Abstract
The Cdc14 gene of Phytophthora infestans is transcribed specifically during sporulation, with no mRNA detectable in vegetative hyphae, and is required for sporangium development. To unravel the mechanisms regulating its transcription, mutated Cdc14 promoters plus chimeras of selected Cdc14 sequences and a minimal promoter were tested in stable transformants. This revealed that a tandem repeat of three copies of the motif CTYAAC, located between 67 and 90 nucleotides (nt) upstream of the major transcription start site, is sufficient to determine sporulation-specific expression. All three repeats need to be present for activity, suggesting that they bind a transcription factor through a cooperative mechanism. Electrophoretic mobility shift assays indicated that the CTYAAC repeats are specifically bound by a protein in nuclear extracts. Evidence was also obtained for a second region within the promoter that activates Cdc14 transcription during sporulation which does not involve those repeats. The CTYAAC motif also affects the specificity of transcription initiation. Wild-type Cdc14 is transcribed from a major start site and minor site(s) located about 100 nt upstream of the major site. However, stepwise mutations through the CTYAAC triad caused a graded shift to the upstream sites, as did mutating bases surrounding the major start site; transcripts initiated from the upstream site remained sporulation specific. Replacing the Cdc14 initiation region with the Inr-like region of the constitutive Piexo1 gene had no apparent effect on the pattern of transcription. Therefore, this study reports the first motif determining sporulation-induced transcription in oomycetes and helps define oomycete core promoters.
Spores enable the spread of many important pathogens on a continental or global scale (4, 37). One of the most significant plant pathogens is the oomycete Phytophthora infestans, the agent of potato late blight, which is responsible for billions of dollars of losses worldwide each year (11). The main inoculum for late blight are asexual sporangia, which form upon the termini of specialized hyphae called sporangiophores. Sporangia are easily spread by wind or water to new potential host plants, where they can germinate directly or by releasing zoospores (21). Various environmental and physiological factors are known to favor sporulation in Phytophthora, including nutrient depletion, high humidity, high oxygen, and low carbon dioxide (34). However, little is known at the molecular level of how such signals are translated into the cellular responses that cause sporulation. Studies of genes induced during sporulation, including analyses of promoter motifs and transcription factors responsible for stage-specific expression, will advance our understanding of spore development and may lead to new ways to control disease.
Recent advances have begun to shed light on molecular aspects of Phytophthora spore biology, including the identification of genes induced at stages of development and the use of silencing to test function (1, 3, 14, 26, 28, 29, 41). One such gene is the protein phosphatase Cdc14. Orthologs play a range of roles that include regulating exit from mitosis in Saccharomyces cerevisiae (20), septation in Schizosaccharomyces pombe (13), cytokinesis in Caenorhabditis elegans (15), and the centrosome cycle in humans (27). Cdc14 of P. infestans complements a cdc14ts defect in budding yeast, indicating its potential role in regulating mitosis (1). However, while its nonoomycete orthologs are transcribed throughout the life cycle, Cdc14 of P. infestans is expressed only during sporulation. Its transcripts appear early during sporulation, persist in sporangia and zoospores, and disappear soon after zoospore cysts germinate. Silencing of Cdc14 blocks sporulation, indicating its essential role in spore development (1, 43).
While insight into the role of Cdc14 in Phytophthora is now available, little is known about the mechanisms regulating its expression. Moreover, in general very little information is available about the basic transcriptional machinery of oomycetes. Functional studies demonstrate that promoters from most nonoomycetes, including true fungi, have little activity in Phytophthora (23). This is not surprising, since oomycetes reside on a branch of the eukaryotic tree distant from that of traditional model systems, close to brown algae and diatoms (2). Phytophthora promoters appear to lack a TATA box; however, an Inr-like motif has been reported in some promoters (31). The size of promoters appears to be modest (<500 nucleotides [nt]), based on the small average size of intergenic regions in Phytophthora genomes. Based on our understanding of transcriptional regulation in other eukaryotes (35, 44), it is expected that the expression of Cdc14 in P. infestans is regulated by the interplay of both constitutive and developmentally controlled regulators that bind the core promoter and distal sites.
With goals of improving our general understanding of both oomycete transcription and how sporulation-specific expression is regulated, here we describe the structure and activity of the Cdc14 promoter from P. infestans. Functional analyses of deleted, mutated, and chimeric promoters indicated that at least two independent and apparently redundant mechanisms activate transcription during sporulation. Detailed analyses of one of these identified a putative transcription factor binding site comprised of tandem repeats of a 6-nt motif that is conserved in related species. Complex interactions between this site, the core promoter, and others regulate the specificity of initiation.
MATERIALS AND METHODS
Growth and manipulation of P. infestans.
Isolate 1306 was grown on rye A agar at 18°C. Sporangia and nonsporulating hyphae were obtained as described elsewhere (23). In brief, sporangia were released from 8- to 10-day cultures by flooding plates with water and rubbing with a glass rod. Nonsporulating hyphae were from submerged cultures of clarified rye broth inoculated with 104 sporangia/ml. Stable transformants were obtained from isolate 1306 using the protoplast method and stained histochemically for the activity of β-glucuronidase (GUS) (22).
Plasmid construction.
Reporter constructs are based on pOGUS (8), which contains a promoterless GUS gene and npt for G418 selection. Unless specified otherwise, promoter fragments were ligated into ApaI and ClaI sites upstream of GUS. Inserts generated by PCR were obtained using Platinum Taq DNA polymerase high-fidelity (Invitrogen, Carlsbad, CA) and the primers listed in Table 1. The integrity of cloned sequences was confirmed by sequencing. Most plasmids described in the following paragraphs were then transformed into P. infestans, although some were only used as templates for constructing other plasmids.
TABLE 1.
