Abstract
Cytotoxic necrotizing factor type 1 (CNF1) and CNF2 are highly homologous toxins that are produced by certain pathogenic strains of Escherichia coli. These 1,014-amino-acid toxins catalyze the deamidation of a specific glutamine residue in RhoA, Rac1, and Cdc42 and consist of a putative N-terminal binding domain, a transmembrane region, and a C-terminal catalytic domain. To define the regions of CNF1 that are responsible for binding of the toxin to its cellular receptor, the laminin receptor precursor protein (LRP), a series of CNF1 truncated toxins were characterized and assessed for toxin binding. In particular, three truncated toxins, ΔN63, ΔN545, and ΔC469, retained conformational integrity and in vitro enzymatic activity and were immunologically reactive against a panel of anti-CNF1 monoclonal antibodies (MAbs). Based on a comparison of these truncated toxins with wild-type CNF1 and CNF2 in LRP and HEp-2 cell binding assays and in MAb and LRP competitive binding inhibition assays and based on the results of confocal microscopy, we concluded that CNF1 contains two major binding regions: one located within the N terminus, which contained amino acids 135 to 164, and one which resided in the C terminus and included amino acids 683 to 730. The data further indicate that CNF1 can bind to an additional receptor(s) on HEp-2 cells and that LRP can also serve as a cellular receptor for CNF2.
Cytotoxic necrotizing factor type 1 (CNF1) is produced by many strains of uropathogenic Escherichia coli (UPEC), which are agents that are responsible for the majority of uncomplicated urinary tract infections (9). CNF1 is a 115-kDa cytoplasmic protein that is a member of a family of toxins that target small GTPases. Specifically, CNF1 deamidates glutamine 63 of RhoA and glutamine 61 of Rac1 and Cdc42, modifications that result in constitutive activation of these small GTPases (1). This activation leads to the formation of stress fibers and focal adhesions (RhoA), lamellipodia (Rac1), and filopodia (Cdc42) in CNF1-intoxicated cells and ultimately results in rearrangement of the cytoskeleton (12, 18, 28). Phenotypically, CNF1 causes multinucleation of various tissue culture cells (8, 11) but can also be cytotoxic against certain cell lines, including Swiss 3T3 and 5637 bladder cells (19, 22). In vivo, CNF1 evokes necrosis when it is injected intradermally into rabbit skin (4). Moreover, members of our laboratory, in collaboration with colleagues, demonstrated that in two animal systems CNF1 expression contributes to the virulence of UPEC strains. In a rat model of acute prostatitis, we found that intraurethral infection with a CNF1-positive strain leads to a significantly enhanced inflammatory response compared to that elicited by an isogenic, CNF1-negative mutant, even when the bacterial counts are equivalent (26). Similarly, in a mouse model of urinary tract infection, the production of CNF1 by UPEC strains results in higher bacterial counts and increased inflammation compared to the results for cnf1 isogenic mutants, in part due to the capacity of the toxin to alter the phagocytic and killing activities of murine polymorphonuclear leukocytes (7, 27).
CNF1 is composed of an N-terminal binding domain that also contains a region postulated to be responsible for toxin translocation and a C-terminal enzymatic domain (2). Previous work by Fabbri and colleagues localized the binding domain of CNF1 to the first 190 amino acids of the toxin (10). In that seminal study, the investigators concluded that hydrophilic amino acids that span residues 53 to 75 play a role in the association of CNF1 with its then-unidentified cellular receptor. However, a synthetic peptide of this region failed to inhibit holotoxin binding. Therefore, the authors considered the possibility that the conformational structure of a larger portion of the CNF1 N terminus was required for receptor binding (10).
Recently, the receptor for CNF1 produced by an E. coli K1 meningitis strain was identified as the 37-kDa laminin receptor precursor protein (LRP) present in human brain microvascular endothelial cells (HBMECs) (5). The natural ligand of LRP is laminin, a ubiquitous substance present in all eukaryotic cells. The wide cellular distribution of LRP may in part explain the high degree of LRP sequence conservation among many organisms (20, 31). Through a process that is not well understood, LRP can dimerize to form the mature 67-kDa laminin receptor protein (15). Moreover, recently, Kim et al. demonstrated that an E. coli K1 strain that expresses CNF1 can be internalized by HBMECs after the toxin binds to the mature laminin receptor (16). Both the precursor and mature forms of the laminin receptor are expressed on the surface of eukaryotic cells and are therefore accessible to CNF1 released from bacteria; however, it is unclear whether CNF1 binds preferentially to one of the two receptor forms. After CNF1 attaches to susceptible target cells, the toxin is internalized via clathrin-dependent and -independent mechanisms and subsequently moves to late endosomes, where it is released into the cytosol after vacuole acidification (6).
The N-terminal binding domain of CNF1 shares homology with two other bacterial toxins: CNF2 and Pasteurella multocida toxin (PMT) (23). CNF1 and CNF2 share 85% amino acid identity and 90% similarity over the entire length of the toxins (14, 32). Although the receptor for CNF2 has not been identified, the high degree of sequence similarity between CNF1 and CNF2 suggests that these toxins may share the same or a related receptor. CNF1 and PMT share only 24% homology in their N termini, and the greatest level of amino acid conservation occurs between the regions considered to be responsible for toxin translocation (amino acids 250 to 530 of PMT and amino acids 200 to 465 of CNF1) (24). Moreover, it has recently been suggested that vimentin, a component of type III intermediate filaments, serves as the receptor for PMT (29).
In this investigation, we analyzed a series of CNF1 truncated toxins to determine the precise regions of CNF1 that directly bind to HEp-2 cells and to LRP. In this work, two domains of CNF1 that are important for binding to LRP were identified: an N-terminal binding domain that contains amino acids 135 to 164 and a C-terminal domain that includes amino acids 683 to 730. Additionally, we observed binding of CNF1 to HEp-2 cells in the presence of saturating amounts of LRP, a finding that suggests that there is a second receptor for CNF1 on these cells. Lastly, we addressed whether LRP can serve as a cellular receptor for CNF2 and showed that LRP can function in that capacity.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and protein purification.
