Abstract
Mycobacterium tuberculosis Rv3676 encodes a cyclic AMP (cAMP) receptor-like protein (CRPMt) that has been implicated in global gene regulation and may play an important role during tuberculosis infection. The CRPMt ortholog in Mycobacterium bovis BCG, CRPBCG, is dysfunctional in an Escherichia coli CRP competition assay and has been proposed as a potential source of M. bovis BCG's attenuation. We compared CRPBCG and CRPMt in vitro and in vivo, in M. bovis BCG and M. tuberculosis, to evaluate CRPBCG's potential function in a mycobacterial system. Both proteins formed dimers in mycobacterial lysates, bound to the same target DNA sequences, and were similarly affected by the presence of cAMP in DNA binding assays. However, CRPMt and CRPBCG differed in their relative affinities for specific DNA target sequences and in their susceptibilities to protease digestion. Surprisingly, CRPBCG DNA binding activity was stronger than that of CRPMt both in vitro and in vivo, as measured by electrophoretic mobility shift and chromatin immunoprecipitation assays. Nutrient starvation-associated regulation of several CRPMt regulon members also differed between M. bovis BCG and M. tuberculosis. We conclude that CRPBCG is a functional cAMP-responsive DNA binding protein with an in vivo DNA binding profile in M. bovis BCG similar to that of CRPMt in M. tuberculosis. However, biologically significant functional differences may exist between CRPBCG and CRPMt with respect to gene regulation, and this issue warrants further study.
Tuberculosis (TB) remains a global epidemic, with one-third of the world's population infected, 9 million active cases, and over 2 million deaths annually (3). Increased drug resistance and a lethal synergism with human immunodeficiency virus exacerbate the morbidity and mortality already associated with Mycobacterium tuberculosis infection (7, 9). The molecular pathogenesis of M. tuberculosis is poorly understood, and better characterization of the mechanisms by which the tubercle bacillus responds to its host environment is needed if we are to control this deadly infection.
Cyclic AMP (cAMP), generated by adenylate cyclase, is an important signaling molecule in many bacterial species and eukaryotic cells. cAMP-mediated gene regulation in Escherichia coli, a process which requires interaction with the cAMP receptor protein (CRP), has been well defined (16). Additional roles for cAMP in metabolism have been reported for many pathogens (2, 6, 8, 13, 18, 19, 23, 30, 36), and recent studies increasingly point to an important role for cAMP signaling in microbial pathogenesis. Vfr, a CRP homolog in Pseudomonas aeruginosa, coordinates the expression of a variety of virulence-associated factors including type IV pili and the type III secretion system (35). Mutation of a cyclase-encoding gene (cya) in Vibrio vulnificus resulted in attenuation of the bacterium, with a 100-fold increase in the 50% lethal dose for mice (15). Pathogenic fungi also use the highly conserved cAMP signal transduction pathway to regulate cellular differentiation and pathogenesis (20, 21, 31).
The potential significance of cAMP signaling in M. tuberculosis pathogenesis was first explored in an early study that reported elevated levels of cAMP in the cytoplasms of macrophages infected with M. microti (24). Those authors correlated such increased cAMP levels with impaired phagosome-lysosome fusion. However, little more work was done on this topic until an in silico study identified genes for 15 adenylate cyclases and 10 cNMP-binding proteins in the M. tuberculosis genome (27). At least 10 of these adenylate cyclases are functional, as demonstrated by in vitro biochemical assays (33). It was recently shown that cAMP signaling is likely to be an important global regulator of gene regulation in M. tuberculosis and also that Rv3676 (CRPMt) is a CRP-like cAMP-responsive transcription factor with a putative regulon of more than 100 genes (4, 32, 34). Deletion of Rv3676 caused impaired growth in laboratory medium, in bone marrow-derived macrophages, and in a mouse model of TB (32).
CRPMt and its ortholog in M. bovis are identical. However, the Mycobacterium bovis BCG Pasteur ortholog, which we refer to as CRPBCG, contains two amino acid differences, L47P and E178K, relative to the CRPMt sequence. It was recently reported that the L47P change resulted in loss of cAMP binding and that the E178K substitution caused a DNA binding defect in an E. coli model system (34). The authors proposed a role for CRPBCG in M. bovis BCG's attenuation based on these results. However, potential differences between CRPMt and CRPBCG function have not been examined biochemically or in TB complex mycobacteria. Such studies are a critical first step in the assignment of a role for this CRP-like protein in the attenuation of M. bovis BCG.
We compared various functional aspects of CRPMt and CRPBCG in vitro and in vivo in M. tuberculosis and M. bovis BCG by use of members of CRPMt's putative regulon (4). We report here that CRPBCG is a fully functional DNA binding protein with enhanced activity relative to CRPMt, both in vitro and in vivo. However, differences were found between CRPMt's and CRPBCG's overall structural conformations, and the starvation-associated regulation of several CRPMt regulon members differed between M. tuberculosis and M. bovis BCG. The implications of these findings for CRPBCG's potential as an attenuating factor in M. bovis BCG are discussed.
MATERIALS AND METHODS
Bacterial strains and culture.
M. tuberculosis H37Rv and recombinant M. bovis BCG (Pasteur strain; Trudeau Institute) were grown in Middlebrook 7H9 medium supplemented with 0.5% glycerol, 10% oleic acid-albumin-dextrose-catalase, 0.05% Tween 80 as previously described (10) or on Middlebrook 7H10 agar (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase and 0.01% cycloheximide. Fresh cultures were inoculated from frozen stocks for every experiment. Bacteria were typically used in late log phase, at 7 days of growth. Cultures were grown under shaking conditions and under ambient air. E. coli strains were grown in Luria-Bertani broth or on Luria-Bertani agar plates. Kanamycin at 25 μg/ml was added for recombinant strains. All cultures were grown at 37°C.
Nutrient starvation conditions.
For some experiments, bacteria were subjected to nutrient starvation conditions, as previously described by others (5). Briefly, M. tuberculosis or M. bovis BCG was grown in 7H9 broth with supplements for 6 days with shaking in tissue culture flasks. The culture was split into two aliquots and harvested by centrifugation. One aliquot was resuspended into phosphate-buffered saline (PBS), and the other aliquot was resuspended in the same volume of fresh 7H9 broth with supplements. After agitation for another 24 h, bacteria were harvested, and RNA was prepared for reverse transcriptase PCR (RT-PCR) to assess the expression of selected genes. Primers for these experiments are shown in Table 1.
TABLE 1.
