Abstract
The intracellular survival of the bacterial pathogen Chlamydia trachomatis depends on protein synthesis by the microbe soon after internalization. Pharmacologic inhibition of bacterial translation inhibits early trafficking of the parasitophorous vacuole (inclusion) to the microtubule-organizing center (MTOC) and promotes its fusion with lysosomes, which is normally blocked by Chlamydia. Depletion of cellular tryptophan pools by gamma interferon-inducible indoleamine-2,3-dioxygenase (IDO) is believed to be the major innate immune mechanism controlling C. trachomatis infection in human cells, an action to which the bacteria can respond by converting into a nonreplicating but highly reactivatable persistent state. However, whether severe IDO-mediated tryptophan starvation can be sufficient to fully arrest the chlamydial life cycle and thereby counteract the onset of persistence is unknown. Here we demonstrate that at low exogenous tryptophan concentrations a substantial fraction of C. trachomatis bacteria fail to traffic to the MTOC or to switch into the conventional persistent state in gamma interferon-induced human cells. The organisms stay scattered in the cell periphery, do not retain infectivity, and display only low transcriptional activity. Importantly, the rate at which these aberrant Chlamydia bacteria become reactivated upon replenishment of cellular tryptophan pools is substantially lower. Thus, severe tryptophan depletion in cells with high IDO activity affects chlamydial development more rigorously than previously described.
The obligate intracellular pathogen Chlamydia trachomatis is a gram-negative bacterium that causes severe infections of the eye and the urogenital tract. C. trachomatis serovars A to C are the leading cause of preventable blindness worldwide, whereas serovars D to K are responsible for the majority of sexually transmitted infections in the United States, causing serious health problems, such as pelvic inflammatory disease, ectopic pregnancy, salpingitis, and infertility (7). Serovars L1 to L3 share the unique ability to pass through epithelia, after which they disseminate, invade, and destroy lymphatic tissue and cause lymphogranuloma venereum (22). In 2005 almost 1 million new C. trachomatis infections were reported to the Centers for Disease Control and Prevention; the number of reported cases is increasing yearly, but 1 million cases still is much below the estimated 4 million new cases per year in the United States (9, 33). The associated annual costs for treatment exceed 2.4 billion dollars (33).
Chlamydia species have a biphasic life cycle (for a review, see reference 7) that is initiated by the endocytic uptake into the host cell of an elementary body (EB), the infectious but metabolically inactive form of the bacterium. The chlamydial vacuole, which is termed an inclusion, shows no apparent acquisition of lysosomal markers (28). Interestingly, the ability to inhibit fusion with the endosomal system is independent of chlamydial protein synthesis during the early phase of the life cycle but requires bacterial translation at later stages (28, 29). Upon entry, the organism exploits the host's intracellular trafficking machinery by recruiting the microtubule motor protein dynein to the outer surface of the vacuole, which drives the migration of the inclusion along the microtubule network towards the microtubule-organizing center (MTOC), where it eventually resides in a perinuclear position (14). If a cell becomes multiply infected, fusion of the individual inclusions occurs. Importantly, bacterial translation is essential for both intracellular trafficking (14) and late inhibition of lysosomal fusion (29), demonstrating that these processes are actively controlled by Chlamydia. Differentiation of the EB into the metabolically active form of the bacterium, the so-called reticulate body (RB), is probably initiated early after cell entry and is a prerequisite for Chlamydia to start multiplication. Twenty-four to 72 hours after entry the organisms convert back into EBs, which eventually are released through host cell lysis or extrusion.
The immune response against C. trachomatis strongly depends on the inflammatory cytokine gamma interferon (IFN-γ), which highly induces the cytosolic tryptophan-degrading enzyme indoleamine-2,3-dioxygenase (IDO) (27). In human cells IDO-mediated tryptophan starvation seems to be the major innate immune mechanism controlling C. trachomatis growth (27). In turn, the organism responds to limiting cellular tryptophan concentrations by converting into an aberrant nondividing persistent form (2) that can be very efficiently reactivated and proceeds with the normal life cycle upon withdrawal of the persistence-inducing stimulus (3, 5). Although it has been shown that IDO can degrade culture medium-derived tryptophan into concentrations in the nanomolar range in vitro (34), very little is known about cellular tryptophan levels that are established by IDO in the context of an infection in vivo. However, one recent study showed that in Toxoplasma gondii-infected mice the tryptophan concentration in lung tissue dramatically dropped from ∼30 μM to ∼2 μM, an effect that was entirely dependent on IFN-γ signaling (11). This suggests that IDO-mediated depletion of cellular tryptophan pools is very efficient also in vivo. Moreover, many cells lack high-affinity tryptophan import systems and are in turn predominantly, if not exclusively, dependent on the neutral amino acid transporter system L for import of tryptophan (25, 31, 32). However, considering the relatively low substrate affinity in combination with the extensive substrate promiscuity of this permease, tryptophan uptake would be greatly reduced at very low exogenous concentrations as other amino acids compete for transport (24, 31). Consequently, cells that depend on system L for tryptophan import would likely undergo dramatic depletion of their tryptophan pools upon IFN-γ-mediated induction of IDO.
These issues prompted us to focus on the development of C. trachomatis under severe tryptophan starvation conditions. We show here that at very low tryptophan levels a substantial fraction of bacteria fail to convert properly into the persistent state. Both trafficking of the organism to the MTOC and fusion of multiple inclusions in the cell are effectively suppressed under these conditions, suggesting that there is early developmental arrest. The aberrant bacteria are scattered in the cell periphery, where they neither form infectious progeny nor retain their infectivity. Most importantly, this fraction of bacteria is characterized by a significantly lower rate of reactivation upon replenishment of the tryptophan pools.
In summary, we provide evidence that moderate and severe tryptophan starvation differentially affect C. trachomatis. Scattered bacterial forms that characteristically arise when the tryptophan concentration is very low display altered early development, are morphologically distinct from typical persistent giant RBs, and show a substantially lower rate of reactivation.
MATERIALS AND METHODS
Cells, bacteria, reagents, and antibodies.
HeLa M cells were grown in Iscove's modified Dulbecco's medium-10% fetal calf serum (FCS) (Gibco) supplemented with nonessential amino acids (Gibco) and penicillin/streptomycin (Gibco). C. trachomatis serovar L1 was propagated in HeLa cells grown in antibiotic-free Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS.
Fluorescein isothiocyanate (FITC)-labeled anti-Chlamydia lipopolysaccharide (LPS) antibodies were purchased from Argene. These antibodies did not stain uninfected cells, confirming their specificity (data not shown). Mouse monoclonal antibody TUB 2.1 recognizes human β-tubulin and was purchased from Sigma. Alexa-546-coupled anti-mouse immunoglobulin G and Topro-3 iodide were purchased from Molecular Probes. IFN-γ was purchased from Peprotech.
Infections, flow cytometry, and immunofluorescence.
A total of 1 × 106 cells per well were seeded overnight in a six-well plate. The next day, the cells were washed with phosphate-buffered saline (PBS), and the medium was replaced with 5 ml antibiotic-free custom-synthesized tryptophan-negative high-glucose DME H-21 (UCSF Cell Culture Facilities) supplemented or not supplemented with serum or various amounts of tryptophan. Where indicated below, the medium also contained 200 U/ml (10 ng/ml) IFN-γ. Following a 24-h incubation at 37°C, 7.5 × 106 infectious units (IFUs) of C. trachomatis serovar L1 was applied to the cells in 1 ml serum-free medium on a rocking platform at room temperature for 2.5 h. Next, the cells were extensively washed to remove remaining extracellular bacteria, and the type of medium that had been used for the preinfection period was freshly prepared and added to the cells, which were incubated for another 24 h. Afterwards, the cells were washed with PBS, detached using 1.3 mM EDTA, fixed in 2% formaldehyde in PBS-6.7 mM HEPES, permeabilized with 0.5% saponin (Sigma), and stained with FITC-labeled anti-Chlamydia LPS antibodies. Finally, infected cells were analyzed by flow cytometry using a FACScan system (Becton Dickinson) or by confocal fluorescence microscopy using a Leica TCS SP2 confocal microscope (Leica Microsystems).
IFU titer determination.
