Abstract
In various organisms, thioredoxins are known to be involved in the reduction of protein disulfide bonds and in protecting the cell from oxidative stress. Genes encoding thioredoxins were found by searching the complete genome sequence of the filamentous ascomycete Podospora anserina. Among them, PaTrx1, PaTrx2, and PaTrx3 are predicted to be canonical cytosolic proteins without additional domains. Targeted disruption of PaTrx1, PaTrx2, and PaTrx3 shows that PaTrx1 is the major thioredoxin involved in sulfur metabolism. Deletions have no effect on peroxide resistance; however, data show that either PaTrx1 or PaTrx3 is necessary for sexual reproduction and for the development of the crippled growth cell degeneration (CG), processes that also required the PaMpk1 mitogen-activated protein kinase (MAPK) pathway. Since PaTrx1 PaTrx3 mutants show not an enhancement but rather an impairment in CG, it seems unlikely that PaTrx1 and PaTrx3 thioredoxins participate in the inhibition of this MAPK pathway. Altogether, these results underscore a role for thioredoxins in fungal development.
In eukaryotes, complex networks of signaling pathways control cell differentiation, cell proliferation or programmed cell death. Conserved mitogen-activated protein kinase (MAPK) cascades are among the key players of those pathways. Various extracellular signals, including oxidative stresses, can trigger the activation of MAPK modules. Once initiated, signal transduction proceeds through a sequence of successive phosphorylations. At the top of a module, a MAPK kinase kinase (MAPKKK) is activated by various means, including self-phosphorylation. When activated, the MAPKKK phosphorylates the corresponding MAPK kinase (MAPKK), which in turn phophorylates the MAPK. Phosphorylation of effectors located downstream of the cascade then leads to diverse but specific cell responses. In the filamentous fungus Podospora anserina, activation of the PaASK1/PaMKK1/PaMpk1 module, akin to the yeast Saccharomyces cerevisiae MPK1 MAPK module, is required to signal differentiation processes that include mycelium pigmentation, development of aerial hyphae, and sexual reproduction (14, 15). It is also necessary for hyphal interference, a defense process presented by P. anserina upon meeting a fungal competitor (36). In addition, when activated in an inappropriate manner, namely, at the apexes of the growing hyphae, this MAPK pathway results in the development of the epigenetic crippled growth (CG) cell degeneration phenomenon (37). The CG alteration appears as sectors of poor growth, displaying an overpigmented and female sterile mycelium. Mutant analysis has also uncovered the NADPH oxidase PaNox1 as controlling the same processes (24, 36). NADPH oxidases are enzymes that produce reactive oxygen species (ROS) using cytoplasmic NADPH as an electron donor. In addition, it has been demonstrated that both peroxides and/or superoxides are secreted during vegetative growth, sexual reproduction, ascospore germination, and hyphal interference of P. anserina (24, 36). ROS have also been implicated in ageing of P. anserina. Indeed, the growth of wild-type P. anserina thalli is limited by a process called senescence, and longevity can be conveniently measured in this organism (38). Processes that affect ROS production also alter longevity (23). Taken together, these results point toward an active role of ROS in P. anserina physiology, possibly as second messengers. However, despite numerous examples of ROS-induced developmental processes, it is not well understood how ROS signals are integrated at the molecular level for signaling purposes. Nevertheless, in mammals it has been demonstrated that activation of the tumor necrosis factor alpha-induced apoptosis pathway effected by the ASK1 MAPK pathway depends on the redox status of a thioredoxin (Trx) (33). The reduced form of the Trx protein inhibits the activation of ASK1 MAPKKK through direct interaction with its N terminus. Yet, when oxidized by ROS, the binding of Trx to ASK1 is disrupted, and the subsequent activation of the released MAPKKK leads to transduction of apoptosis signals through activation of Jun N-terminal protein kinase and/or p38 MAPK. More recently, it was established that interaction between Trx and ASK1 promotes ASK1 ubiquitination and degradation (22).
Thioredoxins are ubiquitous small proteins with a redox-active site that have been conserved throughout evolution since they have been described in prokaryotes, as well as in eukaryotes, including, fungi, plants, invertebrates, and vertebrates. As hydrogen donors, thioredoxin systems regulate many metabolic enzymes that form disulfide bonds during their catalytic cycle and can function as scavengers of ROS. Thioredoxins obtain their reducing power from NADPH by way of the thioredoxin reductase. Therefore, in addition to sensing oxidative stress, to which isolated cells or more integrated tissues can be subjected, thioredoxins might also be good sensors of the nutrient state of any given cell. Thus, it is not surprising that deregulation of thioredoxin expression leads to a broad range of phenotypes. Targeted disruption of the mouse cytosolic thioredoxin gene (Txn) causes early embryonic lethality (25), whereas transgenic mice overexpressing the human Trx gene have longer life spans (28). Moreover, when the same Trx gene was overexpressed in human fibrosarcoma cells, cell growth was retarded and senescence was induced. However, resistance to oxidative stress caused by H2O2 or drugs such as paraquat was enhanced (4). The TRX homologous gene, identified in Drosophila melanogaster, is required for female meiosis and early embryonic development (34). Caenorhabditis elegans mutants with a deletion of the allele trx-1, a gene encoding a thioredoxin homologous to the human gene, have reduced life spans and are sensitive to oxidative stress (12, 27).
In plants, thioredoxins also play a fundamental role in plant tolerance to oxidative stress. The Arabidopsis thaliana TRXh5 gene encoding a cytosolic thioredoxin is upregulated in response to oxidative stresses but also during wounding, abscission, and incompatible interactions with pathogens (17). Interestingly, a 28-bp antioxidant responsive cis element has been characterized in the promoter of both a soybean thioredoxin gene that participates in root nodule development and the rice trxh gene that also encodes a cytosolic thioredoxin (21, 42). S. cerevisiae contains two genes encoding a cytoplasmic thioredoxin. They are dispensable, since deletion of either TRX1 or TRX2 has no effect under normal growth conditions (6, 29). However, trx2 mutants are hypersensitive to H2O2 (7). In contrast, simultaneous inactivation of TRX1 and TRX2 leads to a broad range of phenotypes: methionine auxotrophy, impaired cell cycle (i.e., a threefold-longer S phase and a shorter G1 phase), increase in generation time, significantly larger cell sizes (6, 29), constitutive activation of the transcription factor YAP-1 (10), and a drastic defect in vacuole inheritance (43).
To date, very little is known on thioredoxin functions in filamentous fungi, besides the recent identification of a transcript involved in the virulence of the chestnut blight fungus Cryphonectria parasitica, which presents significant similarity with the Neurospora crassa thioredoxin gene (16). In recent years, the discovery that ROS are involved in developmental processes in various filamentous fungi (18, 24, 26, 40, 41) makes thioredoxin genes attractive targets for analysis, especially in the view of the limited data available for these genes in such organisms. The possible crucial role of ROS in signaling many aspects of P. anserina physiology make this fungus an appealing model to study thioredoxins. We have thus undertaken to characterize the role(s) of cytosolic thioredoxins in this species.
MATERIALS AND METHODS
Strain, culture conditions, nucleic acid manipulations, and genetic analyses.
