Abstract
Bordetella pertussis, the causative agent of human whooping cough, or pertussis, is an obligate human pathogen with diverse high-affinity transport systems for the assimilation of iron, a biometal that is essential for growth. Under iron starvation stress conditions, B. pertussis produces the siderophore alcaligin. The alcaligin siderophore gene cluster, consisting of the alcABCDERS and fauA genes, encodes activities required for alcaligin biosynthesis, the export of the siderophore from the cell, the uptake of the ferric alcaligin complex across the outer membrane, and the transcriptional activation of alcaligin system genes by an autogenous mechanism involving alcaligin sensing. The fauA gene encodes a 79-kDa TonB-dependent outer membrane receptor protein required for the uptake and utilization of ferric alcaligin as an iron source. In this study, using mixed-infection competition experiments in a mouse respiratory model, inactivation of the B. pertussis ferric alcaligin receptor protein was found to have a profound impact on in vivo growth and survival of a fauA mutant compared with a coinfecting wild-type strain. The attenuating effect of fauA inactivation was evident early in the course of the infection, suggesting that the contribution of ferric alcaligin transport to the ecological fitness of B. pertussis may be important for adaptation to iron-restricted host conditions that exist at the initial stages of infection. Alcaligin-mediated iron acquisition by B. pertussis may be critical for successful host colonization and establishment of infection.
One consensus that has emerged from global analyses of a diverse assortment of host-pathogen systems is that the most prevalent class of in vivo-expressed genes consists of those that function in nutrient acquisition (34). Furthermore, genes involved in metal acquisition, primarily iron, dominate the nutrient acquisition genes expressed in the host (34). Consequently, microbial iron acquisition gene expression is regarded as a transcriptional signature of the host microenvironment (29, 51). Active sequestration of iron by the host represents a type of nutritional immunity mediated primarily by specific host iron-binding glycoproteins such as lactoferrin in mucosal secretions. The level of freely available iron in extracellular tissue fluids of mammals is approximately 10−18 M, which is well below the 4 × 10−7 to 4 × 10−6 M iron concentration required to support the growth of most microorganisms (19, 59), necessitating the involvement of intensive microbial iron-scavenging mechanisms to acquire iron for growth.
Bordetella pertussis and the other so-called classical Bordetella species, B. bronchiseptica and B. parapertussis, produce the siderophore alcaligin (Fig. 1A) (11, 36). Alcaligin binds ferric iron with a stability constant of 1037 M−1 at physiological pH (38). The genetic organization of the alcaligin siderophore gene cluster (Fig. 1B) is shared by all of the classical Bordetella species (40) as well as by Bordetella holmesii (20). In B. holmesii, the alcaligin genes reside on a genomic island apparently acquired by horizontal transfer from B. pertussis, a process that is thought to have contributed to the emergence of B. holmesii and its adaptation to the human host. The alcABCDER genes encode alcaligin biosynthesis and regulatory functions (4, 8, 25, 26, 31, 43), and the alcS gene specifies a membrane efflux pump involved in alcaligin export (16). Ferric alcaligin uptake requires the 79-kDa TonB-dependent outer membrane receptor protein FauA (13). The fauA gene is expressed as a monocistronic transcript from its own Fur- and iron-repressible promoter (13, 32). Maximal expression of fauA and alcABCDER during iron starvation requires the AlcR regulator with the alcaligin siderophore as the inducer (13, 14).
FIG. 1.
Molecular structure of alcaligin and the spatial organization of the Bordetella alcaligin siderophore system gene cluster. (A) Molecular structure of alcaligin. (B) Genetic map of the alcaligin system gene cluster. Arrows indicate the transcriptional orientations and spatial limits of genes. An arrow representing the 2.2-kb fauA coding sequence is enlarged below, and the DNA region deleted in the construction of the nonpolar ΔfauA2 mutant strain PM11 is indicated by the white segment. The small arrowheads flanking the fauA region deleted in PM11 approximate the positions of oligonucleotide primers used in cPCR to determine strain ratios in bacterial populations recovered from infected mice. The small white arrowhead above the deleted region represents the position of the oligonucleotide probe used in confirmatory colony hybridizations to distinguish fauA+ bacteria from ΔfauA2 mutants in the infection output. A 1-kb scale bar is shown.
Previous studies established the importance of heme utilization for in vivo multiplication and survival of B. pertussis (17) using mixed-infection competition experiments in mice. Competition infection experiments are a preferred means to assess the role of certain genes in microbial virulence (9, 24). The degree of attenuation caused by a given mutation is inferred from the relative change in strain abundance that occurs under the selective pressures of the host environment. Typically, the coinfecting strains are differentially marked with antibiotic resistance markers for identification. In this study, the application of competitive PCR (cPCR) (58) to determine strain ratios obviates the requirement for different antibiotic resistance phenotypes and their potential attenuating effects. The current study was aimed at assessing the impact of ferric alcaligin utilization on B. pertussis multiplication and survival in vivo using a mouse respiratory infection model system.
MATERIALS AND METHODS
Bacterial strains and in vitro growth conditions.
B. pertussis UT25Sm1 (12) and PM11 were grown on Bordet-Gengou (BG) medium (10). Modified Stainer Scholte (SS) broth (50, 53) was used as liquid medium for B. pertussis. Escherichia coli DH5α (Bethesda Research Laboratories, Gaithersburg, MD) was used for routine plasmid propagation and DNA cloning procedures and as the donor in triparental matings. E. coli strain SURE (Stratagene, La Jolla, CA) was employed for the propagation of M13 mp18 bacteriophage derivatives (39), and E. coli host strain CJ236 (30) was used for site-directed mutagenesis procedures. E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates (49). Antibiotics were used at the indicated final concentrations: ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; tetracycline, 15 μg/ml; and nalidixic acid, 35 μg/ml.
Plasmids and genetic methods.