The bacterial strains and plasmids used in this study are listed in Table 1. Plasmid pGEX-2T-LRP (5) was a gift from Kwang Sik Kim (Johns Hopkins University School of Medicine, Baltimore, MD). Plasmids pCNF24 (CNF1), p2CNF (CNF2), pΔN63, pΔN75, pΔN134, pΔN272, pΔN545, pΔC469, and pΔ442 were transformed into E. coli M15(pREP4) and grown in Luria-Bertani broth supplemented with 100 μg/ml ampicillin and 25 μg/ml kanamycin. Expression of CNF1, CNF2, CNF1 truncated proteins, and LRP was induced in cultures grown at 22 to 25°C by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. N-terminal histidine (His)-tagged proteins were purified over HisTrap Ni2+ affinity columns by the fast phase liquid chromatography ÁKTA system using the manufacturer's protocol (GE Healthcare, Piscataway, NJ). Buffer exchange of affinity column-eluted proteins from 250 mM imidazole to 20 mM Tris-HCl (pH 7.8) was done by fast phase liquid chromatography with HiTrap HP desalting columns (GE Healthcare) or by dialysis. The purity of purified toxins was determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting. Briefly, toxins were subjected to SDS-PAGE (4 to 20% gradient; Invitrogen, Carlsbad, CA), and proteins were either transferred to a 0.45-μm nitrocellulose membrane and blocked overnight at 4°C with BLOTTO (Tris-buffered saline with 5% skim milk and 0.05% Tween 20) or fixed and stained with Coomassie blue G-250 (Bio-Rad). The blocked membrane was incubated with goat anti-CNF1 polyclonal antisera (1:5,000), followed by horseradish peroxidase (HRP)-conjugated porcine anti-goat immunoglobulin (IgG) secondary antibodies (1:7,500). Reactive proteins were visualized with ECL Plus (GE Healthcare).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB+lacIqZΔM15::Tn10(Tetr)] | Stratagene |
| M15(pREP4) | NaIs Strs Rifs Δlac-ara-gal-mtl F−recA uvr (pREP4 lacI Kanr) | QIAGEN |
| Cloning vectors | ||
| pBluescript II SK(−) | E. coli phagemid cloning vector (Ampr) | Stratagene |
| pQE30 | E. coli expression vector with six-His tag 5′ of the polylinker (Ampr) | QIAGEN |
| pGEX-2T | E. coli expression vector with glutathione S-transferase tag 5′ of the polylinker (Ampr) | GE Biosciences |
| Plasmids | ||
| pCNF24 | Wild-type cnf1 gene (nucleotides 1 to 3045) amplified from pHLK102 and cloned into BamHI/KpnI sites of pQE30 | 21 |
| p2CNF | Coding region of cnf2 cloned into SacI/KpnI sites of pQE30 | This study |
| pEOSW30 | 3.3-kb BanI/HaeII fragment containing the cnf2 gene cloned into the SmaI site of pK184 | 23 |
| pΔN63 | pCNF24 with deletion of 187 nucleotides from the 5′ end of the cnf1 gene | 21 |
| pΔN75 | 2.8-kb BamHI/KpnI cnf1 fragment (nucleotides 226 to 3045) cloned into pQE30 | 21 |
| pΔN134 | 2.6- kb BamHI/KpnI cnf1 fragment (nucleotides 403 to 3045) cloned into pQE30 | 21 |
| pΔN272 | 2.2-kb BamHI/KpnI cnf1 fragment (nucleotides 817 to 3045) cloned into pQE30 | 21 |
| pΔN545 | 1.4-kb PstI cnf1 fragment (nucleotides 1637 to 3045) cloned into pQE30 | 21 |
| pΔC469 | 1.6-kb BamHI/PstI cnf1 fragment (nucleotides 1 to 1637) cloned into pQE30 | 21 |
| pΔ442 | pCNF24 with an internal in-frame BclI deletion (nucleotides 1115 to 2348) | 21 |
| pGEX-2T-LRP | Full-length lrp gene cloned into BamHI/EcoRI sites of pGEX-2T | 5 |
| pHisLRP | Full-length lrp gene subcloned into BamHI/XmaI sites of pQE30 | This study |
Construction of histidine-tagged LRP and CNF2 expression plasmids.
To subclone the lrp gene into the His-tagged expression vector pQE30 (QIAGEN, Valencia, CA), plasmids pGEX-2T-LRP and pQE30 were digested with XmaI and BamHI, and the 899- and 3-kb fragments, respectively, were purified from a 1% sodium borate agarose gel (Faster Better Media LLC, Hunt Valley, MD) with a QIAEX II gel extraction kit (QIAGEN) used according to the manufacturer's instructions. Purified DNA fragments were ligated and transformed into supercompetent XL1-Blue cells (Stratagene, Cedar Creek, TX) prior to transformation into E. coli M15(pREP4) for protein expression of the 37-kDa LRP.
To subclone the cnf2 gene into pQE30, the full-length gene was amplified from pEOSW30 (23) with primers CNF2F1 (5′ ATGAGCTCATGAACGTT CAATGGCAAC) and CNF2R3045 (5′ ATGGTACCTCAAAAATCTTTTGAAAAAAC ATGC) that were designed to incorporate SacI and KpnI restriction sites (underlined), respectively, at their 5′ ends in order to facilitate directional cloning into pQE30. The cnf2 gene amplicon and pQE30 were each digested with SacI/KpnI and ligated, and the resultant plasmid, p2CNF, was transformed into chemically competent E. coli XL1-Blue cells. For protein purification and examination of toxin activity on HEp-2 cells, p2CNF was subsequently transformed into E. coli M15(pREP4) and lysates were examined as previously described (19).
Cell lines and media.
The HEp-2 cell line, a human laryngeal cell line (ATCC CCL-23), was grown at 37°C with 5% CO2 in Eagle's minimal essential medium with Earle's balanced salt solution (Cambrex, East Rutherford, NJ) supplemented with 10% fetal bovine serum (Invitrogen-Biosource, Carlsbad, CA), 2 mM l-glutamine (Invitrogen), 10 μg/ml gentamicin, 10 U/ml penicillin, and 10 μg/ml streptomycin.
Dot blot analysis of truncated CNF1 toxins.
Dot blotting of native and denatured toxin samples was performed as previously described, with the following modifications (19). Briefly, purified CNF1, CNF2, and truncated CNF1 toxins were diluted in either phosphate-buffered saline (PBS) (native) or 6× SDS-dithiothreitol buffer (denatured) to obtain a final concentration of 50 μg/ml, and 100-μl portions of each sample were then transferred to nitrocellulose membranes with a Minifold microsample filtration manifold (Schleicher & Schuell, Keene, NH). Toxins diluted in denaturation buffer were boiled at 95°C for 5 min prior to transfer. Membranes were washed with 1× PBS, dried, and then blocked overnight at 4°C in BLOTTO. Blots were probed with a panel of CNF1-specific monoclonal antibodies (MAbs) (21), as well as polyclonal anti-CNF1 sera (22), followed by incubation with HRP-conjugated goat anti-mouse IgG or porcine anti-goat IgG secondary antibodies (Roche, Indianapolis, IN). Reactive proteins were visualized with diaminobenzamidine (Sigma, St. Louis, MO).
LRP receptor binding ELISA.
Vinyl alphanumeric 96-well U-bottom enzyme-linked immunosorbent assay (ELISA) plates (Thermo Fisher Scientific, Waltham, MA) were coated with purified native LRP in 1× PBS at a concentration of 200 μg/ml (20 μg/well), and the plates were incubated overnight at 4°C. Wells were washed with 1× PBS-0.1% Tween 20 (PBS-T) and then blocked with PBS that contained 3% bovine serum albumin (BSA) for 2 h at 37°C. Once blocked, wells were again washed with PBS-T, and purified toxins were added at a concentration of 50 μg/ml (5 μg/well). Plates were incubated for 2 h at 37°C, washed with PBS-T, and then sequentially incubated with goat anti-CNF1 polyclonal sera (1:5,000) for 1 h at 37°C and with an anti-goat IgG HRP-conjugated antibody (1:7,500; Roche) for 1 h at room temperature. Color development was achieved with the 3,3′,5,5′-tetramethylbenzidine substrate (Bio-Rad, Hercules, CA) used according to the manufacturer's instructions, and plates were read with an ELx800 plate reader (BioTek Instruments, Winooski, VT) at 405 nm. To ensure equal binding of the antisera to each toxin, a concurrent ELISA was performed as described above in the absence of LRP.
HEp-2 receptor binding ELISA.