Oligonucleotides used in this study
| Purpose and primer name | Sequence (5′ to 3′) |
|---|---|
| Employed for cloning | |
| 105f | TATAGGGCCCCGCATTCTTCGACTTCTT |
| 260f | TATAGGGCCCACGCACTATTTTGGGTTTC |
| 67BamHI | CAGTGGATCCGCACCTTTCGACCGCATCC |
| 67f | TATAGGGCCCGCACCTTTCGACCGCATCC |
| 75f | TATAGGGCCCAGCCTCAACGCACCCTTTC |
| 81f | TATAGGGCCCCTCAACAGCCTCAACGCACC |
| 868f | TATAGGGCCCTTTCTACCTTCTGCCGAG |
| 868r | GGTGGTATCGATGCTGAAGAGATGGAGGTG |
| 9067f | CGTTGAGGCTGTTGAGACGGTTAAGGGGCC |
| 9067r | CCTTAACCGTCTCAACAGCCTCAACGGGCC |
| 90f | TATAGGGCCCCTTAACCGTCTCAACAGCCT |
| 90BamHI | CAGTGGATCCAAGTCGAAGAATGCGAGAGT |
| CHP8F | CCTTAACCGTCTCAACAGAAGACCAGGGCC |
| CHP8R | CTGGTCTTCTGTTGAGACGGTTAAGGGGCC |
| M1f | CTCGGTTGAATATAGACCATGGTCCCGTGATCCAG |
| M1M6M7f | AGACCACTAAGACCATACAAGTTCGACCGCATCCC |
| M1M6M7r | TGCGGTCGAACCTTGTATGGTCTTAGTGGTCACGG |
| M1r | TCACGGGACCATGGTCTATATTCAACCGAGGCTGC |
| M2f | CCGTGATCCATGTCGGACAGCTCGCATTCTTCG |
| M2r | AGAATGCGAGCTGTCCGACATGGATCACGGGACCC |
| M3f | TGATTCACTCAGATACGGAGTCGACTTCTTAACC |
| M3r | AAGAAGTCGACTCCGTATCTAGTGAATCACTGG |
| M4f | CTCGCATTCTGATCAGGAGGAACCGTCTCAACAGC |
| M4r | TGAGACGGTTCCTCCTGATCAGAATGCGAGAGTGA |
| M5f | GGCTGTTGAGCATTGGAAGAAGTCGAAGAAT |
| M5r | TCGACTTCTTCCAATGCTCAACAGCCTCAAC |
| M6f | TCTTAACCGTAGACCACTAATCAACGCACCTTTCG |
| M6r | GGTGCGTTGATTAGTGGTCTAAGAAGTCG |
| M7f | TCAACAGCCTACCATACAAGTTCGACCGCATCCC |
| M7r | TGCGGTCGAACTTGTATGGTAGGCTGTTGAGACGG |
| M8r | CGTTGAGGCTTGGTCTACGTGGCCTAAGTCG |
| BLOCK1F | GGTTGAAGCGAGACCAGGGTCCCGTGATCCA |
| BLOCK1R | CACGGGACCCTGGTCTCGCTTCAACCGAGGC |
| BLOCK2F | TCTTCGACTTAGGCCACGTCTCAACAGCCTC |
| BLOCK2R | TGTTGAGACGTGGCCTAAGTCGAAGAATGCG |
| BLOCK3F | TAGGCCACGTAGACCAAGCCTCAACGCACGT |
| BLOCK3R | CGTTGAGGCTTGGTCTACGTGGCCTAAGTCG |
| BLOCK4F | TAGACCAAGCAGACCAGCACCTTTCGACCGC |
| BLOCK4R | CGAAAGGTCGTGGTCTGCTTGGTCTACGTGG |
| TSSf1 | TAAACCCCCTCTCTCATTTCCGCATTTGCTACCAGGCACCCCGTT |
| TSSf2 | TAAACCACCCGACTCGATGGGGACTATATTTACCAGGCACCCCGTT |
| TSSr1 | GGTGCCTGGTAGCAAATGCGGAAATGAGAGAGGGGGTGGTTTAAAGGT |
| TSSr2 | GGTGCCTGGTAAATATAGTCCCCATCGAGTCGGGTGGTTTAAAGGT |
| Used for EMSAs | |
| WT-S | ACTTCTTAACCGTCTCAACAGCCTCAACGCACC |
| WT-AS | GGTGCGTTGAGGCTGTTGAGACGGTTAAGAAGT |
| MUT-S | ACTTAGGCCACGTAGACCAAGCAGACCAGCACC |
| MUT-AS | GGTGCTGGTCTGCTTGGTCTACGTCGTGGCCTAAGT |
| NSP-S | CGAGGTCAGAGCGGACGAGACTCGAGGGTGGTAA |
| NSP-AS | TTACCACCCTCGAGTCTCGTCCGCTCTGACCTCG |
The 868-nt intact Cdc14 promoter was amplified using primers 868f and 868r and cloned into pOGUS to form plasmid p868; primer 868r represents the 3′ end of all promoter fragments and matches the +71 to +88 region of Cdc14, with +1 representing the transcription start site (TSS). To clone promoter fragments deleted at their 5′ ends, the 868-nt promoter was either digested with restriction sites within the promoter (HincII, BclI, or XbaI) followed by blunt ending, ClaI digestion, and ligation into suitably prepared pOGUS, or truncated by PCR. In the case of the latter, truncated promoters were generated using a common primer, 868r, along with primers 260f, 105f, 90f, 81f, 75f, and 67f. Internal deletions of the −90 to −67 region involved combining PCR products generated using primer pairs 868f/90BamHI and 868r/67BamHI, which were digested with BamHI, ligated, amplified using 868f and 868r, digested with ApaI and ClaI, and cloned into pOGUS.
Site-directed mutagenesis involved a combined PCR method (40) in which A was changed to C, C to A, G to T, and T to G. To mutate block M1, for example, PCR was performed using primer pairs 868f/M1r and M1f/868r, and then the two reaction mixtures were combined and amplified with 868f and 868r, digested with ApaI and ClaI, and cloned into pOGUS. This strategy was also used to mutate blocks M2 to M7 by using primer pairs 868f/M2r and M2f/868r, 868f/M3r and 868r/M3f, 868f/M4r and 868r/M4f, 868f/M5r and 868r/M5f, 868f/M6r and 868r/M6f, and 868f/M7r and 868r/M7f, respectively. Double and triple mutations were generated using mutated promoters as template. A similar strategy was used to mutate the four CTYAAC motifs, using a succession of amplifications that involved first performing two PCRs with primer pairs BLOCK1F/868r and BLOCK1R/260f, followed by amplification with 260f/868r; then reactions with BLOCK2F/868r and BLOCK2R/260f, followed by amplification with 260f/868r; then BLOCK3F/868r and BLOCK3R/260f reactions, followed by a 260f/868r reaction; and then BLOCK1F/868r and BLOCK1R/260f reactions, followed by a 260f/868r reaction. The region encompassing the TSS (nt −4 to +13) was mutated using primer pairs 105f/TSSr2 and TSSf2/868r, yielding pInrΔ-4/+13. To swap the Inr-like region of the Cdc14 and Piexo1 promoters, PCR was performed using primer pairs TSSf1/868r and TSSr1/105f, yielding pInrSwap. The chimeric promoter p24Cdc::74NIF was generated by annealing oligonucleotides 9067f and 9067r and cloning the resulting double-stranded DNA via ApaI-compatible overhangs into ApaI-digested p74NIFS, a derivative of pOGUS containing a 74-nt minimal promoter from the NIFS gene. p17Cdc::74NIF was made by annealing oligonucleotides CHP8F and CHP8R, which were ligated into pOGUS.