Primers used in this study
| Assay type and gene namea | Primer | Oligonucleotide sequenceb |
|---|---|---|
| EMSA and ChIP | ||
| Rv3857c | KM1218 | F: GGATCCTAGCCAACCTGATACGTGCC |
| KM1219 | R: GGATCCGAGGGCACAGTTCATC | |
| serC-Rv0885 | KM1229 | F: GGATCCTATGCGCGTCCTGTCCATTC |
| KM1230 | R: GGATCCTGGTCGGCCATGCCATCAG | |
| frdA | KM1260 | F: GGATCCGTAAACACAATCGCAGAACTGG |
| KM1261 | R: GGATCCATAACCACGATGTTGTGTTGGG | |
| Rv0019c | KM1262 | F: GGATCCTCGTCCGCATGCACTGAACC |
| KM1263 | R: GGATCCGTCAGTTGCAGTACCAACC | |
| PE15 | KM1266 | F: GGATCCTATCTCGCTGCCGTGTAGC |
| KM1267 | R: GGATCCGGAACGACTCGCAACGTCAC | |
| Rv0145 | KM1270 | F: GGATCCACCTGACCGAGACCTAAC |
| KM1271 | R: GGATCCTCGCGAGGATGGTAACGAGG | |
| Rv0950c-sucC | KM1601 | F: GGATCCAACTCCTTGGCTTGATACTC |
| KM1602 | R: GGATCCGAGGTGTGCGAATCGCTG | |
| Rv1158c-Rv1159 | KM1641 | F: GGATCCTAGCGATCGCCGAGCGCACC |
| KM1642 | R: GGATCCGCGGACAAACGTCCAGATGG | |
| sigA internal fragment | KM1309 | F: GATGACCGAGCTTAGCGAGC |
| KM1310 | R: CGTAGGTGGAGAACTTGTACC | |
| RT-PCR | ||
| Rv3676 | KM859 | F: ACGGCCGAGAAAACCTGTTAACC |
| KM860 | R: GACCCGCAATGCGACCTTCC | |
| 16S rRNA | KM886 | F: GCGAACGGGTGAGTAACACG |
| KM887 | R: TGAAAGAGGTTTACAACCCG | |
| Rv0145 | KM1272 | F: ATAGCTGCCGGATTCGATGG |
| KM1273 | R: GGCTCGTGGTAGGTCAACG | |
| frdA | KM1274 | F: GCGATAGCCGAAACCAATCC |
| KM1275 | R: GTGAAATCCCGTCTTGTCGG | |
| Rv0019c | KM1276 | F: TCATCTGGTCCGTGCTACGG |
| KM1277 | R: ATCCTAGATCTTCGACGTACC | |
| PE15 | KM1280 | F: CTGCCATCGAAGCAGTGACC |
| KM1281 | R: CCACCGCTGAGATACGACGC | |
| Rv0885 | KM1282 | F: CGAGTACGTGGAAATGCTGG |
| KM1283 | R: GAACGTGTAGTTCATCAGGC | |
| serC | KM1286 | F: GGCTATGAGGTGATACTGGG |
| KM1287 | R: GGCGTCGATGACGACCAAGG | |
| sucC | KM1288 | F: TTCGCCAAGCACAACGTGCC |
| KM1289 | R: TATCGCTAGCCTCAGCGACC | |
| sigA | KM1309 | F: GATGACCGAGCTTAGCGAGC |
| KM1310 | R: CGTAGGTGGAGAACTTGTACC | |
| Rv1158c | KM1643 | F: GGTATCGCTCACGCAGATCC |
| KM1644 | R: CGTCAGTCCTGGAAAGGTGG | |
| Rv0950c | KM1675 | F: GTGCTGACCGCTCATGCATC |
| KM1676 | R: GCGTTAGCCAGGTCGATACC | |
| Rv1159 | KM1677 | F: CATTTTCGGTGGTCGTGTTCC |
| KM1678 | R: GCAGCAGGCCGAACAACTCG | |
| Rv3857c | KM1679 | F: CTCGGATTTGATACGAAACC |
| KM1680 | R: CAGTTGCCCGAAGACGACCC |
The target sequence for each gene is the intergenic region for EMSA and the ChIP assay and an internal region for RT-PCR, according to the complete sequence of M. tuberculosis H37Rv on the TubercuList World Wide Web server (http://genolist.pasteur.fr/TubercuList/).
Oligonucleotide sequences are from 5′ to 3′. F, forward primer; R, reverse primer.
Expression and purification of CRPMt and CRPBCG.
Expression and purification of CRPMt have been reported previously (4). Expression of CRPBCG was by the same procedure, except that M. bovis BCG DNA was used as template for PCR amplification of the encoding DNA. The DNA was cloned into pET28a(+) vector (Novagen) between EcoRI and HindIII to generate pMBC570. The sequence was verified and maintained in E. coli BL21(DE3). The expression of CRPMt and CRPBCG was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. The His-tagged CRPMt and CRPBCG were purified using a HisTrap column (Amersham Biosciences) as previously reported (4). Protein concentration was measured with a NanoOrange protein quantitation kit (Molecular Probes) and diluted to 2 mg/ml before storage in aliquots at −70°C.
Preparation of polyclonal antibody and Western blot analysis.
Two New Zealand White rabbits were immunized subcutaneously with 1 mg purified His-CRPMt emulsified 1:1 with Titermax Gold (Sigma) and boosted initially and again 2 weeks later with the same amount of protein and adjuvant. The specificity of serum was confirmed by Western blotting with His-CRPMt and extract of M. bovis BCG as the antigen. Extracts of M. tuberculosis and M. bovis BCG were mixed with sample buffer and subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were blotted onto polyvinylidene difluoride membranes and sequentially probed with anti-CRPMt serum and with a peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibody (Jackson ImmunoResearch Laboratories). Peroxidase detection was carried out with the ECL Western blotting detection reagent and analysis system (Amersham Biosciences).
Cross-linking of CRPMt and CRPBCG by glutaraldehyde.
Cross-linking of purified His-tagged CRPMt and CRPBCG was modified from a previously reported method (22). CRPMt and CRPBCG were diluted in cross-linking buffer (50 mM sodium phosphate, pH 7.4, 20% glycerol, 5 mM MgCl2) to 310 nM and were then incubated with glutaraldehyde at a final concentration of 7 mM for 1 h at room temperature. The reactions were quenched by the addition of SDS-PAGE sample buffer, and a portion of each was separated on a 12% SDS-PAGE gel. The proteins were transferred onto a polyvinylidene difluoride membrane and visualized with anti-CRPMt antibody.
Proteolysis of CRPMt and CRPBCG with trypsin.
The effects of 100 μM cAMP on CRPMt's and CRPBCG's conformation were compared by incubation of 5 μg of CRPMt and CRPBCG with 0.2, 0.1, or 0.02 μg trypsin (Sigma) in a 20-μl reaction mixture for 10 min at 37°C, as described by others (17). One-half of the digested protein was separated on a 15% SDS-PAGE gel. AMP (100 μM) was used as a control for cAMP.