Approximately 5 × 104 HEp-2 cells/well were seeded in 96-well plates and incubated overnight at 37°C with 5% CO2. The following day, cells were fixed with 2% formalin for 20 min at room temperature, and then wells were washed with PBS-T prior to blocking for 1 h at 37°C with 1× PBS-3% BSA. Once blocked, wells were washed with PBS-T and purified toxins were added at a concentration of 50 μg/ml (5 μg/well). Controls without toxin were included to determine the degree of nonspecific antibody binding to cells. Plates were incubated for 1 h at 37°C and washed, and then the wells were sequentially incubated with goat anti-CNF1 polyclonal sera (1:5,000) for 1 h at 37°C and with anti-goat HRP-conjugated IgG (1:7,500; Roche) for 1 h at room temperature. Color development was achieved with the 3,3′,5,5′-tetramethylbenzidine substrate (Bio-Rad) used according to the manufacturer's instructions, and the reaction was stopped by addition of 1 N H2SO4. To prevent interference from HEp-2 cell monolayers, samples (100 μl) were transferred to a 96-well plate, after which they were read with an ELx800 plate reader (BioTek Instruments) at 405 nm.
Binding inhibition assays.
Preliminary binding experiments were initially done to determine the concentrations of CNF1, CNF2, and ΔN545 that yielded approximately equivalent absorbance values in the HEp-2 receptor binding ELISA. Based on the information obtained, CNF1 (2.5 μg/well), CNF2 (5 μg/well), and ΔN545 (2.5 μg/well) were incubated with 5 μg of either CNF-neutralizing MAb BF8 (IgA) or NG8 (IgG2a) or a mixture of both MAbs for 2 h at 37°C. As controls, the toxins were also incubated with 5 μg of isotype-matched control non-CNF1-neutralizing MAb GC2 (IgG2a) and NOS-E1, a commercially available anti-nitric oxidase synthase IgA MAb (Sigma), as well as irrelevant non-isotype-matched MAbs CA6 (anti-CNF1, IgG1) and 13C4 (anti-Shiga toxin type 1 [Stx1], IgG1) (Hycult, The Netherlands). After incubation, the toxin-MAb samples were added to fixed and blocked HEp-2 cells, and the assay was performed using the receptor binding ELISA protocol described above. As an additional control to ensure that MAbs BF8 and NG8 did not interfere with the capacity of the polyclonal antisera to recognize the toxins, CNF1, CNF2, and ΔN545 were incubated with BF8 or NG8 or both as described above for 2 h at 37°C. The toxin-antibody mixtures were then permitted to bind to wells of an ELISA plate overnight at 4°C. The plates were washed with TBS-T, and the assay was done using the receptor binding ELISA protocol described above. For competitive inhibition of binding with exogenous LRP, CNF1 and ΔN545 at the concentrations described above were preincubated with 200 μg/ml of purified LRP for 2 h at 37°C, and the samples then were used in HEp-2 receptor binding ELISAs. Paired, two-tailed t tests were used to determine whether there were statistically significant differences between untreated toxin samples and toxins that had been preincubated with CNF1 MAbs or exogenous LRP.
In vitro deamidation of Rho GTPases.
Deamidation of RhoA, Rac1, and Cdc42 was done as described previously (19). Briefly, a mixture containing GTPase and toxin at a 20:1 molar ratio was incubated in deamidation buffer (50 mM NaCl, 50 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride) for 2.5 h at 37°C. Untreated GTPases served as negative controls. After toxin treatment, samples were concentrated by addition of 10% trichloroacetic acid and stored overnight at 4°C. Precipitated proteins were pelleted, washed with acetone, air dried, and resuspended in 20 mM Tris-HCl (pH 7.4). Samples were subjected to SDS-PAGE with 12% acrylamide, and proteins were transferred to 0.45-μm nitrocellulose membranes. Membranes were blocked overnight at 4°C in BLOTTO and then probed either with MAbs against RhoA (1:1,500; Santa Cruz Biotechnology, Santa Cruz, CA), Rac1 (1:5,000; Upstate Biotechnology), or Cdc42 (1:1,000; Santa Cruz Biotechnology) or with a rabbit polyclonal antisera raised against the deamidated RhoA peptide (1:2,000) (19, 32). Reactive proteins were visualized after incubation with HRP-conjugated goat anti-mouse IgG (1:2,000, 1:7,500, and 1:3,000 for RhoA, Rac1, and Cdc42, respectively; Roche) or donkey anti-rabbit IgG (1:3,000; GE Healthcare), followed by detection with ECL Plus (GE Healthcare). The pixel density of total and modified RhoA was analyzed with NIH ImageJ 1.34S software (http://rsb.info.nih.gov/ij/). The percent modification was calculated as described previously (19) using the values derived from ImageJ and the following formula: [(modified RhoA value/total RhoA value) × 100%] − [(RhoA control-modified RhoA value/RhoA control- total RhoA value) × 100%].
Colocalization of CNF1 and LRP by immunofluorescence.
Immunofluorescence experiments were done as follows. HEp-2 cells were seeded in eight-well glass or Permanox chamber slides at a concentration of approximately 5 × 106 cells/well and incubated for 24 h at 37°C with 5% CO2. Slides were then chilled to 4°C and washed with cold Hanks’ balanced salt solution (Invitrogen/Gibco) prior to addition of purified CNF1, CNF2, or ΔN545 which had been diluted to obtain a final concentration of 12 μg/well in cold binding buffer without maltose (13, 22). Control wells received binding buffer in the absence of toxin. Cells were incubated with toxin for 1 h at 4°C, washed with PBS, and then fixed with 2% formalin for 20 min at room temperature. After fixation, wells were washed and blocked with 1% BSA in PBS overnight at 4°C. The following day, wells were sequentially incubated for 1 h at 4°C with goat anti-CNF1 polyclonal antisera (1:500), followed by AlexaFluor 488-conjugated chicken anti-goat IgG (1:300; Invitrogen-Molecular Probes), rabbit anti-LRP polyclonal antisera (1:500; Abcam, Cambridge, MA), and AlexaFluor 555-conjugated donkey anti-rabbit IgG (1:300; Invitrogen-Molecular Probes). Prior to examination, slides were treated with Fluoromount G (Southern Biotech, Birmingham, AL) and coverslips were applied. Cells were visualized at a magnification of ×100, and confocal images were taken with a Zeiss LSM Pascal confocal microscope and processed with LSM Image 5 Browser software.
RESULTS
Evaluation of CNF1 truncated toxins.