IFUs were released from infected cells by ultrasound using an ice-cooled cup horn Sonifier (three treatments at 85-W for 20 s; Branson Digital Sonifier; Branson) before confluent HeLa cell layers were incubated with a 10-fold dilution series of sonicates derived from equal numbers of cells for 2 h. The freshly infected cells were incubated at 37°C for 24 h in DMEM containing 10% FCS and 1 μg/ml cycloheximide, fixed, and stained with FITC-labeled anti-Chlamydia LPS antibodies. The percentage of infected cells in each sample was determined by flow cytometry.
Analysis of chlamydial cluster formation.
Cells were cultivated 24 h before infection as well as after infection in tryptophan-negative DMEM supplemented or not supplemented with 200 U/ml (10 ng/ml) IFN-γ and 10% serum or the indicated amount of tryptophan. Infection was carried out on 12-mm glass coverslips at a multiplicity of infection (MOI) of 30 at room temperature for 2 h. Cells were fixed and permeabilized by 10 min of incubation with cold methanol at −20°C, followed by 1 min of incubation with cold acetone on ice. Antibodies were applied in PBS-1% bovine serum albumin for 1 h at dilutions recommended by the supplier. Samples were extensively washed and mounted in ProLong Gold antifade reagent (Invitrogen) for immunofluorescence analysis. At least 1,000 cells per sample were blind counted to determine the percentage of cells harboring a perinuclear bacterial cluster.
Reactivation assay.
A total of 1.5 × 106 cells prestarved for 24 h in tryptophan-negative medium containing 200 U/ml (10 ng/ml) IFN-γ were infected at an MOI of 1 or 5 with C. trachomatis and cultivated for another 24 h with 0, 3, or 6 μM tryptophan in the presence of 200 U/ml (10 ng/ml) IFN-γ. Induction of persistence with 6 μM tryptophan under these conditions was confirmed in a separate experiment (data not shown). Next, to reactivate viable bacteria, the medium was exchanged against IFN-γ-free regular DMEM-10% FCS for 24 h. Following this, cells were harvested and analyzed as described above.
Reverse transcription (RT)-PCR and real-time PCR.
Bacterial RNA was isolated from equal numbers of Chlamydia-infected cells by standard Trizol-chloroform extraction and was further purified using an RNeasy mini kit (QIAGEN), and DNA contamination was removed with a Turbo DNAfree kit (Ambion). cDNA was prepared using a First Strand synthesis kit (Stratagene) in combination with random primers. The actin, incG, trpB, and 16S rRNA genes were detected with a standard Taq-driven PCR (annealing temperature, 65°C; 27 cycles) using primer pairs 5′-GGCATCGTGATGGACTCCG-3′/5′-GCTGGAAGGTGGACAGCGA-3′, 5′-TTTGTTTGCGGTGTTTGCTTCAGG-3′/5′-CGAATGCCCCAGAAACACTATCTCC-3′, 5′-ATGCTCTTGGTCAGTGTTTGC TTGC-3′/5′-GCCCAGTCCTCCCCCTTCCAC-3′, and 5′-GCTGCGGTAATACGGAGGGTGC-3′/5′-AACCCAACACCTCACGGCACG-3′, respectively. PCR products were sequenced. To exclude the possibility that the PCR amplification was derived from bacterial genomic DNA contamination rather than from mRNA, all PCR analyses included controls performed with an additional “mock cDNA” sample to which no reverse transcriptase had been added. Standard SYBR green-based real-time PCR (Applied Biosystems) was carried out with a Stratagene Mx3000P Lightcycler using the same primer pairs. Each reaction was set up in duplicate.
RESULTS
Severe tryptophan starvation gives rise to aberrant scattered C. trachomatis forms in human IFN-γ-induced cells.
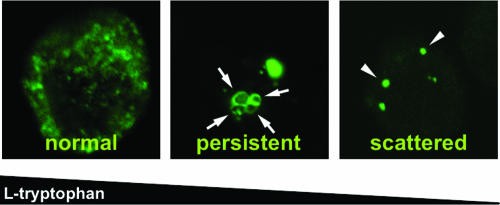
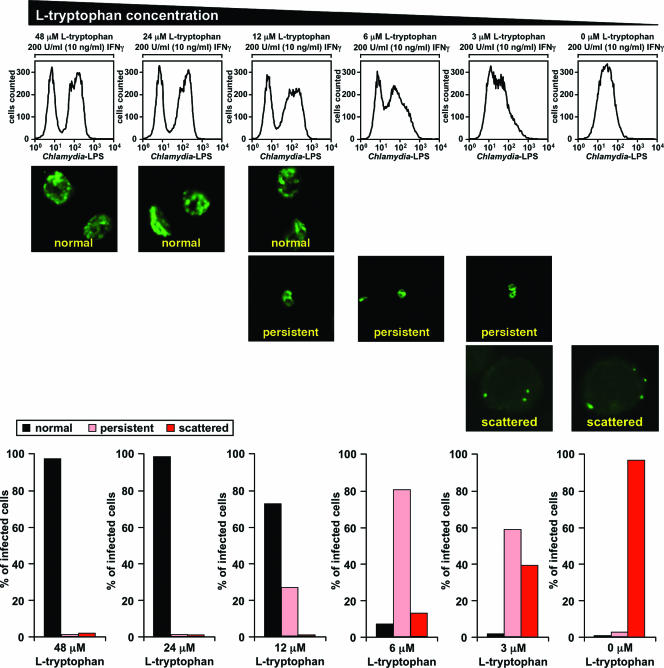
Immunofluorescence has been successfully used to study persistence-associated changes in the morphology of Chlamydia inclusions (3, 12). Therefore, we used this technique coupled with flow cytometry as a high-throughput approach to monitor chlamydial growth and development in IFN-γ-induced human cells in the presence of various concentrations of exogenous tryptophan. To this end, we incubated HeLa M cells in serum-free medium containing 200 U/ml (10 ng/ml) IFN-γ plus the indicated amounts of tryptophan 24 h before and 24 h after infection with C. trachomatis serovar L1. Note that the indicated tryptophan concentrations represented the levels of the amino acid at the start of the experiment and that the levels were expected to drop during IDO-mediated breakdown. The cells were then harvested, fixed, intracellularly stained with anti-Chlamydia LPS antibodies, and analyzed by examining immunofluorescence (Fig. 1 and 2, middle panel) or by flow cytometry (Fig. 2, top panel). To quantify the immunofluorescence results, inclusions were classified into three clearly distinguishable categories according to their morphology (Fig. 1 and 2, lower panel). In line with the results of Gieffers and coworkers (12), large Chlamydia vacuoles displaying a high degree of granularity were considered “normal” inclusions, whereas small but intensely bright inclusions containing giant RBs and lacking granularity were recognized as “persistent” inclusions (Fig. 1, left and middle panels). Consistent with the previous finding that provision of excess tryptophan can rescue the growth of C. trachomatis in IFN-γ-stimulated HeLa cells (8), only “normal” inclusions could be found with 24 to 48 μM exogenous tryptophan (Fig. 2, columns 1 and 2). However, with 12 μM tryptophan normal inclusions started to disappear, and such inclusions were almost completely absent with 6 μM tryptophan when the “persistent” population peaked (Fig. 2, columns 3 and 4). These changes could not be readily seen when the correlating fluorescence-activated cell sorting (FACS) histograms were examined, indicating that flow cytometry is not capable of distinguishing between “normal” and “persistent” inclusions (Fig. 2, top panel, columns 1 to 4).
FIG. 1.
Inclusion morphologies observed with different tryptophan concentrations. HeLa M cells were incubated in the presence of 200 U/ml (10 ng/ml) IFN-γ with a high (80 μM) (left panel) or moderate (6 μM) (middle panel) exogenous tryptophan concentration or without tryptophan (right panel) for 24 h before and after infection with C. trachomatis serovar L1. The cells were stained intracellularly with FITC-labeled anti-Chlamydia LPS antibodies and analyzed by immunofluorescence. The arrows indicate individual giant RBs in conventional persistent inclusions (middle panel), while the arrowheads indicate dot-shaped scattered bacteria (right panel).
FIG. 2.