Electrophoretic mobility shift assays (EMSA).
PCR forward primer was labeled with [γ-33P]ATP (MP Biomedicals) by use of T4 DNA polynucleotide kinase (New England Biolabs). DNA fragments were then labeled by PCR, and about 0.05 pmol DNA probe was used in each 10-μl binding reaction mixture, as described previously (4). Samples were loaded on an 8% nondenaturing polyacrylamide gel and run for 2 to 3 h at 14 V/cm. Gels were vacuum dried, exposed on a phosphor screen, scanned with a Storm 860 PhosporImager (Molecular Dynamics), and analyzed with ImageQuant software (Molecular Dynamics).
DNase I footprinting.
A 156-bp DNA fragment containing the serC-Rv0885 intergenic region was PCR amplified with primers KM1299 (GCTGGTACCGGGAATTATCC) and KM1300 (TGCCATCAGGGTAGTGAGGG). Either KM1299 or KM1300 was end labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (MP Biomedicals) to generate a single end-labeled probe. Binding reactions were performed in a final volume of 100 μl containing approximately 0.25 pmol DNA probe, 100 nM of either CRPMt or CRPBCG in binding buffer (10 mM Tris-HCl [pH 8.0], 50 mM KCl, 1 mM EDTA, 50 μg/ml bovine serum albumin, 1 mM dithiothreitol, 0.05% nonionic P-40 detergent, 100 μM cAMP) and 20 μg/ml poly(dI-dC) as a nonspecific competitor. Binding reactions were performed for 30 min at room temperature and were then subjected to DNase I digestion in 10 mM MgCl2, 5 mM CaCl2 for 1 min. Digestion was terminated by the addition of 200 μl stop solution (1% SDS, 200 mM NaCl, 20 mM EDTA, and 20 μg/ml salmon sperm DNA). Samples were extracted once with phenol-chloroform, and DNA was precipitated with ethanol, dried, and resuspended in formamide loading dye. Samples were loaded on a 7 M urea-6% polyacrylamide sequencing gel. A dideoxynucleotide sequencing ladder was generated using a Thermo Sequenase cycle sequencing kit (USB) with the same end-labeled primer as was used for the footprinting.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were carried out by a method modified from the literature (12, 29). M. tuberculosis and M. bovis BCG were grown until late log phase. Paraformaldehyde was added to a final concentration of 4% for M. tuberculosis, and formaldehyde was added to a final concentration of 1% for M. bovis BCG, for 30 min at room temperature. Cells were harvested, washed twice in PBS containing 1% Tween 80, and then resuspended in 1 ml lysis buffer containing 0.5% SDS, 50 mM Tris-HCl (pH 8.0), 10 mM EDTA (pH 8.0), 50 μl protease inhibitor cocktail (Sigma), and 10 mg lysozyme. After incubation for 20 min at 37°C, bacterial DNA was sheared by sonication as previously reported, except that the sonication time was increased to 10 min at setting 8 (28). Cell debris was removed by centrifugation, and the supernatant was diluted 1:5 in IP buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% Triton X-100) for use as the input sample. A 1-ml aliquot of the input sample was incubated overnight at 4°C on a nutator with 10 μl of antiserum or the preimmune serum from the same rabbit. Protein A-agarose beads (50 μl; Boehringer Mannheim) was added and incubated for 1 h at room temperature on a nutator, followed by four washes with IP buffer and two elutions in 150 μl of elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, pH 8.0, and 1% SDS). The eluate was incubated overnight at 65°C, followed by the addition of proteinase K to 100 μg/ml and incubation for 2 h at 37°C. Immunoprecipitated DNA was recovered by ethanol precipitation after phenol-chloroform extraction and used for PCR amplification of selected intergenic regions (30 cycles). An internal fragment of sigA amplified using KM1309 and KM1310 (Table 1) was used to normalize the amount of DNA between CRP-immune and preimmune IP samples.
RNA extraction and RT-PCR.
Total RNA extraction from cultures was carried out as described by others (25). After isopropanol precipitation, RNA was treated twice with RNase-free DNase I (RNase Free DNase set; QIAGEN) on an RNeasy mini kit column (QIAGEN). The RNA concentration was determined spectrophotometrically with a biophotometer (Eppendorf) at 260 nm. cDNA was prepared and amplified as previously described (11). Control reactions were performed against 16S RNA (1) by use of cDNA dilutions of 10−4 or 10−5 and against sigA (26) by use of cDNA diluted 10−2. PCR of 16S RNA and of sigA was also performed using total RNA without reverse transcription to ensure the absence of DNA contamination. PCR was run for 30 cycles. Gels were scanned with a FluorImager595 (Molecular Dynamics) and analyzed with ImageQuant software.
Statistical analysis.
A two-tailed t test was performed using GraphPad InStat version 3.0b for Macintosh (GraphPad Software, CA) to determine the significance of binding and expression differences between genes in M. tuberculosis and M. bovis BCG. P values of <0.05 were considered significant.
RESULTS
Dimerization of CRPMt and CRPBCG.
Many transcriptional regulators form oligomers when bound with their DNA ligands. An example is E. coli CRP, which binds specific DNA targets as a dimer (16). CRPMt recognizes a CRP-like palindromic core sequence, suggesting that it also functions as a dimer in M. tuberculosis.
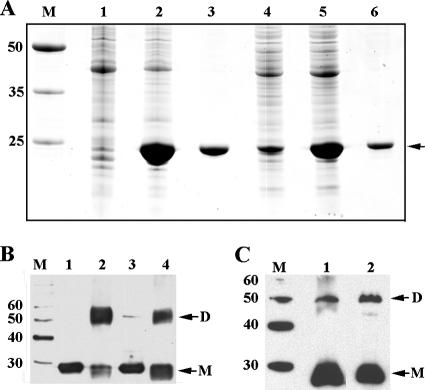
His-tagged CRPMt and CRPBCG were overexpressed in E. coli and then purified for in vitro analyses. Dimer formation was examined by glutaraldehyde cross-linking and Western blot analysis using anti-CRPMt serum (Fig. 1). Untreated proteins migrated predominantly as monomers during SDS-PAGE, but bands corresponding in mobility to dimers were present for both CRPMt and CRPBCG following glutaraldehyde treatment (Fig. 1B). At high protein concentrations, CRPBCG, but not CRPMt, also produced a faint higher-molecular-weight band and large aggregates that failed to enter the gel (not shown). Bands corresponding in size to both dimers and monomers were also observed for native bacterial protein extracts from both M. tuberculosis and M. bovis BCG (Fig. 1C). These results indicate that native expression and dimerization of these CRP-like proteins are similar in M. tuberculosis and M. bovis BCG.