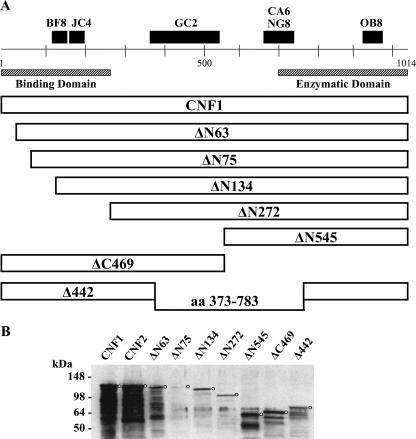
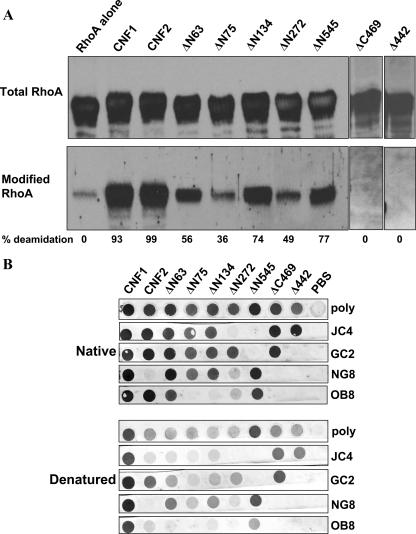
Prior to their use in binding domain studies, a series of truncated CNF1 toxins (Fig. 1A) were assessed for purity (Fig. 1B), activity, and conformational integrity. When toxin preparations were analyzed in Coomassie blue-stained gels (data not shown) and in Western blots, most truncated toxins (2 μg/well) were easily detectable with polyclonal anti-CNF1 sera and considered relatively pure; the only exception was ΔN75 (Fig. 1B), which showed weak immunoreactivity. Breakdown products were apparent for many of the toxin preparations; however, this finding coincides with previous data from our laboratory which indicate that CNF1 at high concentrations has a tendency to degrade during the purification process despite the inclusion of protease inhibitors (Fig. 1B) (22). To reduce the potential for toxin degradation and to ensure that the protein concentrations were the concentrations of primarily full-length wild-type and mutant toxins, all toxin preparations were flash frozen in small aliquots immediately after protein purification and thawed just prior to use in assays. Mutant toxins were initially examined to determine their capacity to multinucleate HEp-2 cells, a hallmark phenotype of CNF1 intoxication; however, none of the truncated toxins evoked such a response (data not shown). Given that multinucleation reflects a series of steps that begins with toxin binding to the target cell and ends with deamidation of the small cytoplasmic GTPase targets, RhoA, Rac1, and Cdc42, the mutant toxins were further evaluated for retention of enzymatic activity. To determine whether the mutants were enzymatically active in vitro, the capacity of each protein to deamidate RhoA, Rac1, and Cdc42 was tested. As shown in Fig. 2A, toxins with various portions of the N terminus deleted still retained the capacity to deamidate RhoA but not to the same extent as that demonstrated by wild-type CNF1 or CNF2. Similarly, the N-terminal truncation mutants also retained the capacity to deamidate Rac1 and Cdc42, but at levels below those obtained with wild-type toxins (data not shown). The ΔN75 and ΔN272 mutants were the least enzymatically active of the toxins (Fig. 2A); nevertheless, the extent to which these proteins deamidated RhoA was greater than the background activity (reactivity of RhoA without toxin). It is noteworthy that mutant toxin Δ442, which has a deletion of amino acids 320 to 783 but still harbors the putative binding and enzymatic regions of the toxin, showed only background levels of deamidation activity despite the presence of the catalytic domain (Fig. 2A). As expected, ΔC469, which is devoid of the entire catalytic domain, was not enzymatically active (Fig. 2A). Somewhat surprising was the fact that deletions in the N terminus of CNF1 resulted in reduced enzymatic activity of the toxin, a finding which has not been previously reported. These results may suggest an additional role of the N terminus of CNF1; perhaps this domain of CNF1 interacts directly with RhoA, as has been shown to be the case with the N terminus of YopT, a cysteine protease that cleaves small GTPases (30).
FIG. 1.
(A) Schematic diagram of CNF1 truncated toxins, with the regions and epitopes recognized by several CNF MAbs indicated at the top. MAbs JC4 and NG8 can neutralize only CNF1, while MAb BF8 can neutralize both CNF1 and CNF2. aa, amino acids. (B) Western blots of purified wild-type and truncated CNF1 toxins. Toxins (2 μg of CNF2 and the CNF1 truncated toxins and 500 ng of CNF1) were applied to SDS-PAGE gels and transferred to nitrocellulose membranes. Blots were probed with anti-CNF1 polyclonal antisera, followed by an HRP-conjugated secondary antibody and the ECL substrate. Circles indicate the position of each full-length toxin.
FIG. 2.
Enzymatic activity and conformation of CNF1 truncated toxins. (A) Western blot analysis of RhoA incubated in the presence or absence of purified toxins. Membranes were probed with a MAb against RhoA (top panel) or with antisera that recognize only the deamidated form of the GTPase (bottom panel), and reactive proteins were detected with appropriate HRP-conjugated secondary antibodies and the ECL substrate. The pixel density of total and modified RhoA was used to calculate the percent modification, and the results are indicated below the blots. Background values for the RhoA control were subtracted from each of the sample values. (B) Purified toxins (5 μg/well) were either applied directly to nitrocellulose membranes (upper panels) or denatured (with SDS, dithiothreitol, and boiling) prior to application (lower panels). As indicated on the right, blots were probed with either CNF1 polyclonal antisera (poly) or a panel of CNF1 MAbs, and reactive proteins were visualized with HRP-conjugated secondary antibodies and the diaminobenzamidine substrate.
Finally, to evaluate conformational structure and integrity, the truncated toxins were examined in native and denatured dot blots probed with a panel of anti-CNF MAbs (21). As shown in Fig. 2B, CNF1 reacted with all MAbs as expected, and CNF2 was detected by all MAbs except the previously identified CNF1-specific MAb NG8 (24). Furthermore, truncated toxins ΔN63, ΔN545, and ΔC469 all produced the expected reactivity patterns under native conditions but showed partial or complete abrogation of immunoreactivity when they were examined under denaturing conditions; these results suggest that conformational integrity was retained by these molecules. It is noteworthy that mutant toxins ΔN75, ΔN134, and Δ442 generated the expected reactivity patterns with all the MAbs except MAb OB8. The latter finding suggests that the conformational integrity of the distal portion of the C terminus of these toxins may have been comprised by the deletion. In addition, mutant toxin ΔN272 showed weak reactivity with MAbs NG8 and OB8, which also suggested that the conformational structure of the C terminus of this molecule was not maintained. Taken together, the dot blot and enzymatic analyses indicate that three (ΔN63, ΔN545, and ΔC469) of the seven truncated mutants maintained proper structural conformation and, with the exception of toxin ΔC469, which was devoid of the catalytic site, retained enzymatic activity.
Binding of CNF1 and the CNF1 truncated mutants to HEp-2 cells.
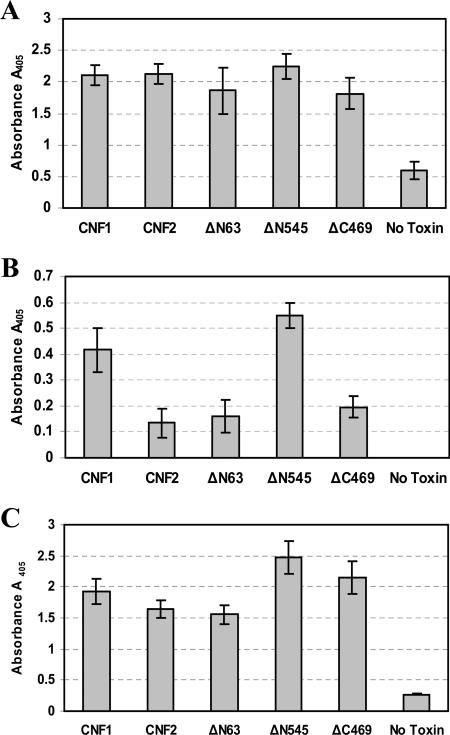
Prior to assessment of whether the properly folded and enzymatically active CNF1 mutant toxins ΔN63 and ΔN545, as well as the conformationally intact toxin ΔC469, retained the capacity to bind to HEp-2 cells, each toxin was used as a coating antigen in an indirect ELISA to establish the optimal dilution of polyclonal anti-CNF1 sera to use for comparable detection of all toxins. As shown in Fig. 3A, when used at a dilution of 1:5,000, the polyclonal anti-CNF1 sera were equally effective for detection of all toxins and produced similar signal levels with the molecules. These signals were considered to be CNF1 specific because minimal reactivity was observed in the absence of toxin. This indirect ELISA therefore served as an internal positive control and was performed concurrently with binding ELISAs. To ascertain the capacity of each toxin to bind to HEp-2 cells, a receptor binding ELISA was devised in which toxin was added to wells of fixed HEp-2 cell monolayers and bound toxin was subsequently detected with an optimal dilution of anti-CNF1 polyclonal sera. As shown in Fig. 3B, both wild-type and truncated toxins bound to fixed HEp-2 cells, but striking differences in the efficiency of binding were observed. Most notable was the degree of binding of the N-terminal truncated mutant ΔN545. This toxin, which contains the catalytic domain of CNF1, showed considerably better binding than CNF2 and, while the data were not statistically significant, also showed slightly greater binding than wild-type CNF1. Together, these results suggest that a HEp-2 binding site is located within the catalytic domain of the toxin and that this site is potentially masked by the natural conformation of the holotoxin. Mutant toxin ΔC469, which contains the putative binding domain of CNF1, also bound to HEp-2 cells but only to the same extent as CNF2 and the deletion mutant ΔN63. The latter observation indicates that it is possible that CNF1 harbors two HEp-2 cell binding regions, one within the N terminus, as previously described, and the other within the distal portion of the molecule. In addition, it appeared that the capacity of CNF2 to bind to HEp-2 cells was significantly reduced (P = 0.047, as determined by a t test) compared to the capacity of CNF1, a difference which occurred despite the fact that these two toxins share 90% amino acid similarity.
