Abstract
Salmonella enterica is an important diarrheal pathogen, and infections may involve severe systemic sequelae depending on serovar- and host-specific factors. The molecular mechanisms underlying translocation of host-restricted and -specific serovars of S. enterica from the intestines to distal organs are ill defined. By surgical cannulation of lymph and blood vessels draining the distal ileum in cattle, S. enterica serovar Dublin was observed to translocate predominantly via mesenteric lymph nodes to efferent lymphatics in a manner that correlates with systemic virulence, since the fowl typhoid-associated serovar Gallinarum translocated at a significantly lower level. While both S. enterica serovars Dublin and Gallinarum were intracellular while in the intestinal mucosa and associated with major histocompatibility complex class II-positive cells, the bacteria were predominantly extracellular within efferent lymph. Screening of a library of signature-tagged serovar Dublin mutants following oral inoculation of calves defined the role of 36 virulence-associated loci in enteric and systemic phases of infection. The number and proportion of tagged clones reaching the liver and spleen early after oral infection were identical to the values in efferent lymph, implying that this may be a relevant mode of dissemination. Coinfection studies confirmed that lymphatic translocation requires the function of type III secretion system 1 (T3SS-1) but, remarkably, not T3SS-2. This is the first description of the mode and genetics of systemic translocation of serovar Dublin in its natural host.
Salmonella enterica is a facultative intracellular pathogen of animals and humans. More than 2,400 serovars exist, and these can be divided into three broad classes on the basis of their host specificity (reviewed in reference 37); ubiquitous serovars (e.g., S. enterica serovar Typhimurium) produce acute but self-limiting enteritis in a broad range of hosts, whereas host-specific serovars (e.g., S. enterica serovar Gallinarum) are associated with severe systemic disease in a single species, which may not involve diarrhea. Host-restricted serovars are primarily associated with systemic disease in one host but may cause disease in a limited number of other species (e.g., S. enterica serovar Dublin in cattle and humans) (reviewed in reference 39). The molecular basis of the differential virulence, tissue tropism, and host range of S. enterica serovars is poorly defined.
Genome-wide mutagenesis has indicated that enteric and systemic virulence of Salmonella in mice and cattle is influenced by Salmonella pathogenicity island 1 (SPI-1) and SPI-2, which, respectively, encode type III secretion system 1 (T3SS-1) and T3SS-2 (2, 17, 22, 26, 36; reviewed in reference 16). These systems inject bacterial proteins into host cells which subvert cellular pathways to the benefit of the pathogen (reviewed in reference 14). T3SS-1 promotes invasion of intestinal M cells and enterocytes by rearrangement of the subcortical actin cytoskeleton, whereas T3SS-2 facilitates the replication of intracellular bacteria within Salmonella-containing vacuoles. Targeted mutagenesis has confirmed the role of T3SS-2 during systemic infection, for example, for serovar Dublin in calves (2). In contrast, T3SS-1 primarily influences the enteric phase of infection, with structural components being required for invasion (42) and induction of intestinal inflammatory and secretory responses (40, 41) in a bovine ligated ileal loop model. T3SS-1-secreted effectors act in concert to promote these events (21, 44). Experiments using cattle and streptomycin-pretreated mice indicate that T3SS-2 also plays a role in the enteric phase of infection (2, 8, 26, 36). Consistent with this, SPI-2 gene expression is induced in the intestinal lumen (5). It has been suggested that early induction of T3SS-2 prepares Salmonella to persist in macrophages during translocation to distal sites; however, little evidence exists in support of this.
Salmonella may penetrate the intestinal epithelium by a variety of routes. Following oral inoculation of mice, S. enterica serovar Typhimurium has been observed to invade and destroy M cells located in the follicle-associated epithelium in a T3SS-1-dependent manner (20, 30). Intestinal villous M cells dispersed in the mucosa (18) and solitary intestinal lymphoid tissue (15) have also been observed to take up Salmonella. Invasive Salmonella can also penetrate enterocytes to gain access to the subepithelial space (12, 35), and localized damage at the site of entry as a result of host or bacterial processes may facilitate translocation via interstitial spaces. In addition, there is evidence for an alternative route of translocation since noninvasive serovar Typhimurium mutants can still reach the spleen following oral dosing of mice (13). This process is believed to involve T3SS-1-independent transport of bacteria across the epithelium via CD18+ phagocytic cells, such as dendritic cells (DCs) and/or tissue macrophages (38). DCs can open tight junctions between epithelial cells, project dendrites into the intestinal lumen to sample Salmonella (27, 31), and migrate via blood or lymph to distal sites. It was recently reported that serovar Typhimurium promotes phagocyte motility by a process dependent on the SPI-2 gene srfH, and this was correlated with increased systemic dissemination in mice (43). The relative importance of these routes and the role of T3SSs are not fully understood and may vary depending on the serovar and host. Here, we characterize the mode and genetics of systemic translocation of the host-restricted S. enterica serovar Dublin in its natural bovine host.