FIG. 1.
Expression and dimerization of CRPMt and CRPBCG. (A) CRPMt and CRPBCG were overexpressed in E. coli and purified with a HisTrap column (Amersham Biosciences). Lanes: M, broad-range protein molecular weight markers (Promega); 1 to 3, CRPMt; 4 to 6, CRPBCG. Lanes 1 and 4 are lysates of uninduced bacteria; lanes 2 and 5 are lysates of bacteria induced with IPTG; lanes 3 and 6 are purified proteins. (B) Glutaraldehyde cross-linking of purified CRPMt and CRPBCG analyzed by Western blotting. Lanes: M, MagicMark XP Western protein standards (Invitrogen); 1 and 2, CRPMt; 3 and 4, CRPBCG. Proteins in lanes 2 and 4 were cross-linked with glutaraldehyde. (C) Dimerization in bacterial lysates of M. tuberculosis (lane 1) and M. bovis BCG (lane 2) analyzed by Western blotting. Lane M, MagicMark XP Western protein standards. Note that the overexpressed proteins in panel B contain a His tag of about 4.4 kDa. Therefore, the antiserum-recognized bands in panel B and panel C differ in size.
Conformational differences between CRPMt and CRPBCG.
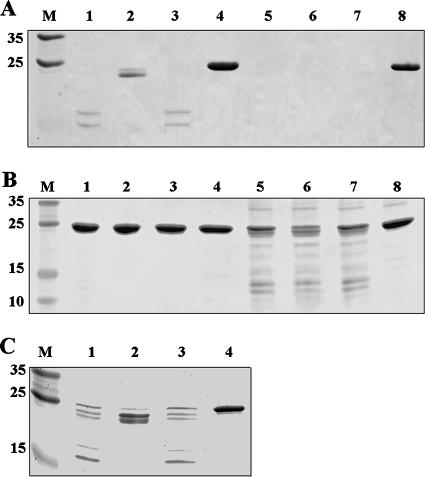
We previously reported that CRPMt changed its conformation upon binding with cAMP (4). Here we evaluated both CRPMt and CRPBCG by limited proteolysis with trypsin to compare their interactions with cAMP. CRPBCG was more sensitive than CRPMt to trypsin digestion, whether or not cAMP was present (Fig. 2). CRPMt showed partial protection from trypsin digestion by cAMP when 5 μg of protein was treated with 0.2 μg trypsin (Fig. 2A), similar to our previous result (4). However, CRPBCG was completely digested and could not be evaluated for cAMP binding in these conditions (Fig. 2A). CRPBCG still showed substantial cleavage when the quantity of trypsin was reduced to 0.02 μg, but CRPMt exhibited no obvious proteolysis under these conditions (Fig. 2B). CRPBCG showed a banding pattern in the presence of cAMP only slightly different from what was seen with trypsin only or AMP controls (Fig. 2B), which differed from the results obtained with CRPMt (Fig. 2A and C). These results indicate that CRPMt and CRPBCG differ in their overall sensitivities to trypsin and possibly in their interactions with cAMP.
FIG. 2.
Evidence of allosteric alteration of CRPMt and CRPBCG by cAMP. CRPMt and CRPBCG were digested with 0.2 μg (A), 0.02 μg (B), or 0.1 μg (C) trypsin. Lanes: M, broad-range molecular markers (Promega); 1 to 4, CRPMt; 5 to 8, CRPBCG. Lanes 4 and 8 are undigested controls. Lanes 1 to 3 and 5 to 7 were digested with trypsin; 100 μM cAMP was supplemented in lanes 2 and 6; 100 μM AMP was supplemented in lanes 3 and 7. Figures are representative of at least two independent experiments.
CRPMt and CRPBCG have similar DNA binding specificities.
DNA binding of CRPMt and CRPBCG was tested with the serC-Rv0885 intergenic DNA used as a model sequence. Our previous results indicated that CRPMt binds to this sequence specifically, and with enhancement in the presence of 100 μM of cAMP (4). In the present study, CRPBCG showed higher DNA binding affinity than did CRPMt in either the absence (Fig. 3A) or presence (Fig. 3B) of 100 μM cAMP. Addition of cAMP enhanced the DNA binding of CRPBCG to the same extent as seen for CRPMt (1.5- to 2-fold) (Fig. 3C). However, CRPBCG was also able to form a higher-molecular-mass protein-DNA complex that was most prominent in the presence of cAMP (Fig. 3B and C). These results indicate that CRPBCG is fully competent for DNA binding, although its binding properties are not identical to those of CRPMt.
FIG. 3.
Comparison of DNA binding between CRPMt and CRPBCG, and the effects of cAMP on CRPBCG binding. (A) Binding of CRPMt and CRPBCG with the serC-Rv0885 intergenic region in the absence of cAMP. Lanes: 1, free probe; 2 to 6, CRPMt; 7 to 11, CRPBCG. Lanes 2 to 6 and 7 to 11 are in the presence of 0.35 nM, 0.7 nM, 1.7 nM, 3.5 nM, and 7 nM purified protein, respectively. (B) Comparison of CRPMt's and CRPBCG's binding with the serC-Rv0885 intergenic region in the presence of 100 μM cAMP. Probe and protein dilutions are the same as the lanes in panel A. (C) CRPBCG's binding with the serC-Rv0885 intergenic region in the absence (lanes 1 to 6) or in the presence (lanes 7 to 12) of cAMP. Lanes 1 and 7 are free probes. Lanes 2 to 6 and 8 to 12 are in the presence of 0.35 nM, 0.7 nM, 1.7 nM, 3.5 nM and 7 nM purified protein, respectively. DNA binding data are plotted to the right of each panel. For each panel, quantitation of band intensity was done using ImageQuant software, and the percent DNA bound was calculated as the intensity of each shifted band divided by the intensity of the unshifted band in the probe-only sample (lane 1). The apparent dissociation constant (kd) (37) in nM is also indicated.
CRPMt's and CRPBCG's DNA binding specificities were further explored by DNase I footprinting of the serC-Rv0885 intergenic region. The same sequence was protected from digestion with DNase I, in the presence of either CRPMt or CRPBCG (Fig. 4). This protected region is consistent with the binding motif that we identified previously (4), but it includes asymmetric flanking sequences as well (Fig. 4).
FIG. 4.

