Abstract
A single-chain variable fragment (scFv) antibody library against Bordetella pertussis was constructed using M13 phage display. The library was enriched for phages surface displaying functional scFv by biopanning against B. pertussis immobilized on polystyrene plates. Two hundred eighty-eight individual clones from the enriched library were screened for binding to B. pertussis cells, filamentous hemagglutinin (FHA), and pertactin (PRN) in enzyme-linked immunosorbent assays (ELISAs). Based on the binding ability, the clones were put into eight groups. The scFv DNA inserts from the 288 clones were digested with BstOI, and 18 unique restriction patterns, named types 1 to 18, were found. Eight clones (types 1 to 7 and 18) were selected for further testing against FHA, PRN, and B. pertussis by ELISA. The results showed that types 1, 5, 7, and 18 bound strongly to B. pertussis cells as well as FHA and PRN. Type 3 bound strongly to the cells and FHA but weakly to PRN. Types 4 and 6 bound FHA only, and type 2 did not bind to the cells or antigens. The ability of the eight clones to inhibit B. pertussis from binding to HEp-2 cells was assayed. Types 1, 5, and 7, but not the remaining clones, inhibited the adherence of B. pertussis to HEp-2 cells. The scFvs were sequenced, and the deduced amino acid sequence showed that the scFvs were different antibodies. Maltose-binding protein (MBP) fusion proteins composed of three different regions of FHA (heparin-binding domain, carbohydrate recognition domain, and the RGD triplet motif) were constructed. The three fusion proteins and Mal85 (MBP-FHA type I domain) were used to map the binding sites for scFvs of types 1, 5, and 7 by ELISA. The results showed that all three scFvs bound to the heparin-binding domain fusion protein but not the other fusion proteins. BALB/c mice who received recombinant phage-treated B. pertussis had reduced bacterial counts in the nasal cavity, trachea, and lungs compared to the control groups.
Pertussis is a highly contagious acute respiratory disease that is more serious in infants (24, 33). Despite widespread immunization with acellular and inactivated whole-cell vaccines, many countries still report epidemic outbreaks (3), and the disease afflicts 40 million and kills 400,000 individuals annually (33).
Bordetella pertussis, the etiological agent of pertussis, expresses a plethora of virulence factors that are critical to its pathogenesis. The virulence factors include multiple toxins that inhibit host defenses and cause damage to local tissues. Adhesins, such as filamentous hemagglutinin (FHA), are also important since they facilitate adherence and initiate the infection process (4, 8, 21, 22, 23, 26, 32). There has been considerable debate as to whether FHA can confer protection against B. pertussis infection. Previous work by Sato et al. (28) and Sato and Sato (27) questioned the protective efficacy of FHA. Cherry et al. (7) and Storsaeter et al. (31) showed no correlation between the presence of anti-FHA antibodies and protection against household exposure to B. pertussis. However, anti-FHA antibodies were found in sera from convalescent patients and vaccinated infants (20). In animal models, a number of laboratories, including ours, showed that antibodies to FHA or FHA fragments are protective (1, 5, 6, 8, 11, 29, 30).
Currently, two different commercial types of vaccines against pertussis are available, inactivated whole-cell vaccines and the more expensive acellular vaccines that are composed of one to five defined purified antigens. All acellular vaccines contain pertussis toxoid; FHA is included in most acellular vaccines, while pertactin (PRN) and fimbriae make up the remaining components of some acellular vaccines. Both the acellular and whole-cell vaccines require multiple doses to achieve adequate immune responses and fail to provide long-lasting immunity. The vaccines offer little protections for newborns and those who are immunocompromised.
Passive immunization has the potential to provide a solution to this problem. However, the production of antibodies for passive immunization is very expensive, and often, it is difficult to quantify the protective antibodies within the preparations. The use of single-chain variable fragment (scFv) antibodies can be employed to avoid these problems. Single-chain antibodies are recombinant antibodies that have the variable light and heavy chains fused together with a glycine/serine linker sequence and can be produced by bacteria inexpensively. scFv has been surface expressed by the commensal bacteria Lactobacillus (12) and Streptococcus gordonii (9) as a new strategy to prevent dental caries.
Our laboratory is interested in using S. gordonii as a live oral vaccine vehicle against respiratory diseases (15). S. gordonii is a gram-positive bacterium that arrives in the oral cavity of infants as young as 6 months of age and remains as normal oral flora throughout life. We have previously shown that S. gordonii can serve as a host to express vaccine antigens (14, 18, 19), and mucosal immune responses could be generated in oral colonization studies in mice (17). S. gordonii can potentially be genetically engineered to secrete scFv to bathe the mucosa in the oral-respiratory tract for passive immunization. In this paper, we report the construction and characterization of scFv antibodies against B. pertussis as a first step in achieving the goal of passive immunity against B. pertussis.
MATERIALS AND METHODS
Bacteria and growth conditions.
B. pertussis Tohama I was grown on Bordet Gengou plates supplemented with 15% horse blood in a moist environment at 37°C. Escherichia coli was grown aerobically with shaking at 200 rpm at 37°C in super broth medium (1% MOPS [morpholinepropanesulfonic acid], 3% tryptone, and 2% yeast extract [wt/vol]) or rich broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl, and 0.2% glucose [wt/vol]). Antibiotics, when needed, were added to the medium at 100 μg/ml ampicillin, 10 μg/ml tetracycline, 70 μg/ml kanamycin, 50 μg/ml spectinomycin, and 34 μg/ml chloramphenicol. All antibiotics were purchased from Sigma-Aldrich Chemical Co. (Oakville, Ontario, Canada). For the cultivation of E. coli on plates, Luria-Bertani agar (1% tryptone, 0.5% yeast extract, 1% NaCl, and 1.5% agar [wt/vol]) was used.
Immunization and total RNA isolation.