RNA analysis.
RNA was extracted and blotted as described previously (23) using probes for the full open reading frames of Cdc14 and GUS. Most analyses involved 1.2% agarose gels electrophoresed until the bromophenol blue tracking dye had moved about 9 cm, with higher-resolution separations employing 1.0% gels run until the dye moved at least 12 cm. Signals were detected by phosphorimager analysis. 5′-rapid amplification of cDNA ends (RACE) was performed with a kit from Invitrogen using reverse transcription at 50°C. Products were cloned in pGEMT-Easy (Promega, Madison, WI) and sequenced.
Promoter alignments.
The sequence of the P. infestans Cdc14 promoter was obtained from a bacterial artificial chromosome clone. Phytophthora ramorum, Phytophthora sojae, and Phytophthora capsici sequences were obtained from the U.S. Department of Energy-Joint Genome Institute (http://www.jgi.doe.gov). Alignments were performed using DIALIGN and default parameters.
Nuclear extracts.
The nuclear extracts were prepared using sporangia from 56 9-day-old, 150-mm agar cultures. Sporangia were resuspended in buffer A [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) pH 7.0, 10 mM MgCl2, 25% (vol/vol) glycerol, 10 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride (PMSF)]. Homogenization was performed on ice using 0.5-mm glass beads in a Bead-Beater (Biospec Products, Bartlesville, OK), employing three strokes of 30 s, each separated by a 2-min cooling period. Subsequent steps were performed at 4°C. An equal volume of buffer A containing 1% Triton X-100 was then added and incubated for 10 min. Nuclei were pelleted at 8,000 × g for 5 min, washed twice with 25 ml buffer A, and resuspended in a 1.5× pellet volume of buffer B (10% glycerol, 15 mM HEPES, pH 7.9, 0.5 M KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol [DTT], 1% Triton X-100) to which was added 0.5 mM PMSF, a protease inhibitor cocktail, 4.75 volumes of water, and 1.25 volumes of 3 M KCl (dropwise), and then the sample was stirred gently for 30 min. After centrifugation for 30 min at 16,000 × g, the supernatant was dialyzed against buffer C (15 mM HEPES pH 7.9, 100 mM KCl, 1 mM EDTA, 2 mM DTT, 0.1 mM PMSF, and 15% glycerol), clarified by centrifugation at 16,000 × g for 30 min, and applied to a column containing 1 ml of heparin-agarose (Amersham Biosciences, Piscataway, NJ) equilibrated with buffer C. The column was washed with buffer C, and elution was performed in the same buffer containing 0.3 M KCl.
Mobility shift assays.
Double-stranded probes were generated by annealing complementary oligonucleotides, purified by nondenaturing polyacrylamide gel electrophoresis, and end labeled with [γ-32P]ATP using T4 polynucleotide kinase. These included sense- and antisense-specific oligonucleotides (WT-S and WT-AS) (Table 1), those with mutated CTYAAC motifs (MUT-S and MUT-AS), and nonspecific oligonucleotides based on the NIFC1 promoter (NS-S and NS-AS). A typical binding reaction mixture contained 1.6 ng of labeled DNA, 1 μg poly(dI-dC), 15 mM HEPES pH 7.9, 25 mM MgCl2, 100 mM KCl, 15% glycerol, 1 mM DTT, and 5 μg of nuclear protein in 15 μl. For competition assays, binding reaction mixtures were preincubated with unlabeled fragment for 30 min on ice and then incubated for a further 30 min with labeled probe on ice. Samples were electrophoresed on 4.5% acrylamide in 0.5× Tris-borate-EDTA for 3 h at room temperature, and the gel was dried and analyzed with a phosphorimager.
RESULTS
Cdc14 has major and minor transcription start sites.
Cdc14 was described previously as encoding a 1.5-kb sporulation-specific transcript (1). Based on 5′-RACE analysis, its 5′ terminus was mapped to a site, designated as +1, located 89 nt upstream of the start codon. This site in which transcription begins, TCAGA+1TC, is similar to the metazoan Initiator (Inr) consensus sequence, YYANWYY (35).
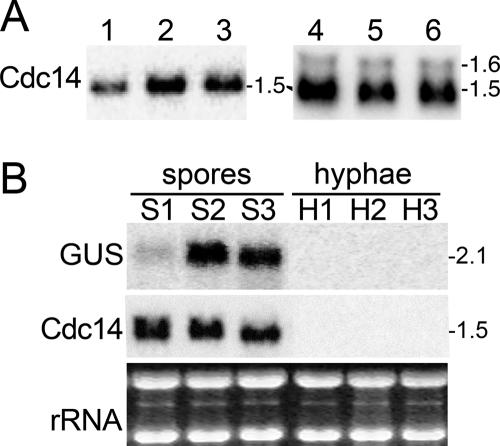
Subsequent studies using higher-resolution gels showed that Cdc14 also encodes a slightly larger RNA of about 1.6 kb (Fig. 1A); the presence of the minor transcript is noted here to aid in the interpretation of promoter mutation studies described later, some of which shift initiation from the major to minor site(s). Phosphorimager analysis indicated that the ratio of major to minor transcripts is typically greater than 20:1. To map these minor start sites, 5′-RACE was performed using primers just upstream of the major start site. Based on sequencing 10 RACE clones, typical 5′ termini of these minor transcripts were at −92, −101, and −109. Due to the absence of ATG motifs in the 200 nt upstream of the +1 position, all major and minor transcripts should encode the same protein.
FIG. 1.