DNase I footprinting of CRPMt's and CRPBCG's binding sites in the serC-Rv0885 intergenic region. The −155/+1 DNA region upstream of the serC translational start site was labeled at the −155 end (A) or the +1 end (B) and then digested with DNase I in the absence of protein (−) or in the presence of 100 nM of either CRPMt (H37Rv) or CRPBCG (BCG). A dideoxynucleotide sequencing ladder, run in parallel with the footprinting reactions (data not shown), was used to determine the sequence of the protected region. The black bars indicate the previously reported CRPTB binding motif (TGTGAN6TCACA).
Together, EMSA, DNase I footprinting, and limited proteolysis results indicate that CRPBCG retains the ability to interact with both DNA and cAMP at a functional level, despite its conformational differences with CRPMt.
CRPMt and CRPBCG bind similar target sequences with differing affinities.
CRPMt's and CRPBCG's DNA binding interactions were further examined, with a broader range of DNA sequences, in vitro and in vivo. In vitro binding was measured by EMSA, while in vivo binding was assessed by ChIP assays that used the same intergenic DNA sequences. Intergenic regions of serC-Rv0885, Rv0950c-sucC (Rv0951), and Rv1158c-Rv1159, as well as regions upstream of PE15 (Rv1386), frdA (Rv1552), Rv3857c, and Rv0019c, were selected from the CRPMt's list of putative binding sequences (4). Another sequence upstream of Rv0145 which binds with CRPMt but was not included in that previous study was also tested. All of these intergenic DNA sequences are identical in M. bovis BCG and M. tuberculosis H37Rv, except that Rv3857c has one base difference 164.5 bp downstream from the binding motif axis, and frdA has two nucleotide substitutions located 136.5 and 175.5 bp upstream of the binding motif axis. The H37Rv sequences were used for all experiments.
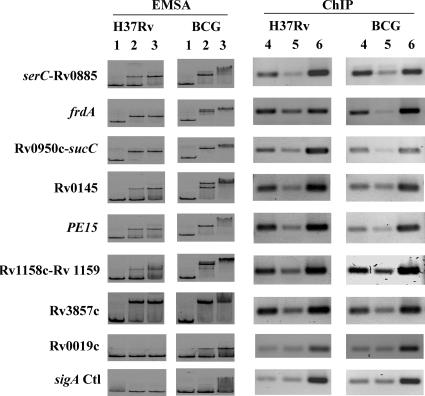
All putative binding sequences tested using EMSA, except Rv0019c, showed clear binding with both CRPMt and CRPBCG (Fig. 5). The Rv0019c intergenic region bound CRPBCG with very low affinity and failed to bind at all with CRPMt at molarities up to 35 nM. Neither CRPMt nor CRPBCG bound to a mycobacterial sigA internal fragment, which was used as a negative control. The affinity of CRPBCG for DNA was slightly higher than that of CRPMt, and in some cases, (e.g., serC-Rv0885, frdA, Rv0145, Rv1158c-Rv1159), the mobility of CRPBCG-DNA complex bands was supershifted in the presence of 3.5 nM or more protein (Fig. 5). This phenomenon was not observed with CRPMt (Fig. 5), even at protein concentrations up to 3.5 μM (data not shown).
FIG. 5.
In vitro and in vivo binding of CRPMt and CRPBCG with selected intergenic regions. Lanes 1 to 3, EMSA of CRPMt (H37Rv) and CRPBCG (BCG) with selected intergenic probes amplified from M. tuberculosis H37Rv. No cAMP was supplemented in the binding buffer. Lane 1 is free probe. Protein-DNA interaction is shown with 3.5 nM (lane 2) and 35 nM (lane 3) protein. The intergenic regions of genes tested are labeled to the left. serC, sucC, and Rv1158c are divergent from Rv0885, Rv0951, and Rv1159, respectively. Note that the size of the protein-DNA complex varies according to the probe size of different genes. Lanes 4 to 6, ChIP in M. tuberculosis and M. bovis BCG. Lane 4 shows PCR from immunoprecipitation with anti-CRPMt serum, and lane 5 is mock precipitation with preimmune serum prepared from the same rabbit. Lane 6 shows PCR control results from input DNA. The intergenic regions of genes tested (except the sigA control, which is in the open reading frame [ORF]) are listed to the left. Note that the results are pooled from different gels, so that bands of different genes are not quantitatively comparable. The ChIP results represent three biological repeats.
Bacterial protein-DNA complexes were cross-linked in culture suspension and analyzed by ChIP to evaluate DNA binding in vivo. A parallel sample using preimmune serum from the same rabbit was run as a background reference control, and an internal sigA DNA sequence was used for PCR normalization. All DNA sequences that showed in vitro binding by EMSA were also enriched by ChIP in both M. tuberculosis and M. bovis BCG lysates (Fig. 5). Rv0019c, which was negative by EMSA, was not enriched by ChIP. Two sequences, frdA and Rv0950c-sucC, reproducibly showed stronger binding with CRPBCG than with CRPMt (Fig. 5; Table 2). These results indicate that although CRPMt and CRPBCG bind similar regulatory sequences in vivo, they differ in their relative affinities for certain DNA sequences.
TABLE 2.
Quantitation of ChIP assay resultsa
| DNA amplifiedb | Immune/preimmune ratio (mean ± SEM) forc:
|
P value (H37Rv vs BCG)d | |
|---|---|---|---|
| H37Rv | BCG | ||
| serC-Rv0885 | 2.6 ± 0.6 | 2.8 ± 0.2 | 0.76 |
| frdA | 1.6 ± 0.1 | 4.5 ± 0.1 | 0.001 |
| Rv0950c-sucC | 1.7 ± 0.1 | 4.1 ± 0.1 | 0.0009 |
| Rv0145 | 2.3 ± 0.1 | 2.0 ± 0.7 | 0.65 |
| PE15 | 2.3 ± 0.4 | 2.1 ± 0.2 | 0.65 |
| Rv1158c-Rv1159 | 1.5 ± 0.2 | 1.9 ± 0.2 | 0.30 |
| Rv3857c | 1.7 ± 0.1 | 2.1 ± 0.4 | 0.38 |
| Rv0019c | 0.9 ± 0.1 | 1.1 ± 0 | - |
PCR bands were scanned and analyzed with ImageQuant software.
DNA amplified from the intergenic regions of divergent genes or upstream of others. sigA was amplified from within its ORF.
Ratio of bands amplified from immunoprecipitated immune serum to bands amplified from the preimmune serum control. The results are presented as means ± standard errors of the means of three biological repeats.
P values from H37Rv versus BCG comparisons were compared using a two-tailed t test. Comparisons with P values of <0.05 are shown in bold. It was not possible to calculate a P value for Rv0019c (indicated by the dash), due to the 0 value for the standard error of the means.
Gene regulation of CRPMt regulon members under nutrient starvation conditions.