MATERIALS AND METHODS
Bacterial strains.
S. enterica serovars Dublin 3246 and Gallinarum SG9 (29) and derivatives were cultured in Luria-Bertani (LB) medium supplemented as appropriate with ampicillin ([Amp] 100 μg ml−1), kanamycin ([Kan] 50 μg ml−1), or nalidixic acid ([Nal] 20 μg ml−1). Fully virulent signature-tagged mutants of serovar Dublin 3246 (mutants 27E4, 27E2, and 27F11) and mutants lacking sipD (26C8) and sseD (29D11) were isolated from a library screened by intravenous inoculation of calves (2). Signature-tagged mutants of serovar Gallinarum SG9 (mutants 12D10, 12B6, and 14C3) that were fully virulent in chicks were a kind gift from J. Olsen (Royal Veterinary and Agricultural University, Frederiksberg C, Denmark).
Experimental animals.
Animal experiments were conducted according to the requirements of the Animal (Scientific Procedures) Act 1986 (license 30/1998) with the approval of the local ethical review committee. Friesian bull calves aged 25 to 30 days were reared, housed, and confirmed to be culture negative for Salmonella as described previously (29).
Surgical cannulation.
A laparotomy was performed under terminal anesthesia, ca. 80 cm of the distal ileal loop was ligated, and an efferent lymphatic vessel draining a suitable mesenteric lymph node (MLN) was dissected and cannulated as described previously (29). Venules draining the distal ileal mucosa were also cannulated, and jugular blood was sampled every 2 h. Preinfection blood and lymph were collected, and the loop was inoculated with 3 × 1010 to 1 × 1011 CFU of strain 3246 or SG9 or 1:1 mixes of strain 3246 and isogenic mutants. Lymph and blood were collected continuously in 2-h intervals for 12 h, and bacteria were enumerated by plating serial dilutions on MacConkey agar. Gentamicin-protected bacteria were enumerated following incubation of the sample with 100 μg ml−1 gentamicin for 30 min at 37°C, followed by centrifugation (200 × g for 3 min at room temperature) and two washes with phosphate-buffered saline (PBS). This method was previously shown to kill extracellular Salmonella organisms without affecting the viability of intracellular bacteria (3). All samples were mixed with 1% (vol/vol) Triton X-100 (9:1 ratio) prior to plating on MacConkey agar and overnight incubation at 37°C.
Immunohistochemistry.
Biopsies of ileal mucosa were fixed in 4% (wt/vol) paraformaldehyde in PBS 12 h after inoculation of ligated ileal loops with approximately 3 × 1010 CFU of Salmonella or 24 h after oral inoculation with approximately 1 × 109 CFU of Salmonella. Two calves were used for each combination of serovar and inoculation route. Agar-embedded lymph was prepared by mixing 1 ml of lymph to 1 ml of molten 10% (wt/vol) agar in PBS containing 4% (wt/vol) paraformaldehyde, followed by centrifugation at 200 × g for 3 min. Sections of tissue (30 to 50 μm) and agar-embedded lymph (100 μm) were cut using a vibrating microtome. These were permeabilized in 0.1% saponin in PBS, immunolabeled, and imaged by confocal laser scanning microscopy as described previously (24). The antibodies are described in the figure legends and were obtained from Molecular Probes (Leiden, The Netherlands) unless stated otherwise. Two sections of each sample were analyzed, together with a cyto-centrifuged preparation from lymph.
Construction of signature-tagged serovar Dublin mutants.