FIG. 3.
HEp-2 and LRP receptor binding ELISAs. (A) Indirect ELISA that served as an internal control for subsequent binding ELISAs. Purified toxins (5 μg/well) were used as coating antigens and detected with CNF1 polyclonal antisera at a dilution (1:5,000) that allowed comparable signals from all toxins assayed. The data are the averages of two experiments done in triplicate. (B) HEp-2 receptor binding ELISA in which purified toxins (5 μg/well) were added to fixed HEp-2 cells for 1 h and bound toxin was detected with anti-CNF1 polyclonal sera. The data are the averages of two experiments done in triplicate. (C) LRP binding ELISA in which purified toxins were incubated in LRP-coated ELISA plate wells and bound toxin was subsequently detected with anti-CNF1 polyclonal sera. The data are the averages of two experiments done in triplicate. In all panels, the error bars indicate the standard deviation above and below the mean. Also, the values for wells containing toxin but no anti-CNF1 sera served as background controls and were subtracted from all sample values.
Localization of the region of CNF1 that binds LRP.
Recently, LRP was identified as a cellular receptor for CNF1. To identify the region(s) of CNF1 that binds to LRP, a receptor binding ELISA was designed in which purified LRP, coated on the wells of an ELISA plate, was incubated with CNF1, CNF2, or the CNF1 truncated mutants and bound toxin was subsequently detected with anti-CNF1 polyclonal sera at a dilution previously determined to yield similar signal levels for toxins (Fig. 3A). As shown in Fig. 3C, all truncated toxins, as well as CNF1 and CNF2, appeared to specifically bind to purified LRP and minimal levels of signal were observed in the absence of toxin. Similar to what was demonstrated in HEp-2 receptor binding assays, mutant toxin ΔN545 showed the highest levels of binding to LRP. This observation supports the possibility that there is a binding site within this region that is fully exposed only in the absence of the N terminus of CNF1. Mutant toxins ΔN63 and ΔC469 also bound to LRP, which coincides with previous reports that indicated that a binding domain is present within the N terminus of CNF1. Finally, these results showed that CNF2 could bind to LRP at a level similar to that of CNF1, a finding which suggests that LRP may serve as a receptor for both toxins.
Inhibition of toxin binding to HEp-2 cells.
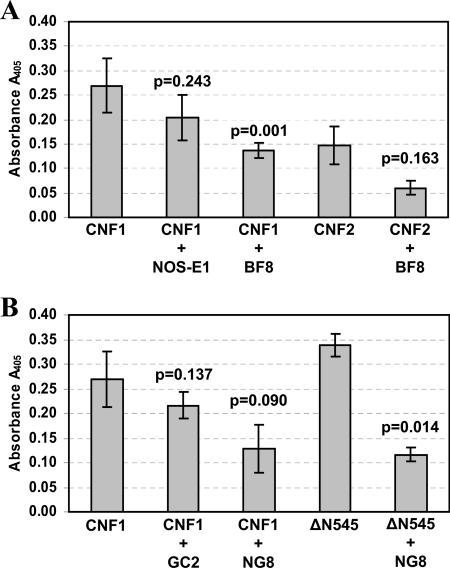
To begin to more precisely define the location of the CNF1 binding domains identified with the HEp-2 and LRP binding ELISAs, a series of competitive binding inhibition studies were carried out. Specifically, wild-type CNF1, CNF2, and mutant toxin ΔN545 were examined to determine their capacities to bind to HEp-2 cells in the presence of anti-CNF1 MAbs or exogenous LRP. To appropriately assess the degree of binding of CNF1, CNF2, and ΔN545 to HEp-2 cells, the toxin concentration required to produce comparable absorbance signals in the HEp-2 receptor binding ELISA was first established for all three toxins (data not shown). In addition, each toxin was tested to ensure that there was similar recognition by both MAb BF8 and MAb NG8 in either an indirect ELISA (MAb BF8) (data not shown) or a dot blot analysis (MAb NG8) (Fig. 2B). Furthermore, an indirect ELISA was used to ensure that binding of either BF8 or NG8 or both MAbs did not interfere with the capacity of the anti-CNF1 polyclonal sera to recognize the toxin (data not shown). Based on this information, appropriate amounts of each toxin were then incubated with 5 μg of either CNF1-neutralizing MAb BF8 or NG8, non-CNF1-neutralizing MAb GC2 or CA6, or irrelevant MAbs (Stx1 MAb 13C4 and NOS-E1) prior to evaluation in the HEp-2 receptor binding ELISA. The regions recognized by these CNF1 MAbs have been previously mapped to amino acids 135 to 164 for BF8, 373 to 546 for GC2, and 683 to 730 for NG8 and CA6 (21) (Fig. 1).
As shown in Fig. 4A, incubation of CNF1 with MAb BF8 significantly reduced binding of the toxin to fixed HEp-2 cell monolayers (P = 0.001). The inhibition of binding appeared to be a specific property of this MAb as coincubation with an isotype-matched control MAb (anti-nitric oxide synthase clone NOS-E1) did not alter the capacity of the toxin to bind to cells (P = 0.243). These findings indicate that the capacity of MAb BF8 to neutralize CNF1 is the direct result of inhibition of toxin binding and that amino acids 135 to 164 of CNF1 are required for this binding activity. Although the results are not statistically significant (P = 0.090), MAb NG8 also appeared to possess the capacity to reduce the binding of CNF1 to HEp-2 cells compared to the effect produced by isotype-matched control MAb GC2 (P = 0.137). Similarly, two irrelevant MAbs, anti-CNF1 MAb CA6 and anti-Stx1 MAb 13C4, which blocks Stx1 engagement with its cellular receptor globotriaosylceramide (29a), also did not inhibit the binding of CNF1 to HEp-2 cells (P = 0.404 and P = 0.779, respectively) (data not shown). While MAb NG8 showed some effect on wild-type CNF1 binding to fixed HEp-2 cells, this MAb was found to significantly diminish the capacity of the N-terminally truncated toxin ΔN545 to bind to cells (P = 0.014). Together with data from the previous CNF1 binding experiments, these data suggest that a second binding site that contains the NG8 epitope (amino acids 683 to 730) is located within the C terminus of the molecule and that this binding site is at least partially masked in the holotoxin because MAb NG8 reduced wild-type CNF1 binding only to a limited degree. Finally, when CNF1 was incubated in the presence of both MAbs BF8 and NG8, a marked reduction in toxin binding was apparent (P = 0.034) (data not shown); however, complete inhibition of binding was not observed (average A405 for CNF1 with BF8 and NG8, 0.162 ± 0.061). The latter finding suggests the possibility that there are other binding sites within CNF1 that can participate to a minor extent in adherence to HEp-2 cells and that these regions are distinct from those which show reactivity to MAbs BF8 and NG8. Alternatively, the full extent of binding inhibition exhibited by these MAbs may be limited by the sensitivity of the assay.