Plasmid pGEM3Z (Promega, Madison, WI) and bacteriophage vector M13 mp18 (39) were used to construct recombinant plasmids and bacteriophages. Plasmid pRK2013 (23) provided transfer functions in triparental matings. Plasmid pSS1129 (54) was used in the construction of B. pertussis mutant PM11. General genetic techniques were performed as described previously (49). DNA and protein sequence analysis was done using Lasergene version 5.53 software (DNASTAR, Inc., Madison, WI). Conjugal transfer of plasmids was performed as described previously (12), and transformation of E. coli strains used standard methods. Synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Nucleotide sequencing was performed by the University of Minnesota BioMedical Genomics Center.
Construction of B. pertussis ΔfauA2 mutant strain PM11.
A nonpolar in-frame B. pertussis ΔfauA2 deletion mutation was generated by in vitro site-directed mutagenesis (33) as described previously (13) for the construction of B. bronchiseptica ΔfauA2 strain BRM18 except using the cloned fauA region of B. pertussis UT25 (22) as the starting DNA template. The mutation was confirmed by nucleotide sequencing, delivered to the chromosome of B. pertussis strain UT25 Sm1 using allelic exchange plasmid pSS1129, and verified by PCR mapping using oligonucleotide primers 5′-GACACTCTCGCCACGCGAAAC-3′ (upstream primer fauA12) and 5′-CGTGATGGGCACGGAGATGTC-3′ (downstream primer fauA13) as described previously (13).
Iron source utilization assays.
Iron source utilization ability was assessed using a radial diffusion method developed previously for B. pertussis (56). Alcaligin was purified as previously described (11). Bovine hemin chloride (Sigma) was used as the positive-control iron source.
Preparation of mixed suspensions with various strain ratios for cPCR analysis.
Mixed suspensions of B. pertussis UT25Sm1 and PM11 used to establish the correlation between fauA allele-specific DNA copy number ratios and CFU ratios were prepared by culturing the strains separately in SS broth to the mid-exponential growth phase, collecting the bacterial cells by centrifugation, and resuspending them in SS broth at an optical density at 600 nm of 1.00. Mixed suspensions with various estimated mutant fractions ranging from 0.00 to 1.00 were prepared. Actual CFU ratios in the mixtures were extrapolated from CFU counting of the individual strain suspensions. DNA was prepared for cPCR analysis by a rapid boiling method that involved suspending bacterial cells recovered from 100 μl of a mixed-strain suspension in 100 μl H2O, boiling for 5 min, and removal of cell debris by centrifugation.
In vitro growth competition.
B. pertussis strains UT25Sm1 and PM11 were subcultured from BG medium to SS broth at an initial optical density at 600 nm of 0.1 and grown at 37°C with shaking (300 rpm) for 24 h. Bacteria were recovered and suspended in phosphate-buffered saline to approximately 2 × 108 CFU/ml based on optical densities. Equal volumes of UT25Sm1 and PM11 suspensions were combined (CFU ratio of 1:1) and used to inoculate iron-replete SS broth versus iron-replete SS broth supplemented with 25 μg/ml ethylenediamine-di-[(o-hydroxyphenyl)acetic acid] (EDDA) chelator (iron-restricted SS broth) to conditionally restrict iron bioavailability. Cultures were sampled at 0, 8, 24, and 48 h for total CFU enumeration by standard plate counting on BG medium (in triplicate). Bacterial DNA was prepared from culture samples by the rapid boiling method. Relative copy numbers of mutant ΔfauA2 and wild-type fauA+ DNA in the inoculum and in output samples were determined in triplicate by cPCR and fluorescence image analysis as detailed in the cPCR analysis described above in Materials and Methods. For each culture condition, the competitive index (CI) at each time point was calculated as the mutant (normalized for molar equivalence)/wild-type DNA fluorescence intensity peak area ratio for the output divided by the mutant (normalized)/wild-type DNA fluorescence intensity peak area ratio determined for the input inoculum.
Preparation of inoculum for mixed-infection competition experiments in mice.
The inoculum was prepared as described previously (17), except using strains UT25Sm1 and PM11, and the total CFU were enumerated by plate counting on BG medium. The strain ratio in the input was determined by cPCR using fauA-specific primers and DNA prepared from the input suspension.
In vivo mixed-infection competition.
All research involving experimental animals was performed in accordance with federal guidelines and institutional policies. Twenty-five female BALB/cAnNHsd mice (10 to 20 g) (Harlan Sprague Dawley, Inc.) were sedated by isoflurane inhalation and infected intranasally with ∼2 × 106 CFU of a 1:1 mixture of UT25Sm1 and PM11 in a 10-μl volume. At 3 days, 7 days, 14 days, and 21 days postinfection, five mice were euthanized (six mice were taken on day 14), and respiratory tissue (lungs and trachea) homogenates were plated onto BG medium for total CFU enumeration. Total DNA was isolated from the homogenates, and the ratio of mutant to wild-type bacteria was determined by cPCR using fauA-specific primers.
Preparation of total DNA from mouse respiratory tissue homogenates.
DNA was recovered from tissue homogenates essentially as described previously (55). Control DNA samples were prepared from uninfected mouse respiratory tissues. For the inoculum, a scaled-down version of the same procedure was used for isolation of genomic DNA from a 100-μl volume of the suspension.
Competitive PCR analysis.