Severe tryptophan starvation gives rise to aberrant scattered C. trachomatis forms in human IFN-γ-treated cells. A total of 1.5 × 106 HeLa M cells were incubated for 24 h in tryptophan-free medium supplemented with the indicated amounts of tryptophan and 200 U/ml (10 ng/ml) IFN-γ, infected with 7.5 × 106 IFUs of C. trachomatis serovar L1 for 2.5 h, and cultivated for another 24 h in the same medium used for the preinfection incubation. Cells were detached, fixed, stained with FITC-labeled anti-Chlamydia LPS antibodies, and analyzed by flow cytometry (upper panel) or immunofluorescence (middle panel). Inclusion types detected by immunofluorescence were classified according to morphological characteristics shown in Fig. 1: normal (large granular vacuoles), persistent (smaller, nongranular, intensely bright vacuoles harboring giant RBs), and scattered (dot-shaped inclusions mostly spread in the cell periphery). Cells harboring both scattered and persistent forms simultaneously were included in only the persistent category.
As in vivo tissue concentrations of tryptophan of ∼2 μM have been observed in a murine infection model (11), which are concentrations at which tryptophan import into cells lacking a high-affinity transport system would be practically eliminated (31), we were also interested in the development of Chlamydia with lower concentrations of this essential amino acid. Most strikingly, with an exogenous tryptophan concentration of 6 μM a third population of inclusions appeared; this population was very prominent when the tryptophan concentration was 3 μM, and these inclusions were the only type of inclusions when tryptophan was not included in the assay mixture (Fig. 2, lower panel, columns 4 to 6). In the corresponding cells Chlamydia vacuoles were seen as dots mainly scattered in the cell periphery (Fig. 1, right panel, and Fig. 2, middle panel, columns 4 to 6). The organisms did not seem to replicate, and their inclusions failed to fuse. We noted that not only normal chlamydial development but also the conversion of microbes into the giant forms characteristic of the persistent state (2, 12, 16) was completely suppressed in this fraction of bacteria. The observed drastic growth defect was also reflected in the correlated FACS histograms, which displayed a dramatic reduction in the average fluorescence of infected cells (Fig. 2, top panel, columns 5 and 6).
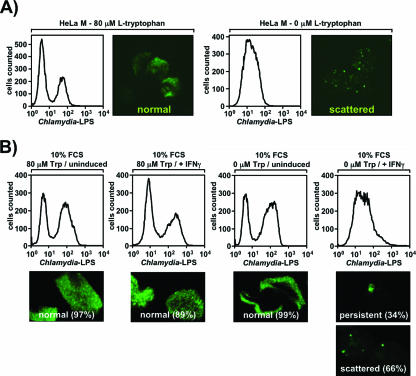
Interestingly, the appearance of “scattered” Chlamydia inclusions did not require the presence of IFN-γ (Fig. 3A) when cells were cultured in the absence of tryptophan, demonstrating that depletion of this essential amino acid alone is sufficient to suppress the organism. As IDO is not detectable in non-IFN-γ-treated HeLa cells, this also argues against a role for tryptophan metabolites in the induction of the “scattered” state. In line with this, exogenous provision of culture medium with even 1 mM of the tryptophan metabolite l-kynurenine failed to induce “scattered” organisms (data not shown). Consequently, it is IDO-mediated severe tryptophan depletion rather than IDO-mediated generation of tryptophan metabolites that drives the bacteria into the “scattered” state in IFN-γ-induced cells. “Scattered” Chlamydia forms were also detected in analogous experiments in which IFN-γ-stimulated cells were grown in medium that included 10% serum instead of a tryptophan supplement, showing that this effect is not a result of serum starvation (Fig. 3B, column 4). Again, the severe developmental block could also be observed by flow cytometry. It is worth noting that under these conditions two-thirds of the cells harbored “scattered” bacteria, whereas in non-IFN-γ-treated cells virtually all inclusions were normal (Fig. 3B, column 3), underscoring the high activity of IDO in the cytokine-stimulated cells. However, based on our experience, the use of serum as a tryptophan source introduces substantial variation in experiments depending on the FCS batch used, which likely reflects different levels of free tryptophan in sera from different suppliers (data not shown). The fact that scattered Chlamydia cells have not been described so far in previous studies (2) may be the result of the use of relatively tryptophan-rich FCS in the experiments. Thus, we are confident that supplementation of serum-free tryptophan-negative medium with defined amounts of this amino acid is the most accurate and reproducible way of monitoring Chlamydia development, particularly with low tryptophan concentrations.
FIG. 3.
IFN-γ treatment or serum starvation is not essential for generation of scattered Chlamydia. (A) IFN-γ is not essential for generation of scattered Chlamydia. Chlamydia-infected cells were analyzed as described in the legend to Fig. 1 except that no IFN-γ was added at any time. Two different tryptophan concentrations (80 μM [left panel] and 0 μM [right panel]) were used. (B) Serum starvation is not required for generation of scattered Chlamydia. Chlamydia-infected cells were analyzed as described in the legend to Fig. 1 except that all samples contained 10% FCS plus the indicated amounts of added tryptophan; 200 U/ml (10 ng/ml) IFN-γ was added where indicated. The frequencies of the inclusion types in infected cells are expressed as percentages.
In summary, we show here that severe tryptophan starvation causes drastic developmental and growth defects in C. trachomatis that efficiently suppress the conversion of the organism into the long-lived persistent form.
Tryptophan depletion impairs chlamydial trafficking to the MTOC.
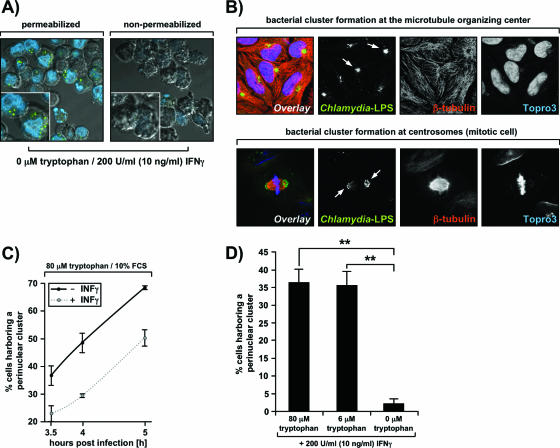
Two distinct scenarios may account for the observation of scattered Chlamydia forms in IFN-γ-induced cells grown in the presence of tryptophan concentrations of ≤6 μM: either the entry of the bacteria into prestarved HeLa M cells may be inhibited or the trafficking of the bacteria to the MTOC may be blocked. In the first scenario the bacteria would attach extracellularly to the surface of the host cells, whereas in the second scenario they would reside intracellularly beneath the plasma membrane. To distinguish between these possibilities, we stained infected cells grown without tryptophan simultaneously with Chlamydia-specific antibodies and Topro-3 under either permeabilizing or nonpermeabilizing conditions (Fig. 4A). Topro-3 is a membrane-impermeant DNA-binding dye that served as a permeabilization control in this experiment. Nonpermeabilized cells did not take up the dye and were not stained with anti-Chlamydia antibodies at all (Fig. 4A, right panel). This showed that the bacteria were in fact intracellular (Fig. 4A, left panel) and that the entry step was not significantly affected under these conditions.
FIG. 4.
