbles (Fig. 6B). This result suggests that Sst4p and Hse1p function at the same point in the sporulation process. Hse1p-GFP was coimmunoprecipitated with Sst4p-HA-FLAG from the cell lysate when we used an anti-HA antibody, confirming that Sst4p and Hse1p form a complex in vivo (Fig. 6C). Consistent with the coimmunoprecipitation, Hse1p-GFP colocalized with Sst4p-RFP during vegetative growth and sporulation (Fig. 6D).
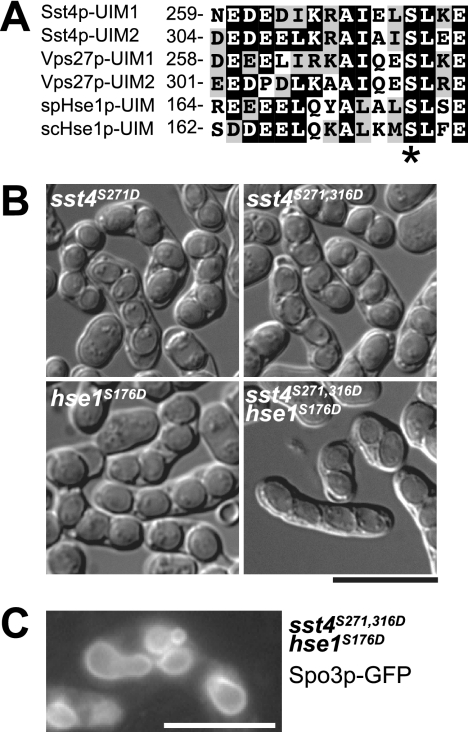
It has been shown that the primary role of the Vps27p/Hse1p complex in S. cerevisiae is to bind to ubiquitinated cargo proteins on the limiting membrane of the endosomes that recruit the ESCRT complexes (26). To test the possibility that binding of the Sst4p/Hse1p complex to ubiquitinated proteins was important for sporulation, we introduced serine-to-aspartate mutations into two UIM domains of Sst4p and a UIM domain of Hse1p (Fig. 7A). This mutation in the conserved amino acid residue has been shown to reduce the ubiquitin-binding activity of the UIM domain in vitro and to be required for proper sorting of endosomal cargo proteins in vivo (54). Interestingly, none of the strains carrying single UIM mutations exhibited defects in sporulation, whereas the sst4S271,316D hse1S176D mutant, in which all three UIMs were mutated, formed sst4Δ hse1Δ-like oval-shaped spores (Fig. 7B and Table 3). Moreover, the forespore membrane in this strain was accompanied by bubbles (Fig. 7C). These results suggest that the UIM domains in the Sst4p/Hse1p complex are important for sporulation and that all three of the UIM domains need not be intact for its function.
FIG. 7.
UIM domains of Sst4p/Hse1p are important for sporulation. (A) Amino acid sequence alignment of UIM domains of Sst4p (S. pombe), Vps27p (S. cerevisiae), spHse1p (S. pombe), and scHse1p (S. cerevisiae). The number to the left of each sequence denotes the amino acid position of the first residue. The conserved serine residues are indicated by an asterisk. (B) Sporulation of UIM domain mutants. Sporulation of strains MI578 (sst4S271D), MI577 (sst4S271,316D), MI545 (hse1S176D), and MI596 (sst4S271,316D hse1S176D) was induced for 48 h. (C) Cells mutated in all three UIM domains form forespore membranes with bubbles. Sporulation of strain MI596 carrying plasmid pAL(Spo3p-GFP) was induced for 14 h. Bars, 10 μm.
DISCUSSION
In this study, we investigated the role of Sst4p, a Vps27p/Hrs homolog, during the sporulation process. sst4Δ cells formed oval-shaped spores with low viability. Fluorescence and electron microscopic analyses revealed that the extension of the forespore membrane was slightly retarded and that bubbles were frequently formed, a defect also observed in pik3Δ cells. Sst4p was localized in a punctate pattern adjacent to the vacuoles and to the forespore membranes, and this localization required both the PtdIns(3)P binding activity of its FYVE domain and the presence of PtdIns(3)P.