The strains were all derived from the S strain ensuring a homogeneous genetic background (30). The Δmus51 strain (kindly provided by Carole Sellem) is a strain lacking the mus51 subunit of the complex involved in end joining of broken DNA fragments. DNA integration in this strain proceeds almost exclusively by homologous recombination. Standard culture conditions, media, and genetic methods for P. anserina were as described previously (31; http://podospora.igmors.u-psud.fr). M2 medium is a minimal medium in which carbon is supplied as dextrin and nitrogen as urea. The methods used for nucleic acid extraction and manipulation have been described (2, 19). Transformation of P. anserina protoplasts was carried out as described previously (3). When necessary, the M2 medium was supplemented with 20 μg of methionine/ml.
Cloning and sequencing of the thioredoxin genes.
The present study was started before the availability of the complete genome sequence of P. anserina. To isolate the P. anserina thioredoxin genes, a PCR was performed in 50-μl reaction volume containing 100 ng of P. anserina genomic DNA, 25 pmol of each primer thiored1 (5′-TGGTGYGGNCCNTGYAARGC-3′) and thiored2 (5′-ARRAANGTNGGCATNGC-3′), 200 μM concentrations of each deoxynucleoside triphosphate, 1.5 U of Taq polymerase (Takara), and buffer supplied by the manufacturer under the following conditions: an initial denaturation step of 2 min at 94°C, followed by two cycles of denaturation for 1 min at 94°C, annealing for 1 min at 37°C, and elongation for 1 min at 72°C. This was the followed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C. Two PCR products, 150 and 250 bp, were gel purified and sequenced. Sequencing revealed that each product coded part of thioredoxin genes that were designated PaTrx1 and PaTrx2. To retrieve the complete genes, two pairs of primers were designed to screen a P. anserina cosmid genomic DNA library from strain S (kindly provided by M. Dequard-Chablat). TRX1-3 (5′-CGACGTTGACGCCTCGGCCG-3′) and TRX1-6 (5′-CTCAGCCGAACCCTCGGCAC-3′) amplified a 150-bp fragment of the PaTrx1 gene and TRX2-1 (5′-CCACATCACGCCCTGCTTTC-3′) and TRX2-4 (5′-GGGTTGGCGCCCATGAGCTCG-3′) amplified a 250-bp fragment of the PaTrx2 gene. Cosmid 9F1 containing the entire PaTrx1 coding sequence (CDS) and cosmid 12B36 containing the entire PaTr2 CDS were recovered. The insert of cosmids 9F1 and 12B36 were used to sequence the entire 577 bp of PaTrx1 CDS and the entire 493 bp of PaTrx2 CDS, respectively. Following the availability of the genome sequence, additional genes were detected by sequence comparison using BLAST (1) with the S. cerevisiae thioredoxin as a query (see Results).
Deletion of PaTrx1, PaTrx2, and PaTrx3 genes.
To construct the Δtrx1::hygR-null allele, an approximately 6.5-kb XhoI fragment containing the PaTrx1 gene was released from cosmid 9F1 and cloned into the unique XhoI site of the pBluescript KS(+) vector (Stratagene) to create the pKStrx1 plasmid. This plasmid was hydrolyzed by XhoI and HindIII to generate a 3-kb fragment corresponding to the 5′ PaTrx1 flanking sequence. A 3′ 1.2-kb PaTrx1 flanking sequence was PCR amplified using the following pair of primers TRX1R (5′-AAATCGATTGTCTCTGGCTTGTGTGGCGC-3′) and M13-20 (5′-GTAAAACGACGGCCAGT-3′) with the pKStrx1 plasmid as a template. The PCR-amplified fragment was cloned into the pGEM-T vector (Promega). One of the resulting plasmids that showed proper orientation of the insert was hydrolyzed by using ApaI and XhoI to generate a 1.2-kb fragment that was then ligated with both the pBC-hygro (35) previously cleaved by ApaI and HindIII and the 3-kb XhoI/HindIII 5′ fragment. From this three-partner ligation experiment, the Δtrx1::hygR plasmid harboring a complete deletion of the PaTrx1 gene was retrieved. Transformation of the P. anserina wild-type S strain was performed with 10 μg of the Δtrx1::hygR plasmid previously linearized at the unique XhoI site to generate homologous recombination ends. A total of 151 hygromycin B-resistant clones were obtained. Some of them showed an altered vegetative growth on regular M2 medium. PCR analysis and Southern blot (see Fig. S1 in the supplemental material) verified that in these transformants, homologous recombination had replaced the PaTrx1 wild-type allele by the Δtrx1::hygR mutant allele. One mutant denoted ΔPaTrx1 was selected for further studies.
To construct the Δtrx2::phleoR-null allele, an ∼6-kb KpnI fragment containing the PaTrx2 gene was released from cosmid 12B36 and circularized by intramolecular ligation. Circular DNA served as a template for PCR amplification using the pair of primer pair TRX2F (5′-TTATCGATTGGGGGTTGTGGTTGTGAAGG-3′) and TRX2R (5′-TTATCGATTGGGAGCGGTGCTGGTGCTAG-3′); the underlined nucleotides correspond to the addition of a ClaI restriction site for cloning purposes. A 5.5-kb fragment was PCR amplified by using the TRX2-TRX2R pair of primers, hydrolyzed by ClaI, and cloned into the unique ClaI site of the pBC-phleo plasmid (35). The resulting 437-bp deletion encompasses the start codon of the PaTrx2 gene, whose native CDS is 493 bp long. Transformation of the P. anserina wild-type S strain was performed with 10 μg of the Δtrx2::phleoR plasmid previously linearized at the unique KpnI site to generate homologous recombination ends. A total of 48 phleomycin-resistant clones were obtained. They were first screened by PCR using the TRX2-1 (5′-CCACATCACGCCCTGCTTTC-3′) and TRX2-4 (5′-GGGTTGGGGCCATGAGCTCG-3′) primers to search for internal deletions of the PaTrx2 gene, and candidate mutants were selected. PCR analysis and Southern blot (see Fig. S1 in the supplemental material) verified that homologous recombination had correctly replaced the PaTrx2 wild-type allele by the Δtrx2::phleoR mutant allele in these candidates. One such mutant, denoted ΔPaTrx2, was selected for further studies.
To delete PaTrx3, the hygromycin B resistance cassette of the pBC-hygro vector (35) was amplified by PCR with the oligonucleotides trx3-1 (5′-TACACGACTCTCTCCCAAAGACCATTACCAATTTCGAAATGGCCGGAAGCG AGAAGA-3′) and trx3-2 (5′-GAATCTCTACAGTCCACCACCATCTCCAGGCCATTACTCGGCACTCTTTGCTGCTTGG-3′). This permitted the recovery of the PCR1 DNA fragment that contained the hygromycin B resistance gene flanked on either side with 50 bp from the region bordering the PaTrx3 coding sequence. However, when used in transformation experiments with the Δmus51 strain, this construct did not permit us to correctly replace the PaTrx3 gene with the ΔPaTrx3 deleted allele. The 5′ bordering region was increased to 200 bp by PCR with genomic DNA as a template and PCR1 and the oligonucleotide trx3-5 (5′-GCTTTCCCAGGGGGTTAG-3′) as primers. This yielded PCR2, a 5′-extended product that was further extended in the 3′ region by PCR with genomic DNA as a template and PCR2 and oligonucleotide trx3-6 (5′-GCTTCCCTCTCCGTGACA-3′) as primers. This last PCR resulted in a DNA fragment containing the hygromycin B resistance gene flanked by 200 bp bordering the PaTrx3 coding sequence. The PCR product was introduced into the Δmus51 strain by transformation, and two independent hygromycin B-resistant clones were obtained. PCR analysis and Southern blot (see Fig. S1 in the supplemental material) verified that they had the correct gene replacement. One mutant denoted ΔPaTrx3 was selected for further studies. It was crossed with wild type to segregate out the Δmus51 mutation.