Thirty-six uniquely tagged mini-Tn5Km2 mutants of serovar Dublin 3246 with insertions in known or putative virulence-associated genes were created by λRed recombinase-mediated integration of linear PCR products. The PCR products mostly comprised mini-Tn5Km2 insertions with 150 to 200 bp flanking DNA and were amplified from a library of serovar Typhimurium 4/74 mutants previously screened in calves, chickens, and pigs (6, 26). Additional mutants were identified from the 4/74 library by hierarchical PCR using transposon- and gene-specific primers. Products were digested with DpnI, purified using spin columns, and electroporated into serovar Dublin 3246 harboring the λRed helper plasmid pKD46 (11), and Kanr Nalr Amps transformants were selected at 37°C. Mutations in sodC1, sodC2, and slyA were constructed by amplification of transposons with compatible signature tags by PCR using primers which incorporate 40-nucleotide gene-specific homology extensions, designed to replace an internal portion of the gene with the transposon. Insertion sites were confirmed by PCR, and sequence analysis and the data are available from the authors on request.
Screening of signature-tagged mutants following oral inoculation of calves.
Forty-two Salmonella mutants (see Tables 1 and 2) were inoculated into LB broth with Kan and Nal and incubated overnight at 37°C. The mutants were pooled, and an aliquot was removed for preparation of “input pool” genomic DNA, as described previously (17). Calves were orally inoculated with the pool (1.6 × 1010 CFU for calves A to C and 3.6 × 108 CFU for calves D and E) in 20 ml of antacid [5% (wt/vol) Mg(SiO3)3, 5% (wt/vol) NaHCO3, and 5% (wt/vol) MgCO3 in H2O]. Calves were anesthetized at 11 h (A to C) or 4 days (D and E) postinoculation, and the distal ileal loop was exteriorized but not ligated. Efferent lymph vessels were cannulated, and lymph was collected in 2-h intervals at 12 to 24 h postinoculation (A to C) and for 2 h at 4 days postinoculation (D and E). Lymph samples and homogenates of tissue collected at postmortem examination (ileal mucosa, MLNs, liver, and spleen) were plated onto MacConkey agar with Kan and Nal to isolate “output pool” bacteria. For each site, ca. 2,000 to 8,000 colonies were pooled for preparation of output pool genomic DNA. Amplification of labeled tags from input and output pools and dot blot hybridizations were performed as described previously (26).
TABLE 1.
Mean attenuation scores of signature-tagged serovar Dublin 3246 mutants in efferent lymph samples at intervals following oral inoculation of calves
| Mutant | Mean attenuation score in efferent lymph at the indicated time postinoculationa
|
||||||
|---|---|---|---|---|---|---|---|
| 12-14 h | 14-16 h | 16-18 h | 18-20 h | 20-22 h | 22-24 h | 4 days | |
| T3SS-1 structural and translocon components | |||||||
| prgHb | 0 | 0 | 0 | 0.3 | 0.3 | 0.3 | 0 |
| sipDb | 0 | 1 | 0.7 | 0.3 | 0.3 | 0.3 | 0 |
| spaR | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| sipC | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| sipA | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T3SS-1-secreted effectors | |||||||
| sptP | 3 | 3 | 2.7 | 3 | 2 | 3 | 3 |