Wild-type and mutant fauA target sequences were coamplified by cPCR using primers that flank the ΔfauA2 mutation and direct the production of a 430-bp product from the wild-type fauA allele and a 175-bp product from the ΔfauA2 allele (the same primer pair used to map the fauA deletion mutation in PM11). PCR conditions used a solution containing 20 mM Tris-HCl (pH 8.8)-2 mM MgSO4-10 mM KCl-10 mM (NH4)2SO4-0.1% Triton X-100-100 μg/ml nuclease-free bovine serum albumin-5% dimethyl sulfoxide-1 mM deoxynucleoside triphosphates-500 nM each fauA-specific oligonucleotide primer and 0.5 U Pfu Turbo DNA polymerase (Stratagene) with 1 μl DNA sample in a 20-μl reaction volume. The PCR program used 35 cycles of denaturation at 96°C, primer annealing at 64°C, and extension at 72°C for 45 s unless otherwise indicated. PCR products were resolved on 5% polyacrylamide gels, stained with 0.5 μg ethidium bromide/ml, and imaged under UV transillumination using a FOTO/Analyst Archiver electronic photodocumentation system equipped with a charge-coupled-device camera (Fotodyne Inc., Hartland, WI). Quantitative image analysis used ImageJ version 1.36b software (http://rsb.info.nih.gov/ij). Fluorescence intensity peak areas of PCR product bands were normalized for the molar difference in ethidium bromide dye binding by multiplying the peak areas of the ΔfauA2-derived PCR products by 2.457, the wild-type product/mutant product size ratio (430 bp/175 bp). The mutant (normalized)-to-wild-type PCR product peak area ratio corresponds to the ΔfauA2/fauA+ DNA copy number ratio in the tissue sample. Since fauA is a single-copy gene, the mutant-to-wild-type DNA copy number ratios were used to derive the CI values.
In preliminary studies of the influence of PCR cycle number and amplification phase on PCR product molar ratios, DNA purified from a mixed-strain suspension of UT25Sm1 and PM11 was used as the template in 10 replicate PCRs. After every three cycles, a reaction tube was removed from the thermocycler and held on ice until completion of the amplification program. The products from each reaction were quantitated by fluorescence image analysis of ethidium bromide-stained gels.
Bacterial colony hybridization.
The PM11/UT25Sm1 strain ratios in the input suspension and the output from the infected mice were estimated by colony hybridization. Approximately 100 bacterial colonies from the inoculum suspension or tissue homogenate CFU plates for each mouse were patched onto BG medium along with PM11 and UT25Sm1 controls and cultured for 2 to 3 days at 37°C. Growth patches were lifted onto nitrocellulose filters and probed by DNA hybridization (28) using a 32P-end-labeled oligonucleotide, 5′-CCTGAGCCAATGGAACTATG-3′, specific for DNA sequences deleted in the ΔfauA2 mutant strain PM11. Mutant-to-wild-type strain ratios were calculated using the formula (T − W)/W, where T is the total number of colonies, W is the number of colonies producing positive hybridization signals, and T − W was deduced to be the number of mutant colonies.
Statistical methods.
CI values were calculated as the ΔfauA2/fauA+ DNA copy number ratio in mouse samples divided by the ΔfauA2/fauA+ DNA copy number ratio in the inoculum. For the in vivo studies, the mean CI at each time point is the mean of five independent mouse infections. A Student's t test was used to determine whether the mean CI at each time point differed significantly from the hypothesized mean value of 1.00 (the predicted mean CI if there was no difference in fitness between the two strains used in mixed infections or in vitro cultures) and whether the change in the mean CI between consecutive time points was significant (hypothesized mean CI difference of 0.00 between each time point). Probabilities (P values) of ≤0.001 were considered to be significant.
For estimates of the mutant-to-wild-type strain ratio in the infection output by colony hybridization, a random sample size of 100 colonies per mouse, associated with a maximum statistical margin of error of ±9.8% at the 95% confidence level, was accepted as an estimate of the true relative prevalence of strains UT25Sm1 and PM11 in the output population. Covariation analysis compared mean CI values by cPCR at each time point, with mean CI values based on colony hybridization results. Statistical analyses used Statview version 4.51 software and the Analysis Toolpak add-in of Microsoft Excel 2004 for Mac software, with assistance provided by the Statistical Consulting Service of the University of Minnesota School of Statistics.
RESULTS
B. pertussis ΔfauA2 mutant PM11 is defective in the utilization of ferric alcaligin as a nutritional iron source.
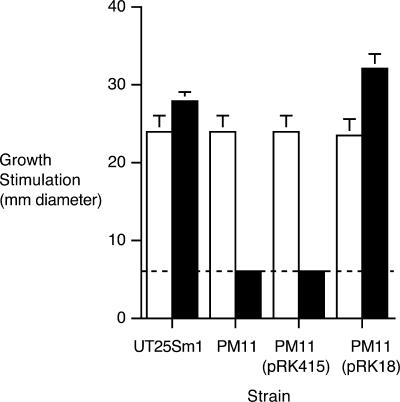
B. pertussis PM11 carries a 255-bp in-frame deletion mutation in fauA, which encodes the TonB-dependent outer membrane receptor for the ferric alcaligin siderophore complex (Fig. 1). PM11 showed no detectable growth stimulation by alcaligin in iron source utilization bioassays compared with the fauA+ strain UT25Sm1 (Fig. 2) but was unaffected in its ability to utilize hemin, a control iron source. The B. pertussis ΔfauA2 mutant phenotype is similar to that previously reported for a B. bronchiseptica ΔfauA2 mutant (13). FauA production and the wild-type ferric alcaligin utilization phenotype were restored to PM11 by trans complementation using plasmid pRK18 (13), which carries a 3.8-kb fauA+ chromosomal DNA fragment of B. pertussis strain UT25 (Fig. 2 and data not shown). FauA is not required for alcaligin production, and fauA mutants do not hyperproduce alcaligin; PM11 produces and exports alcaligin at normal levels (data not shown).
FIG. 2.
Ferric alcaligin utilization assays. Bars indicate mean diameters in millimeters (n = 3) (±1 standard deviation) of growth stimulation zones surrounding wells filled with iron source solutions. Relevant genotypes of B. pertussis indicator strains used are as follows: UT25Sm1, fauA+; PM11, ΔfauA2; PM11(pRK415), ΔfauA2 (plasmid vector control); and PM11(pRK18), ΔfauA2/fauA+ (fauA+ in trans). The dashed line corresponds to the 6-mm diameter of the sample well containing the iron source solution. Filled bars, alcaligin (125 μM); open bars, hemin chloride (100 μM) (positive control iron source).
Coamplification of ΔfauA2 and fauA+ alleles by cPCR provides a reliable measure of the PM11/UT25Sm1 strain ratio in a mixed bacterial population.