Complementation of the ΔPaTrx1 mutant and overexpression of PaTrx2.
The complete PaTrx1 and PaTrx2 CDS were PCR amplified by using the primer pair TRX1F3 (5′-ATTCTAGATAGTTCCTCCCGCTAC-3′) and TRX1R3 (5′-ATTCTAGAAGCGGCCAACCGTTTG-3′) and primer pair TRX2F1 (5′-ATTCTAGATAAACCAAGAGCCTTC-3′) and TRX2R1 (5′-ATTCTAGATTATCTGCATGCAAGC-3′), respectively; the underlined nucleotides correspond to the XbaI restriction site added for cloning purpose. The 759-bp PaTrx1 PCR-amplified fragment and the 599-bp PaTrx2 PCR-amplified fragment were cloned into the pGEM-T vector and sequenced. Plasmids carrying an insert without mutation were hydrolyzed by XbaI. The resulting fragments were cloned at the unique XbaI restriction site of the pBCH-A plasmid (15). This plasmid contains the promoter of the AS4 gene, which is highly and constitutively expressed throughout the life cycle (39). pBCH-ATrx1-3, containing the PaTrx1 native CDS downstream of the AS4 promoter, and pBCH-ATrx2-16, containing the PaTrx2 native CDS downstream of the AS4 promoter, were selected after restriction analyses. Transformation of the P. anserina wild-type S strain was performed with 10 μg of either the pBCH-ATrx1-3 plasmid or the pBCH-ATrx2-16 plasmid. Twelve independent hygromycin B-resistant clones were obtained after transformation with pBCH-ATrx1-3, and two independent hygromycin B-resistant clones were obtained after transformation with pBCH-ATrx2-16. Both types of transformants were crossed with a ΔPaTrx1 strain of compatible mating type. In the progeny of these crosses, strains carrying both the ΔPaTrx1 allele and either the pBCH-ATrx1-3 transgene or the pBCH-ATrx2-16 transgene were selected and assayed for methionine prototrophy.
Phenotype analysis.
Fertility was measured as described previously (31), except that the growth medium was M2. Longevity was measured on at least 18 cultures as described previously (39). The production of superoxides and peroxides was assayed as described previously (24). Hyphal interference, including peroxide production and cell death, was assayed as described previously (36).
Western blot analysis.
Phosphorylation of PaMpk1 was evaluated as described previously (14). Anti-phospho-p42/p44 antibody 9101 from Cell Signaling Technology was used as a primary antibody.
Database searches.
Databases were searched with the BLAST program using the default values (1). Several CDS were detected in the P. anserina and N. crassa databanks (http://podospora.igmors.u-psud.fr and http://www.broad.mit.edu/annotation/fungi/fgi/, respectively). These were compared to the Swiss-Prot databanks, and CDS that matched thioredoxin with an e-value ≤10−10 were kept as bona fide thioredoxin. This analysis also yielded CDS for disulfide isomerases, which are related to thioredoxins. Moreover, additional CDS with a thioredoxin fold could be detected both in the P. anserina and in the N. crassa genomes, but with e-values too high to consider them as coding for typical thioredoxins. These may code for highly divergent thioredoxin-like proteins.
RESULTS
Thioredoxin and thioredoxin-like proteins of P. anserina.
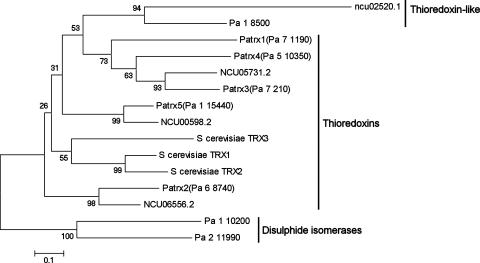
The complete P. anserina predicted CDS set available at http://podospora.igmors.u-psud.fr was searched with the BLAST program using the S. cerevisiae thioredoxins as a query. Six CDS potentially coding for thioredoxins were detected: Pa_7_210, Pa_5_10350, Pa_7_1190, Pa_6_8740, Pa_1_15440, and Pa_1_8500. A BLAST comparison with the Swiss-Prot database as a target revealed that Pa_7_210, Pa_5_10350, Pa_7_1190, Pa_6_8740, and Pa_1_15440 were most similar to thioredoxins, while Pa_1_8500 was most similar to mammal thioredoxin-like proteins (20). Pa_7_1190, Pa_6_8740, Pa_7_210, Pa_5_10350, and Pa_1_15440 were thus named PaTrx1, PaTrx2, PaTrx3, PaTrx4, and PaTrx5, respectively. A similar search in the complete CDS set of N. crassa available at http://mips.gsf.de/projects/fungi/neurospora.html revealed only four CDS: NCU05731.2, NCU06556.2, NCU00598.2, and NCU02520.1. To gain insight into the evolution of these thioredoxin CDS, a phylogenetic comparison was made between these 10 CDS and the TRX1, TRX2, and TRX3 proteins of S. cerevisiae (Fig. 1). Two putative P. anserina disulfide isomerases were included as an outgroup. On the phylogenetic tree, one can see that the two additional thioredoxins of P. anserina result from the triplication of one ancestral gene to yield PaTrx1, PaTrx3, and PaTrx4, while only one gene, NCU05731.2, is present in N. crassa. Apart from this triplicated set, all of the other CDS had a single ortholog in P. anserina and N. crassa. Because of the small size of these proteins, the bootstrap values associated with many branches were low, preventing the recovery of phylogenetic relationship between the isoforms with statistically significant support. Nevertheless, careful analysis of the protein comparison (see Fig. S2 in the supplemental material) revealed the following points. First, the three S. cerevisiae proteins (TRX1, TRX2, and TRX3), the two P. anserina proteins (PaTrx2 and PaTrx5), and the two N. crassa proteins (NCU06556.2 and NCU00598.2) all present near the C terminus of the protein a GANPXAL motif, suggesting that they all derived from the same ancestral gene. Interestingly, S. cerevisiae TRX3 is located in the mitochondria and thus possibly is derived from a cytosolic form. Second, no obvious mitochondrial thioredoxin is present in the genome of P. anserina and N. crassa. Indeed, only P. anserina PaTrx4 presents a potential targeting presequence, but analysis of the probable localization of this protein with PSORT yielded the secretory pathway as the most probable fate of PaTrx4 with high confidence, since 24 of the 26 closest neighbor presequences are those of secreted proteins. In addition, this PaTrx4 thioredoxin lacks the canonical CGPC motif required for activity (9). It is replaced by a CPPC motif, which may either abolish or impair activity. Third, for the couple of orthologous proteins PaTrx5 and NCU00598.2, the thioredoxin domain is followed by a C-terminal extension of about 70 amino acids, suggesting that these isoforms may have an additional activity. This 70 amino acids extension is conserved in other sordariomycete fungi.