| avrA | 2.3 | 2.3 | 2.7 | 2.3 | 2.3 | 2.7 | 3 |
| sopA | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| sopB | 1.7 | 2 | 2 | 2.3 | 2.3 | 2 | 3 |
| sopE2 | 2.7 | 3 | 2.7 | 3 | 2 | 2.7 | 3 |
| T3SS-2 structural and translocon components | |||||||
| sseDb | 3 | 3 | 3 | 3 | 2.7 | 2.7 | 1 |
| ssaJ | 2.3 | 2.7 | 2 | 2 | 1.3 | 2.7 | 0 |
| sseC | 1.7 | 2 | 2 | 2.3 | 1.7 | 2 | 0 |
| sseD | 3 | 3 | 3 | 3 | 2.3 | 3 | 2 |
| sseF | 3 | 2.7 | 2.3 | 3 | 2.7 | 2.7 | 2 |
| T3SS-2-secreted effectors | |||||||
| sseI | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| sifA | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| sifB | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| sseK1 | 2.7 | 3 | 3 | 3 | 3 | 3 | 2 |
| SPI-3 genes | |||||||
| mgtC | 3 | 3 | 3 | 3 | 3 | 3 | 2 |
| rmbA | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| SPI-4 genes | |||||||
| siiD | 2 | 1.3 | 1.3 | 2.3 | 2 | 1.7 | 1 |
| siiE | 0.7 | 0.3 | 1 | 1.3 | 1.3 | 1.7 | 0 |
| siiF | 1 | 1.3 | 1.3 | 1.7 | 1.3 | 1 | 1 |
| SPI-5 genes | |||||||
| pipB | 3 | 3 | 2.7 | 3 | 2.7 | 3 | 3 |
| pipC | 3 | 2.7 | 3 | 3 | 3 | 3 | 3 |
| pipD | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Virulence plasmid genes | |||||||
| spvB | 2 | 2.7 | 1.3 | 2 | 1.3 | 2 | 0 |
| spvR | 1.7 | 2 | 2 | 1.7 | 1.3 | 2 | 0 |
| Other virulence genes | |||||||
| virK | 3 | 2.3 | 2.3 | 2.7 | 2.3 | 2.7 | 3 |
| somA | 2.3 | 2 | 2.3 | 2 | 2.3 | 2 | 2 |
| pmrAB | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| mig-14 | 3 | 2.7 | 2.7 | 2.7 | 3 | 3 | 0 |
| sodC1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| sodC2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| slyA | 2.7 | 2.3 | 2.7 | 2.7 | 2 | 2.3 | 2 |
| Virulent serovar Gallinarum controls | |||||||
| 12D10 | 1.3 | 1.3 | 0.7 | 0.7 | 1 | 0 | 0 |
| 12B6 | 1 | 0.7 | 0.7 | 0 | 0.3 | 0 | 0 |
| 14C3 | 0.7 | 0.7 | 0.3 | 0 | 0 | 0 | 0 |
| Virulent serovar Dublin controls | |||||||
| 27E4 | 3 | 3 | 3 | 2.7 | 3 | 3 | 3 |
| 27E2 | 3 | 3 | 3 | 2.7 | 3 | 3 | 3 |
| 27F11 | 3 | 2.7 | 3 | 3 | 2.7 | 3 | 3 |
Values represent the mean score per mutant of duplicate blots from 3 calves (12- to 24-h samples) or 2 calves (4-day samples) according to the following scale: 0, no signal; 1, ca. one-third of input signal; 2, ca. two-thirds of input signal; 3, signal equivalent to input.
As tested in CI experiments.
TABLE 2.
Mean attenuation scores of signature-tagged serovar Dublin 3246 mutants in tissue samples following oral inoculation of calves
| Mutant | Mean attenuation score in tissue at the indicated time postinoculationa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Ileal mucosa
|
MLN
|
Liver
|
Spleen
|
|||||
| 24 h | 4 days | 24 h | 4 days | 24 h | 4 days | 24 h | 4 days | |
| T3SS-1 structural and translocon components | ||||||||
| prgHb | 0.7 | 0.5 | 0.3 | 0 | 0.3 | 0 | 0 | 0 |
| sipDb | 0.7 | 0.5 | 0 | 0.5 | 0.3 | 0.5 | 0 | 0 |
| spaR | 0 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| sipC | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 | 0 |
| sipA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T3SS-1-secreted effectors | ||||||||
| sptP | 3 | 1.5 | 2.7 | 1 | 2.7 | 1 | 2.3 | 2.5 |
| avrA | 2.3 | 3 | 2.7 | 2.5 | 2 | 2.5 | 2 | 3 |