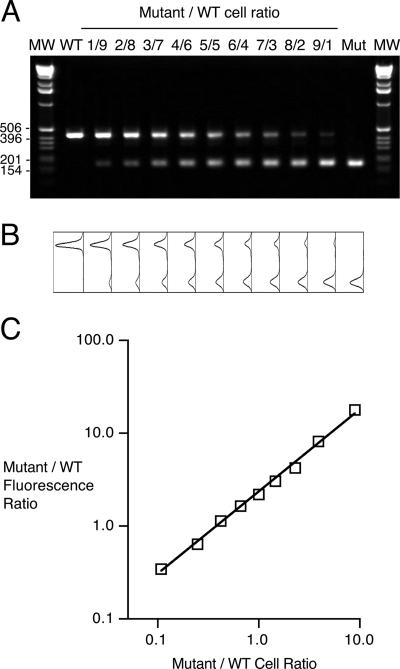
Mixed-infection competition experiments rely on the ability to measure changes in the coinfecting strain ratios. To determine whether cPCR could be used to monitor strain ratios using unmarked strains, the correlation between the fauA allele-specific DNA copy number ratio determined by cPCR and the actual CFU ratio in a mixed bacterial population was examined in vitro using defined mixtures of PM11 and UT25Sm1. Quantitative image analysis of ethidium bromide-stained electrophoretic gels (Fig. 3A) confirmed that the molar ratio of ΔfauA2 to fauA+ DNA copies varied directly as the PM11/UT25Sm1 cell ratio for these mixed-strain populations (Fig. 3B). The relationship between the fluorescence peak area ratios and the actual CFU ratios for the strain mixtures yielded a correlation coefficient r of 0.999.
FIG. 3.
Relationship between ΔfauA2/fauA+ PCR product DNA molar ratios and cell ratios in mixed bacterial suspensions. (A) Ethidium bromide-stained gel showing PCR products generated by coamplification of ΔfauA2 and fauA+ DNA copies in mixed bacterial suspensions with various PM11/UT25Sm1 cell ratios (1/9 [1 volume of mutant suspension combined with 9 volumes of wild-type suspension], 2/8 [2 volumes of mutant suspension combined with 8 volumes of wild-type suspension], etc.). Lanes labeled WT and Mut show PCR products generated from pure suspensions of UT25Sm1 and PM11 alone, respectively. MW, size markers with indicated sizes in bp. (B) Plots showing raw fluorescence peaks associated with the corresponding gel lanes in A. (C) Mutant-to-wild-type (WT) fluorescence ratio as a function of the mutant-to-wild-type cell ratio in mixed strain suspensions. Fluorescence intensity peak areas were normalized for molar differences in the fluorescent dye-binding capacities of the wild-type- and mutant-specific products as described in Materials and Methods. The line represents the best-fit line (r = 0.999).
Influence of PCR cycle number and amplification phase on the ΔfauA2/fauA+ DNA copy number ratio determined by cPCR.
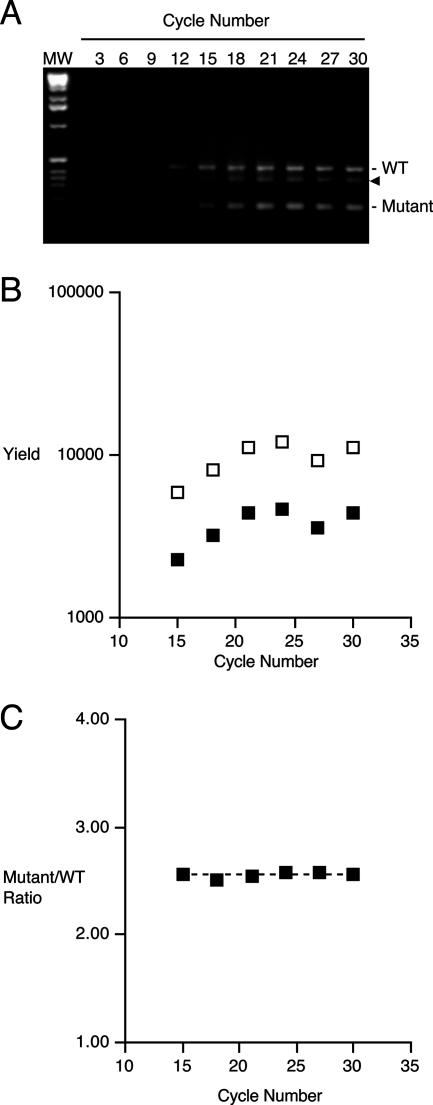
Allele-specific PCR product yields and molar DNA product ratios were determined for replicate cPCR reactions prepared using a mixed-strain DNA template and subjected to amplification programs varying from 3 to 30 cycles, encompassing the exponential and plateau phases of PCR (Fig. 4). The 430-bp fauA+- and 175-bp ΔfauA2-derived PCR products were visibly detectable in ethidium bromide-stained electrophoretic gels after 15 cycles (Fig. 4A). Product yields increased exponentially until 21 cycles (Fig. 4B), at which stage the amplification program entered the plateau phase, and no further increase in product yield was apparent in any reaction. Variations were observed and are included in the data shown to exemplify some useful features of cPCR. For example, the reduced product yields for the reaction mixture subjected to 27 cycles (Fig. 4B) are probably due to pipetting error during template addition to the reaction or to sample loss during gel loading. In addition, an unknown DNA species of intermediate size, thought to represent a ΔfauA2/fauA+ DNA heteroduplex, was detected in products from this reaction set (Fig. 4A). It is notable that despite these variations, the ΔfauA2/fauA+ PCR product DNA molar ratio was constant at 2.56 ± 0.02 (mean ± standard deviation for the reactions undergoing 15 to 30 cycles) (n = 6) (Fig. 4C). The reproducibility of this value confirms that the DNA copy number ratio determined by cPCR is dependent only on the DNA copy number ratio in the initial mixture and is independent of PCR cycle number and amplification phase, obviating the need to restrict the cPCR analysis to any particular amplification phase.
FIG. 4.