FIG. 1.
Phylogenetic tree of the proteins indicated produced by using the neighbor-joining method with 1,000 replication for bootstrap. Analysis was performed with the thioredoxin domain underlined on Fig. S2 in the supplemental material. Similar trees were obtained with the maximum-parsimony and minimum evolution algorithms. The GenBank accession numbers for PaTrx1 is AY077728 and that for PaTrx2 is AY077731.
A search of the P. anserina EST database revealed that the five thioredoxin genes and the thioredoxin-like genes are expressed (see Table S1 in the supplemental material). In summary, the genome of P. anserina contains one thioredoxin-like protein and five putative thioredoxins. Among these, three are canonical thioredoxins (PaTrx1, PaTrx2, and PaTrx3) with a probable cytosolic location, one may be secreted and possibly inactive (PaTrx4), and one possesses an additional domain at its C terminus (PaTrx5). PaTrx1, PaTrx3, and the potentially secreted PaTrx4 derive from a triplication or two successive duplications of an ancestral gene, present in a single copy in the genome of N. crassa. As a first step in the analysis of the role of thioredoxins in P. anserina phylogeny, the genes encoding the canonical cytosolic thioredoxins PaTrx1, PaTrx2, and PaTrx3 were deleted.
PaTrx1 deletion causes partial methionine auxotrophy, whereas PaTrx2 and PaTrx3 deletions do not.
To investigate the role of PaTrx1, PaTrx2, and PaTrx3, three mutants harboring ΔPaTrx1, ΔPaTrx2, or ΔPaTrx3 null alleles were generated, respectively. To achieve this, defective alleles were constructed in vitro by replacing the entire thioredoxin CDS with the hygromycin B or phleomycin resistance selectable marker (see Materials and Methods). To allow substitution of the native alleles by the deleted alleles in vivo, the corresponding constructs were introduced into P. anserina by transformation. In each case, after appropriate PCR and Southern blot analysis (see Fig. S1 in the supplemental material), several independent resistant transformants showing homologous integration of the defective allele at the proper locus were recovered. Because of the functional redundancy that the PaTrx1, PaTrx2, and PaTrx3 genes might display, the phenotypes of all of the double mutants as well as that of the triple ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutant were examined.
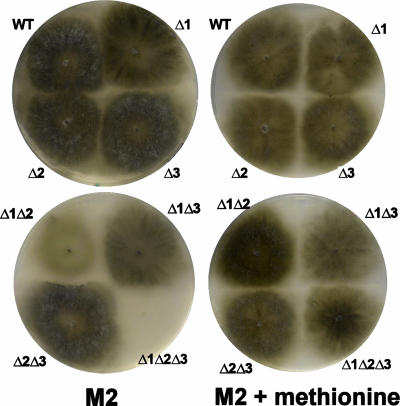
The single ΔPaTrx2 and ΔPaTrx3 mutants did not display any obvious vegetative phenotype under standard growth conditions, whereas inactivation of the PaTrx1 gene resulted in partial methionine auxotrophy, since on M2 medium lacking methionine the mycelium of the ΔPaTrx1 mutants produced fewer aerial hyphae (Fig. 2), was female sterile, and grew slightly slower than wild-type mycelium (Table 1). The addition of 20 mg of methionine/ml to the medium was sufficient to restore wild-type vegetative growth and fertility in the ΔPaTrx1 mutants (Fig. 2). In the double ΔPaTrx1 ΔPaTrx2 and ΔPaTrx1 ΔPaTrx3 mutants the auxotrophy was exacerbated (their mycelia were thinner than that of ΔPaTrx1 [Fig. 2]), whereas the double ΔPaTrx2 ΔPaTrx3 mutants remained prototrophic for methionine. Increase in methionine auxotrophy was particularly clear in the triple ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutants since growth was slowed down (Table 1) and the mycelium was very spindly (Fig. 2). To further confirm that methionine auxotrophy is due to lack of thioredoxin, the PaTrx1 CDS was placed under the control of the highly expressed AS4 gene (39), and the resulting chimeric gene was introduced into the genome of the ΔPaTrx1 mutant. This mutant regained methionine prototrophy, showing that the phenotype is indeed due to the lack of PaTrx1. To circumvent methionine auxotrophy, all subsequent experiments were performed in medium supplemented with methionine (except when indicated). On this methionine-supplemented medium, the wild-type strain behaves as on nonsupplemented medium for all of the phenotypes investigated below.
FIG. 2.
Mycelium morphology of strains with the thioredoxin genes deleted on M2 and on M2 supplemented with methionine. WT, wild type; Δ1, ΔPaTrx1, Δ2: ΔPaTrx2; Δ3, ΔPaTrx3.
TABLE 1.
Effect of thioredoxin deletions during P. anserina life cycle
| Category | Parameter | Straina
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | Δ1 | Δ2 | Δ3 | Δ1Δ2 | Δ1Δ3 | Δ2Δ3 | Δ1Δ2Δ3 | surT2 Δ1Δ3 | ||
| Methionine auxotrophy | Growth rate (mm/day) on medium lacking methionine | 7 | 6 | 7 | 7 | 6 | 6 | 7 | 5 | 7 |
| Vegetative growth on medium supplemented with methionine | Morphology | WT | WT | WT | WT | WT | Flat and dark | WT | Flat and dark | WT |
| Longevity (mean cm ± SD) | 9.3 ± 1.0 | 16.8 ± 3.7 | 8.6 ± 1.4 | 9.5 ± 1.2 | >27 | >27 | 12.0 ± 1.0 | >27 | NT | |
| CG on M2 | No | No | No | No | No | No | No | No | No | |
| CG on M2 +YE | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No | |
| H2O2 inhibition dose (%)b | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
| H2O2 lethal dose (%)c | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | |
| Sexual reproduction | Perithecium production | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No |
WT, wild-type strain; Δ1, ΔPaTrx1 strain; Δ2, ΔPaTrx2 strain; Δ3, ΔPaTrx3 strain; YE, yeast extract; flat, fewer aerial hyphae than WT; dark, more pigmented than WT. NT, not tested.
That is, the dose at which the growth rate of the indicated strains starts to decrease to 90% of that on medium not supplemented with H2O2. The same doses promote the same effects on medium lacking methionine or on medium supplemented with cysteine.
That is, the dose at which the indicated strain does not present any sign of growth after 2 days of incubation.
Effect of thioredoxin deletions during vegetative growth.