A 5-week-old BALB/c mouse (Charles River Laboratory, St. Constant, Quebec, Canada) was immunized by intraperitoneal injections of 0.1 ml of the commercial pertussis vaccine Quandracel (Sanofi Pasteur, Toronto, Ontario, Canada) on days 1, 28, 35, and 44. The amounts of antigens in each injection were 4 μg pertussis toxoid, 4 μg FHA, 1 μg fimbriae, 0.6 μg PRN, 3 limit-of-flocculation units of diphtheria toxoid, 1 limit-of-flocculation unit of tetanus toxoid, and inactivated poliomyelitis viruses (8, 1, and 6.4 D-antigen units of poliovirus types 1, 2, and 3, respectively). The animal was euthanized on day 53, and the spleen was removed, immediately minced, and placed into 1 ml of TRIzol (Invitrogen Life Technologies, Burlington, Ontario, Canada). The sample was incubated for 5 min at room temperature and centrifuged at 2,500 × g for 10 min. The supernatant fluid was vortexed with 0.1 ml of 1-bromo-3-chloro-propane (Sigma-Aldrich) for 15 s and incubated at room temperature for 15 min. The mixture was then centrifuged at 17,500 × g for 15 min, and the upper aqueous layer was saved. Total RNA was precipitated from the aqueous layer by the addition of 0.5 ml of isopropanol and incubation at room temperature for 15 min. The precipitated RNA was recovered by centrifugation at 17,500 × g for 15 min, washed with 75% ethanol, and redissolved in RNase-free water. The amount of RNA was determined by absorbance measurements at 260 nm. A total of 655 μg of RNA was obtained.
Phage display library construction.
cDNA was obtained from 20 μg total RNA using oligo(dT) as the primer and Superscript II (Invitrogen) according to the manufacturer's instructions. The phage display library was constructed from the cDNA according to procedures described previously by Barbas et al. (2). Briefly, the variable light-chain (VL) and heavy-chain (VH) DNA fragments were amplified by PCR using mixed primers as described previously by Barbas et al. (2). The resulting 0.4-kb VL and VH DNA fragments were gel purified and used in overlapping PCR to produce the scFv antibody DNA. The resulting 0.8-kb scFv fragments were gel purified and ligated into the SfiI sites of the phagemid pComb3H. The ligated DNA was transformed into electrocompetent E. coli XL1-Blue cells to obtain the library. Recombinant phages were prepared from the library with the helper phage VCSM13. The phage library obtained was precipitated with polyethylene glycol and stored in 2 ml of Tris-buffered saline (TBS) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl) containing 1% (wt/vol) bovine serum albumin (BSA) at 4°C.
Biopanning.
To enrich phages that surface displayed the antipertussis scFv, the phage library was biopanned against B. pertussis whole cells. Two wells of a 96-well polystyrene microtiter plate were each coated with 1 × 109 B. pertussis cells in phosphate-buffered saline (PBS) containing 0.125% glutaraldehyde for 30 min at room temperature. The wells were blocked with 3% BSA in TBS for 1 h at 37°C, and 100 μl of the phages was added to each well. The plate was incubated at 37°C for 2 h and washed five times with TBS containing 0.5% Tween 20. The adherent phages were eluted with 100 μl of 0.1 M glycine-HCl (pH 2.2). The eluate was neutralized immediately with 3 μl of 2 M Tris base. The eluated phages were grown in E. coli XL1-Blue with VCSM13, and the phage preparation was obtained by polyethylene glycol precipitation. The phages were panned a total of four times as described above. The final phage preparation was used for individual clone screening.
Phage preparation.
For the initial screening of clones in enzyme-linked immunosorbent assays (ELISAs), phage particles were prepared as follows. Super broth medium (3 ml) was inoculated with a single colony and grown for 4 h at 37°C with shaking at 200 rpm. Thirty microliters of VCSM13 helper phage (1012 PFU/ml) was added. The culture was incubated for another 2 h, at which time kanamycin was added and left to shake overnight. The following day, the culture was split in half (1.5 ml each half) and centrifuged at 2,800 × g for 15 min at 4°C. The supernatant (2 × 1.2 ml) was saved, and 300 μl of 5× polyethylene glycol-NaCl (20% polyethylene glycol 8000 and 15% NaCl [wt/vol]) was added. The mixture was incubated on ice for 1 h to precipitate the phage particles. After precipitation, the solution was centrifuged at 10,000 × g for 5 min at 4°C. The two phage pellets were resuspended in a total of 100 μl of 1% (wt/vol) BSA in TBS and used without determining the titer. For large-scale phage preparations to be used in quantitative ELISA, 100 ml of culture was used, and the phages were resuspended in 2 ml of 1% BSA in TBS. Phage titer was determined by plating serially diluted phage and E. coli XL1-Blue cells onto agar plates (2).
For testing the binding specificity of the recombinant phages type 1 (T1), T5, and T7 or parent phage 3H to maltose binding protein (MBP)-FHA fragment fusion proteins, the phages were biotinylated. Biotinylation was needed to avoid the use of secondary antibodies, which were found to cross-react with the RGD-containing FHA fragment. Phage particles (ca. 5 × 1012 PFU) resuspended in 0.5 ml of PBS were incubated with 500 μg of biotinamidohexanoic acid 3-sulfo-N-hydroxysuccinide ester (Sigma-Aldrich) at room temperature with gentle mixing. After 45 min, the mixture was applied to a PD-10 Sephadex G-25 column (10 ml; GE Healthcare Bio-Sciences, Inc., Baie d'Urfe, Quebec, Canada), and the phage particles eluted with PBS in the void volume were collected. Phage titer was determined by plating, and phages were confirmed to have been biotinylated by reactions to avidin-alkaline phosphatase (Sigma-Aldrich) by ELISA.
ELISA.