The differing affinities of CRPMt and CRPBCG for some regulatory sequences suggested that members of the CRP regulon might be regulated to different extents in M. tuberculosis and M. bovis BCG. We previously observed that a high percentage of the putative CRPMt regulon candidates were associated with starvation in another published study (4, 5). Both anaerobic and starved cultures have been used as models to study the molecular basis of dormancy (14), and the CRPMt regulon has been implicated in the persistence pathway (32).
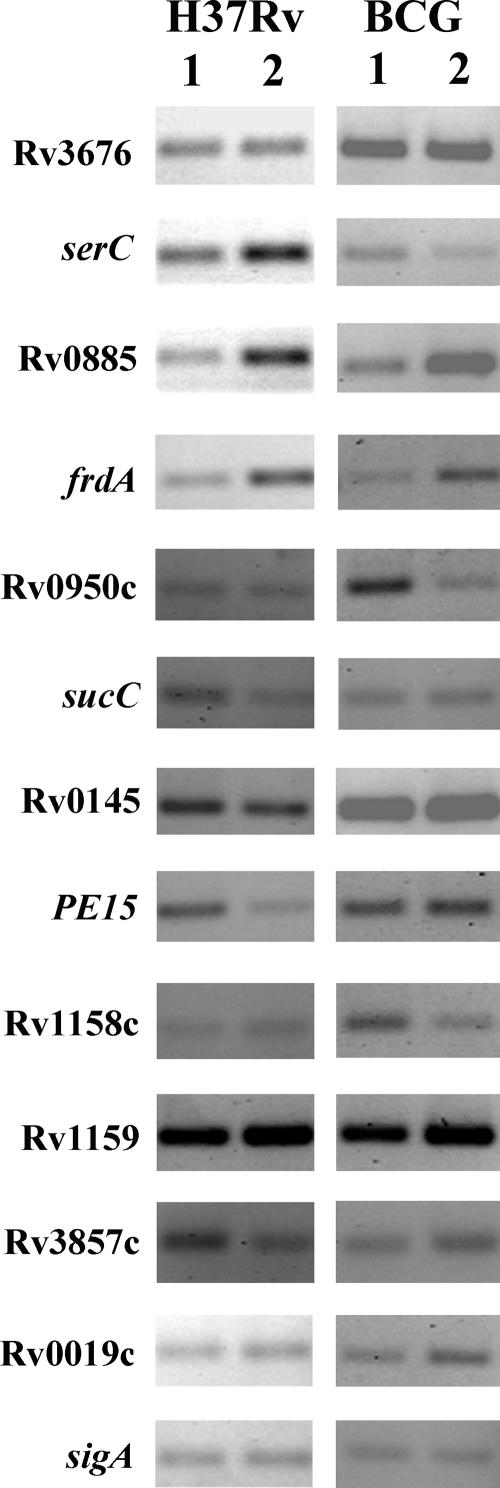
We compared expression levels of putative regulon gene members by semiquantitative RT-PCR in M. tuberculosis and M. bovis BCG subjected to starvation versus nutrient-rich conditions (Fig. 6; Table 3). Rv0885, frdA, Rv0950c, Rv0145, PE15, and Rv1158c were up-regulated at least 1.5-fold in response to starvation in at least one of the bacteria. frdA was similarly up-regulated, while Rv0950c was down-regulated, in both species. Three genes, Rv0145, PE15, and Rv1158c, showed statistically significant differences in their regulatory patterns between M. tuberculosis and M. bovis BCG. Rv0885 was generally more strongly regulated in M. tuberculosis than M. bovis BCG, but the difference was not statistically significant. These results indicate that the gene regulatory responses of M. tuberculosis and M. bovis BCG to starvation are not identical. Further studies are needed to determine the extent of these regulatory differences and whether they are mediated by the CRP-like proteins in TB complex bacteria.
FIG. 6.
Transcription of selected genes after starvation of M. tuberculosis and M. bovis BCG for 24 h in PBS. RT-PCR with RNA prepared from M. tuberculosis H37Rv and M. bovis BCG after 24 h of starvation in PBS. Lanes: 1, 7H9 broth control; 2, starvation samples. The genes tested are listed to the left. sigA is a control for normalizing the cDNA level. Note that the results are pooled from PCRs with different concentrations of cDNA. Therefore, the bands are not comparable between different genes. The figure is a representative of three independent experimental repeats.
TABLE 3.
Quantitation of RT-PCR resultsa
| ORF | Gene name | Fold (mean ± SEM) induction by starvationb
|
P value (H37Rv vs BCG)c | |
|---|---|---|---|---|
| H37Rv | BCG | |||
| Rv3676 | 1.03 ± 0.07 | 0.93 ± 0.03 | 0.25 | |
| Rv0884c | serC | 1.00 ± 0.20 | 0.67 ± 0.07 | 0.19 |
| Rv0885 | 2.30 ± 0.60 | 1.27 ± 0.20 | 0.18 | |
| Rv1552 | frdA | 2.70 ± 0.42 | 2.30 ± 0.25 | 0.46 |
| Rv0950c | 0.40 ± 0.10 | 0.30 ± 0.06 | 0.44 | |
| Rv0951 | sucC | 0.70 ± 0.10 | 0.97 ± 0.03 | 0.06 |
| Rv0145 | 0.53 ± 0.12 | 0.90 ± 0.06 | 0.05 | |
| Rv1386 | PE15 | 0.53 ± 0.12 | 0.97 ± 0.03 | 0.03 |
| Rv1158c | 1.30 ± 0.20 | 0.53 ± 0.07 | 0.02 | |
| Rv1159 | 0.83 ± 0.09 | 1.13 ± 0.20 | 0.24 | |
| Rv3857c | 0.97 ± 0.32 | 1.17 ± 0.07 | 0.57 | |
| Rv0019c | 0.97 ± 0.17 | 1.07 ± 0.20 | 0.72 | |
Gels were scanned and analyzed with ImageQuant software.
Results were expressed as means ± standard errors of the means of three biological repeats and normalized by the induction (n-fold) of sigA.
P values for H37Rv versus BCG comparisons were compared using a two-tailed t test. P values relating to significant species-related differences in response to starvation are shown in bold.
DISCUSSION
Allosteric and functional comparison of CRPMt and CRPBCG.
CRPMt is an established CRP-like gene regulator in M. tuberculosis (4, 32). However, a recent report indicated that two amino acid changes alter CRPBCG's function relative to CRPMt, making CRPBCG unable to compete with CRP for DNA and/or cAMP binding in an E. coli functional model (34). This finding raised the possibility that this cAMP-responsive transcription regulator is inactive or defective in M. bovis BCG, possibly contributing to BCG's attenuation.