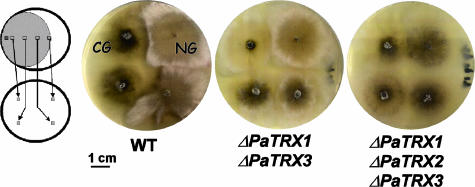
Growth of the thioredoxin mutants on methionine-supplemented medium was similar to the wild-type P. anserina, except for the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 strains, whose mycelium presented fewer aerial hyphae and was more pigmented (Fig. 2). This phenotype was also seen on M2 medium supplemented with 5 g of yeast extract/liter, suggesting that it did not result from the requirement of any additional metabolite. The longevity of the ΔPaTrx1 strain was increased almost twofold (Table 1). The longevity of strains carrying additional deletions in combination with ΔPaTrx1 was further increased since most cultures had not died when the experiment was terminated (at which stage the cultures had grown more than three times longer than wild-type cultures). Only two strains were affected in their ability to present CG. In wild-type strains, CG was never observed on M2 medium or on M2 medium supplemented with methionine but is readily observed on medium supplemented with yeast extract (8). None of the mutant strains exhibited CG on M2 medium supplemented with methionine, and apart from the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 strains, all exhibited CG as did the wild-type strain on M2 medium supplemented with yeast extract. However, the ΔPaTrx1 ΔPaTrx3 strains presented a CG that was greatly reduced, and the ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 strains presented no CG whatsoever (Fig. 3). The lack of CG can be due either to the inability of these strains to build the cytoplasmic and infectious C element responsible for the growth alteration or to restriction of its propagation during growth (8). To differentiate between these two possibilities, contamination experiments were conducted between the ΔPaTrx1 ΔPaTrx3 and the ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 strains as the donor and the AS6-5 strain as the receptor under conditions previously used to differentiate between the two possibilities (8, 14, 15, 24). The recovery of sectors of altered growth in the receptor strain after contamination allowed us to conclude that although CG was either reduced or absent, the C element was still made during stationary phase by both donor strains.
FIG. 3.
Lack of CG in the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 strains. On the left is depicted the experimental setup. A culture is grown for 7 days on M2 medium supplemented with methionine, and explants taken at various distances from the growing edge are replicated onto M2 medium supplemented with yeast extract. The C element responsible for CG is induced in the stationary phase and triggers CG in explants taken from the part of the thallus that is in the stationary phase. In contrast, explants taken from the growing edge yield mycelia in normal growth (NG) conditions. CG is visible in wild type grown on M2 plus yeast extract as a flat and spindly growth with pigment accumulation (8, 37). In the ΔPaTrx1 ΔPaTrx3 mutant, growth impairment is much reduced and visible as a slightly flatter mycelium. In the ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutant no discernible CG was observed. Note that the mycelium from this strain grows more slowly, produces fewer aerial hyphae, and accumulates more pigment on M2 supplemented with yeast extract than normal-growing wild-type strains.
The recovery of CG sectors in contamination experiments strongly suggested that the ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 triple mutant strain was able to undergo anastomosis. To verify this directly, we checked whether heterokaryons could be obtained by mixing mycelium from the partial methionine auxotroph ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutant with mycelium from the leu1-1 mutant that is auxotrophic for leucine. Upon plating on M2 medium, the mixed mycelia were able to grow like the wild type, indicating that prototrophic heterokaryons had formed. Similar data were obtained with the ΔPaTrx1 ΔPaTrx2 and ΔPaTrx1 ΔPaTrx3 mutants. Therefore, cell fusion is not compromised in strains with the thioredoxin genes deleted. Similarly, hyphal interference occurred as in the wild type in all single-, double-, and triple-mutant strains (data not shown). Finally, resistance to peroxide was not affected in any of the mutants (Table 1).
Effect of thioredoxin deletions during sexual reproduction.
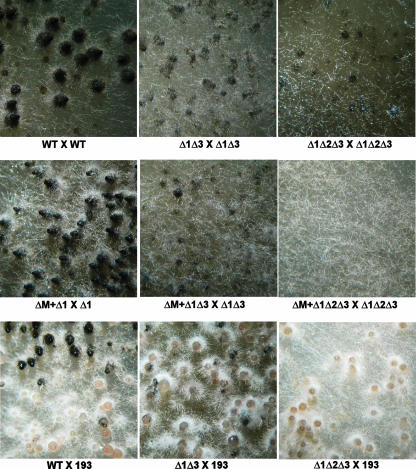
The fertility of the thioredoxin mutants was similar to that of wild-type mycelium except for the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutants. These two mutants were fertile as males but were sterile as females in homozygous crosses (Fig. 4) and also in crosses with wild-type fungus. However, ascospores with the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 genotypes (as well as all of the other combinations of Trx gene deletion) germinated like wild-type fungus. The fertility defect was more pronounced in the ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutant since the ΔPaTrx1 ΔPaTrx3 produced small fruiting bodies with an envelope, sometimes differentiating a neck and a centrum, but without asci, whereas only tiny abortive fructifications were generated by the ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutant. Since the ΔPaTrx1 ΔPaTrx2 mutants are auxotrophic for methionine but fertile on medium supplemented with methionine (Fig. 2), the sexual defect exhibited by ΔPaTrx1 ΔPaTrx3 was likely not due to methionine auxotrophy per se. To confirm this, a homozygous cross of the methionine auxotroph met2 (kindly provided by A. Sainsard-Chanet) was set up on M2 medium supplemented with methionine. This mutant was fully fertile on such medium, ruling out the effect of methionine auxtrophy in the sterility of the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutants (see Fig. S3 in the supplemental material). Homokaryotic spores were recovered from the progeny of homozygous met2 crosses. Thalli derived from these ascospores all showed methionine auxotrophy.
FIG. 4.
Effect of thioredoxin deletions during sexual reproduction. (Top row) Fertility defect of the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 strains. mat+/mat− heterokaryotic mycelia of the indicated genotypes were incubated at 27°C for 1 week, at which time the perithecia are fully mature and starting to eject ascospores in wild-type strains. Incubation times of more than 3 weeks did not permit further maturation of perithecia in the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutants. (Middle row) The fertility defect of the indicated mat+/mat− heterokaryons was not corrected when Δmat nuclei were added in the heterokaryons. (Bottom row) The perithecial envelope of the thioredoxin mutants/193 heterokaryon derived mostly or exclusively from the 193 parent, showing that expression of PaTrx1 or PaTrx3 was required in the perithecial envelope.
To further define at which step the thioredoxins are needed, the following trikaryotic strains were constructed: Δmat/ΔPaTrx1 ΔPaTrx2 ΔPaTrx3mat+/ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mat− and Δmat/ΔPaTrx1 ΔPaTrx3mat+/ΔPaTrx1 ΔPaTrx3 mat−. No rescue of fertility was observed in these trikaryotic strains (Fig. 4). The Δmat nuclei lack the mating type locus are thus unable to participate in fertilization; however, they are fully competent to provide hyphae for the generation of the envelope, provided the dikaryon resulting from fertilization is able to send the signal required for the development of the envelope (11). The absence of fruiting bodies in such trikaryons suggests that the ΔPaTrx1 ΔPaTrx2 ΔPaTrx3mat+/ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mat− and ΔPaTrx1 ΔPaTrx3mat+/ΔPaTrx1 ΔPaTrx3 mat− dikaryotic strains are impaired in their ability to signal the development of the perithecial envelope. The presence of either Patrx1 or PaTrx3 is thus essential within the dikaryons.