Influence of PCR cycle number and amplification program phase on fauA allele-specific PCR product yields and ΔfauA2/fauA+ PCR product molar ratios in cPCR analysis of a prepared mixture of PM11 and UT25Sm1. (A) Ethidium bromide-stained gel showing coamplified fauA+ and ΔfauA2-specific PCR products produced using a mixed-strain DNA sample and various cycle numbers of a PCR amplification program. Positions of the wild-type (WT) and mutant (Mutant) products are indicated. The arrowhead marks the position of the putative mutant/wild-type DNA heteroduplex species. MW, size markers. (B) Yield of ΔfauA2- and fauA+-specific PCR products at various cycle numbers determined by quantitative image analysis of the gel shown in A and normalized for their molar differences in fluorescence dye-binding capacities. Filled squares, fluorescence peak areas for 430-bp wild-type products; open squares, fluorescence peak areas (normalized) for 175-bp mutant products. (C) Mutant (normalized)/wild-type fluorescence peak area ratios at various cycle numbers representing the exponential and plateau phases of the amplification program calculated from product yields shown in B. The dashed line represents the mean mutant (normalized)/wild-type molar ratio of 2.56.
B. pertussis ΔfauA2 mutant PM11 has no growth defect in vitro.
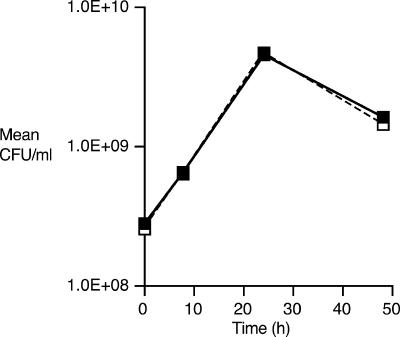
B. pertussis UT25Sm1 and PM11 were cultured in parallel in iron-replete SS broth (36 μM iron), and growth was monitored by optical density and CFU counting. The strains exhibited comparable growth with respect to duration of lag phase, exponential growth rate, maximal growth levels at stationary phase, and rate of decline (Fig. 5), indicating that the mutant has no fundamental growth defect in vitro.
FIG. 5.
In vitro growth of wild-type and ΔfauA2 B. pertussis strains in iron-replete SS broth. Growth of fauA+ strain UT25Sm1 and ΔfauA2 strain PM11 cultured separately in standard SS broth for 48 h is shown as mean CFU/ml (n = 3) (± standard deviation). Filled symbols and solid line, UT25Sm1; open symbols and dashed line, PM11.
PM11 exhibits reduced fitness relative to the fauA+ parent strain UT25Sm1 when cocultured in vitro under conditions that restrict iron availability.
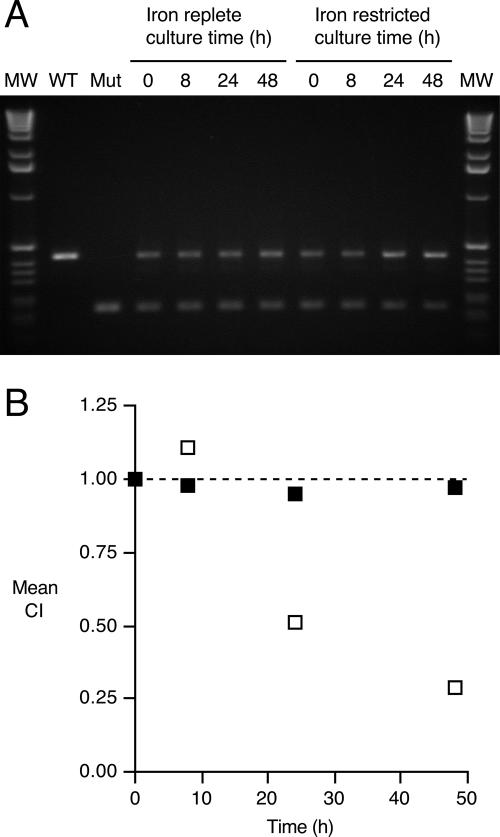
PM11 and UT25Sm1 were cocultured in iron-replete SS broth versus iron-restricted SS broth, and strain ratios were determined by cPCR at 0, 8, 24, and 48 h. The relative yields of ΔfauA2- and fauA+-specific PCR products are shown on the gel pictured in Fig. 6A. CI values (Fig. 6B) derived by quantitative image analysis of a representative gel (Fig. 6A) determined that the mutant-to-wild-type product molar ratio in the iron-replete culture remained virtually unchanged from the initial ratio, indicating that PM11 has equivalent fitness to its wild-type parent when cocultured under iron-replete conditions. In contrast, PM11 had a relative growth disadvantage compared to UT25Sm1 under iron-restricted culture conditions with EDDA-bound iron. The mean CI values for the iron-restricted cultures decreased to 0.51 by 24 h and 0.29 by 48 h, indicating that the inactivation of fauA imposes a fitness cost on B. pertussis PM11 that is evident under conditions that require the production and transport of ferric alcaligin for iron assimilation.
FIG. 6.
In vitro growth competition between wild-type and ΔfauA2 B. pertussis strains in iron-replete and iron-restricted SS broth. (A) Ethidium bromide-stained gel showing relative yield of 430-bp fauA+- and 175-bp ΔfauA2-specific PCR products coamplified from DNA prepared from mixed-strain cultures in iron-replete and iron-restricted SS broth. Mut, mutant; WT, wild type; MW, size markers. (B) Mean CI values (n = 3) (y error bars representing ±1 standard deviation are hidden by the symbols) derived from ΔfauA2/fauA+ DNA copy number ratios determined by fluorescence image analysis. Filled squares, iron-replete conditions; open squares, iron-restricted conditions. The dashed line (y value of 1.00) corresponds to the hypothesized CI value if there was no difference in fitness between the strains cocultured under the specified conditions.
FauA function contributes to in vivo fitness of B. pertussis during primary respiratory infection in mice.