| sopA | 3 | 3 | 3 | 3 | 3 | 2.5 | 2.7 | 2.5 |
| sopB | 2.7 | 3 | 2.7 | 3 | 2 | 1.5 | 1.3 | 1.5 |
| sopE2 | 3 | 3 | 3 | 3 | 2.3 | 1.5 | 2.3 | 3 |
| T3SS-2 structural and translocon components | ||||||||
| sseDb | 3 | 1.5 | 2.7 | 1 | 2.3 | 1 | 2 | 1 |
| ssaJ | 3 | 0.5 | 2 | 0 | 3 | 0 | 2 | 0 |
| sseC | 2.3 | 1 | 1.7 | 0 | 1 | 0 | 0.7 | 0 |
| sseD | 3 | 1.5 | 2.7 | 1.5 | 3 | 1 | 2.7 | 2 |
| sseF | 3 | 2 | 2.7 | 2 | 1.7 | 1 | 1.7 | 0 |
| T3SS-2-secreted effectors | ||||||||
| sseI | 3 | 3 | 3 | 3 | 3 | 2.5 | 3 | 3 |
| sifA | 3 | 2 | 3 | 2 | 2.7 | 2 | 2.7 | 1 |
| sifB | 3 | 3 | 3 | 3 | 3 | 2.5 | 3 | 3 |
| sseK1 | 3 | 3 | 2.7 | 2.5 | 2 | 3 | 2.7 | 2.5 |
| SPI-3 genes | ||||||||
| mgtC | 3 | 2.5 | 3 | 1 | 3 | 0.5 | 3 | 0 |
| rmbA | 3 | 3 | 3 | 3 | 2.7 | 3 | 3 | 2.5 |
| SPI-4 genes | ||||||||
| siiD | 2 | 1 | 2 | 1 | 2.7 | 2.5 | 1.3 | 3 |
| siiE | 1.7 | 1 | 2.3 | 1 | 1 | 2.5 | 2.3 | 2 |
| siiF | 1.3 | 1.5 | 1.7 | 1 | 0.7 | 2 | 2.3 | 2.5 |
| SPI-5 genes | ||||||||
| pipB | 3 | 3 | 2.7 | 3 | 2 | 3 | 3 | 3 |
| pipC | 3 | 3 | 3 | 2.5 | 3 | 2 | 2.3 | 3 |
| pipD | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Virulence plasmid genes | ||||||||
| spvB | 3 | 1.5 | 2.3 | 2 | 2 | 0.5 | 1.7 | 0 |
| spvR | 2.3 | 3 | 2 | 2.5 | 1 | 0.5 | 1.7 | 0 |
| Other virulence genes | ||||||||
| virK | 3 | 2 | 2.3 | 2 | 2.7 | 2 | 2.3 | 2 |
| somA | 2.3 | 2 | 2.3 | 2.5 | 1.7 | 0.5 | 1.7 | 1 |
| pmrAB | 3 | 3 | 3 | 3 | 2.7 | 2.5 | 2.7 | 2.5 |
| mig-14 | 3 | 2 | 3 | 1.5 | 2 | 0.5 | 1.7 | 1 |
| sodC1 | 3 | 3 | 3 | 2.5 | 3 | 2.5 | 2.7 | 2.5 |
| sodC2 | 3 | 3 | 3 | 2.5 | 3 | 2.5 | 2.7 | 2.5 |
| slyA | 2.7 | 2 | 2.7 | 1 | 2.3 | 1.5 | 1.7 | 1.5 |
| Virulent serovar Gallinarum controls | ||||||||
| 12D10 | 0 | 0 | 0.3 | 0 | 0 | 0 | 0 | 0 |
| 12B6 | 0.3 | 0 | 0.3 | 0 | 0 | 0 | 0 | 0 |
| 14C3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Virulent serovar Dublin controls | ||||||||
| 27E4 | 3 | 3 | 3 | 3 | 2.7 | 3 | 2.3 | 3 |
| 27E2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2.5 |
| 27F11 | 3 | 3 | 3 | 3 | 2.7 | 3 | 3 | 3 |
Values represent the mean score per mutant of duplicate blots from 3 calves (24-h samples) or 2 calves (4-day samples), according to the following scale: 0, no signal; 1, ca. one-third of input signal; 2, ca. two-thirds of input signal; 3, signal equivalent to input.
As tested in CI experiments.
Determination of CIs.
Salmonella strains were grown for 24 h at 25°C in LB broth, subcultured 1:3 into LB broth, and incubated at 37°C for 90 min to induce expression of T3SS-1. Wild-type (wt) and mutant strains were mixed in equal numbers (ca. 3 × 1010 CFU) in a total volume of 50 ml and injected into ligated ileal loops (three calves per pair of strains). The wt and mutant bacteria were enumerated by plating serial dilutions of the inoculum, lymph, or tissue homogenates on MacConkey agar with Nal and with Nal plus Kan. Competitive indices (CIs) were calculated as the ratio of mutant to wt in the output pool divided by the ratio of mutant to wt in the inoculum. The data were analyzed by a paired t test (Minitab Inc., State College, PA).
RESULTS
Serovar Dublin translocates from bovine MLNs via efferent lymph in a predominantly cell-free niche.