The 193mat+/ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mat− and 193mat+/ΔPaTrx1 ΔPaTrx3 mat− dikaryotic strains were then constructed. 193mat+/ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mat− yielded colorless perithecia, and 193mat+/ΔPaTrx1 ΔPaTrx3 mat− yielded mostly colorless perithecia, along with few perithecia containing areas of pigmented hyphae (Fig. 4). 193 is a mutation located in a polyketide synthase gene acting at the first step of pigment biosynthesis (5). The 193 mutant allele promotes lack of pigmentation at all stages of the life cycle. The perithecia obtained in the 193mat+/ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mat− and 193mat+/ΔPaTrx1 ΔPaTrx3 mat− dikaryotic strains have thus an envelope that results exclusively or essentially from hyphae with 193 nuclei, respectively. These results support the previous observation made in Δmat trikaryotic strains and strongly suggest that the PaTrx1 or PaTrx3 proteins are needed in the envelope for the proper development of fruiting bodies.
The fertility defect of the ΔPaTrx1 ΔPaTrx3 mutants is not due to a defect in the PaMpk1 MAPK cascade.
A severe defect in perithecium production is observed in mutants of the PaMpk1 cascade (14, 15, 24). We thus investigated whether PaMpk1 is correctly phosphorylated and located in the nuclei in the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutants. As seen in Fig. S4 in the supplemental material, phosphorylation and nuclear localization are not affected in either mutant.
Overexpression of PaTrx2 suppresses methionine auxotrophy but not the fertility defect.
From these experiments, it appears that PaTrx1 is critical for methionine prototrophy and that either PaTrx1 or PaTrx3 is needed for fertility. Both of these thioredoxins are derived from the same ancestral gene (Fig. 1), which leaves open a physiological role for PaTrx2. To assay whether PaTrx2 shares some redundancy with PaTrx1 or PaTrx3, it was overexpressed. The PaTrx2 CDS was placed under the control of the AS4 promoter, and the AS4-PaTrx2 chimeric gene was introduced into the ΔPaTrx1 mutant. The resulting AS4-PaTrx2 ΔPaTrx1 strains were prototrophic for methionine, showing that high levels of PaTrx2 can functionally replace PaTrx1 in sulfur metabolism (29). The AS4-PaTrx2 ΔPaTrx1 strain was then crossed with ΔPatrx3, and AS4-PaTrx2 ΔPaTrx1 ΔPatrx3 strains were obtained in the progeny. These strains remained sterile (Fig. 5) and presented a weak CG, as does the ΔPaTrx1 ΔPatrx3 strain. This shows that overexpression of PaTrx2 cannot replace the lack of both these proteins.
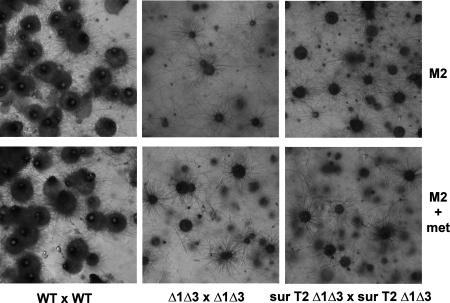
FIG. 5.
Lack of fertility on M2 medium and M2 medium supplemented with methionine of strains lacking PaTrx1/PaTrx3 and overexpressing PaTrx2 (surT2 Δ1Δ3). mat+/mat− heterokaryotic strains of the indicated genotypes were incubated for 10 days on M2 medium or M2 medium supplemented with 20 μg of methionine/ml, at which time the wild type produces fully mature perithecia, whereas strains with PaTrx1 and PaTrx3 deleted do not produce mature fructifications even if PaTrx2 is overexpressed. A longer incubation time does not result in perithecium maturation in these mutants.
DISCUSSION
The thioredoxin system in P. anserina seems to comprise five genes coding for thioredoxin and one gene coding for a thioredoxin-like protein. Nevertheless, additional genes coding highly divergent thioredoxins may be present. Among the bona fide thioredoxin genes, three seem to code for cytosolic proteins (PaTrx1, PaTrx2, and PaTrx3) and none codes for a mitochondrial protein. One is possibly secreted and possibly inactive (PaTrx4), and one is fused with a domain conserved in sordariomycetes but whose function is unknown. Interestingly, PaTrx1, PaTrx3, and the potentially secreted PaTrx4 seem to result from the triplication of an ancestral gene, which is present as a single copy in the related fungus N. crassa. Deletion of the three cytosolic canonical thioredoxin genes allowed us to better define their role in relation to the P. anserina life cycle. Clearly, PaTrx1 plays the predominant role; however, differences in the roles of PaTrx1/PaTrx3 versus PaTrx2 can be defined, especially during sexual reproduction and CG development.
Most prominent is methionine auxotrophy promoted by the deletion of PaTrx1. It has been shown in S. cerevisiae that methionine auxotrophy is due to the absence of reduction of the 3′-phosphoadenosine 5′-phosphosulfate reductase (PAPS) by Trx1 or Trx2 cytosolic thioredoxins. PAPS converts 3′-phosphoadenosine 5′-phosphosulfate to sulfite. Since glutaredoxins can also perform this reaction, methionine auxotrophy is seen only in the quadruple mutant lacking thioredoxins and glutaredoxins (29). A similar perturbation of the PAPS redox status could also explain the methionine auxotrophy of the P. anserina ΔPaTrx1 mutants. Interestingly, the methionine auxotrophy phenotype is more severe in double and triple mutants, indicating that PaTrx1 can be substituted for by PaTrx2 or PaTrx3, although inefficiently. This observation suggests that the P. anserina thioredoxins might be functionally less redundant than the S. cerevisiae enzymes. This is in line with the fact the S. cerevisiae proteins closely resemble PaTrx2 (Fig. 1) but are more distantly related to PaTrx1/PaTrx3. However, overexpression of PaTrx2 in a ΔPaTrx1 ΔPaTrx3 background leads to the restoration of methionine prototrophy.
The severity of methionine auxotrophy is correlated with an increase in life span (Table 1). It is known from previous analyses that many genes influence longevity in P. anserina (32), including the leu1-1 gene involved in leucine biosynthesis. At the present time the connection between auxotrophy and life span modification is not clear, especially because longevity is measured on supplemented medium. However, an imbalance in the pool of cytosolic amino acids may perturb efficient translation, which has been shown to be a major determinant of life span (38). Alternatively, ROS are hypothesized to modulate life span (23), and tampering with thioredoxins may modify ROS homeostasis, although we were not able to detect a modification of peroxide resistance in the thioredoxin mutants. Remarkably, in yeast and in mammalian cell cultures, aging is expressed through radically different aspects but is also controlled by thioredoxins (12, 27-29). It remains to be determined whether the underlying molecular mechanisms involved are conserved in all of these organisms.