Transcription from the Cdc14 promoter of P. infestans. (A) Expression of native Cdc14. RNA was electrophoresed under standard (1.2% agarose; lanes 1 to 3) or extended resolution (1.0% agarose; lanes 4 to 6) conditions, blotted, and hybridized with a probe for Cdc14. Using the latter parameters, a band of about 1.6 kb in addition to the predominant 1.5-kb band can be discerned. The band(s) detected is from the native Cdc14 gene, even though the strains portrayed are transformants containing various promoters fused to GUS; similar patterns are detected in the wild-type progenitor strain 1306 (not shown). (B) Expression of the GUS transgene from the Cdc14 promoter. RNA from three independent transformants containing plasmid p868 was electrophoresed in 1.2% agarose and hybridized with a probe for GUS or the Cdc14 open reading frame. Samples are from sporangia (spores; S1, S2, and S3) and nonsporulating hyphae (H1, H2, and H3). An ethidium bromide-stained image is shown as a loading control. Due to position effects or copy number variation, lower activity is displayed by the transformant in lane S1.
Strategy for use of the GUS reporter system for promoter analysis.
As will be detailed later, altered promoters were tested in stable transformants of P. infestans to identify sequences within Cdc14 responsible for its regulation. This employed plasmids containing wild-type, chimeric, or mutagenized promoters driving the GUS gene, in addition to the nptII marker for resistance to G418. We reported previously that GUS fused to an 868-nt Cdc14 promoter exhibited sporulation-specific expression based on histochemical staining (1).
During the present study, it became evident that histochemical staining was not entirely reliable for assessing promoter function due to limitations of the Phytophthora transformation system. Since neither the site of integration nor transgene copy number can be controlled, position effects are significant (22). Therefore, the intensity of staining varies widely between transformants, with some exhibiting no expression. For example, of 698 transformants generated using 17 “functional” promoter plasmids, the fraction of GUS-positive transformants ranged from 21 to 93%, averaging 52%; in most cases, experiment-to-experiment variation in the fraction of transformants showing GUS staining from the same promoter plasmid was not very different from that observed between plasmids. Moreover, RNA blot analyses of transformants revealed cases where the GUS transcript was too large to have been initiated within Cdc14 sequences; in such cases, expression is likely due to cryptic promoters in the plasmid backbone, chromosomal DNA flanking the integration site, or vector rearrangement.
Consequently, histochemical staining was used only for preliminary screenings of transformants for promoter activity, with final assessments derived from blot analyses of RNA from sporangia and nonsporulating hyphae. For example, of 51 transformants obtained using an 868-nt Cdc14 promoter fused to GUS (plasmid p868), 18 exhibited activity based on histochemical staining. Illustrated in Fig. 1B is an RNA blot of three of these GUS-positive strains; each produces a sporulation-specific transcript of 2.1 kb, which is consistent with initiation at the normal site within Cdc14 sequences (i.e., the major +1 start site) (Fig. 2). This indicates that the 868-nt region contains a functional promoter. Conversely, in cases where an altered promoter fragment seemed to lack function based on GUS staining, RNA blot analysis was performed on multiple transformants before concluding that promoter activity was absent. RNA blots from transformants lacking the GUS transcript were also hybridized with a probe for the Cdc14 open reading frame to eliminate the possibilities that a Cdc14 binding transcription factor had been titrated out due to high transgene copy number or that transformants were otherwise incapable of expressing sporulation-induced promoters.
FIG. 2.
Alignment of Cdc14 promoters from four species of Phytophthora. Sequences are from P. infestans, P. sojae, P. ramorum, and P. capsici (top to bottom), are numbered with respect to the major transcription start site in P. infestans (bent arrow at + 1), and terminate at the ATG start codon. M1 to M7 represent the conserved blocks described in Results, and triangles are drawn below the four CTYAAC motifs. The original alignments were performed using 750-nt promoters, but to save space only the more conserved regions (from −155 to +91 in P. infestans) are shown.
Interspecific promoter comparisons reveal conserved regions.
An alignment of 750 nt of Cdc14 promoter sequences from P. capsici, P. infestans, P. ramorum, and P. sojae revealed several zones of conservation. Most are within 155 nt of the TSS of the P. infestans gene, as shown in Fig. 2. These include the area flanking the TSS and multiple regions centered about 100 nt upstream of the TSS, which are labeled as blocks M1 to M7; as will be described later, these represent regions subjected to site-directed mutagenesis and are not proposed to delimit distinct functional motifs. Interestingly, four copies of a sequence having a consensus in P. infestans of CTYAAC are in the conserved regions, once within M1 and three times in the interval spanning M4 to M7. This consensus is mostly conserved in the other Phytophthora species and lacks a significant hit to known eukaryotic transcription factor binding sites in databases such as TRANSFAC and JASPAR. Data to be presented later indicate that sporulation-specific expression is conferred by a combination of three of these 6-nt elements, which also contribute to the specificity of transcription initiation.
Mutating individual conserved blocks does not abolish sporulation-specific activity.
To test whether they overlap transcription factor binding sites, blocks M1 to M7 were mutated within the 868-nt promoter and evaluated in stable transformants by GUS staining followed by RNA blot analysis. As illustrated in Fig. 3A, this entailed using site-directed mutagenesis to alter blocks individually (M2, M3, M4, and M5) or in combination (M1 with M6 and M1 with M6 and M7). None of these changes prevented sporulation-specific expression. For example, of 46 transformants obtained with p868mM4, which contains mutations in block M4, 12 exhibited GUS histochemical staining. When four were tested in RNA blot analysis, all displayed a sporulation-specific GUS transcript of approximately 2.1 kb, as illustrated for two representative transformants in Fig. 3B. Similar results were obtained for the other plasmids (Fig. 3A and B). Such findings indicate that binding sites for the relevant transcription factors are outside the blocks or that redundancy exists between blocks.
FIG. 3.
Site-directed mutagenesis of conserved blocks in the Cdc14 promoter. (A) Schematic of promoter-GUS fusion, with the M1 to M7 and TSS blocks indicated by rectangles. Solid and open rectangles represent native and mutagenized sequences, respectively, and the bent arrow indicates position +1. To save space, unmutagenized 5′ regions of the rest of the 868-nt promoters are not shown. In pInrSwap, the TSS block was replaced by an equivalent region from Piexo1. All eight plasmids confer sporulation-induced expression in transformants. (B) RNA blots obtained from 1.2% agarose gels were hybridized with a probe for GUS, using RNA from sporangia (S1 and S2) or hyphae (H1 and H2) of two representative transformants containing each plasmid. Ethidium bromide-stained images are shown as loading controls.