Chlamydial trafficking to the MTOC requires tryptophan. (A) Aberrant scattered Chlamydia forms are intracellular. Cells were incubated with 200 U/ml (10 ng/ml) IFN-γ in serum-free tryptophan-negative DMEM 24 h before and after infection with C. trachomatis. Subsequently, cells were split into two samples and stained with FITC-labeled anti-Chlamydia LPS antibodies (green) and Topro-3 (blue) under either permeabilizing (0.5% saponin) (left panel) or nonpermeablizing (right panel) conditions. Cells were analyzed by confocal microscopy, and fluorescent signals were overlaid onto a differential interference contrast image to visualize all cells. (B) C. trachomatis inclusions migrate to the MTOC soon after entry into the cell. Cells grown on coverslips were cultivated in DMEM-10% FCS for 24 h before and 5 h after infection with C. trachomatis at an MOI of 30. Next, the cells were fixed in methanol-acetone, stained with antibodies against Chlamydia LPS (green) and β-tubulin (red) and with Topro-3 (blue) and analyzed by confocal fluorescence microscopy. The arrows indicate bacterial clusters formed at the MTOC in nonmitotic cells (top panels) and at the centrosomes of a mitotic cell (bottom panels). (C) IFN-γ delays C. trachomatis trafficking to the MTOC: time course analysis of chlamydial cluster formation at the MTOC. The experimental setup was similar to that used for panel B, with the modification that cells were treated (dotted line) or not treated (solid line) with 200 U/ml (10 ng/ml) IFN-γ before and after infection. The percentage of cells harboring a bacterial perinuclear cluster at 3.5, 4, or 5 h postinfection was determined by blind counting of FITC-labeled anti-Chlamydia LPS-stained samples. The error bars indicate the standard deviation from the mean of two independent experiments, and in each experiment at least 1,000 cells were blind counted. (D) Tryptophan is essential for chlamydial trafficking. Chlamydial cluster formation at the MTOC of IFN-γ-induced cells at 5 h postinfection was analyzed as described above for panel B except that the media used contained 200 U/ml (10 ng/ml) IFN-γ, a defined tryptophan concentration, and no serum. The error bars indicate the standard deviation from the mean of three independent experiments, and in each experiment at least 1,000 cells were blind counted. The differences between cluster formation with 0 and 6 μM tryptophan (two asterisks; P < 0.01) and between cluster formation with 0 and 80 μM tryptophan (two asterisks; P < 0.01) are statistically significant, as assessed by a one-way analysis of variance test with the Dunnett posttest.
Consequently, our findings point towards a chlamydial trafficking defect in cells in which tryptophan pools have been depleted through the action of IFN-γ-inducible IDO. To test this hypothesis, we infected cells at a high MOI (30) and monitored the subsequent formation of perinuclear bacterial clusters. In the presence of high tryptophan levels most cells contained a bacterial cluster that colocalized with the MTOC at 5 h postinfection (Fig. 4B, upper row showing nonmitotic cells and lower row showing mitotic cells), indicating that the migration of inclusions along the microtubule network occurred properly in these cells. However, treatment of the cells with IFN-γ significantly delayed chlamydial trafficking even when the initial exogenous tryptophan concentration was >80 μM (80 μM medium-derived tryptophan plus serum tryptophan), as revealed when the formation of bacterial clusters was assessed in a time course experiment (Fig. 4C). We noted that under these conditions chlamydial growth appeared to be fairly normal in IFN-γ-induced cells (Fig. 3B, column 2), indicating that the delay does not have detrimental consequences for the organism. Next, we analyzed bacterial migration towards the MTOC in the presence of defined tryptophan concentrations that either allow normal growth (80 μM), induce persistence (6 μM), or give rise to aberrant scattered forms (0 μM) in the maximal number of IFN-γ-induced cells (Fig. 2, columns 4 and 6). Surprisingly, a 13-fold reduction in exogenous tryptophan levels (6 μM versus 80 μM) had hardly any effect on chlamydial trafficking, whereas the absence of tryptophan stalled this process (Fig. 4D).
Together, the results show that mild tryptophan depletion delays trafficking of intracellular C. trachomatis to the MTOC, whereas severe tryptophan starvation blocks trafficking of intracellular C. trachomatis to the MTOC.
Aberrant scattered Chlamydia forms do not retain infectivity and display low transcriptional activity.
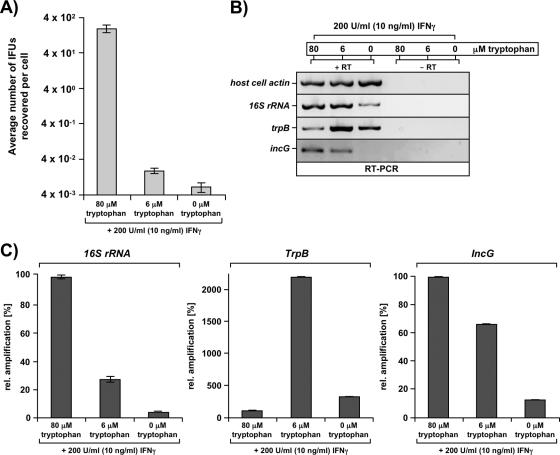
Morphologically visible transition of infectious, metabolically inactive EBs into noninfectious, metabolically active RBs requires bacterial translation (29) and is preceded by modification of the inclusion membrane through insertion of bacterial proteins, as well as early migration to the MTOC. Thus, the observed chlamydial trafficking defects with limiting tryptophan concentrations suggest that the development of the organism is quickly arrested upon cell entry. This prompted us to ask whether aberrant scattered Chlamydia bacteria would stay in the cell periphery as inert, transcriptionally silent forms that retain their infectivity. Therefore, we assessed the number of IFUs recoverable from IFN-γ-induced cells that harbored mainly normal (80 μM tryptophan), persistent (6 μM tryptophan), or scattered (0 μM tryptophan) inclusions. Twenty-four hours postinfection, cells grown in the presence of excess tryptophan contained on average ∼200 infectious EBs, whereas 4-orders-of-magnitude-fewer IFUs were recovered from cells harboring persistent RBs (Fig. 5A). Strikingly, infected cells grown in the absence of tryptophan yielded even lower numbers of IFUs than of persistent RBs, clearly showing that the bacteria do not retain their infectivity in the “scattered state” (Fig. 5A). Differences in the number of recoverable IFUs between scattered Chlamydia and persistent Chlamydia may reflect a somewhat decreased survival rate in the scattered population.
FIG. 5.
Aberrant scattered Chlamydia forms do not retain infectivity and display low transcriptional activity. (A) Scattered Chlamydia bacteria are not infectious. IFUs were released by sonication from equal numbers of IFN-γ-induced cells grown in the presence of defined tryptophan concentrations that allow normal bacterial growth (80 μM) or persistence (6 μM) or give rise to aberrant scattered Chlamydia bacteria (0 μM). IFU titers were determined by infecting confluent HeLa cell layers with a dilution series of the sonicate and are expressed as the average number of IFUs recovered per cell. The results of one representative experiment of two independent experiments are shown. (B and C) Scattered Chlamydia bacteria display low transcriptional activity. RNA was extracted from 5 × 105 infected cells that had undergone the indicated treatment. Conventional RT-PCR (B) or real-time PCR (C) was carried out using primers specific for the host cell actin, bacterial 16S rRNA, trpB, or incG gene. Amplification from mRNA rather than contaminating genomic DNA was confirmed using a mock cDNA synthesis reaction mixture lacking reverse transcriptase (RT) (right three lanes in panel B). All real-time PCRs were performed in duplicate, and the error bars show the deviations between the two samples.
However, since the majority of scattered Chlamydia bacteria appear to remain alive, as they can be reactivated (see below), we analyzed whether these bacteria are transcriptionally active. To this end, we extracted RNA from infected IFN-γ-stimulated cells that had been incubated in the presence of 80, 6, or 0 μM tryptophan, both preinfection and postinfection, and examined the expression of three well-characterized bacterial genes, the 16S rRNA, trpB and incG genes, by conventional semiquantitative RT-PCR (Fig. 5B) and quantitative real-time PCR (Fig. 5C). It is important to note that the absence or presence of tryptophan did not affect the infection efficiency in our experimental system. Isolation of RNA from equal numbers of cells was confirmed by a host cell actin gene-specific RT-PCR (Fig. 5B, row 1). Consistent with the findings of Ouellette and coworkers (23), we found that the 16S rRNA gene levels were reduced in persistent bacteria compared with normal growing bacteria (Fig. 5C, left panel). In cells harboring scattered Chlamydia bacteria, however, even lower levels of the 16S rRNA gene were detected (Fig. 5B, row 2, and Fig. 5C, left panel), indicating either that these bacterial forms have drastically lower average transcriptional activity or that a large fraction of these bacteria does not survive (or is transcriptionally inert) under severe tryptophan starvation conditions. Next, we probed for expression of trpB, a gene that is directly regulated by tryptophan levels through derepression of the promoter in the absence of the amino acid (1). As expected, we found substantial induction of the gene in persistent bacteria compared with normal growing bacteria (Fig. 5B, row 3, lanes 1 and 2; Fig. 5C, middle panel). Most interestingly, we also saw increased levels of trpB in scattered Chlamydia bacteria (Fig. 5B, row 3, lanes 1 and 3; Fig. 5C, middle panel), albeit to a minor extent, which could reflect active induction of the gene by either a low tryptophan concentration or a constitutively permissive promoter due to impaired trpR repressor protein synthesis. In either case, this result shows that at least some aberrant scattered Chlamydia bacteria are not completely silent in transcription and still respond to environmental conditions. Finally, we investigated the expression of the chlamydial incG gene, a prototype early gene whose expression is initiated within the first 2 h upon internalization (30). incG appeared to be expressed at somewhat reduced levels in persistent bacteria compared with normal growing bacteria (Fig. 5B, row 4, lanes 1 and 2; Fig. 5C, right panel), and again, scattered Chlamydia forms followed the trend observed for persistent RBs, but the reduction in the PCR signal was even more pronounced (Fig. 5B, lane 3).