Based on these results, we conclude that Sst4p functions downstream of Pik3p during sporulation. We have already shown that two PX domain proteins that are constituents of the retromer complex, Vps5p and Vps17p, also function downstream of Pik3p during sporulation (28). pik3Δ cells show a pleiotropic phenotype in spore membrane formation: formation of small spore membranes because of inefficient forespore membrane extension, disoriented extension of the forespore membrane, failure of closure of the leading edges, and formation of bubbles. Among these, formation of small spore membranes is also found in vps5Δ and vps17Δ mutants; therefore, these proteins may be responsible for the Pik3p-mediated retrograde trafficking to the Golgi apparatus, which, in turn, is required for efficient anterograde membrane flux to the forespore membrane. In contrast, Sst4p may function at a later stage of forespore membrane formation, specifically when the leading edge closes, because the forespore membrane extends but is arrested specifically at this stage in sst4Δ cells. Thus, it is likely that Pik3p and its product, PtdIns(3)P, play multiple roles during sporulation, regulating these downstream effectors with different timings to contribute to the development and closure of the forespore membrane.
We also found that Hse1p functions in cooperation with Sst4p. Class E vps genes are required for sporulation, as all of the mutants tested so far form abnormal spores. Among them, the phenotypes of the hse1Δ mutant were very similar to those of the sst4Δ mutant, forming oval-shaped spores without affecting the spore numbers in each ascus. Hse1p colocalized and was coimmunoprecipitated with Sst4p, suggesting that Hse1p and Sst4p may form a complex. Other mutants often formed asci with less than four spores; however, some of them (sst6/vps23Δ, vps28Δ, vps20Δ, and sst2/AMSHΔ) formed oval-shaped spores when they succeeded in generating four spores. Introduction of the sst4Δ mutation into these mutants gave no additional phenotype (M. Iida, unpublished). These results suggest that the Sst4p/Hse1p complex may function at a specific point, whereas other proteins may have some additional roles that are responsible for regulating spore number. Further studies are necessary for understanding the roles of these additional proteins. That these other vps and sst mutants show more severe defects than sst4Δ, despite having defects in the MVB pathway similar to those of sst4Δ (22), argues against the possibility that the abnormal morphology of the spores in sst4Δ cells is a secondary effect of inefficient bulk degradation of the preexisting proteins.
The UIM domains of Sst4p and Hse1p may be important because the mutations in all three UIM domains (two in Sst4p and one in Hse1p) trigger the formation of oval-shaped spores. We have shown previously that Sst4p is required for sorting of Ub-GFP-CPS into vacuoles via the MVB pathway (23), suggesting the possibility that the Sst4p/Hse1p complex binds to ubiquitinated cargoes in S. pombe. Indeed, we detected some ubiquitinated proteins that coimmunoprecipitated with Sst4p in specific genetic backgrounds, such as hse1Δ, sst6Δ, and sst2Δ (M. Iida, unpublished). In S. cerevisiae, mutations in one of the UIM domains in Vps27p result in impaired sorting of biosynthetic and endocytic cargoes into MVBs (54). This difference in the requirements for the UIM domains in the two yeasts implies that the functions of the Sst4p/Hse1p complex and the Vps27p/Hse1p complex could be somewhat different or that some aspect of the machinery of sorting to the forespore membrane might not be identical to that of the MVB sorting pathway. Nonetheless, our results demonstrate the importance of UIM domains for sporulation, which implies the existence of some ubiquitinated cargo proteins that are sorted to the forespore membranes via the Sst4p/Hse1p-mediated pathway.
sst4Δ cells form oval-shaped, immature spores. Transition of the shapes of the spores from ovals to spheres has been considered to depend on spore wall synthesis (55). Although the involvement of a number of wall enzymes and materials that contribute to spore wall development has been known (2, 17, 24, 29, 30, 36, 59), no particular material responsible for the shaping process has been identified, and little is known about the mechanisms for sorting and deposition of these proteins. For S. cerevisiae, in contrast, it has been shown that at least some enzymes are redirected from the plasma membrane to the spore membrane by endocytosis (33). It is interesting to speculate that Sst4p and Hse1p are involved in the sorting of spore wall enzymes or other proteins required for the normal spore morphology to the forespore membrane.