Whether the conserved block surrounding the TSS (−4 to +13) contributes to sporulation specificity was also assessed. This likely represents the core promoter, since it contains a sequence resembling the metazoan Inr element, although it differs from the oomycete Inr-containing consensus proposed previously (GCTCATTYYNCAWTTTNYY [31]). Because of its potential divergence from the oomycete Inr consensus, and since some core promoters are known to contribute to developmental regulation (10, 24), testing its potential contribution to sporulation-specific transcription was required. Therefore, the −4 to +13 interval was exchanged with the TSS region of the Piexo1 gene of P. infestans, which is not sporulation specific and has <50% sequence identity with the Cdc14 TSS region (31). The resulting plasmid, pInrSwap, resulted in the normal pattern of sporulation-specific expression of a 2.1-kb band (Fig. 3). Therefore, sequences surrounding the Cdc14 TSS are not specific for sporulation-specific transcription.
A 24-nt region containing three CTYAAC motifs determines sporulation-specific expression.
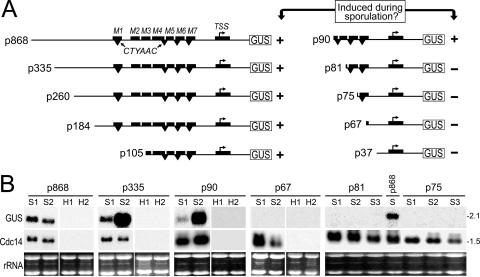
In parallel with mutagenesis of the blocks described above, 5′ deletion analysis was performed and indicated that a determinant of sporulation-specific transcription resides between −90 and −67 (Fig. 4). This is because plasmids p335, p260, p184, p105, and p90 (containing Cdc14 sequences from the ATG codon to 335, 260, 184, 105, and 90 nt upstream of the TSS, respectively) exhibited sporulation-specific expression based on histochemical staining and RNA blot analysis, while p67 and p37 lacked activity. In the case of p90, for example, four of five transformants examined by RNA blot analysis exhibited a sporulation-specific 2.1-kb transcript (as illustrated for two representative transformants in Fig. 4B). In contrast, for p67 no expression was detected based on analyzing RNA from 12 transformants; data from two representative strains are shown in Fig. 4B.
FIG. 4.
5′ deletion analysis of the Cdc14 promoter. (A) Schematic of promoter constructs, with M1 to M7 and TSS blocks indicated by rectangles above the center line and CTYAAC motifs represented by inverted triangles below the line. Plasmid names indicate the size of the promoter upstream of the TSS (bent arrow; position +1), i.e., p335 contains DNA up to nucleotide −335. Whether plasmids confer sporulation-induced expression is indicated by plus or minus signs. (B) RNA from sporangia (S) or hyphae (H) of representative transformants containing each plasmid, electrophoresed on 1.2% gels. Filters were probed for GUS, stripped, and then hybridized with the Cdc14 open reading frame as a control. Hyphae from p81- and p75-containing transformants were also negative for GUS RNA (not shown). Ethidium bromide-stained images are shown as controls.
Inspection of the apparently critical 24-nt interval between −90 and −67 revealed that it contained three of the CTYAAC repeats described earlier. Finer 5′ deletions were therefore constructed to test whether one, two, or all three repeats were required. Plasmids p75 and p81, which include one and two CTYAAC repeats, respectively, did not display sporulation-specific activity (Fig. 4). This suggests that all three repeats are important, possibly due to cooperative binding of a transcription factor.
A repeat-containing region confers spore-specific expression to a minimal promoter.
The 24-nt region containing the three CTYAAC motifs was placed upstream of a minimal promoter to further test its role (Fig. 5). This employed a 74-nt minimal promoter from the NIFS gene of P. infestans, which was developed by deleting sequences upstream of its TSS. As illustrated for representative transformants in Fig. 5B, the 74-nt promoter lacks expression in spores or hyphae (p74Nif). A transformant expressing GUS from a fully functional 363-nt NIFS promoter is shown as a control (p363Nif). In contrast, a chimera of the minimal promoter and the −90 to −67 region of Cdc14 exhibited sporulation-specific expression (p24Cdc::74Nif) (Fig. 5C). This confirms the presence of a sporulation-specific determinant within the 24-nt region.
FIG. 5.
Use of a chimeric promoter to prove function of the −67 to −90 region. (A) Schematic of plasmids, showing the TSS (rectangle) from the NIFS gene and CTYAAC motifs (inverted triangles, with filled and open triangles representing wild-type and mutagenized sequences). Whether plasmids confer sporulation-induced expression is indicated by plus or minus signs. (B) Blot of RNA from sporangia (S) or hyphae (H) of representative transformants containing p74Nif, performed under high-resolution (1.0% agarose) conditions. Filters were hybridized with a probe for GUS, stripped, and hybridized with the Cdc14 open reading frame as a control. The ethidium bromide-stained image is shown as a control. (C) The same experiment as shown in panel B, except that transformants contained p24Cdc::74Nif. The promoter in p17Cdc::74Nif showed no activity based on blot analysis of RNA from hyphae and sporangia (not shown).
Tests of a modification of this chimeric promoter suggested that all three CTYAAC motifs within the 24-nt region are required for sporulation-specific expression. This involved site-directed mutagenesis of the −67 to −74 region to remove the 3′ motif while maintaining the distance of the other two motifs from the 24-nt region to the TSS. This eliminated sporulation-specific expression (p17Cdc::74Nif) (Fig. 5A). For example, no evidence for appropriate expression was revealed from histochemical staining of 109 transformants and RNA blot analysis of selected transformants (not shown). Combined with the earlier observation that 5′ deletion constructs having only the second and third motif (p81 [Fig. 4]) or third motif (p75) lack activity, it follows that the region spanning all three CTYAAC motifs is needed for function.
A nuclear protein binds the CTYAAC repeat region.
Electrophoretic mobility shift assays (EMSAs) identified a binding activity specific to the CTYAAC motif. Using a radiolabeled double-stranded probe centered on the three repeats, corresponding to bases −94 to −62, a shifted band was detected (Fig. 6). Binding was competed by the unlabeled probe (“self”), but not by a DNA fragment derived from the promoter of another gene (nonself) or a version of the −94 to −62 region in which CTYAAC motifs were altered (mutated self).
FIG. 6.
EMSA using labeled probe representing the −94 to −62 region of Cdc14 and nuclear protein from sporangia. Labels denote a specific protein-DNA complex (SP), nonspecific complex (NS), and free probe (FP). Lane 1, no extract; lane 2, extract and probe; lanes 3 to 14, reaction mixtures with increasing amounts of unlabeled competitor DNA (1×, 10×, 100×, 200×). Competitors are the DNA used as the labeled probe (self), DNA derived from another gene (nonself), and the −94 to −62 region in which the CTYAAC motifs were changed to other bases (mutant self).