FIG. 4.
Inhibition of toxin binding to HEp-2 cells by CNF1 MAbs. (A) Wild-type CNF1 and CNF2 were incubated with either 5 μg of MAb BF8 (IgA) or the isotype-matched control MAb NOS-E1 and then assessed to determine their capacity to bind to HEp-2 cells in a receptor binding ELISA. (B) Wild-type CNF1 and ΔN545 were incubated with either 5 μg of MAb NG8 (IgG2a) or the isotype-matched control MAb GC2 and assessed to determine their capacity to bind HEp-2 cells in a receptor binding ELISA. The data are the cumulative averages of triplicate readings from eight independent experiments for CNF1 and from two independent experiments for all other toxins; the error bars indicate the standard deviation above and below the mean. Data were analyzed by paired, two-tailed t tests, and P values are indicated. The values for wells containing toxin but no anti-CNF1 polyclonal sera served as background controls and were subtracted from all sample values (average background control value, 0.689 ± 0.33).
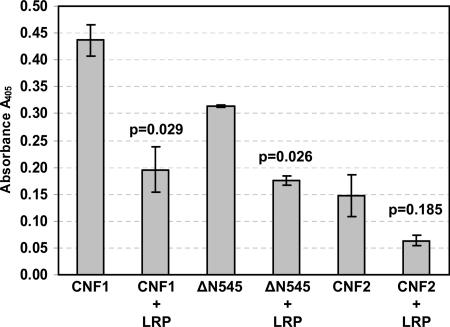
To establish whether the binding of CNF1 and the truncated toxin ΔN545 to HEp-2 cells was through LRP, a second set of competitive binding inhibition experiments was performed, in which toxins were coincubated with exogenous LRP (20 μg) prior to addition to fixed HEp-2 cell monolayers. As expected, exposure of CNF1 or ΔN545 to exogenous LRP significantly inhibited the binding of either toxin to HEp-2 cells (Fig. 5) (P = 0.029 and P = 0.026, respectively). These findings indicate that CNF1 can use surface-exposed LRP on HEp-2 cells as a receptor and that one site required for interaction with LRP is located within the C terminus of the toxin. Furthermore, because toxin binding was not completely abrogated by exogenous LRP, even when it was used at saturating concentrations (data not shown), the results also suggest that there is an additional receptor for CNF1 on HEp-2 cells.
FIG. 5.
Inhibition of toxin binding to HEp-2 cells by exogenous LRP. Wild-type CNF1, CNF2, and ΔN545 were incubated with 20 μg of LRP prior to addition to HEp-2 cells. The data are the averages of two independent experiments performed in triplicate, and the error bars indicate the standard deviation above and below the mean. Data were analyzed by paired, two-tailed t tests, and P values are indicated. The values for wells containing toxin but no anti-CNF1 polyclonal sera served as background controls and were subtracted from all sample values (average background control value, 0.790 ± 0.046).
Results from our laboratory indicate that CNF2 can also bind to and intoxicate HEp-2 cells but not to the same extent as CNF1 (K. Grande, S. Rasmussen, K. Meysick, and A. D. O'Brien, unpublished data). To determine whether CNF1 and CNF2 share common binding regions, CNF2 was examined for its binding to fixed HEp-2 cell monolayers after exposure to either MAb BF8, a cross-reactive CNF1 MAb that can partially neutralize CNF2 activity (21), or exogenous LRP. As shown in Fig. 4A and 5, the presence of MAb BF8 as well as LRP diminished CNF2 binding to HEp-2 cells; however, in neither case was the reduction in binding statistically significant compared to the results for untreated toxin. Based on these results, it appears that the CNF2 epitope recognized by MAb BF8 partially contributes to toxin binding. Furthermore, the data suggest that there are unique CNF2 binding sites or possibly a secondary CNF2 receptor on HEp-2 cells in addition to LRP.
Colocalization of LRP and toxin on HEp-2 cells by immunofluorescence.
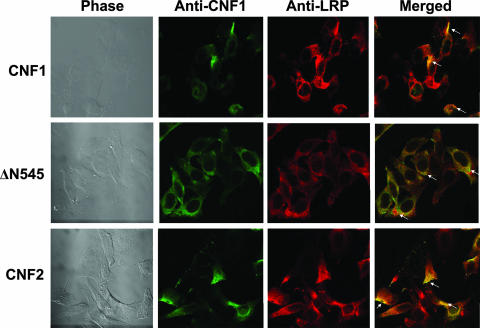
As a final means by which to confirm the LRP binding regions of CNF1, full-length CNF1 and the truncated CNF1 toxin ΔN545, as well as CNF2, were used in immunofluorescence colocalization experiments with HEp-2 cells. For these studies, each toxin was allowed to bind to HEp-2 cells at 4°C prior to fixation and subsequent incubation with polyclonal anti-CNF1 sera, anti-LRP sera, and differentially labeled fluorescent secondary antibodies. As shown in Fig. 6, CNF1, ΔN545, and CNF2 all colocalized with LRP. As toxins were not applied at saturating concentrations, some unbound LRP on cells was also apparent in merged images.
FIG. 6.
Colocalization of CNF1 and LRP on HEp-2 cells. CNF1, ΔN545, and CNF2 were bound to HEp-2 cells, and cells were subsequently stained with polyclonal anti-CNF1 sera, anti-LRP sera, and the corresponding fluorescently labeled secondary antibodies to allow visualization of toxin (green; Alexa 488) and LRP (red; Alexa 555). Confocal microscopy was used to demonstrate colocalization of the toxin and LRP receptor, as indicated by a yellow color (arrows) in the merged panels. Images were taken at a magnification of ×100.
DISCUSSION
Only one region of CNF1, its enzymatic C-terminal domain, has been crystallized (3). Thus, other functions attributed to the structure of this Rho GTPase-activating toxin have remained largely undefined. Prior to this work, only two published reports focused on identification of CNF1 domains required for eukaryotic cell binding (10, 17). Furthermore, much of the previous work on the delineation of CNF1 binding domains was hindered by the fact that the CNF1 receptor had not been identified. However, recent work that indicates that the 37-kDa LRP can serve as a receptor for CNF1 on HBMECs (5) has provided new information that can be applied to CNF1 structure-function analyses. In this study, recombinant LRP was used to help define CNF1 cell binding domains, and here we report the involvement of two new regions of the toxin necessary for binding to LRP and HEp-2 cells.
One domain necessary for complete CNF1 binding to HEp-2 cells is in the N terminus of the toxin. Based on the results of a competitive binding inhibition assay, it appears that amino acids 135 to 164, which comprise the reactive epitope for CNF1-neutralizing MAb BF8 (21), are required for toxin binding. The location of this binding site is consistent with a previous report which indicated that the first 190 amino acids of CNF1 were required for receptor binding (10). Although this previous study proposed that amino acids 53 to 75 are critical for cell binding, this region did not appear to be essential for toxin interaction with the target cell because a peptide that contained these residues did not inhibit binding (10). Based on the latter finding, Fabbri and colleagues suggested that the overall conformation of the N terminus may be necessary for full toxin binding. In part, our data support this concept because the truncated mutant ΔN63 displayed reduced binding to HEp-2 cells; however, the fact that mutant toxin ΔC469, which contains the complete N terminus of CNF1, showed a similar reduction in binding may indicate that the overall conformation of the holotoxin is essential for complete toxin binding to HEp-2 cells. It did not appear that deletion of various portions of CNF1 had as dramatic effect on the binding of toxin to purified LRP. One explanation for this apparently discrepant result is that the available binding regions on purified LRP are likely to be different from those on LRP expressed on the surface of eukaryotic cells.