A clear difference in the roles of PaTrx1/PaTrx3 versus PaTrx2 can be seen during sexual reproduction and CG development. Indeed, deletions of both PaTrx1 and PaTrx3 result in defects in fruiting body formation and in restriction of CG. Additional deletion of PaTrx2 results in more severe phenotypes, but overexpression of PaTrx2 does not compensate for the fertility and CG defects. Therefore, a protein belonging to the PaTrx1/PaTrx3 tandem is essential for both processes, and this tandem cannot be substituted for by PaTrx2. The physiological role of PaTrx2 is unclear since deletion of this gene does not result in any modified phenotype. It is possible that it substitutes for the other thioredoxins during vegetative growth when P. anserina meets special conditions. At the present time, the exact cellular activity of PaTrx1/PaTrx3 during development is unknown. In mammals, a thioredoxin is involved in the activation of the ASK1 MAPKKK by ROS (33). The Trx thioredoxin binds to the nonphosphorylated and inactive ASK1 MAPKKK protein. Upon activation by ROS, thioredoxin releases ASK1, which, following an unknown stimulus, self-activates by autophosphorylation. In P. anserina, we described the PaMpk1 MAPK cascade necessary for perithecium production, CG, and hyphal interference (14, 15). The requirement of the PaNox1 NADPH oxidase in the same processes (14, 24) suggested that the MAPKKK of this cascade could also be activated by ROS. If a thioredoxin, playing a role similar to that of the mammalian Trx, is present in P. anserina, the PaMpk1 cascade is expected to be constitutively activated in the corresponding lack of function mutant. This should result in the facilitation of CG, as we have previously shown by overexpressing PaASK1 and PaMpk1 (14, 15). On the contrary, we observed that deletions of the PaTrx1 and PaTrx3 genes result in an impairment of CG, indicating that thioredoxins do not act as in mammals by repressing the PaASK1 MAPKKK. Moreover, since we were not able to show any difference in phosphorylation and nuclear localization for PaMpk1 in these thioredoxin mutants, the involvement of the PaTrx1/PaTrx3 tandem into production of perithecia is not likely mediated by the PaMpk1 cascade. This is in accordance with the effects of the deletions on sexual reproduction, CG and hyphal interference. Indeed, first, thioredoxins are dispensable for hyphal interference, whereas the MAPK module and PaNox1 are not (36). Second, mutants of the thioredoxin genes do not impair the formation of the C cytoplasmic and infectious factor responsible for CG, whereas mutations in the genes coding MAPKKK, MAPKK, MAPK, and NADPH oxidase abolish production of the C factor (14, 15, 24). The effect of the thioredoxin mutations is to impair C propagation, as do some of the previously described mutants affecting CG (8). One can hypothesize that these mutants trigger a constitutive stress, known to restrict the propagation of C (8, 37). The same explanation can be proposed in the case of the thioredoxin mutants. Third and finally, the steps at which the thioredoxins, the MAPK PaMpk1, and the PaNox1 NADPH oxidase are required during fructification development are different. The MAPK PaMpk1 and NADPH oxidase PaNox1 are required to build the envelope, which originates from the female parent, and are dispensable in the dikaryon resulting from fertilization (11), whereas thioredoxins are required both in the dikaryons and in the envelope of perithecia. A general disturbance of physiology, perhaps due to a compromised redox status, could account for the failure of the ΔPaTrx1 ΔPaTrx3 and ΔPaTrx1 ΔPaTrx2 ΔPaTrx3 mutants to complete sexual reproduction. Alternatively, thioredoxin could possibly participate in the activation or inactivation of some proteins required for sexual reproduction by directly interacting with them, as shown for the YAP1 transcription factor in S. cerevisiae (10). We recently proposed a model, in which ROS involved in signaling perithecium development by PaMpk1 are secreted outside the cells (11), which might be in line with the lack of a direct role of thioredoxins in this pathway. We have undertaken a genetic approach to identify the extracellular effectors of the PaMpk1 pathway (8).
In summary, in the filamentous fungus P. anserina, the three cytosolic thioredoxins PaTrx1, PaTrx2, and PaTrx3 have a broad role in the control of sulfur metabolism, sexual reproduction, and cell degeneration. The different isoforms seem to harbor more specialized functions than in the yeast S. cerevisiae, reflecting the fact that these isoforms belong to two different families and that P. anserina has a more complex life cycle with more regulatory steps to go through. Finally, thioredoxins do not regulate the PaMpk1 MAPK pathway as direct inhibitors. Thus, they seem to act differently from the mammalian thioredoxin Trx known to activate the ASK1 MAPK to signal apoptosis. However, it remains to be tested whether the thioredoxins participate in the activation of the two other MAPKs present in the genome of P. anserina (13). While this manuscript was under revision, M. Thön et al. (41a) published a study demonstrating that deletion of the TrxA gene of Aspergillus nidulans inhibits the maturation of fruiting bodies, indicating that the role(s) of thioredoxins during elaboration of fructifications may have been conserved during filamentous ascomycete evolution.
Supplementary Material
Acknowledgments
We thank Hervé Lalucque, who contributed to the initiation of this study; Sylvain Brun for critical reading of the manuscript; Sylvie François and Magali Prigent for expert technical assistance; and Anne-Lise Haenni for correcting the manuscript.
This study was supported by ANR grant ANR-05-Blan-0385-02.
Footnotes
Published ahead of print on 12 October 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Strul (ed.). 1987. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 3.Brygoo, Y., and R. Debuchy. 1985. Transformation by integration in Podospora anserina. I. Methodology and phenomenology. Mol. Gen. Genet. 200:128-131. [Google Scholar]
- 4.Byun, H. S., E. W. Cho, J. S. Kim, M. S. Moon, J. J. Yum, K. C. Kim, and I. G. Kim. 2005. Thioredoxin overexpression in HT-1080 cells induced cellular senescence and sensitization to gamma radiation. FEBS Lett. 579:4055-4062. [DOI] [PubMed] [Google Scholar]
- 5.Coppin, E., and P. Silar. 2007. Identification of PaPKS1, a polyketide synthase involved in melanin formation and its use as a genetic tool in Podospora anserina. Mycol. Res. 111:901-908. [DOI] [PubMed] [Google Scholar]
- 6.Gan, Z. R. 1991. Yeast thioredoxin genes. J. Biol. Chem. 266:1692-1696. [PubMed] [Google Scholar]
- 7.Garrido, E. O., and C. M. Grant. 2002. Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol. Microbiol. 43:993-1003. [DOI] [PubMed] [Google Scholar]
- 8.Haedens, V., F. Malagnac, and P. Silar. 2005. Genetic control of an epigenetic cell degeneration syndrome in Podospora anserina. Fungal Genet. Biol. 42:564-577. [DOI] [PubMed] [Google Scholar]
- 9.Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264:13963-13966. [PubMed] [Google Scholar]
- 10.Izawa, S., K. Maeda, K. Sugiyama, J. Mano, Y. Inoue, and A. Kimura. 1999. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 274:28459-28465. [DOI] [PubMed] [Google Scholar]
- 11.Jamet-Vierny, C., M. Prigent, and P. Silar. 2007. IDC1, a Pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet. Biol. doi: 10.1016/j.fgb.2007.1004.1005. [DOI] [PubMed]
- 12.Jee, C., L. Vanoaica, J. Lee, B. J. Park, and J. Ahnn. 2005. Thioredoxin is related to life span regulation and oxidative stress response in Caenorhabditis elegans. Genes Cells 10:1203-1210. [DOI] [PubMed] [Google Scholar]
- 13.Kicka, S. 2005. Ph.D. thesis. Université de Paris 7-Denis Diderot, Orsay, France.