Evidence for an independent sporulation-inducing motif.
While the analyses of 5′ deletion and chimeric promoter plasmids described above indicated that the 24-nt region between −90 and −67 is sufficient for conferring expression during sporulation, in combination with a core promoter, it should be remembered that plasmids in which portions of that interval were mutated still resulted in expression (p868mM4, p868mM5, p868mM1M6, and p868mM1M6M7) (Fig. 3). This suggests that a separate region can independently direct transcription during sporulation.
This was confirmed by testing p868Δ90/67, in which the 24-nt region was deleted within the 868-nt promoter, and p260m4MOTIF, in which the four CTYAAC regions were altered by site-directed mutagenesis within the 260-bp promoter (Fig. 7). Both plasmids conferred sporulation-specific activity, as illustrated for representative transformants in Fig. 7B and C. Since p260m4MOTIF lacks sequences more than 260 nt upstream of the major Cdc14 TSS, it follows that the second sporulation-inducing site is between nucleotides −260 and −90. Interestingly, GUS mRNAs from these plasmids have a size of about 2.2 kb, which is slightly larger than those from plasmids having an intact −67 to −90 region; this is addressed in more detail in the next section.
FIG. 7.
Mutagenesis of CTYAAC motifs and the TSS. (A) Schematic of plasmids showing the M1 to M7 and TSS blocks (rectangles) and CTYAAC motifs (inverted triangles). Whether plasmids confer sporulation-induced expression is indicated by plus or minus signs. Filled and open triangles represent wild-type and mutagenized sequences. To save space, the 5′ ends of the map are not to scale. (B) Blot of RNA from sporangia (S) or hyphae (H) of representative transformants expressing p260m4MOTIF, plus transformants containing p868 and p90 as controls, probed for GUS after electrophoresis under high-resolution conditions (1.0% agarose). (C) The same experiment as in panel B, except that transformants contained p868Δ90/67, in which the three CTYAAC motifs were mutagenized. (D) Blot of RNA from sporangia of transformants containing the indicated plasmids, showing a progressive increase in the size of the GUS transcript with increasing numbers of mutations in the CTYAAC motifs. (E) The same experiment as in panel B, except that transformants contained pInrΔ-4/+13, in which the conserved block containing the TSS was removed. A transformant containing p105 is presented for comparison, showing the position of the band originating from the major start site; in a shorter exposure, its pattern resembles that of p105 in panel D, which is the same transformant. Cdc14-hybridized and ethidium bromide-stained panels are presented as controls.
Combined roles of TSS and CTYAAC motifs in start site determination.
As noted earlier, Cdc14 transcripts initiate at both the major site defined as the +1 TSS in Fig. 2 and later figures and a minor site(s) about 100 nt further upstream. Some data indicate that the choice of initiation site is determined by the −67 to −90 region, and likely by the CTYAAC motifs. In particular, when RNA from transformants expressing GUS from promoters containing progressively fewer copies of CTYAAC was examined on gels, the average transcript size shifted higher. As illustrated in Fig. 7D, GUS RNA from “normal” promoters, such as those in p184, p260, or p335, is mostly 2.1 kb, RNA from a promoter mutated in blocks M1 and M6 is distributed between 2.1 and 2.2 kb (p868mM1M6), and RNA from a promoter altered in M1, M6, and M7 is mostly 2.2 kb (p868mM1M6M7). As described earlier, p260m4MOTIF, which contains mutations specifically in the CTYAAC motifs, yields only the 2.2-kb transcript. This indicates that the 6-nt motifs contribute to the specificity of transcription initiation, and without such motifs a greater fraction of transcripts initiate at upstream sites. p105 yields only the 2.1-kb transcript, since it is too small to include the upstream initiation site.
As shown in Fig. 7E for plasmid pInrΔ-4/+13, mutating the region containing the major TSS eliminated the 2.1-kb band; however, transformants bearing that plasmid still produce a weak 2.2-kb band which was sporulation specific. This helps confirm the nature of the upstream start site(s) and their ability to be induced during sporulation. In addition, the loss of the 2.1-kb band from transformants expressing pInrΔ-4/+13 confirms that the −4 to +13 region most likely represents the core promoter. This was not previously obvious, since it matches poorly the Inr-like element defined for oomycetes by McLeod et al. (31), deviating from the consensus at 7 of the 15 core positions. Also, the start site defined by 5′-RACE (TCAGA+1TC) is two bases downstream from the start site predicted using the metazoan Inr consensus (YYA+1NWYY) (35).
DISCUSSION
In plant and animal pathogens, understanding the transcriptional networks that regulate spore formation is of considerable interest due to their essential roles in dissemination and infection. In Aspergillus, Fusarium, and Magnaporthe, for example, the identification of cis-elements and transcription factors participating in sporulation has led to a better understanding of that important pathway in ascomycetes (12, 19, 36, 42). To help reveal the mechanisms of sporulation in an oomycete, we analyzed the promoter of the Cdc14 gene of P. infestans, which is transcribed early during sporulation and required for development. The promoter was discovered to be relatively complex, as it contains multiple initiation sites and two independent modules that activate sporulation-specific expression. One module, represented by three repeated motifs, also influences TSS determination in concert with an Inr-like sequence that is predicted to be part of the core promoter.
The functional complexity of the promoter is consistent with the relatively high degree of sequence conservation observed between the four species of Phytophthora. The approach of comparing promoters, commonly termed phylogenetic footprinting, identifies candidate sites for binding transcription factors (5). Of the 200 nucleotides upstream of the TSS, 38% are unchanged between the four species of Phytophthora; by comparison, Cdc14 coding regions have 72% identity. Therefore, it seems likely that the promoter binds a number of proteins. Conserved sites other than the CTYAAC motifs may play a variety of roles, such as providing contextual information to the sporulation-specific factors or regulating chromatin assembly. Some sites may also have quantitative influences on transcription, although this was not addressed due to the complications of position and copy number effects. We and others previously reported the use of transient-expression systems to quantitate promoter activity in Phytophthora (23, 31), but this is not useful for studying a developmentally regulated promoter.