Thus, in all cases analyzed five- to sevenfold-lower mRNA levels were found in scattered bacteria than in persistent bacteria. This probably reflected overall reduced transcriptional and metabolic activity in the scattered population. Thus, differences on the transcriptional level between the two Chlamydia forms may be primarily quantitative rather than qualitative.
Severe tryptophan depletion decreases the reactivation rate of Chlamydia.
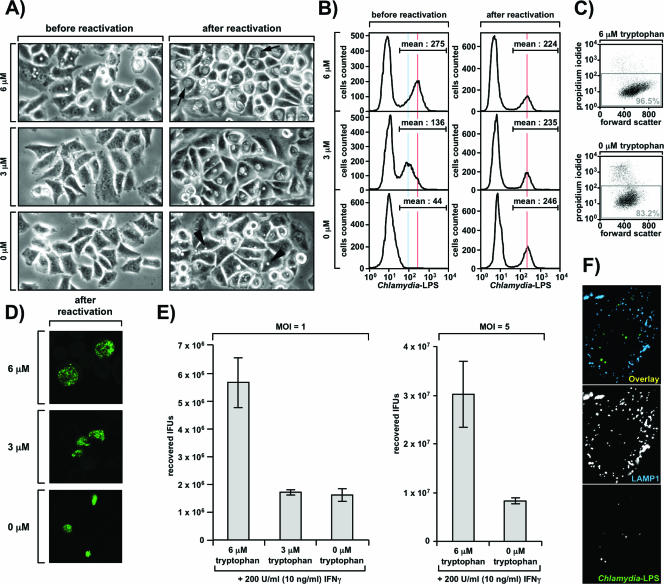
One hallmark of the persistent state is its reversibility, which allows rapid reactivation of the organism upon withdrawal of the persistence-inducing stimulus (3, 5). Therefore, it was important to determine whether scattered Chlamydia bacteria would also survive in the cell and continue with their developmental life cycle upon restoration of favorable growth conditions. To answer this question, we monitored chlamydial survival with low tryptophan concentrations in reactivation assays using light and confocal fluorescence microscopy (Fig. 6A and 6D), flow cytometry (Fig. 6B), and determination of IFU titers (Fig. 6E). To this end, we incubated prestarved Chlamydia-infected (MOI, 1) cells for 24 h under conditions that allow the onset of persistence (6 μM tryptophan) or induce the scattered state (0 μM tryptophan). Additionally, we included an intermediate tryptophan concentration, 3 μM, in this experiment. As expected, small inclusions typical of the persistent state were readily seen by light microscopy in the sample that had received 6 μM tryptophan (Fig. 6A, upper left panel). However, cells cultivated with 3 μM tryptophan displayed only a few very tiny inclusions, and cells grown in the absence of tryptophan did not show any obvious inclusions by light microscopy (Fig. 6A, middle and lower left panels). This documents our repeated observation that in contrast to persistent inclusions, scattered Chlamydia bacteria are not visible at the resolution provided by light microscopy. Consistent with these findings and the established correlation between inclusion brightness and low exogenous tryptophan levels (Fig. 2, right upper panels), flow cytometry using Chlamydia LPS-specific antibodies revealed intense bacterial staining with 6 μM tryptophan, diminished staining with 3 μM tryptophan, and no detectable staining with 0 μM tryptophan (Fig. 6B, left panels). Again, this demonstrates that scattered Chlamydia bacteria are also not detectable by FACS analysis (compare Fig. 6B with Fig. 2, rightmost histogram in the upper panel). To assess the viability of tryptophan-starved host cells under the conditions used, we performed propidium iodide staining with a sample that was mock infected but was otherwise treated as described above. As shown in Fig. 6C, the vast majority of cells excluded the dye, irrespective of whether the cells had been incubated with 0 or 6 μM tryptophan, indicating that the conditions used largely preserved cell viability.
FIG. 6.
Severe tryptophan depletion decreases the reactivation rate of Chlamydia. (A) Analysis of chlamydial reactivation by light microscopy. Prestarved, IFN-γ-stimulated HeLa M cells were incubated postinfection (MOI, 1) for 24 h with 6, 3, or 0 μM tryptophan plus 200 U/ml (10 ng/ml) IFN-γ. Light microscopic images were obtained either directly (left panels) or after reactivation of the same cells after an additional 24 h of incubation in the presence of excess tryptophan (without IFN-γ) (right panels). The arrows indicate large inclusions formed after recovery of persistent RBs. The arrowheads indicate very small inclusions formed after reactivation of scattered bacteria. (B and D) Analysis of chlamydial reactivation by flow cytometry (B) or immunofluorescence (D). Cells were treated as described in the legend to panel A, harvested, stained with anti-Chlamydia LPS antibodies, and analyzed by flow cytometry or confocal microscopy. The blue and red lines in panel B highlight distinct peak fluorescence intensities in the infected population. (C) Viability of HeLa M cells under the reactivation assay conditions. Mock-infected cells otherwise treated as described in the legend to panel A (using only 0 and 6 μM tryptophan) were detached, washed, and stained with propidium iodide to distinguish dead cells (permeabilized) from live cells (in the gray rectangle). (E) Analysis of chlamydial reactivation by IFU titer determination. Either cells were treated as described in the legend to panel A (left panel) or an identical experiment was performed with a higher MOI (MOI, 5) (right panel). Following reactivation, cells were processed for IFU titer determination as described in the legend to Fig. 4A. (F) Scattered Chlamydia bacteria do not reside in lysosomes. HeLa M cells were seeded on coverslips, incubated with 0 μM tryptophan and 200 U/ml (10 ng/ml) IFN-γ 24 h before and after infection (MOI, 5), fixed in methanol-acetone, stained with antibodies against Chlamydia-LPS (FITC conjugated) and LAMP1 (Alexa-647 conjugated), and analyzed by confocal microscopy.
Next, to allow reactivation of intracellular bacteria, the medium of the infected cultures was replaced with tryptophan-sufficient DMEM containing 10% FCS and lacking IFN-γ. Twenty-four hours later the cells were analyzed by light microscopy (Fig. 6A, right panels), flow cytometry (Fig. 6B, right panels), and immunofluorescence (Fig. 6D). As shown in Fig. 6B (right panels), bacteria from all three samples could be clearly reactivated, irrespective of whether they had originally been cultivated with 0, 3, or 6 μM tryptophan, suggesting that, in principle, both persistent and scattered Chlamydia bacteria have the ability to continue their normal life cycle when favorable growth conditions are restored. This is in line with our transcriptional data, which also suggested that at least a fraction of the scattered bacterial population is alive, even when tryptophan depletion is extremely harsh (Fig. 5B and 5C). In particular, persistent RBs were very efficiently reactivated and gave rise to large granular inclusions containing rapidly multiplying bacteria (Fig. 6A, upper panel, and Fig. 6D, upper panel). However, cells initially grown with 3 μM tryptophan harbored visibly smaller vacuoles upon reactivation (Fig. 6A, right middle panel, and Fig. 6D, middle panel), indicating that the ability of the pathogen to continue development was compromised under these conditions. Most strikingly, reactivated inclusions that were derived exclusively from scattered Chlamydia bacteria (0 μM tryptophan) were all extremely small, demonstrating that this bacterial fraction is substantially impaired in the reactivation process (Fig. 6A, right lower panel, and Fig. 6D, lower panel). Surprisingly, these differences could not be detected by flow cytometry (Fig. 6B, right panels) as smaller inclusions interestingly displayed greater brightness, an effect also readily noticed in the immunofluorescence analysis shown in Fig. 6D.