- 14.Kicka, S., C. Bonnet, A. K. Sobering, L. P. Ganesan, and P. Silar. 2006. A mitotically inheritable unit containing a MAP kinase module. Proc. Natl. Acad. Sci. USA 36:13445-13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kicka, S., and P. Silar. 2004. PaASK1, a mitogen-activated protein kinase kinase kinase that controls cell degeneration and cell differentiation in Podospora anserina. Genetics 166:1241-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, J. H., and D. H. Kim. 2006. Cloning and characterization of a thioredoxin gene, CpTrx1, from the chestnut blight fungus Cryphonectria parasitica. J. Microbiol. 44:556-561. [PubMed] [Google Scholar]
- 17.Laloi, C., D. Mestres-Ortega, Y. Marco, Y. Meyer, and J. P. Reichheld. 2004. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 134:1006-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lara-Ortiz, T., H. Riveros-Rosas, and J. Aguirre. 2003. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50:1241-1255. [DOI] [PubMed] [Google Scholar]
- 19.Lecellier, G., and P. Silar. 1994. Rapid methods for nucleic acids extraction from petri dish grown mycelia. Curr. Genet. 25:122-123. [DOI] [PubMed] [Google Scholar]
- 20.Lee, K. K., M. Murakawa, S. Takahashi, S. Tsubuki, S. Kawashima, K. Sakamaki, and S. Yonehara. 1998. Purification, molecular cloning, and characterization of TRP32, a novel thioredoxin-related mammalian protein of 32 kDa. J. Biol. Chem. 273:19160-19166. [DOI] [PubMed] [Google Scholar]
- 21.Lee, M. Y., K. H. Shin, Y. K. Kim, J. Y. Suh, Y. Y. Gu, M. R. Kim, Y. S. Hur, O. Son, J. S. Kim, E. Song, M. S. Lee, K. H. Nam, K. H. Hwang, M. K. Sung, H. J. Kim, J. Y. Chun, M. Park, T. I. Ahn, C. B. Hong, S. H. Lee, H. J. Park, J. S. Park, D. P. Verma, and C. I. Cheon. 2005. Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol. 139:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y., and W. Min. 2002. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ. Res. 90:1259-1266. [DOI] [PubMed] [Google Scholar]
- 23.Lorin, S., E. Dufour, and A. Sainsard-Chanet. 2006. Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina. Biochim. Biophys. Acta 1757:604-610. [DOI] [PubMed] [Google Scholar]
- 24.Malagnac, F., H. Lalucque, G. Lepere, and P. Silar. 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41:982-997. [DOI] [PubMed] [Google Scholar]
- 25.Matsui, M., M. Oshima, H. Oshima, K. Takaku, T. Maruyama, J. Yodoi, and M. M. Taketo. 1996. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev. Biol. 178:179-185. [DOI] [PubMed] [Google Scholar]
- 26.Michan, S., F. Lledias, and W. Hansberg. 2003. Asexual development is increased in Neurospora crassa cat-3-null mutant strains. Eukaryot. Cell 2:798-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miranda-Vizuete, A., J. C. Gonzalez, G. Gahmon, J. Burghoorn, P. Navas, and P. Swoboda. 2006. Lifespan decrease in a Caenorhabditis elegans mutant lacking TRX-1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 580:484-490. [DOI] [PubMed] [Google Scholar]
- 28.Mitsui, A., J. Hamuro, H. Nakamura, N. Kondo, Y. Hirabayashi, S. Ishizaki-Koizumi, T. Hirakawa, T. Inoue, and J. Yodoi. 2002. Overexpression of human thioredoxin in transgenic mice controls oxidative stress and life span. Antioxid. Redox Signal. 4:693-696. [DOI] [PubMed] [Google Scholar]
- 29.Muller, E. G. 1991. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J. Biol. Chem. 266:9194-9202. [PubMed] [Google Scholar]
- 30.Rizet, G. 1952. Les phénomènes de barrage chez Podospora anserina. I. Analyse génétique des barrages entre souches S and s. Rev. Cytol. Biol. Veg. 13:51-92. [Google Scholar]
- 31.Rizet, G., and C. Engelmann. 1949. Contribution à l'étude génétique d'un ascomycète tétrasporé: Podospora anserina (Ces.) Rehm. Rev. Cytol. Biol. Veg. 11:201-304. [Google Scholar]
- 32.Rossignol, M., and P. Silar. 1996. Genes that control longevity in Podospora anserina. Mech. Ageing Dev. 90:183-193. [DOI] [PubMed] [Google Scholar]
- 33.Saitoh, M., H. Nishitoh, M. Fujii, K. Takeda, K. Tobiume, Y. Sawada, M. Kawabata, K. Miyazono, and H. Ichijo. 1998. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17:2596-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salz, H. K., T. W. Flickinger, E. Mittendorf, A. Pellicena-Palle, J. P. Petschek, and E. B. Albrecht. 1994. The Drosophila maternal effect locus deadhead encodes a thioredoxin homolog required for female meiosis and early embryonic development. Genetics 136:1075-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silar, P. 1995. Two new easy-to-use vectors for transformations. Fungal Genet. Newsl. 42:73. [Google Scholar]
- 36.Silar, P. 2005. Peroxide accumulation and cell death in filamentous fungi induced by contact with a contestant. Mycol. Res. 109:137-149. [DOI] [PubMed] [Google Scholar]
- 37.Silar, P., V. Haedens, M. Rossignol, and H. Lalucque. 1999. Propagation of a novel cytoplasmic, infectious and deleterious determinant is controlled by translational accuracy in Podospora anserina. Genetics 151:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silar, P., H. Lalucque, and C. Vierny. 2001. Cell degeneration in the model system Podospora anserina. Biogerontology 2:1-17. [DOI] [PubMed] [Google Scholar]
- 39.Silar, P., and M. Picard. 1994. Increased longevity of EF-1 alpha high-fidelity mutants in Podospora anserina. J. Mol. Biol. 235:231-236. [DOI] [PubMed] [Google Scholar]
- 40.Takemoto, D., A. Tanaka, and B. Scott. 2006. A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell 18:2807-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, A., M. J. Christensen, D. Takemoto, P. Park, and B. Scott. 2006. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell 18:1052-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Thön, M., Q. Al-Abdallah, P. Hortschansky, and A. A. Brakhage. 2007. The thioredoxin system of the filamentous fungus Aspergillus nidulans: impact on development and oxidative stress response. J. Biol. Chem. 282:27259-27269. [DOI] [PubMed] [Google Scholar]
- 42.Tsukamoto, S., S. Morita, E. Hirano, H. Yokoi, T. Masumura, and K. Tanaka. 2005. A novel cis-element that is responsive to oxidative stress regulates three antioxidant defense genes in rice. Plant Physiol. 137:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, Z., and W. Wickner. 1996. Thioredoxin is required for vacuole inheritance in Saccharomyces cerevisiae. J. Cell Biol. 132:787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.