While the three 6-nt repeats exist within a region of overall interspecies conservation, the motifs are not identical in or between those species. For example, based on motifs in the −67 to −90 region, the P. infestans consensus is CTYAAC, while consensus sequences within P. capsici, P. ramorum, and P. sojae are CYCAAC, CTCAAC, and CKCAAC, respectively. This may mean that not all bases within the 6-mer are absolutely required for binding the cognate transcription factor, which is the case for many such regulators (5, 32). Alternatively, considering that Cdc14 has only been studied in P. infestans, there could be divergence in precisely how the orthologs are regulated in the other species.
An interesting feature of the architecture of the Cdc14 promoter is that three copies of the CTYAAC motif are required for function. In other systems, multiple copies of binding sites enable either a graded transcriptional response or an all-or-none pattern (6, 9). Cdc14 appears to fit the latter model, with the repeats likely to either synergistically strengthen the attachment of the regulator to DNA or the interaction between the regulator and RNA polymerase complex (7). A search of the P. infestans genome (www.broad.mit.edu) for three closely spaced CTYAAC motifs upstream of predicted genes did not match any other loci, suggesting that this is a unique site, but this is a tentative finding since gene models are still in a preliminary state. Nevertheless, CTYAAC motifs were not detected in the promoters of 10 sporulation-induced genes for which the automated gene models were verified (unpublished results).
Although this study focused on the CTYAAC-containing activator site, another significant aspect of the promoter is the presence of at least one additional site upstream of position −90 that apparently also induces transcription in spores. Precisely defining this second motif is beyond the scope of this study; however, both sites could play important roles. For example, sporulation is regulated by a number of physiological and environmental inputs, such as nutritional status and humidity, that potentially may act through the two sites (34). Alternatively, the two regulatory sites may act at different stages of development, since Cdc14 transcripts persist from early sporulation until cyst germination (1). Genes regulated by multiple inputs via distinct promoter modules are common in prokaryotes and eukaryotes, as are promoters containing functionally redundant transcription factor sites (16, 18, 25).
Possible redundancy is also seen in the choice of transcription initiation sites within Cdc14, with major and minor sites identified by RNA blot analysis and 5′-RACE. This is notable since oomycete genes have been generally reported to have a single TSS, although in fact detailed analyses have been performed for few genes (31). Heterogeneous start sites are also common in many nonoomycetes. Vertebrate promoters, for example, display either focused initiation in which RNA synthesis starts within a narrow region of several nucleotides or initiation is dispersed over 50 nt or more (24, 39). Also, the HIS3 gene of Saccharomyces cerevisiae initiates transcription from multiple sites directed by different TATA boxes, where one confers constitutive expression and the other responds to amino acid starvation (38). Studies of such TATA-containing promoters indicate that TFIIB and RNA polymerase II dictate the TSS (35). Most oomycete genes lack a TATA box, but many including Cdc14 contain an Inr-like sequence near the major TSS. The Inr itself may not entirely determine the TSS, since in some species it has been shown to form an unstable complex with basal transcription factors (35). Eliminating CTYAAC motifs from Cdc14 reduced initiation from the major site, implicating the cognate transcription factor as a participant in TSS determination. Presumably, the CTYAAC binding factor helps to anchor the RNA polymerase complex to the major TSS. Whether having multiple start sites in native Cdc14 is biologically significant is unknown, but this could be part of a mechanism to diversify its temporal or spatial expression pattern by using both the strong core promoter and possible weak core promoters further upstream (17, 30).
Much remains to be learned about core promoters in oomycetes. Previous analyses of 35 genes identified a partially conserved 19-nt region, which was later discovered to have an Inr-like domain and a downstream motif named the flanking promoter region (31, 33). At the onset of this study, it was not obvious if the conserved block flanking the major TSS of Cdc14 represents its major core promoter, since it is not a good match to the 19-nt consensus. Nevertheless, our functional data suggest that this is the main core promoter, since plasmid pΔINR-4/+13 no longer exhibits the normal major TSS. Through further analyses of genome sequences and in combination with functional studies, it should be possible to better understand the diversity in oomycete core promoters, if they work in combination with other core promoter elements, as seen for metazoan TATA-Inr and Inr-DPE promoters (35), and how they interact with other features within the promoter, such as stage-specific determinants. Purification of the protein(s) that binds the CTYAAC motif should also lead to a better understanding of transcription in oomycetes and enable research into the upstream signaling pathways that activate sporulation.
Acknowledgments
This work was supported by an award to H.S.J. from the U.S. Department of Agriculture National Research Initiative.
We thank S. Roy for helpful discussions.
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Ah-Fong, A., and H. S. Judelson. 2003. Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus-like oomycete Phytophthora infestans. Mol. Microbiol. 50:487-494. [DOI] [PubMed] [Google Scholar]
- 2.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. [DOI] [PubMed] [Google Scholar]
- 3.Blanco, F. A., and H. S. Judelson. 2005. A bZIP transcription factor from Phytophthora interacts with a protein kinase and is required for zoospore motility and plant infection. Mol. Microbiol. 56:638-648. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. K., and M. S. Hovmoller. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537-541. [DOI] [PubMed] [Google Scholar]
- 5.Bulyk, M. L. 2003. Computational prediction of transcription-factor binding site locations. Genome Biol. 5:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey, M., Y. S. Lin, M. R. Green, and M. Ptashne. 1990. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345:361-364. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, T. S., and T. G. Cooper. 1993. The Saccharomyces cerevisiae DAL80 repressor protein binds to multiple copies of GATAA-containing sequences (URSGATA). J. Bacteriol. 175:5851-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cvitanich, C., and H. S. Judelson. 2003. A gene expressed during sexual and asexual sporulation in Phytophthora infestans is a member of the Puf family of translational regulators. Eukaryot. Cell 2:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donahue, T. F., R. S. Daves, G. Lucchini, and G. R. Fink. 1983. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell 32:89-98. [DOI] [PubMed] [Google Scholar]