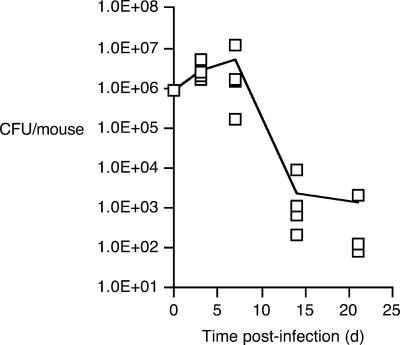
Mice were infected intranasally with 8.77 × 105 CFU of a 1:1 mixture of UT25Sm1 and PM11. Typical of B. pertussis infection kinetics in mice, total bacterial loads (Fig. 7) increased over threefold by 3 days postinfection and reached maximal levels of approximately 4.90 × 106 CFU/mouse at 7 days postinfection. Bacterial loads declined more than 1,000-fold between 7 and 14 days, further decreasing by another twofold between 14 and 21 days. Competitive PCR analysis and the resulting CI values (Fig. 8A and B) revealed a marked shift in the relative abundance of the coinfecting strains, which is indicative of a costly fitness defect in the mutant strain under the selective pressures of the host environment. Mean CI values declined significantly to 0.53 as early as 3 days postinfection (P = 0.0001) and further decreased to 0.12 at 7 days postinfection (P = < 0.0001). This low CI value at 7 days is coincident with peak bacterial loads, the population level at which competition for a limiting nutrient would be predicted to be most acute. The mean CI difference of 0.41 between 3 days and 7 days was also statistically significant (P = < 0.0001). The mean CI did not decrease significantly after the day 7 time point. Respiratory tissue DNA samples from uninfected mice did not yield any detectable fauA-specific PCR products (data not shown). Parallel studies using colony hybridization to estimate the ΔfauA2/fauA+ strain ratios for viable bacteria recovered from infected mice corroborated the ratios based on cPCR; by covariation analysis, a strong correlation between the mean CI values derived using the two different strain detection methods was found to exist (r = 0.948).
FIG. 7.
Total B. pertussis wild-type and mutant CFU recovered from mice in mixed-infection competition studies. Each symbol represents the total CFU recovered from an infected mouse at days 3, 7, 14, and 21; CFU were recovered from only three of the five mice sampled at day 21. The line indicates the mean CFU/mouse as a function of time postinfection (days).
FIG. 8.
PCR product yields and CI values based on cPCR analysis of mouse tissue homogenates in mixed-infection competition studies. (A) Ethidium bromide-stained gel images showing relative yields of 430-bp fauA+- and 175-bp ΔfauA2-specific PCR products coamplified from DNA extracted from infected mouse respiratory tract tissues. Each gel lane represents a single infected mouse. Products generated using the input strain mixture are also shown. (B) CI values based on ΔfauA2/fauA+ fluorescence peak area ratios determined by cPCR. Relative DNA copy number yields of fauA+- and ΔfauA2-specific PCR products were determined by fluorescence image analysis of the stained gels shown in A. Each symbol represents the CI calculated for a single mouse at a given time point, and the line represents the mean CI value. The dashed line (y value of 1.00) corresponds to the hypothesized CI value if there was no difference in fitness between the strains in this mouse model of respiratory infection.
These results indicate that ferric alcaligin utilization contributes to the ecological fitness of B. pertussis during primary respiratory infection in mice and that the ability to assimilate host iron using the alcaligin siderophore system likely plays a significant role in the early stages of host colonization.
DISCUSSION
The ability to overcome host defenses that actively restrict iron availability for invading microbes is an elemental trait of a successful mucosal pathogen (19, 59, 60). B. pertussis colonizes ciliated cells of the upper respiratory tract epithelium, where it multiplies, causing mild to severe local injury and diverse systemic effects. The classical bordetellae have multiple systems for iron retrieval (15, 18) including the three genetically characterized systems for utilization of the native alcaligin siderophore, the enterobactin xenosiderophore, and heme (1, 4, 5, 6, 56). In vitro studies have established that these pathogens also have the ability to utilize several additional catecholate and hydroxamate class xenosiderophores (3, 37, 45). The classical bordetellae have genes for a number of TonB-dependent iron receptors of unknown ligand specificity (6, 7, 40, 45), implying that their iron-scavenging potential is even more diverse than is currently recognized. An important feature shared by the iron retrieval systems for alcaligin, enterobactin, and heme is that each system is up-regulated in response to its cognate iron source (2, 14, 57). This ability to sense and respond to particular iron sources suggests that the ability to selectively deploy different iron retrieval systems contributes to successful in vivo growth.
Changes in host niche conditions over the course of infection are proposed to alter the array of potential iron sources available to B. pertussis. Upon initial colonization, it is conceivable that B. pertussis would exploit lactoferrin in mucosal secretions using its native siderophore alcaligin. Utilization of human lactoferrin and transferrin as iron sources by B. pertussis and B. bronchiseptica in vitro (46, 47) has been shown not to require direct bacterial cell contact with the iron-binding proteins (1, 27), implicating a siderophore in this process. Other siderophores produced by respiratory commensals or transient colonizers could also supply iron to B. pertussis, provided that B. pertussis produces the functions required for their uptake and utilization. As infection progresses, the inflammation and damaging effects of Bordetella toxins may disrupt the epithelial barrier and allow iron-loaded cellular and serum components to escape to the mucosal surface (41). Lysis of host cells with the release of their intracellular components might supply yet additional iron sources such as transferrin and heme proteins that could be assimilated by B. pertussis. Once heme sources are released and made available to B. pertussis on the mucosal surface, the primary host iron-withholding defense imposed by lactoferrin is effectively bypassed since lactoferrin cannot bind and withhold heme.
Previously published studies show the importance of iron acquisition in the pathogenesis of B. pertussis and B. bronchiseptica. TonB is required for high-affinity iron transport in B. pertussis and B. bronchiseptica (37, 44), and infection studies in mice found that a B. pertussis tonB exbB mutant strain could not efficiently colonize the lungs (44). A B. pertussis alcR mutant was found to have no significant defect in the ability to colonize the mouse lung (43); however, alcR strains can still produce, transport, and utilize ferric alcaligin, albeit at reduced levels (8). In a different study, a B. bronchiseptica alcaligin-deficient mutant was impaired in the colonization of neonatal swine, and infected animals presented little or none of the nasal pathology normally associated with atrophic rhinitis (48).