To further confirm our results for the reactivation of Chlamydia, we additionally determined IFU titers recovered after reactivation of scattered (0 μM tryptophan) or persistent (6 μM tryptophan) Chlamydia bacteria from cells infected at an MOI of 1 (Fig. 6E, left panel) or 5 (Fig. 6E, right panel). In line with the results shown in Fig. 6A to D, we were able to detect significant numbers of IFUs in all samples, again showing that scattered bacteria can be reactivated in principle (Fig. 6E). However, in both experiments, which differed only in the MOI used, we found >3.5-fold-reduced IFU titers derived from reactivation of scattered Chlamydia compared with the titers obtained for persistent Chlamydia. Interestingly, a recovery defect was also seen with cells grown with 3 μM tryptophan. We noted that at this tryptophan concentration the scattered population was always prominent (Fig. 2) and in some experiments even constituted the only type of inclusion (data not shown). Taken together, these results substantiate the conclusion that severe tryptophan starvation significantly compromises the ability of scattered Chlamydia to become efficiently reactivated.
The lower number of recoverable IFUs observed under these conditions may reflect an impaired rate of recovery or increased bacterial death in this population. Thus, particularly in light of a study showing that translation-incompetent Chlamydia is eventually delivered to lysosomes (29), it was important to determine whether this also applies to the scattered chlamydial fraction. To this end, we seeded cells on coverslips, incubated them 24 h before and after infection (MOI, 5) with 0 μM tryptophan in the presence of IFN-γ, and finally compared the distribution of intracellular scattered bacteria with the distribution of the lysosomal marker LAMP1 by confocal fluorescence microscopy (Fig. 6F). Although we very occasionally detected single bacteria trapped in LAMP1-positive compartments (data not shown), the vast majority of the scattered population did not colocalize with lysosomes (Fig. 6F). This suggests that an impaired rate of reactivation is the more likely explanation for the reduced IFU titers shown in Fig. 6E. In line with this, we did not find a substantial decrease in the percentage of infected cells upon reactivation of scattered Chlamydia (0 μM tryptophan) compared with persistent Chlamydia (6 μM tryptophan), even when an MOI of 1 was used in the experiment (Fig. 6B, right panels). This indicates that even most cells infected with single bacteria still contained viable bacteria 24 h after restoration of favorable growth conditions. Moreover, we could not detect residual scattered bacteria in the cell periphery upon reactivation in any of the samples (data not shown). Overall, these data argue against a scenario where strongly reduced bacterial survival under low-tryptophan conditions causes the observed phenomena, although our data do not exclude the possibility of an additional contribution of such effects. Instead, the major difference between the samples shown in Fig. 6A and 6D appeared to be the inclusion size, indicating that scattered Chlamydia bacteria survive even within a severely tryptophan-starved cellular environment but that their ability to become rapidly reactivated upon replenishment of tryptophan pools is limited.
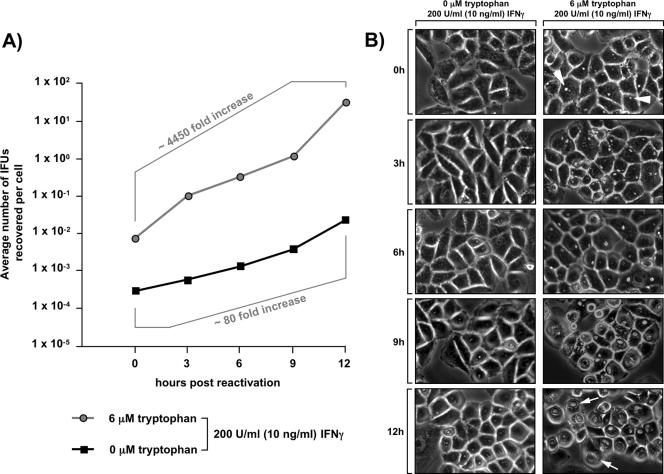
To further substantiate this scenario, we investigated the early reactivation kinetics of persistent or scattered bacteria in a time course assay (Fig. 7A and 7B). Consistent with the potential of scattered Chlamydia to become reactivated when favorable growth conditions are restored (Fig. 6A, 6B, and 6E), the IFU titers for recovering scattered organisms assessed at various time points after addition of tryptophan steadily increased over time, but they were still amplified only ∼80-fold even at 12 h after reactivation (Fig. 7A). In contrast, persistent bacteria displayed a dramatically higher reactivation rate, with ∼4,450-fold amplification of the initial titer within the same time frame. This coincided with the steady transformation of the initially tiny inclusions harboring persistent organisms (Fig. 7B, top right panel) into large inclusions (Fig. 7B, lowest panel on the right) that are typically found in cells in which Chlamydia growth is unrestricted.
FIG. 7.
Scattered Chlamydia bacteria reactivate slower than their persistent counterparts. (A) Analysis of the early chlamydial reactivation kinetics by IFU titer determination. To characterize the recovery of persistent Chlamydia (circles) or scattered Chlamydia (squares), equal numbers of infected cells treated as described in the legend to Fig. 6E (right panel) were subjected to IFU titer determination at 0, 3, 6, 9, or 12 h after restoration of favorable growth conditions. The results are expressed as the average number of IFUs recovered per cell. (B) Analysis of early chlamydial reactivation kinetics by light microscopy: light microscopic images of cells from the experiment whose results are shown in panel A. Photos were taken during the course of reactivation of scattered Chlamydia (left column) or persistent Chlamydia (right column) at the indicated times after restoration of high tryptophan levels. The arrowheads indicate small inclusions formed under conditions that favor the onset of conventional persistence. The arrows indicate large inclusions formed after reactivation of persistent Chlamydia.
The reduced inclusion size (Fig. 6A and 6D) and IFU titers (Fig. 6E) observed with recovering scattered Chlamydia even after 24 h are in keeping with the almost 60-fold-reduced reactivation rate of these forms within the first 12 h compared with that of their persistent counterparts. Thus, severe tryptophan starvation substantially affects the rate at which the bacteria can generate infectious progeny.
DISCUSSION
It is well established that the antichlamydial immune response largely depends on the production of the inflammatory cytokine IFN-γ by CD4+ T cells (7). IFN-γ is known to induce a plethora of downstream genes, many of which exert direct antipathogenic effects or contribute to the function of a proper adaptive and innate immune system. One of the antiparasitic proteins highly induced by the cytokine is the cytosolic enzyme IDO (27), which causes rapid depletion of tryptophan pools in vitro (34) and in vivo (11). Particularly in human cells, starvation of the intruding microbes for tryptophan appears to be the major innate immune mechanism controlling the growth of C. trachomatis (27). Interestingly, Chlamydia species can respond to reduced tryptophan levels by converting into a so-called persistent form, which is specifically adapted to survive within the resulting hostile environment (16). Persistent RBs are atypically large, do not multiply or produce infectious progeny, and display a distinct transcriptional profile (3, 5, 16). Most importantly, they can be rapidly reactivated upon restoration of favorable growth conditions (3, 5).