For the initial screening, a single well was used for each clone. The 96-well polystyrene plates were coated with 100 ng/well of FHA (List Biological Laboratories Inc., Campbell, CA), 100 ng/well of PRN (Sanofi Pasteur, Toronto, Ontario, Canada), or 1 × 109 B. pertussis cells/well. The plates were then blocked for 2 h with 1% BSA in TBS. Following blocking, 25 μl of phages and 50 μl of PBS were added. The plates were incubated overnight at 4°C. Following incubation, the plates were washed, and bound phages were detected with horseradish peroxidase-conjugated anti-M13 monoclonal antibody (1:7,000; GE Healthcare Bio-Sciences). For quantitative ELISA, antigens and B. pertussis were coated in triplicate as described above. Phages (5.6 × 1010 PFU) were added to each well. Phage was detected with the anti-M13 monoclonal antibody followed by the goat anti-mouse immunoglobulin G-alkaline phosphatase conjugate (1:7,500; Sigma-Aldrich). Wells with an A405 reading of 0.1 over the control well (reacted with parent phage) were considered to be positive.
For determining the binding specificity of the T1, T5, and T7 phages, microtiter plates were coated with 100 ng/well of FHA or each of the MBP-FHA fragment fusion proteins (see below for isolation) and blocked with BSA. Biotinylated phages (1.25 × 1011 PFU) were added to each well and detected with avidin-alkaline phosphatase.
PCR and restriction analysis of the initial clones.
The scFv DNA from 288 recombinant phage particles was amplified by PCR using the primer pair SL283/SL284 and Taq DNA polymerase. Primer sequences are listed in Table 1. The PCR products were analyzed on 0.8% agarose gels, and a clear 0.8-kb band was observed for each of the clones (data not shown). The PCR products were digested with BstOI (Promega, Madison, WI) for 3 h at 60°C and analyzed on 2% agarose gels.
TABLE 1.
Primers used in this studya
| Primer | Nucleotide sequence (5′-3′) | Direction |
|---|---|---|
| SL283 | GAGGCGGGGCCCAGGCGGCCGAGCTC | Forward |
| SL284 | GAGCCTGGCCGGCCTGGCCACTAGTG | Reverse |
| SL371 | TATCTAGAGGCGTGGCGCTGCAA | Forward |
| SL372 | TTGAAAGCTTTCACACGGCCGAATCGGC | Reverse |
| SL373 | GTTCTAGACATCAGGGCGTGCTGG | Forward |
| SL374 | AGCTAAGCTTTCAGCCTTTGGTGGCCATGA | Reverse |
| SL390 | ATTCTAGAAAGGTACAGGCCACGCTGTTGAATG | Forward |
| SL391 | AGATAAGCTTTCAGCGATTGGCGTGCAGCGAGTT | Reverse |
Underlined sequences in SL371, SL373, and SL390 are the XbaI restriction sites, while those in SL372, SL374, and SL391 are the HindIII restriction sites.
HEp-2 cell binding assay.
HEp-2 cells (ATCC CCL-23, derived from human larynx carcinoma) were grown in Earle's minimum essential medium (Invitrogen) to confluence at 37°C in a CO2 incubator. Cells were then trypsinized and washed with Hanks balanced salt solution (Invitrogen). Cells were centrifuged at 330 × g for 10 min and resuspended in PBS to a cell density of 2 × 107 cells/ml. B. pertussis cells from Bordet Gengou slants containing 1% proteose peptone were suspended in PBS containing 1% Casamino Acids to a cell density of 5 × 108 CFU/ml. Fifty microliters (2.5 × 107 cells) of B. pertussis cells was mixed with 50 μl containing 3.6 × 1010 phage particles. The reaction mixture was mixed at 4 rpm at 37°C for 1 h. One hundred microliters of HEp-2 cells (2 × 106) was then added to the reaction mixture. The mixture was further mixed for 90 min at 37°C. The mixture was then centrifuged at 110 × g for 4 min, and the top 50 μl of the liquid, containing unbound B. pertussis cells, was serially diluted and plated onto Bordet Gengou plates. Two controls were run alongside this assay using PBS or parent phage in place of the recombinant phages. Parallel to the procedures described above, B. pertussis cells were incubated alone, and the plate counts gave the total B. pertussis count. Plates were counted after 5 days of incubation. The results were expressed as percent nonadherent B. pertussis cells, which was calculated as follows: (cell count from each reaction/total B. pertussis cell count) × 100.
Construction and isolation of MBP-FHA fusion proteins.
The DNAs coding for three regions of FHA (HBD [amino acids 390 to 874], carbohydrate recognition domain [CRD] [amino acids 1101 to 1340], and Arg-Gly-Asp triplet motif [RGD] [amino acids 921 to 1140]) were amplified from B. pertussis chromosomal DNA using the primer pairs SL390/SL391, SL373/SL374, and SL371/SL372, respectively (Table 1). The PCR products, 1,500 bp, 720 bp, and 660 bp, respectively, were each cloned into the pMal-C expression vector (New England BioLabs) using the XbaI and HindIII restriction sites. The cloning created in-frame fusions with the FHA fragments located at the C terminus of MBP. The MBP-HBD and MBP-CRD constructs were maintained in E. coli TB1 cells, and the MBP-RGD construct was maintained in E. coli Rosetta gami 2 cells (Novagen, Inc., Madison, WI). The recombinant E. coli cells were grown in 500 ml of rich broth for 3 h and were induced with 0.12 mM isopropyl-β-d-thiogalactoside for 3 h at 37°C with shaking at 200 rpm. Cells were harvested by centrifugation (10,000 × g) and resuspended in 10 ml of wash buffer (50 mM Tris [pH 7.5], 1 mM EDTA, and 200 mM NaCl) and sonicated (15 10-s bursts at an amplitude of 35 separated by 10-s cooling periods, followed by 15 10-s bursts at an amplitude of 55 separated by 10-s cooling periods). The sonicate was centrifuged at 27,500 × g for 20 min at 4°C. The clear supernatant was applied to a 15-ml amylose column (New England BioLabs) equilibrated with wash buffer, and unwanted proteins were removed with 45 ml of wash buffer. The fusion proteins were eluted with 10 mM maltose in wash buffer as described previously (16). E. coli (pMal85) cells expressing the MBP-FHA type I domain (amino acids 1655 to 2013), kindly provided by C. Locht (Institut Pasteur de Lille) (20), were similarly grown, and the fusion protein Mal85 was isolated as previously described (11).