The second CNF1 cell binding domain identified in this study is in the C terminus of the molecule. The evidence that supports this conclusion includes the observation that the truncated toxin ΔN545 exhibited strong binding to both HEp-2 cells and purified LRP. Since ΔN545 generally showed better binding than CNF1, it is possible that the binding site located within the region may be only partially exposed in the native conformation of the holotoxin and more accessible only when the N terminus of the molecule is removed. Furthermore, based on competitive binding inhibition studies, it appears that this C-terminal binding site associates either directly or indirectly with LRP expressed on the surface of HEp-2 cells. In addition, as both CNF1 and ΔN545 showed significantly diminished binding to HEp-2 cells in the presence of neutralizing MAb NG8, it appears that this reactive epitope (amino acids 683 to 730) (21) may contain the entire toxin binding site or at least encompass a portion of the site.
From the data presented here it appears that CNF1 contains two regions that are important for toxin binding, a finding that is particularly surprising given that one of these regions is located near the C-terminal catalytic domain of the toxin. There are several possible explanations for why CNF1 possesses two cellular binding domains. First, CNF1 may require two binding regions in order to permit maximal binding to surface-exposed LRP. Since the crystal structure has been resolved only for amino acids 720 to 1014 of CNF1 (3), it is not clear how the N terminus of the molecule folds or interacts with its putative receptor. Therefore, it is possible that the binding sites within the N and C termini fold in such a way that they form an LRP binding pocket. A second possible explanation for why CNF1 possesses two binding domains is the presence of a second cellular receptor to which CNF1 can bind. Our finding that toxin binding can still occur even in the presence of high levels of exogenous LRP supports this possibility. Moreover, addition of exogenous LRP has also been shown to block CNF1 binding to the mature laminin receptor (16). Furthermore, Blumenthal et al. (1a) recently showed that treatment of HeLa cells with sodium chlorate, a substance that blocks the synthesis of heparan sulfate proteoglycans (HSPGs), can reduce CNF1 uptake. These authors suggested that CNF1 may bind to a HSPG/LRP complex. Thus, HSPGs may serve as a coreceptor that requires a different portion of the toxin to permit complete binding. The possibility that an HSPG is a secondary or coreceptor along with LRP and the ubiquity of these molecules (6a, 13a, 20, 25) may also help explain the wide range of tissue culture cell types that are susceptible to CNF1 (22). Finally, it is possible that only the N-terminal binding region, which includes amino acids 134 to 164, is required for interaction with LRP or other cellular receptors and that the C-terminal binding region is merely an artifact created by truncation that has led to exposure of a portion of the molecule that is normally masked or inaccessible in the wild-type toxin. However, the fact that the CNF1-neutralizing MAb NG8 not only recognized and blocked the binding of truncated toxin ΔN545 to HEp-2 cells but was equally effective in inhibition of wild-type toxin binding does not support the latter possibility. The data clearly indicate that the NG8 epitope is exposed and accessible in both ΔN545 and the holotoxin. As CNF1 appears to undergo degradation in vitro (22) and potentially in vivo, the presence of a binding site in the C-terminal enzymatic region of the toxin may also provide an alternative mechanism for CNF1 entry into eukaryotic cells and subsequent deamidation of Rho GTPase proteins.
The fact that CNF1 and CNF2 share a high degree of amino acid homology also suggests that these two toxins could possess a common eukaryotic receptor, specifically LRP. Based on the binding and colocalization studies presented here, it does appear that CNF2 can bind to exogenous LRP as well as to surface-bound LRP on HEp-2 cells, but to a lesser extent than that observed with CNF1. However, neither the presence of neutralizing MAb BF8 nor exogenous LRP significantly affected the level of CNF2 that bound to HEp-2 cells. Taken together, these data indicate that CNF2 can use LRP as its receptor ligand but also suggest that there are differences in affinity or avidity to LRP between these two toxins. As it has recently been shown that CNF1 and the highly homologous molecule CNFY of Yersinia pseudotuberculosis bind to different cellular receptors (2), it is not completely unexpected to find that CNF1 and CNF2 show specific differences in LRP binding. These results may also help explain why CNF1 appears to be more potent than CNF2 in multinucleation activity on HEp-2 cells (Grande et al., unpublished data).
In summary, two new regions of CNF1 have been identified that mediate toxin binding to purified preparations of the CNF1 receptor, LRP, as well as to LRP expressed on the surface of HEp-2 cells. One region is located in the N terminus of the molecule and contains amino acids 135 to 164, while the second binding site is located in the C terminus and encompasses amino acids 683 to 730. In addition, we have found that CNF2 can also bind to exogenous LRP and colocalizes with LRP on the surface of HEp-2 cells. The latter result in particular suggests that LRP may also serve as a receptor for CNF2. Finally, it appears that CNF1 toxin-neutralizing MAbs BF8 and NG8 mediate their effect by inhibiting binding of toxin to its LRP receptor.
Acknowledgments
We are grateful to Tom Baginski for his assistance with the confocal microscope and to Cara Olsen for her help with statistical analysis.
This work was supported by grant AI38281 from the National Institutes of Health.
We have no conflicting financial interests. The opinions or assertions in this paper are those of the authors and are not to be construed as the views of the Department of Defense.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 20 August 2007.
REFERENCES
- 1.Aktories, K. 1997. Rho proteins: targets for bacterial toxins. Trends Microbiol. 5:282-288. [DOI] [PubMed] [Google Scholar]
- 1a.Blumenthal, B., C. Hoffmann, K. Aktories, S. Backert, and G. Schmidt. 2007. The cytotoxic necrotizing factors from Yersinia pseudotuberculosis and from Escherichia coli bind to different cellular receptors but take the same route to the cytosol. Infect. Immun. 75:3344-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boquet, P. 2001. The cytotoxic necrotizing factor 1 (CNF1) from Escherichia coli. Toxicon 39:1673-1680. [DOI] [PubMed] [Google Scholar]
- 3.Buetow, L., G. Flatau, K. Chiu, P. Boquet, and P. Ghosh. 2001. Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1. Nat. Struct. Biol. 8:584-588. [DOI] [PubMed] [Google Scholar]
- 4.Caprioli, A., V. Falbo, L. G. Roda, F. M. Ruggeri, and C. Zona. 1983. Partial purification and characterization of an Escherichia coli toxic factor that induces morphological cell alterations. Infect. Immun. 39:1300-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, J. W., S. J. Hong, K. J. Kim, D. Goti, M. F. Stins, S. Shin, V. L. Dawson, T. M. Dawson, and K. S. Kim. 2003. 37-kDa laminin receptor precursor modulates cytotoxic necrotizing factor 1-mediated RhoA activation and bacterial uptake. J. Biol. Chem. 278:16857-16862. [DOI] [PubMed] [Google Scholar]
- 6.Contamin, S., A. Galmiche, A. Doye, G. Flatau, A. Benmerah, and P. Boquet. 2000. The p21 Rho-activating toxin cytotoxic necrotizing factor 1 is endocytosed by a clathrin-independent mechanism and enters the cytosol by an acidic-dependent membrane translocation step. Mol. Biol. Cell 11:1775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Coombe, D. R., and C. W. Kett. 2005. Heparan sulfate-protein interactions: therapeutic potential through structure-function insights. Cell. Mol. Life Sci. 64:410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, J. M., S. B. Rasmussen, and A. D. O'Brien. 2005. Cytotoxic necrotizing factor type 1 production by uropathogenic Escherichia coli modulates polymorphonuclear leukocyte function. Infect. Immun. 73:5301-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donelli, G., and C. Fiorentini. 1992. Cell injury and death caused by bacterial protein toxins. Toxicol. Lett. 64-65(Spec. No.):695-699. [DOI] [PubMed] [Google Scholar]
- 9.Donnenberg, M. S., and R. A. Welch. 1996. Virulence determinants of uropathogenic Escherichia coli, p. 135-174. In H. L. T. Mobley and J. W. Warren (ed.), Urinary tract infections: molecular pathogenesis and clinical management. American Society for Microbiology Press, Washington, DC.