Using mixed-infection competition experiments in a mouse respiratory model, the heme utilization system of B. pertussis was previously shown to be important for multiplication and survival in the host (17). A bhuR strain defective in heme utilization was found to be significantly less fit than the coinfecting wild-type parent strain, as evidenced by a marked reduction in the mean CI value after 7 days postinfection. The timing of this reduction suggests that the limiting resource or host condition responsible for the competitive loss of the mutant strain was not available or active in the host niche at the initiation of infection but was accessible to B. pertussis later in infection. This hypothesis is consistent with the prediction that heme is made available by the action of Bordetella toxic factors on the host primarily during the advanced stages of infection.
In vivo competition results from the current study confirm that ferric alcaligin siderophore utilization contributes fundamentally to the fitness of B. pertussis in the murine respiratory tract and that alcaligin-mediated iron transport is important for B. pertussis adaptation to limiting iron nutritional conditions that exist at the early stages of infection.
In mixed-infection competition experiments, coinfecting strains are usually engineered to express different selectable markers for strain identification. In this study, coinfecting strains differed only by the chromosomal deletion mutation responsible for the ferric alcaligin utilization defect of the mutant. cPCR (21, 35, 42, 52, 58) was used to determine the mutant-to-wild-type strain ratios over the course of infection, and total bacterial loads in infected mice were monitored by CFU counting. Control experiments established a strong positive correlation between CFU ratios based on fauA+-specific colony DNA hybridization and fauA allele-specific target DNA ratios based on cPCR, indicating that the clearance of B. pertussis from infected mouse tissues was associated with a loss of the ability to detect B. pertussis by PCR.
There are several advantages to the application of cPCR in mixed-infection competition experiments. The measure of relative DNA target abundance by cPCR is dependent on the initial ratio of target templates in the samples (21, 52), which remains constant throughout all phases of the amplification program, so it is not necessary to base the strain ratio determinations on data acquired only during the exponential phase of the PCR program. The targets compete for the same primers, there are no priming efficiency differences, and any variable effects due to differences in sample preparation, recovery, or PCR amplification are all internally controlled and affect the yields of wild-type and mutant product equally (52, 58). Perhaps the main advantage of cPCR in mixed-infection competition experiments is that cPCR does not require the use of marked strains, so there is no potential influence of selectable marker expression on virulence.
These findings provide evidence that among the diverse iron retrieval systems of B. pertussis, the alcaligin iron retrieval system has a distinct role in virulence. Alcaligin-mediated iron retrieval is important under host conditions that exist in the early stages of infection and may be essential for the successful colonization of the human respiratory mucosa.
Acknowledgments
We are grateful to Christopher Bingham and the Statistical Consulting Service of the University of Minnesota School of Statistics for advice concerning statistical analyses.
This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
Editor: F. C. Fang
Footnotes
Published ahead of print on 27 August 2007.
REFERENCES
- 1.Agiato, L. A., and D. W. Dyer. 1992. Siderophore production and membrane alterations by Bordetella pertussis in response to iron starvation. Infect. Immun. 60:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. T., and S. K. Armstrong. 2004. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J. Bacteriol. 186:7302-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, M. T., and S. K. Armstrong. 2006. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J. Bacteriol. 188:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong, S. K., and M. O. Clements. 1993. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J. Bacteriol. 175:1144-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 6.Beall, B., and T. Hoenes. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143:135-145. [DOI] [PubMed] [Google Scholar]
- 7.Beall, B. 1998. Two iron-regulated putative ferric siderophore receptor genes in Bordetella bronchiseptica and Bordetella pertussis. Res. Microbiol. 149:189-201. [DOI] [PubMed] [Google Scholar]
- 8.Beaumont, F. C., H. Y. Kang, T. J. Brickman, and S. K. Armstrong. 1998. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J. Bacteriol. 180:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 10.Bordet, J., and O. Gengou. 1906. Le microbe de la coqueluche. Ann. Inst. Pasteur (Paris) 20:731-741. [Google Scholar]
- 11.Brickman, T. J., J. G. Hansel, M. J. Miller, and S. K. Armstrong. 1996. Purification, spectroscopic analysis and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. Biometals 9:191-203. [DOI] [PubMed] [Google Scholar]
- 12.Brickman, T. J., and S. K. Armstrong. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J. Bacteriol. 178:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brickman, T. J., C. K. Vanderpool, and S. K. Armstrong. 2004. Bordetella, p. 311-328. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 16.Brickman, T. J., and S. K. Armstrong. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 187:3650-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brickman, T. J., C. K. Vanderpool, and S. K. Armstrong. 2006. Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect. Immun. 74:1741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brickman, T. J., M. T. Anderson, and S. K. Armstrong. 2007. Bordetella iron transport and virulence. Biometals 20:303-322. [DOI] [PubMed] [Google Scholar]
- 19.Bullen, J. J. 1981. The significance of iron in infection. Rev. Infect. Dis. 3:1127-1138. [DOI] [PubMed] [Google Scholar]
- 20.Diavatopoulos, D. A., C. A. Cummings, H. G. van der Heide, M. van Gent, S. Liew, D. A. Relman, and F. R. Mooi. 2006. Characterization of a highly conserved island in the otherwise divergent Bordetella holmesii and Bordetella pertussis genomes. J. Bacteriol. 188:8385-8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diviacco, S., P. Norio, L. Zentilin, S. Menzo, M. Clementi, G. Biamonti, S. Riva, A. Falaschi, and M. Giacca. 1992. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene 122:313-320. [DOI] [PubMed] [Google Scholar]
- 22.Field, L. H., and C. D. Parker. 1979. Differences observed between fresh isolates of Bordetella pertussis and their laboratory-passaged derivatives, p. 124-132. In C. R. Manclark and J. C. Hill (ed.), International Symposium on Pertussis. Public Health Service, U.S. Department of Health, Education, and Welfare, Washington, DC.