- 10.Duan, Z. J., X. Fang, A. Rohde, H. Han, G. Stamatoyannopoulos, and Q. Li. 2002. Developmental specificity of recruitment of TBP to the TATA box of the human gamma-globin gene. Proc. Natl. Acad. Sci. USA 99:5509-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, J. 1999. Phytophthora: an abiding threat to our crops. Microbiol. Today 26:114-116. [Google Scholar]
- 12.Dutton, J. R., S. Johns, and B. L. Miller. 1997. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16:5710-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteban, V., M. Blanco, N. Cueille, V. Simanis, S. Moreno, and A. Bueno. 2004. A role for the Cdc14-family phosphatase Flp1p at the end of the cell cycle in controlling the rapid degradation of the mitotic inducer Cdc25p in fission yeast. J. Cell Sci. 117:2461-2468. [DOI] [PubMed] [Google Scholar]
- 14.Gaulin, E., A. Jauneau, F. Villalba, M. Rickauer, M. T. Esquerre-Tugaye, and A. Bottin. 2002. The CBEL glycoprotein of Phytophthora parasitica var. nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115:4565-4575. [DOI] [PubMed] [Google Scholar]
- 15.Gruneberg, U., M. Glotzer, A. Gartner, and E. A. Nigg. 2002. The CeCDC-14 phosphatase is required for cytokinesis in the Caenorhabditis elegans embryo. J. Cell Biol. 158:901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309-1320. [DOI] [PubMed] [Google Scholar]
- 18.Huang, M., J. W. Lee, and D. O. Peterson. 1993. Functional redundancy of octamer elements in the mouse mammary tumor virus promoter. Nucleic Acids Res. 21:5235-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iida, Y., T. Ohara, and T. Tsuge. 2007. Identification of genes with changes in transcription levels caused by mutations in conidiation regulator genes REN1 and FoSTUA in Fusarium oxysporum. J. Gen. Plant Pathol. 73:158-167. [Google Scholar]
- 20.Jaspersen, S. L., and D. O. Morgan. 2000. Cdc14 activates Cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 10:615-618. [DOI] [PubMed] [Google Scholar]
- 21.Judelson, H. S., and F. A. Blanco. 2005. The spores of Phytophthora: weapons of the plant destroyer. Nat. Microbiol. Rev. 3:47-58. [DOI] [PubMed] [Google Scholar]
- 22.Judelson, H. S., R. Dudler, C. M. J. Pieterse, S. E. Unkles, and R. W. Michelmore. 1993. Expression and antisense inhibition of transgenes in Phytophthora infestans is modulated by choice of promoter and position effects. Gene 133:63-69. [DOI] [PubMed] [Google Scholar]
- 23.Judelson, H. S., B. M. Tyler, and R. W. Michelmore. 1992. Regulatory sequences for expressing genes in oomycete fungi. Mol. Gen. Genet. 234:138-146. [DOI] [PubMed] [Google Scholar]
- 24.Juven-Gershon, T., J. Y. Hsu, and J. T. Kadonaga. 2006. Perspectives on the RNA polymerase II core promoter. Biochem. Soc. Trans. 34:1047-1050. [DOI] [PubMed] [Google Scholar]
- 25.Keiler, K. C., and L. Shapiro. 2001. Conserved promoter motif is required for cell cycle timing of dnaX transcription in Caulobacter. J. Bacteriol. 183:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, K. S., and H. S. Judelson. 2003. Sporangia-specific gene expression in the oomyceteous phytopathogen Phytophthora infestans. Eukaryot. Cell 2:1376-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krasinska, L., G. de Bettignies, D. Fisher, A. Abrieu, D. Fesquet, and N. Morin. 2007. Regulation of multiple cell cycle events by Cdc14 homologues in vertebrates. Exp. Cell Res. 313:1225-3129. [DOI] [PubMed] [Google Scholar]
- 28.Latijnhouwers, M., and F. Govers. 2003. A Phytophthora infestans G-protein β subunit is involved in sporangium formation. Eukaryot. Cell 2:971-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latijnhouwers, M., W. Ligterink, V. G. Vleeshouwers, P. van West, and F. Govers. 2004. A G-alpha subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans. Mol. Microbiol. 51:925-936. [DOI] [PubMed] [Google Scholar]
- 30.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 31.McLeod, A., C. D. Smart, and W. E. Fry. 2004. Core promoter structure in the oomycete Phytophthora infestans. Eukaryot. Cell 3:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee, S., M. F. Berger, G. Jona, X. S. Wang, D. Muzzey, M. Snyder, R. A. Young, and M. L. Bulyk. 2004. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 36:1331-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pieterse, C. M. J., P. Van West, H. M. Verbakel, P. W. H. M. Brasse, G. C. M. Van Den Berg-Velthuis, and F. Govers. 1994. Structure and genomic organization of the ipiB and ipiO gene clusters of Phytophthora infestans. Gene 138:67-77. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro, O. K. 1983. Physiology of asexual sporulation and spore germination in Phytophthora, p. 55-70. In D. C. Erwin, S. Bartnicki-Garcia, and P. H. Tsao (ed.) Phytophthora, its biology, taxonomy, ecology, and pathology. APS Press, St. Paul, MN.
- 35.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 36.Soanes, D. M., M. J. Kershaw, R. N. Cooley, and N. J. Talbot. 2002. Regulation of the MPG1 hydrophobin gene in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 15:1253-1267. [DOI] [PubMed] [Google Scholar]
- 37.Stevens, D. A. 1995. Coccidioidomycosis. N. Engl. J. Med. 332:1077-1082. [DOI] [PubMed] [Google Scholar]
- 38.Struhl, K. 1986. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol. Cell. Biol. 6:3847-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, Y., H. Taira, T. Tsunoda, J. Mizushima-Sugano, J. Sese, H. Hata, T. Ota, T. Isogai, T. Tanaka, S. Morishita, K. Okubo, Y. Sakaki, Y. Nakamura, A. Suyama, and S. Sugano. 2001. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tani, S., and H. S. Judelson. 2006. Activation of zoosporogenesis-specific genes in Phytophthora infestans involves a 7-nucleotide promoter motif and cold-induced membrane rigidity. Eukaryot. Cell 5:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tani, S., E. Yatzkan, and H. S. Judelson. 2004. Multiple pathways regulate the induction of genes during zoosporogenesis in Phytophthora infestans. Mol. Plant-Microbe Interact. 17:330-337. [DOI] [PubMed] [Google Scholar]
- 42.Vienken, K., and R. Fischer. 2006. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol. Microbiol. 61:544-554. [DOI] [PubMed] [Google Scholar]
- 43.Whisson, S. C., A. O. Avrova, P. van West, and J. T. Jones. 2005. A method for double-stranded RNA-mediated transient gene silencing in Phytophthora infestans. Mol. Plant Pathol. 6:153-163. [DOI] [PubMed] [Google Scholar]
- 44.Wray, G. A., M. W. Hahn, E. Abouheif, J. P. Balhoff, M. Pizer, M. V. Rockman, and L. A. Romano. 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20:1377-1419. [DOI] [PubMed] [Google Scholar]