Pharmacologic inhibition of chlamydial translation stalls early development and intracellular trafficking of the microbe (14), as well as the ability of Chlamydia to block lysosomal fusion with the inclusion (28, 29). Therefore, as C. trachomatis is a natural tryptophan auxotroph (36), we hypothesized that IDO-mediated tryptophan deprivation might have an effect on the organism more drastic than temporarily inducing persistence, provided that it limits the availability of the amino acid sufficiently. However, antibiotic-mediated inhibition of protein synthesis is different from translational suppression, which Chlamydia may encounter upon entry into IFN-γ-stimulated cells. For instance, chloramphenicol, the antibiotic used in the above-mentioned studies, broadly blocks bacterial protein synthesis regardless of individual polypeptides, whereas a reduction in the tryptophan pool size may allow the bacterium to switch to a different translational profile (e.g., synthesize proteins requiring less or no tryptophan). Indeed, it was found that Chlamydia bacteria in the IFN-γ-induced persistent state dampen the production of the tryptophan-rich protein MOMP but continue and even increase the production of tryptophan-free cHSP60 (2, 4). Furthermore, tryptophan-sensitive promoters, like the promoter driving the expression of trpB, may adjust the transcriptional profile to the needs of the bacterium under amino acid deficiency conditions. In short, IDO-mediated tryptophan depletion may allow the organism to respond (and evade), whereas antibiotic-mediated suppression of protein synthesis presumably does not. Also, in contrast to the antibiotic effect of chloramphenicol, a translational block mediated by IDO affects both the intruding microbe and the host cell. Thus, the innate immune defense against intracellular parasites might be impaired per se. In fact, this may be the reason why we could not detect significant colocalization of scattered Chlamydia bacteria with lysosomes 24 h postinfection, as has been reported previously for chloramphenicol-inhibited bacteria (28) (Fig. 6E). However, consistent with data for antibiotic-treated microbes (14, 29), we observed drastic developmental and growth defects in severely tryptophan-starved Chlamydia (Fig. 2 and 4D), indicating that the associated aberrations arise mainly from a block in bacterial translation.
Chlamydial protein synthesis has been shown to be a prerequisite for the association of the host-derived microtubule motor protein dynein with the outer face of the inclusion and consequently for migration of the vacuole to the perinuclear MTOC (14). Hence, the fact that severely tryptophan-starved Chlamydia maintains its location in the cell periphery likely reflects a failure of the microbes to insert a putative dynein receptor into their inclusion membrane. Several nonmutually exclusive scenarios may account for this. Tryptophan starvation may block the synthesis or assembly of the chlamydial type III secretion system, thereby affecting the export of the putative receptor into the host cytosol. However, repression of the bacterial translocation machinery using the newly developed synthetic inhibitor C1 was not observed to stall early bacterial trafficking, although it potently blocked subsequent developmental steps (35). This surprising result argues against involvement of type III secretion in dynein-dependent migration of the bacteria to the MTOC. Alternatively, the translation of the putative dynein receptor itself may be suppressed as a consequence of IDO-mediated abrogation of tryptophan supply to the microbe. Moreover, as our data suggest that scattered Chlamydia bacteria have substantially diminished transcriptional activity compared to that of persistent RBs (Fig. 5B and 5C), the putative dynein receptor may already be insufficiently transcribed. Currently, it is not known which bacterial effectors mediate the early trafficking of the organism. Hence, identification of these effectors remains a major objective for future studies.
Our observation of an altered viable state of Chlamydia at low tryptophan concentrations (Fig. 1 and 2) certainly raises the question of how distinct these bacteria in fact are from conventional persistent RBs. One of the most obvious differences (and in fact this attracted our initial attention) lies in the tiny dot-shaped morphological appearance of the scattered bacterial population, which is radically different from the large characteristic giant forms (Fig. 1) that many Chlamydia species generate in response to environmental stresses as diverse as tryptophan starvation (2, 3, 13), iron depletion (13), glucose deprivation (15), antibiotic exposure (13, 18), viral coinfection (10), phage infection (17), and heat shock (19). The fact that so many different stimuli all lead to remarkably similar morphological outcomes strongly argues that a general advantage of this chlamydial form under unfavorable conditions underlies this common pattern. As a consequence, persistent giant RBs would represent a particularly stress-resistant chlamydial form, well adapted to cope with a hostile environment and prepared to continue with development upon restoration of more favorable growth conditions. Of the numerous indications that support this view, the direct regulation of the trpAB operon by free tryptophan (1) may be one of the most evident. Induction of the corresponding genes when tryptophan levels drop leads to the expression by genital serovars of C. trachomatis of functional tryptophan synthase, an enzyme that is capable of converting indole into tryptophan (8). In this way, persistent RBs may counterbalance and evade IDO-mediated immunity.
In contrast, we showed that in scattered Chlamydia bacteria the induction of trpB is substantially impaired by expression of sevenfold-lower mRNA amounts of this gene than the amounts expressed in persistent RBs (Fig. 5B and 5C). It is tempting to speculate that as a consequence the organisms would be less responsive to indole addition and that their ability to recover in the presence of this compound would be compromised. However, although clearly more genes have to be analyzed to permit a general statement, our data on the transcriptional response argues that the overall rate of transcription might be decreased in scattered Chlamydia compared with persistent Chlamydia (Fig. 5B and 5C). If this is the case, the scattered fraction would likely be much less adapted to environmental stress and may behave suboptimally in various contexts. Moreover, it must be considered that the lower tryptophan levels, at which the peripherally spread bacteria appear, would additionally limit the translational capabilities of the organism and almost necessarily cause alterations in the chlamydial proteome. In line with these considerations, even the early development of the scattered microbes, including their migration to the MTOC, is effectively arrested (Fig. 4D). In contrast, in the context of tryptophan levels that allow the onset of persistence, the bacteria clearly traffic to the perinuclear region (although the movement is somewhat delayed) (Fig. 4C and 4D). Thus, persistent Chlamydia and scattered Chlamydia also undergo differential early development.
Another key feature of persistent Chlamydia bacteria is their ability to be reactivated upon removal of the persistence-inducing stimulus (3, 5). Interestingly, reactivation of the scattered bacterial fraction was also found to be substantially compromised when the early (3 to 12 h postreactivation) rate of generation of infectious progeny (Fig. 7A) and even the size of the inclusions (Fig. 6A and 6D) and the amount of infectious progeny (Fig. 6E) 24 h postreactivation were compared to the data for persistent Chlamydia. Moreover, in a normal infection environmental conditions are unlikely to change as abruptly or dramatically as they do in the in vitro reactivation assay. Consequently, reactivation of scattered Chlamydia occurring in vivo might be even slower and less efficient. In the context of an ongoing cytotoxic T-cell-mediated immune response against intracellular bacteria (26), this may give the microbe a substantial disadvantage, promoting its eventual eradication. When the results are taken together, scattered Chlamydia bacteria are characterized by a fundamentally different morphological appearance, arrested early development, an altered transcriptional profile, and an impaired ability to become reactivated compared to persistent RBs. Consequently, the scattered forms represent a severely compromised bacterial population, which is predicted to have reduced fitness in vivo, whereas persistent RBs are a well-adapted stress-resistant form that may allow the microbe to evade an efficient immune response.
As we observed the appearance of scattered Chlamydia only at very low levels of tryptophan, the in vivo relevance of our findings is dependent on the concentrations that this essential amino acid eventually reaches in infected tissue upon IFN-γ induction. Interestingly, a recent study reported the IFN-γ-dependent reduction of tryptophan levels in explants of term placentas to 2.3 μM, which could be completely blocked by application of the IDO inhibitor 1-methyl-tryptophan (21). Furthermore, Fujigaki and colleagues recently showed that in an infection model tryptophan concentrations dropped dramatically to ∼2 μM in lung tissue of T. gondii-infected mice and that this effect was entirely dependent on IFN-γ signaling (11). This result demonstrates that IDO is enormously potent in vivo. However, as the measured tryptophan concentration represents the average level found in the tissue, minimal concentrations of the amino acid established in local environments are likely to be even below this value. Such local tryptophan-depleted microenvironments have recently been postulated to be generated by the action of macrophages, which possess a unique high-affinity tryptophan import system (31). This highly specific and efficient transport machinery allows the macrophages to import and subsequently degrade tryptophan at very low exogenous concentrations of the amino acid (31). Hence, in infected tissue they could act as a kind of “tryptophan sink,” reducing tryptophan levels below the threshold where import into neighboring cells is efficient. In this context it should be noted that infiltration of macrophages into Chlamydia-infected tissue has indeed been observed in vivo (6). Interestingly, in contrast to macrophages, cells or cell lines having diverse tissue origins seem to use predominantly, if not exclusively, the neutral amino acid transporter system L for uptake of this essential amino acid (25, 31, 32). However, while system L displays substrate affinities in a micromolar to millimolar range (24), it does not show particular selectivity towards tryptophan. Thus, at very low exogenous tryptophan levels other amino acids (present at plasma concentrations of ∼ 50 to 200 μM [20]) would be expected to inhibit competitively tryptophan import via this permease to a significant extent. Consequently, cells that depend on system L for tryptophan uptake would be likely to experience a dramatic depletion of their tryptophan pools upon IFN-γ-mediated IDO induction. Based on these considerations, we believe that the occurrence of the scattered Chlamydia state in vivo is indeed a plausible scenario.