- 23.Figurski, D. H., C. Young, H. C. Schreiner, R. F. Pohlman, D. H. Bechhofer, A. S. Prince, and T. F. D'Amico. 1985. Genetic interactions of broad host-range plasmid RK2: evidence for a complex replication regulon. Basic Life Sci. 30:227-241. [DOI] [PubMed] [Google Scholar]
- 24.Freter, R., B. Allweiss, P. C. O'Brien, S. A. Halstead, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vitro studies. Infect. Immun. 34:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giardina, P. C., L. A. Foster, S. I. Toth, B. A. Roe, and D. W. Dyer. 1995. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene 167:133-136. [DOI] [PubMed] [Google Scholar]
- 26.Giardina, P. C., L. A. Foster, S. I. Toth, B. A. Roe, and D. W. Dyer. 1997. Analysis of the alcABC operon encoding alcaligin biosynthesis enzymes in Bordetella bronchiseptica. Gene 194:19-24. [DOI] [PubMed] [Google Scholar]
- 27.Gorringe, A. R., G. Woods, and A. Robinson. 1990. Growth and siderophore production by Bordetella pertussis under iron-restricted conditions. FEMS Microbiol. Lett. 54:101-105. [DOI] [PubMed] [Google Scholar]
- 28.Grunstein, M., and D. S. Hogness. 1975. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. USA 72:3961-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heithoff, D. M., C. P. Conner, P. C. Hanna, S. M. Julio, U. Hentschel, and M. J. Mahan. 1997. Bacterial infection as assessed by in vivo gene expression. Proc. Natl. Acad. Sci. USA 94:934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyce, C. M., and N. D. Grindley. 1984. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J. Bacteriol. 158:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang, H. Y., T. J. Brickman, F. C. Beaumont, and S. K. Armstrong. 1996. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J. Bacteriol. 178:4877-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang, H. Y., and S. K. Armstrong. 1998. Transcriptional analysis of the Bordetella alcaligin siderophore biosynthesis operon. J. Bacteriol. 180:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahan, M. J., D. M. Heithoff, R. L. Sinsheimer, and D. A. Low. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu. Rev. Genet. 34:139-164. [DOI] [PubMed] [Google Scholar]
- 35.Menzo, S., P. Bagnarelli, M. Giacca, A. Manzin, P. E. Varaldo, and M. Clementi. 1992. Absolute quantitation of viremia in human immunodeficiency virus infection by competitive reverse transcription and polymerase chain reaction. J. Clin. Microbiol. 30:1752-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, C. H., L. A. Foster, D. G. Gerbig, Jr., D. W. Dyer, and B. W. Gibson. 1995. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J. Bacteriol. 177:1116-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson, M. L., and B. Beall. 1999. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology 145:2453-2461. [DOI] [PubMed] [Google Scholar]
- 38.Nishio, T., N. Tanaka, J. Hiratake, Y. Katsube, Y. Ishida, and J. Oda. 1988. Isolation and structure of the novel dihydroxamate siderophore alcaligin. J. Am. Chem. Soc. 110:8733-8734. [Google Scholar]
- 39.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 40.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 41.Persson, C. G., I. Erjefalt, U. Alkner, C. Baumgarten, L. Greiff, B. Gustafsson, A. Luts, U. Pipkorn, F. Sundler, C. Svensson, et al. 1991. Plasma exudation as a first line respiratory mucosal defence. Clin. Exp. Allergy 21:17-24. [DOI] [PubMed] [Google Scholar]
- 42.Piatak, M., Jr., K. C. Luk, B. Williams, and J. D. Lifson. 1993. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques 14:70-81. [PubMed] [Google Scholar]
- 43.Pradel, E., N. Guiso, and C. Locht. 1998. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J. Bacteriol. 180:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pradel, E., N. Guiso, F. D. Menozzi, and C. Locht. 2000. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect. Immun. 68:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pradel, E., and C. Locht. 2001. Expression of the putative siderophore receptor gene bfrZ is controlled by the extracytoplasmic-function sigma factor BupI in Bordetella bronchiseptica. J. Bacteriol. 183:2910-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redhead, K., T. Hill, and H. Chart. 1987. Interaction of lactoferrin and transferrins with the outer membrane of Bordetella pertussis. J. Gen. Microbiol. 133:891-898. [DOI] [PubMed] [Google Scholar]
- 47.Redhead, K., and T. Hill. 1991. Acquisition of iron from transferrin by Bordetella pertussis. FEMS Microbiol. Lett. 61:303-307. [DOI] [PubMed] [Google Scholar]
- 48.Register, K. B., T. F. Ducey, S. L. Brockmeier, and D. W. Dyer. 2001. Reduced virulence of a Bordetella bronchiseptica siderophore mutant in neonatal swine. Infect. Immun. 69:2137-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Schneider, D. R., and C. D. Parker. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoolnik, G. K. 2002. Microarray analysis of bacterial pathogenicity. Adv. Microb. Physiol. 46:1-45. [DOI] [PubMed] [Google Scholar]
- 52.Siebert, P. D., and J. W. Larrick. 1992. Competitive PCR. Nature 359:557-558. [DOI] [PubMed] [Google Scholar]
- 53.Stainer, D. W., and M. J. Scholte. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211-220. [DOI] [PubMed] [Google Scholar]
- 54.Stibitz, S. 1994. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 235:458-465. [DOI] [PubMed] [Google Scholar]
- 55.Strauss, W. M. 1998. Preparation of genomic DNA from mammalian tissue, p. 2.2.1-2.2.3. In R. B. Frederick M. Ausubel, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Hoboken, NJ. [DOI] [PubMed]
- 56.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanderpool, C. K., and S. K. Armstrong. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, A. M., M. V. Doyle, and D. F. Mark. 1989. Quantitation of mRNA by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:9717-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberg, E. D. 1984. Iron withholding: a defense against infection and neoplasia. Physiol. Rev. 64:65-102. [DOI] [PubMed] [Google Scholar]