We found that CRPBCG is a fully functional DNA binding protein in M. bovis BCG, but its pattern of DNA binding is not identical to that of CRPMt in M. tuberculosis. Both CRPMt and CRPBCG were detected as dimers in vitro and in vivo, but the trypsin digestion experiments indicated that the amino acid differences in CRPBCG affected its conformation. The L47P change in CRPBCG deletes one trypsin recognition site, but the E178K change generates another, as determined by PeptideCutter (http://au.expasy.org/tools/peptidecutter/). Therefore, the total number of trypsin recognition sites is the same in each protein, although their positions vary.
It is not clear whether CRPBCG also differs significantly from CRPMt in its functional interactions with cAMP (Fig. 3). The presence of cAMP had very little effect on CRPBCG's sensitivity to trypsin digestion compared with the protection it afforded to CRPMt (Fig. 2). In fact, CRPBCG was slightly more sensitive to trypsin digestion in the presence of cAMP, which is more similar to what is observed with CRP in E. coli than with CRPMt (17). However, the cAMP-associated DNA binding effects observed by EMSA suggest that CRPBCG retains a functional association with cAMP that is not readily detected in the trypsin experiments, due to CRPBCG's increased lability in the presence of protease (Fig. 3). Additional functional studies will be needed to more completely resolve this issue.
In contrast to our expectations based on a previous report (34), we found that CRPBCG shows higher affinity than CRPMt for most target DNA sequences in vitro (Fig. 5), indicating that the conformational changes associated with the L47P and/or E178K substitutions are actually beneficial for binding with DNA. This effect may have biological significance, because in vivo binding by CRPBCG was also significantly enhanced relative to CRPMt in two cases (Fig. 5).
Comparison of in vitro and in vivo DNA binding of CRPMt and CRPBCG.
Our in vitro (EMSA) and in vivo (ChIP) DNA binding results showed good agreement across all eight sequences tested, indicating that EMSA provides a useful predictor for in vivo binding of CRPMt regulon promoters. Seven intergenic sequences from the putative CRPMt regulon showed clear binding by both approaches, thereby supporting our previous in silico prediction of the regulon (4). Nonetheless, these regulon predictions need further optimization. Rv0145 binds CRPMt and CRPBCG in vitro and in vivo, making it a likely regulon member, despite its not being detected in the in silico analysis (4). Rv0145 also regulates in response to starvation in M. tuberculosis (but not M. bovis BCG) but was not previously identified as a member of the starvation regulon (5). In contrast, the regulatory sequence for another putative regulon member predicted in silico, Rv0019c, was not detected in vivo, and it showed only very weak in vitro binding with CRPBCG and no binding with CRPMt (Fig. 5). Rv0019c is also the only previously identified starvation regulon member that failed to be regulated in response to starvation in the present study (5).
Rv0019c and Rv0145 may aid us in better defining the criteria that are most important for binding and in optimizing the CRPMt binding motif. Specifically, alignment of the CRPMt motifs for all DNA sequences used in this study shows that only Rv0019c and Rv0145 have mismatches in both 5-bp halves of the predicted core sequence. Rv0019c has three mismatches, compared with two for Rv0145. None of the remaining sequences had more than one mismatched nucleotide (Fig. 7). Future studies will determine whether it was the total number of mismatches, or the specific nucleotide changes, that caused Rv0019c's lack of activity in this assay.
FIG. 7.
Alignment of CRPMt and CRPBCG binding regions of selected intergenic sequences. Forty-six nucleotides centered on the motif core axis from each intergenic sequence were aligned. Blocks of similar nucleotides are indicated with yellow shading. Identical nucleotides are shown in red, and conserved nucleotides are shown in green. The binding arms in the consensus sequence are indicated with black bars. The protection region from DNase I footprinting in the serC-Rv0885 intergenic sequence is underlined. The sequences for which the experimental results are inconsistent with the previous prediction (4) are indicated with asterisks.
EMSA was a reliable predictor of in vivo DNA binding activity, but the relative affinities of CRPMt and CRPBCG for various DNA sequences in vitro and in vivo did not always coincide (Fig. 5; Table 2). Only two DNA sequences, frdA and Rv0950c-sucC, were bound with more affinity by CRPBCG than CRPMt in vivo, whereas CRPBCG's DNA binding activity was higher for all sequences in vitro (Fig. 5). This is probably because many different factors that can affect binding are included in the complex biological environment but are not present in the defined and “minimalist” in vitro binding conditions used for the EMSA experiments. In this respect, the ChIP approach is likely to be more reliable than EMSA for interpreting differences between CRPMt's and CRPBCG's interactions with different regulatory sequences. Nonetheless, the differing in vivo DNA binding affinities measured for these DNA sequences did not correlate with regulatory differences found in the starvation assay, even though both frdA and Rv0950c were regulated in response to starvation. It is possible that factors other than CRPMt/CRPBCG mediate the starvation response for these genes or that DNA binding affinity is not the primary determinant of regulatory function.
Responses of M. tuberculosis and M. bovis BCG to nutrient starvation.
Twenty-four members of the putative CRPMt regulon (∼21%) (4) were previously identified as being starvation regulated in M. tuberculosis (5). Five of these genes (Rv0885, frdA, Rv0019c, PE15, and Rv1158c) were examined in the present study, and results for all except Rv0019c were consistent with those of a previous microarray analysis (5). In addition, we observed down-regulation of Rv0950c and Rv0145 expression in response to starvation, a finding which has not been reported previously. The reason for these differences in regulation is not clear, but they likely reflect subtle differences in experimental conditions or assay sensitivities between laboratories.
PE15, Rv1158c, and Rv0145 showed differing expression patterns between M. bovis BCG and M. tuberculosis (Table 3; Fig. 6). PE15 and Rv0145 expression was not regulated in M. bovis BCG, while Rv1158c expression was regulated only in M. bovis BCG, indicating differences in the responses of these bacteria to starvation. However, all three genes code for hypothetical proteins of unknown function, making it difficult to assess the possible importance of their dysregulation to M. bovis BCG's attenuation.
In summary, M. tuberculosis and M. bovis BCG contain functional CRP orthologs that behave similarly, but not identically, with respect to DNA binding in vitro and in vivo. Conformational differences may be responsible for these differences in DNA affinity and could also affect the interactions of these CRP-like proteins with RNA polymerase and other gene regulatory factors. Starvation-associated gene regulatory differences between M. bovis BCG and M. tuberculosis may have implications for pathogenesis and warrant further investigation. Further studies are also needed to assess the roles of these CRP orthologs in the gene regulatory responses of M. bovis BCG and M. tuberculosis to starvation and other host-associated environmental conditions, as well as the environmental signals to which these CRP-like global regulators respond.