In summary, the present study describes a novel, yet undefined chlamydial state that appears particularly in severely tryptophan-starved human cells and that is clearly distinct from conventional persistent RBs. Hence, we provide new insights into the biology of C. trachomatis under conditions that the pathogen may encounter during an inflammatory IDO-inducing immune response.
Acknowledgments
We are indebted to W. Yuan for the kind donation of Alexa-647-conjugated LAMP1-specific antibodies and to E. de la Torre for preparation of chlamydial stocks. We thank R. DeMars for providing C. trachomatis serovar L1 and H. Caldwell and K. Swanson for technical advice. We thank D. Stepensky for helpful discussions. E. Hinson and W. Yuan are acknowledged for critically reading the manuscript.
This study was supported by the Howard Hughes Medical Institute (P.C.) and by NIH grant RO1 AI49571 (P.B.K.). Part of the work was supported by an Overseas Research Supporting Grant from the Kangwon National University to S.J.L. We have no financial conflict of interest.
Editor: F. C. Fang
Footnotes
Published ahead of print on 27 August 2007.
REFERENCES
- 1.Akers, J. C., and M. Tan. 2006. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J. Bacteriol. 188:4236-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty, W. L., T. A. Belanger, A. A. Desai, R. P. Morrison, and G. I. Byrne. 1994. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62:3705-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Immunoelectron microscopic quantitation of differential levels of chlamydial proteins in a cell culture model of persistent Chlamydia trachomatis infection. Infect. Immun. 62:4059-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belland, R. J., D. E. Nelson, D. Virok, D. D. Crane, D. Hogan, D. Sturdevant, W. L. Beatty, and H. D. Caldwell. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA 100:15971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilenki, L., S. Wang, J. Yang, Y. Fan, A. G. Joyee, and X. Yang. 2005. NK T cell activation promotes Chlamydia trachomatis infection in vivo. J. Immunol. 175:3197-3206. [DOI] [PubMed] [Google Scholar]
- 7.Brunham, R. C., and J. Rey-Ladino. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 5:149-161. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Investig. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2006. Sexually transmitted disease surveillance 2005 supplement. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, GA.
- 10.Deka, S., J. Vanover, S. Dessus-Babus, J. Whittimore, M. K. Howett, P. B. Wyrick, and R. V. Schoborg. 2006. Chlamydia trachomatis enters a viable but non-cultivable (persistent) state within herpes simplex virus type 2 (HSV-2) co-infected host cells. Cell. Microbiol. 8:149-162. [DOI] [PubMed] [Google Scholar]
- 11.Fujigaki, S., K. Saito, M. Takemura, N. Maekawa, Y. Yamada, H. Wada, and M. Seishima. 2002. l-Tryptophan-l-kynurenine pathway metabolism accelerated by Toxoplasma gondii infection is abolished in gamma interferon-gene-deficient mice: cross-regulation between inducible nitric oxide synthase and indoleamine-2,3-dioxygenase. Infect. Immun. 70:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gieffers, J., J. Rupp, A. Gebert, W. Solbach, and M. Klinger. 2004. First-choice antibiotics at subinhibitory concentrations induce persistence of Chlamydia pneumoniae. Antimicrob. Agents Chemother. 48:1402-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goellner, S., E. Schubert, E. Liebler-Tenorio, H. Hotzel, H. P. Saluz, and K. Sachse. 2006. Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect. Immun. 74:4801-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grieshaber, S. S., N. A. Grieshaber, and T. Hackstadt. 2003. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell Sci. 116:3793-3802. [DOI] [PubMed] [Google Scholar]
- 15.Harper, A., C. I. Pogson, M. L. Jones, and J. H. Pearce. 2000. Chlamydial development is adversely affected by minor changes in amino acid supply, blood plasma amino acid levels, and glucose deprivation. Infect. Immun. 68:1457-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan, R. J., S. A. Mathews, S. Mukhopadhyay, J. T. Summersgill, and P. Timms. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72:1843-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsia, R., H. Ohayon, P. Gounon, A. Dautry-Varsat, and P. M. Bavoil. 2000. Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect. 2:761-772. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, F. W., and D. Hobson. 1977. The effect of penicillin on genital strains of Chlamydia trachomatis in tissue culture. J. Antimicrob. Chemother. 3:49-56. [DOI] [PubMed] [Google Scholar]
- 19.Kahane, S., and M. G. Friedman. 1992. Reversibility of heat shock in Chlamydia trachomatis. FEMS Microbiol. Lett. 76:25-30. [DOI] [PubMed] [Google Scholar]
- 20.Klassen, P., P. Furst, C. Schulz, M. Mazariegos, and N. W. Solomons. 2001. Plasma free amino acid concentrations in healthy Guatemalan adults and in patients with classic dengue. Am. J. Clin. Nutr. 73:647-652. [DOI] [PubMed] [Google Scholar]
- 21.Kudo, Y., C. A. Boyd, I. L. Sargent, and C. W. Redman. 2001. Tryptophan degradation by human placental indoleamine 2,3-dioxygenase regulates lymphocyte proliferation. J. Physiol. 535:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mabey, D., and R. W. Peeling. 2002. Lymphogranuloma venereum. Sex. Transm. Infect. 78:90-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouellette, S. P., T. P. Hatch, Y. M. AbdelRahman, L. A. Rose, R. J. Belland, and G. I. Byrne. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol. Microbiol. 62:1387-1401. [DOI] [PubMed] [Google Scholar]
- 24.Palacin, M., R. Estevez, J. Bertran, and A. Zorzano. 1998. Molecular biology of mammalian plasma membrane amino acid transporters. Physiol. Rev. 78:969-1054. [DOI] [PubMed] [Google Scholar]
- 25.Ritchie, J. W., and P. M. Taylor. 2001. Role of the system L permease LAT1 in amino acid and iodothyronine transport in placenta. Biochem. J. 356:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roan, N. R., and M. N. Starnbach. 2006. Antigen-specific CD8+ T cells respond to Chlamydia trachomatis in the genital mucosa. J. Immunol. 177:7974-7979. [DOI] [PubMed] [Google Scholar]
- 27.Roshick, C., H. Wood, H. D. Caldwell, and G. McClarty. 2006. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect. Immun. 74:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scidmore, M. A., D. D. Rockey, E. R. Fischer, R. A. Heinzen, and T. Hackstadt. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect. Immun. 64:5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scidmore-Carlson, M. A., E. I. Shaw, C. A. Dooley, E. R. Fischer, and T. Hackstadt. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 33:753-765. [DOI] [PubMed] [Google Scholar]
- 31.Seymour, R. L., V. Ganapathy, A. L. Mellor, and D. H. Munn. 2006. A high-affinity, tryptophan-selective amino acid transport system in human macrophages. J. Leukoc. Biol. 80:1320-1327. [DOI] [PubMed] [Google Scholar]
- 32.Shennan, D. B., J. Thomson, M. C. Barber, and M. T. Travers. 2003. Functional and molecular characteristics of system L in human breast cancer cells. Biochim. Biophys. Acta 1611:81-90. [DOI] [PubMed] [Google Scholar]
- 33.Wagenlehner, F. M., K. G. Naber, and W. Weidner. 2006. Chlamydial infections and prostatitis in men. BJU Int. 97:687-690. [DOI] [PubMed] [Google Scholar]
- 34.Werner, E. R., M. Hirsch-Kauffmann, D. Fuchs, A. Hausen, G. Reibnegger, M. Schweiger, and H. Wachter. 1987. Interferon-gamma-induced degradation of tryptophan by human cells in vitro. Biol. Chem. Hoppe-Seyler 368:1407-1412. [DOI] [PubMed] [Google Scholar]
- 35.Wolf, K., H. J. Betts, B. Chellas-Gery, S. Hower, C. N. Linton, and K. A. Fields. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie, G., C. A. Bonner, and R. A. Jensen. 2002. Dynamic diversity of the tryptophan pathway in chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol 3:research0051. [DOI] [PMC free article] [PubMed]