In sst4Δ cells, bubbles emerged from the gaps of the unclosed leading edges of the forespore membranes, suggesting that bubble formation was the consequence of failure in closing the leading edges. Recently, Itadani et al. reported a protein that was localized in the area following the leading edges, type I myosin light chain calmodulin Cam2p (22), along with type I myosin heavy chain Myo1p, F-actin, and a meiotic protein, Meu14p (35, 40, 43). This result strongly suggests that the closure of the leading edges involves the actin-mediated endocytosis machinery, as is the case with cytokinesis of animal and yeast cells. Recent reports suggest the importance of endosome-centered membrane trafficking for cytokinesis in mammalian cells, especially at the very end when the plasma membrane penetrating into the cytosol finally closes (10, 51, 52). Our results that Sst4p and Hse1p localize at intracellular dots which are presumed to be the endosomes and that these proteins are required for the efficient closure of the leading edges might implicate the contribution of endosomal trafficking to the closure of the forespore membranes. Identification of a forespore membrane-targeted protein(s) sorted by the Sst4p/Hse1p-mediated machinery would provide important information for understanding the mechanisms underlying both forespore membrane development and maturation of spores.
Acknowledgments
We thank Takehiko Yoko-o, Hideaki Ishikawa, and Yoshinori Watanabe for providing the plasmids, Taro Nakamura and Chikashi Shimoda for discussions, and Aaron Neiman for reading the manuscript.
This work was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan (to Y.F.).
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Agromayor, M., and J. Martin-Serrano. 2006. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 281:23083-23091. [DOI] [PubMed] [Google Scholar]
- 2.Arellano, M., H. Cartagena-Lirola, M. A. Nasser Hajibagheri, A. Durán, and M. Henar Valdivieso. 2000. Proper ascospore maturation requires the chs1+ chitin synthase gene in Schizosaccharomyces pombe. Mol. Microbiol. 35:79-89. [DOI] [PubMed] [Google Scholar]
- 3.Asao, H., Y. Sasaki, T. Arita, N. Tanaka, K. Endo, H. Kasai, T. Takeshita, Y. Endo, T. Fujita, and K. Sugamura. 1997. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 272:32785-32791. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M. 2005. A protein's final ESCRT. Traffic 6:2-9. [DOI] [PubMed] [Google Scholar]
- 5.Babst, M., T. K. Sato, L. M. Banta, and S. D. Emr. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bache, K. G., C. Raiborg, A. Mehlum, and H. Stenmark. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278:12513-12521. [DOI] [PubMed] [Google Scholar]
- 8.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 9.Balla, T. 2005. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 118:2093-2104. [DOI] [PubMed] [Google Scholar]
- 10.Baluska, F., D. Menzel, and P. W. Barlow. 2006. Cytokinesis in plant and animal cells: endosomes ‘shut the door.’ Dev. Biol. 294:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Bilodeau, P. S., J. L. Urbanowski, S. C. Winistorfer, and R. C. Piper. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534-539. [DOI] [PubMed] [Google Scholar]
- 12.Bilodeau, P. S., S. C. Winistorfer, W. R. Kearney, A. D. Robertson, and R. C. Piper. 2003. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163:237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowers, K., and T. H. Stevens. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744:438-454. [DOI] [PubMed] [Google Scholar]
- 14.Burd, C. G., and S. D. Emr. 1998. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 2:157-162. [DOI] [PubMed] [Google Scholar]
- 15.Burda, P., S. M. Padilla, S. Sarkar, and S. D. Emr. 2002. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115:3889-3900. [DOI] [PubMed] [Google Scholar]
- 16.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortes, J. C., J. Ishiguro, A. Durán, and J. C. Ribas. 2002. Localization of the (1,3)beta-d-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J. Cell Sci. 115:4081-4096. [DOI] [PubMed] [Google Scholar]