Prevention of B. pertussis colonization in mice.
Cohorts of BALB/c mice (female, 5 weeks of age; n = 5) were anesthetized with ketamine and xylazine. The first group of animals was given a mixture of B. pertussis cells plus T1 phage, the second group received B. pertussis cells plus T5 phage, the third group received B. pertussis plus T7 phage, the fourth group received B. pertussis plus a mixture of T1, T5, and T7 phages, the fifth group received B. pertussis cells plus the parent phage (pComb3H), the sixth group received B. pertussis plus a 1/100-diluted antipertussis mouse serum generated against the commercial Quadracel pertussis vaccine (11), and the seventh group received B. pertussis cells plus PBS. The number of B. pertussis cells given was 1 × 108 CFU/animal, and the number of phage particles was 1.7 × 1010 PFU/animal. In group 4, the three phages were each at a concentration 0.57 × 1010 PFU to give a total 1.7 × 1010 PFU. Prior to delivery, the B. pertussis cell mixtures were incubated at 37°C for 30 min with shaking at 116 rpm. The amount of mixture given to each animal was 25 μl, and this was delivered to the nostrils (12.5 μl per nostril) by a micropipette. The animals were euthanized 48 h following the instillation. Swabs were obtained from the nasal cavity and placed into a capped test tube containing 1 ml of PBS. The trachea was excised and placed into 1 ml of PBS. The swabs and trachea were vortexed for 1 min, and serial dilutions were plated onto Bordet Gengou plates to determine bacterial concentrations. The lungs were homogenized in 2 ml of PBS and similarly diluted and plated. The means and standard errors were calculated for the different samples. Significance was calculated using the two-sample t test assuming equal variance, and a P value of ≤0.05 was considered to be significant.
DNA sequencing.
The scFv genes were sequenced using primers SL283 and SL284 (Center for Functional Microbial Genomics and Host Defense, Dalhousie University) (Table 1).
Nucleotide sequence accession numbers.
The GenBank accession numbers for scFv types 1, 2, 4, 5, 6, 7, and 18 are DQ202338, DQ202339, DQ202340, DQ202341, DQ202342, DQ202343, and DQ202344, respectively.
RESULTS
Construction and characterization of an anti-B. pertussis scFv phage display library.
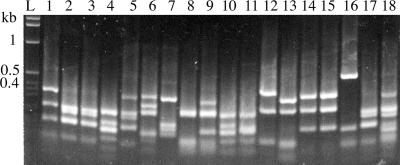
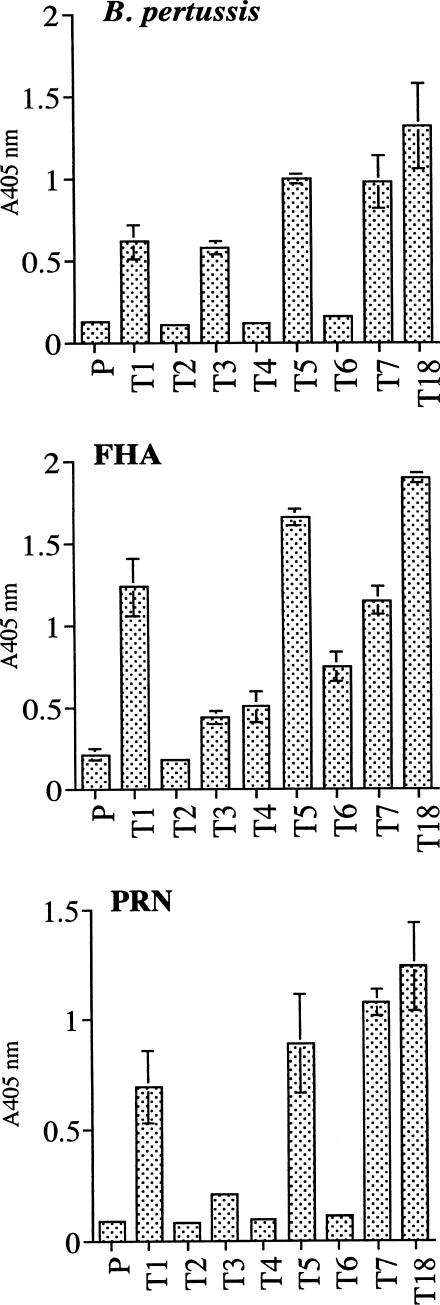
A library containing 3.21 × 108 clones was constructed. From that library, 288 individual clones were grown, and the recombinant phages were tested for binding to B. pertussis, FHA, and PRN by ELISA. Based on the binding ability, the clones were placed into eight distinct groups (groups A to H) (Table 2). The scFv DNA inserts from the 288 clones were PCR amplified and restricted with the enzyme BstOI. The results showed that the clones contained 18 unique banding patterns (Fig. 1), and they were designated types 1 to 18 (Table 2). From the 18 clones, 8 (types 1 to 7 and 18) were selected for further studies. These clones were of particular interest because of their ability to bind B. pertussis cells, and they appeared to be different based on the restriction patterns. Types 1, 2, 3, and 18 were from group A, and types 4, 5, 6, and 7 were from groups B, C, D, and E, respectively. The binding affinity of the eight clones was retested against B. pertussis, FHA, and PRN by ELISA. In this ELISA, the titer of the recombinant phages was standardized to 5.6 × 1010 phage particles per well, and the parent phage (pComb3H) was used as a negative control. The results showed that phages of types 1, 5, 7, and 18 bound strongly to B. pertussis, FHA, and PRN (Fig. 2). Type 3 also bound to B. pertussis, FHA, and PRN, but the binding to PRN was weak. Types 4 and 6 bound to FHA only, and type 2 did not bind to any of the antigens and was likely to be a false positive during the initial screening.