Acknowledgments
We gratefully acknowledge Tachanda Bryant for technical assistance, Lee Ann McCue for sequence analysis, and the Molecular Genetics Core facility of the Wadsworth Center for DNA sequencing.
This work was supported by grants AI4565801 and AI063499 from the National Institutes of Health.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.Alland, D., I. Kramnik, T. R. Weisbrod, L. Otsubo, R. Cerny, L. P. Miller, W. R. Jacobs, Jr., and B. R. Bloom. 1998. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 95:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz, M. A., and A. Wright. 2005. The World Health Organization/International Union Against Tuberculosis and Lung Disease Global Project on Surveillance for Anti-Tuberculosis Drug Resistance: a model for other infectious diseases. Clin. Infect. Dis. 41(Suppl. 4):S258-S262. [DOI] [PubMed] [Google Scholar]
- 4.Bai, G., L. A. McCue, and K. A. McDonough. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 187:7795-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 6.Caler, E. V., R. E. Morty, B. A. Burleigh, and N. W. Andrews. 2000. Dual role of signaling pathways leading to Ca2+ and cyclic AMP elevation in host cell invasion by Trypanosoma cruzi. Infect. Immun. 68:6602-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349-364. [DOI] [PubMed] [Google Scholar]
- 9.Espinal, M. A. 2003. The global situation of MDR-TB. Tuberculosis (Edinburgh) 83:44-51. [DOI] [PubMed] [Google Scholar]
- 10.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gazdik, M. A., and K. A. McDonough. 2005. Identification of cyclic AMP-regulated genes in Mycobacterium tuberculosis complex bacteria under low-oxygen conditions. J. Bacteriol. 187:2681-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grainger, D. C., T. W. Overton, N. Reppas, J. T. Wade, E. Tamai, J. L. Hobman, C. Constantinidou, K. Struhl, G. Church, and S. J. Busby. 2004. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J. Bacteriol. 186:6938-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, A., M. Bouaboula, P. Casellas, J. P. Liautard, and J. Dornand. 2003. Subversion and utilization of the host cell cyclic adenosine 5′-monophosphate/protein kinase A pathway by Brucella during macrophage infection. J. Immunol. 170:5607-5614. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, S., and D. Chatterji. 2005. Stress responses in mycobacteria. IUBMB Life 57:149-159. [DOI] [PubMed] [Google Scholar]
- 15.Kim, Y. R., S. Y. Kim, C. M. Kim, S. E. Lee, and J. H. Rhee. 2005. Essential role of an adenylate cyclase in regulating Vibrio vulnificus virulence. FEMS Microbiol. Lett. 243:497-503. [DOI] [PubMed] [Google Scholar]
- 16.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749-795. [DOI] [PubMed] [Google Scholar]
- 17.Krakow, J. S., and I. Pastan. 1973. Cyclic adenosine monophosphate receptor: loss of cAMP-dependent DNA binding activity after proteolysis in the presence of cyclic adenosine monophosphate. Proc. Natl. Acad. Sci. USA 70:2529-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, N., C. A. D'Souza, and J. W. Kronstad. 2003. Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol. 41:399-427. [DOI] [PubMed] [Google Scholar]
- 19.Li, C. C., D. S. Merrell, A. Camilli, and J. B. Kaper. 2002. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol. Microbiol. 43:1577-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebmann, B., S. Gattung, B. Jahn, and A. A. Brakhage. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269:420-435. [DOI] [PubMed] [Google Scholar]
- 21.Liebmann, B., M. Muller, A. Braun, and A. A. Brakhage. 2004. The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72:5193-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder, J. U., A. Schultz, and J. E. Schultz. 2002. Adenylyl cyclase Rv1264 from Mycobacterium tuberculosis has an autoinhibitory N-terminal domain. J. Biol. Chem. 277:15271-15276. [DOI] [PubMed] [Google Scholar]
- 23.Liu, H. 2002. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 292:299-311. [DOI] [PubMed] [Google Scholar]
- 24.Lowrie, D. B., P. S. Jackett, and N. A. Ratcliffe. 1975. Mycobacterium microti may protect itself from intracellular destruction by releasing cyclic AMP into phagosomes. Nature 254:600-602. [DOI] [PubMed] [Google Scholar]
- 25.Mangan, J. A., K. M. Sole, D. A. Mitchison, and P. D. Butcher. 1997. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 25:675-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 27.McCue, L. A., K. A. McDonough, and C. E. Lawrence. 2000. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10:204-219. [DOI] [PubMed] [Google Scholar]
- 28.McDonough, K. A., M. A. Florczyk, and Y. Kress. 2000. Intracellular passage within macrophages affects the trafficking of virulent tubercle bacilli upon reinfection of other macrophages in a serum-dependent manner. Tuber. Lung Dis. 80:259-271. [DOI] [PubMed] [Google Scholar]
- 29.Molle, V., M. Fujita, S. T. Jensen, P. Eichenberger, J. E. Gonzalez-Pastor, J. S. Liu, and R. Losick. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683-1701. [DOI] [PubMed] [Google Scholar]
- 30.Petersen, S., and G. M. Young. 2002. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70:3665-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pukkila-Worley, R., and J. A. Alspaugh. 2004. Cyclic AMP signaling in Cryptococcus neoformans. FEMS Yeast Res. 4:361-367. [DOI] [PubMed] [Google Scholar]
- 32.Rickman, L., C. Scott, D. M. Hunt, T. Hutchinson, M. C. Menendez, R. Whalan, J. Hinds, M. J. Colston, J. Green, and R. S. Buxton. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56:1274-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shenoy, A. R., and S. S. Visweswariah. 2006. Mycobacterial adenylyl cyclases: biochemical diversity and structural plasticity. FEBS Lett. 580:3344-3352. [DOI] [PubMed] [Google Scholar]
- 34.Spreadbury, C. L., M. J. Pallen, T. Overton, M. A. Behr, S. Mostowy, S. Spiro, S. J. Busby, and J. A. Cole. 2005. Point mutations in the DNA- and cNMP-binding domains of the homologue of the cAMP receptor protein (CRP) in Mycobacterium bovis BCG: implications for the inactivation of a global regulator and strain attenuation. Microbiology 151:547-556. [DOI] [PubMed] [Google Scholar]
- 35.Whitchurch, C. B., S. A. Beatson, J. C. Comolli, T. Jakobsen, J. L. Sargent, J. J. Bertrand, J. West, M. Klausen, L. L. Waite, P. J. Kang, T. Tolker-Nielsen, J. S. Mattick, and J. N. Engel. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 55:1357-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 37.Zeng, X., H. H. Saxild, and R. L. Switzer. 2000. Purification and characterization of the DeoR repressor of Bacillus subtilis. J. Bacteriol. 182:1916-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]