- 18.Dowler, S., G. Kular, and D. R. Alessi. 2002. Protein lipid overlay assay. Sci. STKE 2002:PL6. [DOI] [PubMed] [Google Scholar]
- 19.Egel, R., and M. Egel-Mitani. 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88:127-134. [DOI] [PubMed] [Google Scholar]
- 20.Gaullier, J. M., E. Ronning, D. J. Gillooly, and H. Stenmark. 2000. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J. Biol. Chem. 275:24595-24600. [DOI] [PubMed] [Google Scholar]
- 21.Horazdovsky, B. F., B. A. Davies, M. N. Seaman, S. A. McLaughlin, S. Yoon, and S. D. Emr. 1997. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol. Biol. Cell 8:1529-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itadani, A., T. Nakamura, and C. Shimoda. 2006. Localization of type I myosin and F-actin to the leading edge region of the forespore membrane in Schizosaccharomyces pombe. Cell Struct. Funct. 31:181-195. [DOI] [PubMed] [Google Scholar]
- 23.Iwaki, T., M. Onishi, M. Ikeuchi, A. Kita, R. Sugiura, Y. Giga-Hama, Y. Fukui, and K. Takegawa. 2007. Essential roles of class E Vps proteins for sorting into multivesicular bodies in Schizosaccharomyces pombe. Microbiology 153:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakihara, Y., K. Nabeshima, A. Hirata, and H. Nojima. 2003. Overlapping omt1+ and omt2+ genes are required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 8:547-558. [DOI] [PubMed] [Google Scholar]
- 25.Kashiwazaki, J., T. Nakamura, T. Iwaki, K. Takegawa, and C. Shimoda. 2005. A role for fission yeast Rab GTPase Ypt7p in sporulation. Cell Struct. Funct. 30:43-49. [DOI] [PubMed] [Google Scholar]
- 26.Katzmann, D. J., C. J. Stefan, M. Babst, and S. D. Emr. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura, K., S. Miyake, M. Makuuchi, R. Morita, T. Usui, M. Yoshida, S. Horinouchi, and Y. Fukui. 1995. Phosphatidylinositol-3 kinase in fission yeast: a possible role in stress responses. Biosci. Biotechnol. Biochem. 59:678-682. [DOI] [PubMed] [Google Scholar]
- 28.Koga, T., M. Onishi, Y. Nakamura, A. Hirata, T. Nakamura, C. Shimoda, T. Iwaki, K. Takegawa, and Y. Fukui. 2004. Sorting nexin homologues are targets of phosphatidylinositol 3-phosphate in sporulation of Schizosaccharomyces pombe. Genes Cells 9:561-574. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J., X. Tang, H. Wang, and M. Balasubramanian. 2000. Bgs2p, a 1,3-beta-glucan synthase subunit, is essential for maturation of ascospore wall in Schizosaccharomyces pombe. FEBS Lett. 478:105-108. [DOI] [PubMed] [Google Scholar]
- 30.Martín, V., J. C. Ribas, E. Carnero, A. Durán, and Y. Sánchez. 2000. bgs2+, a sporulation-specific glucan synthase homologue is required for proper ascospore wall maturation in fission yeast. Mol. Microbiol. 38:308-321. [DOI] [PubMed] [Google Scholar]
- 31.McCullough, J., M. J. Clague, and S. Urbé. 2004. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166:487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno, S., A. Klar, and P. Nurse. 1990. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 33.Morishita, M., and J. Engebrecht. 2005. End3p-mediated endocytosis is required for spore wall formation in Saccharomyces cerevisiae. Genetics 170:1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morishita, M., F. Morimoto, K. Kitamura, T. Koga, Y. Fukui, H. Maekawa, I. Yamashita, and C. Shimoda. 2002. Phosphatidylinositol 3-phosphate 5-kinase is required for the cellular response to nutritional starvation and mating pheromone signals in Schizosaccharomyces pombe. Genes Cells 7:199-215. [DOI] [PubMed] [Google Scholar]
- 35.Motegi, F., K. Nakano, C. Kitayama, M. Yamamoto, and I. Mabuchi. 1997. Identification of Myo3, a second type-II myosin heavy chain in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 420:161-166. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, T., H. Abe, A. Hirata, and C. Shimoda. 2004. ADAM family protein Mde10 is essential for development of spore envelopes in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura, T., M. Nakamura-Kubo, A. Hirata, and C. Shimoda. 2001. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding syntaxin-like protein. Mol. Biol. Cell 12:3955-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura-Kubo, M., T. Nakamura, A. Hirata, and C. Shimoda. 