TABLE 2.
Antigen-binding characteristics and BstOI restriction patterns of the 288 clones from the initial screening
| Group | Binding of antigen:
|
Total no. of clones per group | Type(s) of BstOI restriction patterna | ||
|---|---|---|---|---|---|
| B. pertussis | FHA | PRN | |||
| A | Yes | Yes | Yes | 44 | 1, 2, 3, 18 |
| B | Yes | Yes | No | 13 | 1, 3, 4 |
| C | Yes | No | Yes | 33 | 1, 3, 5 |
| D | Yes | No | No | 14 | 1, 3, 6 |
| E | No | No | Yes | 30 | 1, 3, 7 |
| F | No | Yes | No | 12 | 1, 8-15 |
| G | No | Yes | Yes | 7 | 1 |
| H | No | No | No | 135 | NA |
NA, not applicable.
FIG. 1.
BstOI restriction pattern of 18 unique scFv clones. Amplified DNA fragments of different scFv clones were digested using the BstOI restriction enzyme and resolved on a 2% agarose gel. L, 1-kb DNA ladder. Number indicates the type, “T”; hence, lanes 2 to 19 are T1 to T18, respectively.
FIG. 2.
Binding of types 1 to 7 and 18 of scFv-expressing phages to B. pertussis, FHA, and PRN in ELISA. P is the parent phage carrying pComb3H. The number of phage particles used for each phage type was 5.6 × 1010. Results shown are means ± standard deviations (SD) of triplicates. Bars without an error indicate that the SD is too small to be seen.
Inhibition of B. pertussis binding to HEp-2 cells.
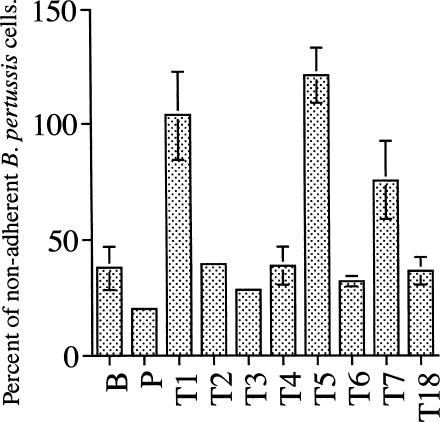
The eight clones were then tested for the ability to prevent B. pertussis binding to HEp-2 cells in an inhibition assay. The results showed that with a ratio of B. pertussis to HEp-2 cells of 10:1, about 35% of B. pertussis cells did not bind to HEp-2 cells (Fig. 3). A similar percentage was observed for the parent phage. However, in the presence of phages of types 1, 5, and 7, the percentage of nonadherent B. pertussis cells increased to 75 to 100%, indicating that the three phages inhibited B. pertussis cells from binding to HEp-2 cells. Interestingly, type 18 did not inhibit the binding of B. pertussis to HEp-2 cells even though it demonstrated strong binding to B. pertussis, FHA, and PRN (Fig. 2). Types 2, 3, 4, and 6 also showed no inhibition.
FIG. 3.
Inhibition of B. pertussis binding to HEp-2 cells by recombinant scFv phages. B is a control in which phage particles have been omitted from the reaction mixture. P is the parent phage not surface expressing any ScFv. All phages were standardized to a titer of 3.6 × 1010 phage particles per reaction. Results shown are means ± SD of triplicates. Percentage of nonadherent B. pertussis cells was calculated as follows: (cell count from each reaction/total B. pertussis count) × 100.
Aligned amino acid sequence of selected clones.
Seven of the eight scFv DNAs were sequenced, and the deduced amino acid sequences were determined (Fig. 4). The results showed the scFv contained the expected variable light- and heavy-chain sequences joined by the glycine-serine linker. Conserved cysteine residues responsible for disulfide bond formation were in the proper positions, as were the conserved glycine markers found in the framework 4 regions of both light and heavy chains. BLAST search results showed that the scFv showed high homology with the variable region of the immunoglobulin chain of mice, further confirming that the scFvs are recombinant antibodies. Close examination of the seven scFv sequences showed that types 1, 2, 5, and 18 were very similar to each other, with the most number of identical amino acid residues particularly in the complementary determining regions (CDRs). Types 4, 6, and 7 had the most diverse amino acid sequences within the CDR. Collectively, the sequences indicate that the seven scFvs are different antibodies.
FIG. 4.
Aligned amino acid sequence of the antipertussis scFv. Sequences shown are VL followed by VH. FR is the framework. Regions in boldface type correspond to the CDR. Underlined residues in the framework correspond to either conserved cysteine residues responsible for disulfide bond formation or conserved glycine markers found in immunoglobulin chains.
scFv types 1, 5, and 7 bound to the HBD-containing fragment of FHA.
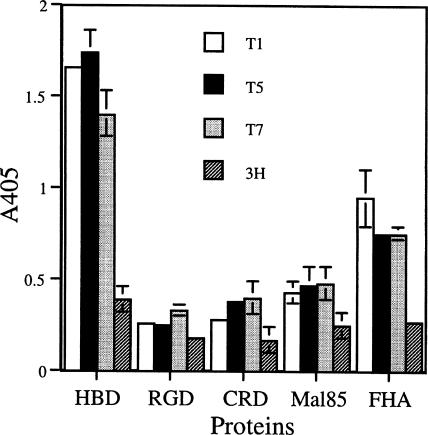
The region on FHA bound by scFv types 1, 5, and 7 was investigated. To help to elucidate this, regions of FHA were constructed as fusions to MBP. The three regions of FHA were HBD, CRD, and RGD, representing the N-terminal portion of FHA. The C-terminal portion of FHA (type I domain) is represented by Mal85. The MBP-FHA fragment fusion proteins were isolated from E. coli lysates, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (13) of the eluates showed the presence of the expected 92-, 66-, 64-, and 85-kDa fusion proteins of the HBD, CRD, RGD, and type I domain of FHA, respectively. The four fusion proteins were used in ELISA to map the binding site for scFv types 1, 5, and 7. As shown in Fig. 5, the three scFvs bound to the HBD fusion but not the other three fusion proteins. As expected, the three recombinant phages also bound to the intact FHA, while the parent phage did not bind to any of the proteins.