- 10.Fabbri, A., M. Gauthier, and P. Boquet. 1999. The 5′ region of cnf1 harbours a translational regulatory mechanism for CNF1 synthesis and encodes the cell-binding domain of the toxin. Mol. Microbiol. 33:108-118. [DOI] [PubMed] [Google Scholar]
- 11.Fiorentini, C., G. Arancia, A. Caprioli, V. Falbo, F. M. Ruggeri, and G. Donelli. 1988. Cytoskeletal changes induced in HEp-2 cells by the cytotoxic necrotizing factor of Escherichia coli. Toxicon 26:1047-1056. [DOI] [PubMed] [Google Scholar]
- 12.Flatau, G., E. Lemichez, M. Gauthier, P. Chardin, S. Paris, C. Fiorentini, and P. Boquet. 1997. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature 387:729-733. [DOI] [PubMed] [Google Scholar]
- 13.Frankel, G., D. C. Candy, P. Everest, and G. Dougan. 1994. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect. Immun. 62:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Häcker, U., K. Nybakken, and N. Perriman. 2005. Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 6:530-541. [DOI] [PubMed] [Google Scholar]
- 14.Horiguchi, Y. 2001. Escherichia coli cytotoxic necrotizing factors and Bordetella dermonecrotic toxin: the dermonecrosis-inducing toxins activating Rho small GTPases. Toxicon 39:1619-1627. [DOI] [PubMed] [Google Scholar]
- 15.Jaseja, M., L. Mergen, K. Gillette, K. Forbes, I. Sehgal, and V. Copie. 2005. Structure-function studies of the functional and binding epitope of the human 37 kDa laminin receptor precursor protein. J. Pept. Res. 66:9-18. [DOI] [PubMed] [Google Scholar]
- 16.Kim, K. J., J. W. Chung, and K. S. Kim. 2005. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J. Biol. Chem. 280:1360-1368. [DOI] [PubMed] [Google Scholar]
- 17.Lemichez, E., G. Flatau, M. Bruzzone, P. Boquet, and M. Gauthier. 1997. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol. Microbiol. 24:1061-1070. [DOI] [PubMed] [Google Scholar]
- 18.Lerm, M., J. Selzer, A. Hoffmeyer, U. R. Rapp, K. Aktories, and G. Schmidt. 1999. Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1: activation of c-Jun N-terminal kinase in HeLa cells. Infect. Immun. 67:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNichol, B. A., S. B. Rasmussen, K. C. Meysick, and A. D. O'Brien. 2006. A single amino acid substitution in the enzymatic domain of cytotoxic necrotizing factor type 1 of Escherichia coli alters the tissue culture phenotype to that of the dermonecrotic toxin of Bordetella spp. Mol. Microbiol. 60:939-950. [DOI] [PubMed] [Google Scholar]
- 20.Mecham, R. P. 1991. Receptors for laminin on mammalian cells. FASEB J. 5:2538-2546. [DOI] [PubMed] [Google Scholar]
- 21.Meysick, K. C., M. Mills, and A. D. O'Brien. 2001. Epitope mapping of monoclonal antibodies capable of neutralizing cytotoxic necrotizing factor type 1 of uropathogenic Escherichia coli. Infect. Immun. 69:2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills, M., K. C. Meysick, and A. D. O'Brien. 2000. Cytotoxic necrotizing factor type 1 of uropathogenic Escherichia coli kills cultured human uroepithelial 5637 cells by an apoptotic mechanism. Infect. Immun. 68:5869-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oswald, E., M. Sugai, A. Labigne, H. C. Wu, C. Fiorentini, P. Boquet, and A. D. O'Brien. 1994. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc. Natl. Acad. Sci. USA 91:3814-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pullinger, G. D., R. Sowdhamini, and A. J. Lax. 2001. Localization of functional domains of the mitogenic toxin of Pasteurella multocida. Infect. Immun. 69:7839-7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieger, R., C. I. Lasmezas, and S. Weiss. 1999. Role of the 37 kDa laminin receptor precursor in the life cycle of prions. Transfus. Clin. Biol. 6:7-16. [DOI] [PubMed] [Google Scholar]
- 26.Rippere-Lampe, K. E., M. Lang, H. Ceri, M. Olson, H. A. Lockman, and A. D. O'Brien. 2001. Cytotoxic necrotizing factor type 1-positive Escherichia coli causes increased inflammation and tissue damage to the prostate in a rat prostatitis model. Infect. Immun. 69:6515-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rippere-Lampe, K. E., A. D. O'Brien, R. Conran, and H. A. Lockman. 2001. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf1) attenuates the virulence of uropathogenic Escherichia coli. Infect. Immun. 69:3954-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt, G., P. Sehr, M. Wilm, J. Selzer, M. Mann, and K. Aktories. 1997. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387:725-729. [DOI] [PubMed] [Google Scholar]
- 29.Shime, H., T. Ohnishi, K. Nagao, K. Oka, T. Takao, and Y. Horiguchi. 2002. Association of Pasteurella multocida toxin with vimentin. Infect. Immun. 70:6460-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Smith, M. J., H. M. Carvalho, A. R. Melton-Celsa, and A. D. O'Brien. 2006. The 13C4 monoclonal antibody that neutralizes Shiga toxin type 1 (Stx1) recognizes three regions on the Stx1 B subunit and prevents Stx1 from binding to its eukaryotic receptor globotriaosylceramide. Infect. Immun. 74:6992-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorg, I., U. M. Goehring, K. Aktories, and G. Schmidt. 2001. Recombinant Yersinia YopT leads to uncoupling of RhoA-effector interaction. Infect. Immun. 69:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorokin, A. V., A. M. Mikhailov, A. V. Kachko, E. V. Protopopova, S. N. Konovalova, M. E. Andrianova, S. V. Netesov, A. N. Kornev, and V. B. Loktev. 2000. Human recombinant laminin-binding protein: isolation, purification, and crystallization. Biochemistry (Moscow) 65:546-553. [PubMed] [Google Scholar]
- 32.Sugai, M., K. Hatazaki, A. Mogami, H. Ohta, S. Y. Peres, F. Herault, Y. Horiguchi, M. Masuda, Y. Ueno, H. Komatsuzawa, H. Suginaka, and E. Oswald. 1999. Cytotoxic necrotizing factor type 2 produced by pathogenic Escherichia coli deamidates a Gln residue in the conserved G-3 domain of the Rho family and preferentially inhibits the GTPase activity of RhoA and Rac1. Infect. Immun. 67:6550-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]