2003. The fission yeast spo14+ gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol. Biol. Cell 14:1109-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakase, Y., T. Nakamura, A. Hirata, S. M. Routt, H. B. Skinner, V. A. Bankaitis, and C. Shimoda. 2001. The Schizosaccharomyces pombe spo20+ gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol. Biol. Cell 12:901-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okuzaki, D., W. Satake, A. Hirata, and H. Nojima. 2003. Fission yeast meu14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J. Cell Sci. 116:2721-2735. [DOI] [PubMed] [Google Scholar]
- 41.Onishi, M., T. Koga, R. Morita, Y. Nakamura, T. Nakamura, C. Shimoda, K. Takegawa, A. Hirata, and Y. Fukui. 2003. Role of phosphatidylinositol 3-phosphate in formation of forespore membrane in Schizosaccharomyces pombe. Yeast 20:193-206. [DOI] [PubMed] [Google Scholar]
- 42.Onishi, M., Y. Nakamura, T. Koga, K. Takegawa, and Y. Fukui. 2003. Isolation of suppressor mutants of phosphatidylinositol 3-phosphate 5-kinase deficient cells in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 67:1772-1779. [DOI] [PubMed] [Google Scholar]
- 43.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell Sci. 111:867-876. [DOI] [PubMed] [Google Scholar]
- 44.Pfeffer, S. R. 2001. Membrane transport: retromer to the rescue. Curr. Biol. 11:R109-R111. [DOI] [PubMed] [Google Scholar]
- 45.Piper, R. C., A. A. Cooper, H. Yang, and T. H. Stevens. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446-455. [DOI] [PubMed] [Google Scholar]
- 47.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rieder, S. E., L. M. Banta, K. Köhrer, J. M. McCaffery, and S. D. Emr. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell 7:985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawano, A., and A. Miyawaki. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28:E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schu, P. V., K. Takegawa, M. J. Fry, J. H. Stack, M. D. Waterfield, and S. D. Emr. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260:88-91. [DOI] [PubMed] [Google Scholar]
- 51.Schweitzer, J. K., E. E. Burke, H. V. Goodson, and C. D'Souza-Schorey. 2005. Endocytosis resumes during late mitosis and is required for cytokinesis. J. Biol. Chem. 280:41628-41635. [DOI] [PubMed] [Google Scholar]
- 52.Schweitzer, J. K., and C. D'Souza-Schorey. 2004. Finishing the job: cytoskeletal and membrane events bring cytokinesis to an end. Exp. Cell Res. 295:1-8. [DOI] [PubMed] [Google Scholar]
- 53.Seaman, M. N., J. M. McCaffery, and S. D. Emr. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih, S. C., D. J. Katzmann, J. D. Schnell, M. Sutanto, S. D. Emr, and L. Hicke. 2002. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4:389-393. [DOI] [PubMed] [Google Scholar]
- 55.Shimoda, C. 2004. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 117:389-396. [DOI] [PubMed] [Google Scholar]
- 56.Stahelin, R. V., F. Long, K. Diraviyam, K. S. Bruzik, D. Murray, and W. Cho. 2002. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 277:26379-26388. [DOI] [PubMed] [Google Scholar]
- 57.Takegawa, K., D. B. DeWald, and S. D. Emr. 1995. Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J. Cell Sci. 108:3745-3756. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka, K., T. Yonekawa, Y. Kawasaki, M. Kai, K. Furuya, M. Iwasaki, H. Murakami, M. Yanagida, and H. Okayama. 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20:3459-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tougan, T., Y. Chiba, Y. Kakihara, A. Hirata, and H. Nojima. 2002. Meu10 is required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 7:217-231. [DOI] [PubMed] [Google Scholar]
- 60.Vanhaesebroeck, B., S. J. Leevers, K. Ahmadi, J. Timms, R. Katso, P. C. Driscoll, R. Woscholski, P. J. Parker, and M. D. Waterfield. 2001. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70:535-602. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe, Y., S. Shinozaki-Yabana, Y. Chikashige, Y. Hiraoka, and M. Yamamoto. 1997. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386:187-190. [DOI] [PubMed] [Google Scholar]