FIG. 5.
Binding of recombinant scFv phages to FHA and the HBD-containing fragment of FHA in ELISA. The proteins used were maltose binding fusion proteins of FHA HBD, FHA RGD fragment (RGD), FHA CRD, or FHA type I domain (Mal85). FHA is the intact native FHA. The results shown are means ± SD of triplicate wells. T1, recombinant phage T1; T5, recombinant phage T5; T7, recombinant phage T7; 3H, parent phage.
Prevention of B. pertussis colonization by T1, T5, and T7 scFv in mice.
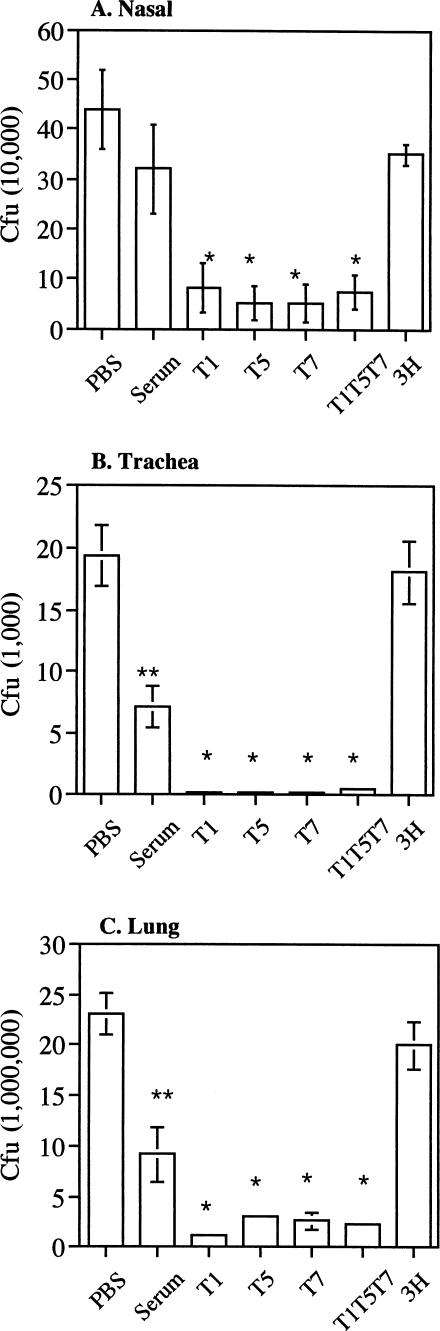
The ability of T1, T5, and T7 phages to neutralize B. pertussis colonization was assessed in a mouse intranasal instillation model. As shown in Fig. 6, the animals that received recombinant phage-treated B. pertussis showed a significant reduction in bacterial counts in the nasal cavity, trachea, and lungs compared to the parent phage-treated group. The levels of recovered B. pertussis between animals that received a single scFv and those that received three scFvs in combination were similar. The level of B. pertussis recovered from the parent phage-treated group was very similar to that of the PBS group. The serum-treated group also showed a reduction in bacterial counts compared to the PBS group, and the reduction was statistically significant for the trachea and lungs. These results indicate that the three recombinant scFvs are capable of neutralizing B. pertussis in the intranasal instillation model.
FIG. 6.
Neutralization of B. pertussis colonization in BALB/c mice. Mice were given B. pertussis premixed with PBS, an antipertussis serum (Serum), recombinant T1, T5, or T7 phage, a mixture of T1, T5, and T7 phages (T1T5T7), or parent M13 phage (3H) via intranasal instillation. Results shown are B. pertussis recovered (mean ± SE) from individual animals (n = 5) 48 h following instillation. Samples without error bars indicate that the errors are very small. * indicates a P value of <0.05 from two-sample t tests between the recombinant phage and parent phage groups. ** indicates a P value of <0.05 from two-sample t tests between the serum-treated and PBS groups.
DISCUSSION
In the present study, an scFv antibody library against B. pertussis antigens was constructed by M13 phage display. The library was biopanned against B. pertussis whole cells. The ELISA screening results showed that 53% (153 clones) of the 288 individual clones displayed reactivity to B. pertussis or its antigens, indicating that enrichment has been achieved via biopanning. Although the biopanning was performed against B. pertussis cells, 47 clones did not react with the cells in ELISA. The reason for this may be attributed to the fact that the phage preparations were not standardized to the same titer before use. Standardization was not performed due to the large number of clones involved, and it was impractical to determine the titers of each preparation. Hence, the lack of reaction to B. pertussis cells may have been due to low titers in these preparations. Therefore, it is possible that many of these 47 clones may appear to be falsely negative against B. pertussis cells during the initial screening. Type 7 is one of those clones.
The restriction analysis of the 288 clones revealed 18 unique banding patterns. However, the diversity of clones might not be limited to 18 types. Restriction analysis, as described previously by Barbas et al. (2), is a method that offers rapid screening of large scFv antibody libraries. The limitation of the method also offers an explanation for the presence of the type 1 pattern in all seven groups. In addition, some patterns appear to be similar (e.g., types 2 and 5); however, slight differences in the patterns are noticeable, and the DNA sequences have significant differences.
The functionality of eight selected clones was tested in a HEp-2 cell-binding assay. The results clearly showed that T1, T5, and T7 prevented the adhesion of B. pertussis to HEp-2 cells, while the remaining five clones did not. Surprisingly, T18 bound strongly to FHA, PRN, and B. pertussis cells but was not able to prevent B. pertussis binding to HEp-2 cells. Perhaps type 18 binds to an epitope that is not involved in adherence.
The ability of scFv types 1, 5, and 7 to neutralize B. pertussis colonization was assessed in a mouse intranasal instillation model. Initial attempts to give the animals scFvs 30 min prior to B. pertussis infection gave inconsistent results for unknown reasons. Since there is a lack of knowledge on scFv stability and host factors that may affect the activity of scFv, we decided to test the biological function of the scFvs following premixing with B. pertussis. The results clearly demonstrate that the three scFvs can greatly reduce the colonization of B. pertussis in the respiratory tract. All three scFvs showed similar degrees of inhibition of colonization, and this may be due to the fact that the three scFvs have the same specificities of binding to the HBD of FHA. This is further supported by the observation that the three scFvs in combination yielded the same degree of inhibition to the individual scFv. It is interesting that each of the recombinant scFvs has a greater effect than the anti-Quadracel serum, which was included as a “positive” control. The serum has an excellent immunoglobulin G titer against FHA (11); however, the exact neutralizing titer is not known. Hence, it is possible that the diluted serum used in the experiment contains a diminished amount of neutralizing antibodies compared to the scFv. The results also showed that the B. pertussis counts in the parent phage-treated group were similar to those in the group that received PBS, indicating that the inhibition by the recombinant phages was specific. The above-described results suggest that antibodies against a specific domain of FHA can abolish B. pertussis colonization in vivo. The effect of this initial abrogation of colonization on the eventual outcome of B. pertussis infection remains to be tested.
The ability of scFvs of types 1, 5, and 7 to bind to both FHA and PRN is intriguing. The DNA sequence clearly showed that the three scFvs are different from one another. It is possible that the scFvs contain one variable fragment against FHA and another against PRN. However, such scFvs normally bind weakly to antigens because optimal antigen binding requires the participation of both VL and VH. Given the observed strong binding to FHA and PRN, types 1, 5, and 7 are unlikely to be composed of variable fragments generated against two different antigens. The most reasonable explanation for the double-antigen-binding phenomenon is that the three scFvs bind to epitopes that are shared between FHA and PRN. The logical candidate epitope is the RGD motif (25), which is present on both PRN and FHA. Surprisingly, the ELISA results showed that all three scFvs bound to the MBP-HBD fusion protein but not the RGD-containing fusion protein. The result suggests that PRN may contain an HBD; however, this remains to be verified. The affinity of the scFvs for HBD may explain the observed in vitro and in vivo results; i.e., the binding to heparan sulfate by B. pertussis has been neutralized by the scFvs. Heparan sulfates are one of the receptors for FHA (10) and are ubiquitous on the epithelial cell surface. In agreement with the observed protection in the animal model, Alonso et al. (1) previously reported that mice immunized with the FHA fragment containing the complete HBD domain (Fha44) were protected against B. pertussis challenge.
In summary, an scFv phage display library against B. pertussis antigens was constructed. Three of the scFvs have affinity for the HBD of FHA and inhibited the binding of B. pertussis to HEp-2 cells. The three scFvs prevented the colonization of B. pertussis in a mouse intranasal instillation model, further indicating that these scFvs have excellent biological activities and are good candidates for further studies in the development of passive immunity against pertussis.
Acknowledgments
We thank Ann MacMillan, Yi-Jing Li, and Annette Morris for their valuable technical assistance. We also thank Carlos Barbas III for providing pComb3H and Jessica Boyd for the gift of the helper phage VCSM13.
This study was supported by the Canadian Institutes of Health Research.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 27 August 2007.
REFERENCES
- 1.Alonso, S., N. Reveneau, K. Pethe, and C. Locht. 2002. Eighty-kilodalton N-terminal moiety of Bordetella pertussis filamentous hemagglutinin: adherence, immunogenicity, and protective role. Infect. Immun. 70:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbas, C. F., III, D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Bass, J. W., and R. R. Wilter. 1994. Return of epidemic pertussis in the United States. Pediatr. Infect. Dis. J. 13:343-345. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, M. J., J. H. Hannah, and E. Leininger. 1991. Adhesion of Bordetella pertussis to sulfatides and to the GalNAcβ4Gal sequence found in glycosphingolipids. J. Biol. Chem. 266:18827-18831. [PubMed] [Google Scholar]
- 5.Cahill, E. S., D. T. O'Hagan, L. Illum, A. Barnard, K. H. G. Mills, and K. Redhead. 1995. Immune responses and protection against Bordetella pertussis infection after intranasal immunization of mice with filamentous haemagglutinin in solution or incorporated in biodegradable microparticles. Vaccine 13:455-462. [DOI] [PubMed] [Google Scholar]
- 6.Cahill, E. S., D. T. O'Hagan, L. Illum, and K. Redhead. 1993. Mice are protected against Bordetella pertussis infection by intra-nasal immunization with filamentous haemagglutinin. FEMS Microbiol. Lett. 107:211-216. [DOI] [PubMed] [Google Scholar]
- 7.Cherry, J. D., J. Gornbein, U. Heininger, and K. Stehr. 1998. A search for serologic correlates of immunity to Bordetella pertussis cough illnesses. Vaccine 16:1901-1916. [DOI] [PubMed] [Google Scholar]
- 8.Coutte, L., S. Alonso, N. Reveneau, E. Willery, B. Quatannens, C. Locht, and F. Jacob-Dubuisson. 2003. Role of adhesin release for mucosal colonization by a bacterial pathogen. J. Exp. Med. 197:735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giomarelli, B., T. Maggi, J., Youson, C. Kelly, and G. Pozzi. 2004. Expression of a functional single-chain Fv antibody on the surface of Streptococcus gordonii. Mol. Biotechnol. 28:105-112. [DOI] [PubMed] [Google Scholar]
- 10.Hannah, J. H., F. D. Menozzi, G. Renauld, C. Locht, and M. J. Brennan. 1994. Sulfated glycoconjugate receptors for the Bordetella pertussis adhesin filamentous hemagglutinin (FHA) and mapping of the heparin-binding domain on FHA. Infect. Immun. 62:5010-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight, J. B., Y. Y. Huang, S. A. Halperin, R. Anderson, A. Morris, A. MacMillan, T. Jones, D. S. Burt, G. Van Nest, and S. F. Lee. 2006. Immunogenicity and protective efficacy of a recombinant filamentous hemagglutinin from Bordetella pertussis. Clin. Exp. Immunol. 144:5543-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruger, C., Y. Hu, Q. Pan, H. Marcotte, A. Hultberg, D. Delwar, P. J. van Dalen, P. H. Pouwels, R. J. Leer, C. G. Kelly, C. van Dollenweerd, J. K. Ma, and L. Hammarstrom. 2002. In situ delivery of passive immunity by lactobacilli producing single-chain antibodies. Nat. Biotechnol. 20:702-706. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Lee, C. W., S. F. Lee, and S. A. Halperin. 2004. Expression and immunogenicity of a recombinant diphtheria toxin fragment A in Streptococcus gordonii. Appl. Environ. Microbiol. 70:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, S. F. 2003. Oral colonization and immune responses to Streptococcus gordonii: potential use as a vector to induce antibody against respiratory pathogens. Curr. Opin. Infect. Dis. 16:231-235. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. F., S. A. Halperin, D. F. Salloum, A. MacMillan, and A. Morris. 2003. Mucosal immunization with a genetically engineered pertussis toxin S1 fragment-cholera toxin subunit B chimeric protein. Infect. Immun. 71:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, S. F., S. A. Halperin, H. Wang, and A. MacArthur. 2002. Oral colonization and immune responses to Streptococcus gordonii expressing a pertussis toxin S1 fragment in mice. FEMS Microbiol. Lett. 208:175-178. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. F., S. A. Halperin, J. B. Knight, and A. Tait. 2002. Purification and immunogenicity of a recombinant Bordetella pertussis S1S3FHA fusion protein expressed by Streptococcus gordonii. Appl. Environ. Microbiol. 68:4253-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, S. F., R. J. March, S. A. Halperin, G. Faulkner, and L. Gao. 1999. Surface expression of a protective recombinant pertussis toxin S1 subunit fragment in Streptococcus gordonii. Infect. Immun. 67:1511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leininger, E., S. Bowen, G. Renauld-Mongenie, J. H. Rouse, F. D. Menozzi, C. Locht, I. Heron, and M. J. Brennan. 1997. Immunodominant domains present on the Bordetella pertussis vaccine component filamentous hemagglutinin. J. Infect. Dis. 175:1423-1431. [DOI] [PubMed] [Google Scholar]
- 21.Leininger, E., M. Roberts, J. G. Kenimer, I. G. Charles, N. Fairweather, P. Novotny, and M. J. Brennan. 1991. Pertactin, an Arg-Gly-Asp-containing Bordetella pertussis surface protein that promotes adherence of mammalian cells. Proc. Natl. Acad. Sci. USA 88:345-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menozzi, F. D., C. Gantiez, and C. Locht. 1991. Interaction of Bordetella pertussis filamentous hemagglutinin with heparin. FEMS Microbiol. Lett. 78:59-64. [DOI] [PubMed] [Google Scholar]
- 23.Menozzi, F. D., R. Mutombo, G. Renauld, C. Gantiez, J. H. Hannah, E. Leininger, M. J. Brennan, and C. Locht. 1994. Heparin-inhibitable lectin activity of the filamentous hemagglutinin adhesin of Bordetella pertussis. Infect. Immun. 62:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills, K. H. G. 2001. Immunity to Bordetella pertussis. Microbes Infect. 3:655-677. [DOI] [PubMed] [Google Scholar]
- 25.Relman, D., E. Tuomanen, S. Falkow, D. T. Golenbock, K. Saukkonen, and S. D. Wright. 1990. Recognition of a bacterial adhesin by an integrin: macrophage CR3 (aMb2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell 61:1375-1382. [DOI] [PubMed] [Google Scholar]
- 26.Relman, D. A., M. Domenghini, E. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato, H., and Y. Sato. 1983. Bordetella pertussis infection in mice: correlation of specific antibodies against two antigens, pertussis toxin and filamentous hemagglutinin, with mouse protectivity in an intracerebral or aerosol challenge system. Infect. Immun. 46:415-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato, Y., K. Izumiya, H. Sato, J. L. Cowell, and C. R. Manclark. 1981. Role of antibody to leukocytosis-promoting factor hemagglutinin and the filamentous hemagglutinin in immunity to pertussis. Infect. Immun. 31:1223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahin, R., M. Leef, J. Eldridge, M. Hudson, and R. Gilley. 1995. Adjuvanticity and protective immunity elicited by Bordetella pertussis antigens encapsulated in poly(dl-lactide-co-glycolide) microspheres. Infect. Immun. 63:1195-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahin, R. D., D. F. Amsbaugh, and M. F. Leef. 1992. Mucosal immunization with filamentous hemagglutinin protects against Bordetella pertussis respiratory infection. Infect. Immun. 60:1482-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storsaeter, J., H. O. Hallander, L. Gustafsson, and P. Olin. 1998. Levels of anti-pertussis antibodies related to protection after household exposure to Bordetella pertussis. Vaccine 16:1907-1916. [DOI] [PubMed] [Google Scholar]
- 32.Tuomanen, E., H. Towbin, G. Rosenfelder, D. Braun, G. Larson, G. Hansson, and R. Hill. 1988. Receptor analogs and monoclonal antibodies that inhibit adherence of Bordetella pertussis to human ciliated respiratory epithelial cells. J. Exp. Med. 168:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 1999. Pertussis vaccines: WHO position paper. Wkly. Epidemiol. Rec. 74:137-142.10355354 [Google Scholar]