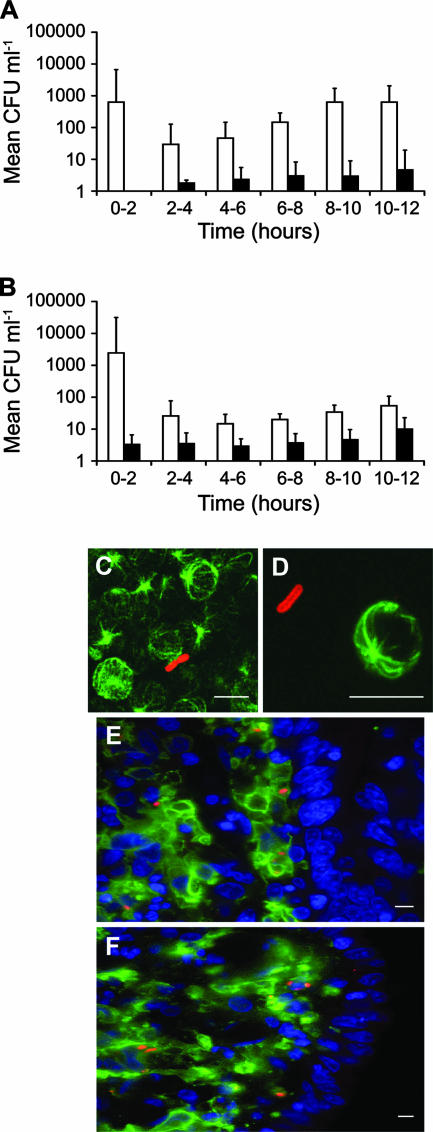
To investigate the route(s) of systemic dissemination of serovar Dublin in cattle, we made use of a novel cannulation model developed in our laboratory (29). Venules and efferent lymph vessels draining the distal ileal loops of calves were cannulated under terminal anesthesia, and blood and efferent lymph were sampled continuously for 12 h after inoculation of the loop with either serovar Dublin 3246 or serovar Gallinarum SG9 (four calves per strain). These serovars have been shown to invade the ileal mucosa and induce enteritis at comparable levels in bovine ligated ileal loops and following oral dosing (29), though serovar Dublin persists within bovine intestinal mucosa in greater numbers. Recovery of both serovars from cannulated venules was lower than the accurate limit of bacterial detection (<102 CFU ml−1). Samples of jugular blood from cannulated calves were intermittently positive by enrichment in some animals as early as 2 h after inoculation. In contrast, by 2 h postinoculation both serovars were present in efferent lymph in high numbers (Fig. 1A and B). At 6 to 12 h, serovar Dublin 3246 was recovered from lymph in significantly higher numbers than serovar Gallinarum SG9 (P value of <0.05) (Fig. 1A and B), consistent with the systemic virulence of strain 3246 and avirulence of strain SG9 following oral dosing of calves (29).
FIG. 1.
Recovery of serovar Dublin 3246 (A) and serovar Gallinarum SG9 (B) from efferent lymph untreated (open bars) or treated (black bars) with gentamicin up to 12 h after inoculation of bovine ileal loops. Each point was derived from the mean (± standard error of the mean) of triplicate lymph samples from four calves per strain. Representative confocal micrographs of cell-free serovar Dublin in a cyto-centrifuged preparation (C) and agar-embedded efferent lymph (D) 12 h after inoculation are shown. Bacteria were labeled with rabbit anti-Salmonella lipopolysaccharide and anti-rabbit immunoglobulin G-AlexaFluor 568 (red). Host cells were labeled with mouse anti-α-tubulin and anti-mouse immunoglobulin G-AlexaFluor 488 (green). Comparable results were obtained at 4, 6, 8, and 10 h after loop inoculation. Serovar Dublin 3246 (E) and serovar Gallinarum SG9 (F) associate with MHC-II+ cells within the lamina propria 12 h after infection of bovine ileal loops. Bacteria were labeled as above. MHC-II+ cells were labeled with CC158 monoclonal antibody (IAH Compton) and anti-mouse immunoglobulin G-AlexaFluor 488 (green). Host cell nuclei (blue) were stained with TO-PRO-3. Scale bars, 10 μm (C and D) and 5 μm (E and F).
Treatment of lymph ex vivo with 100 μg ml−1 gentamicin (a concentration established to kill extracellular, but not intracellular, bacteria) resulted in a 90 to 99% reduction in recoveries of both serovars over the time course (P < 0.01) (Fig. 1A and B), indicating that the majority of bacteria were free in the lymph. The predominantly cell-free location of serovar Dublin in efferent lymph was supported by immunocytochemistry of detergent-permeabilized cyto-centrifuged and agar-embedded lymph (Fig. 1C and D).
The association of Salmonella with cells in the ileal mucosa early after infection was investigated to determine if differences in the magnitude of lymphatic translocation of serovars Dublin and Gallinarum reflect association with different cell types following invasion. Mucosal sections were labeled to detect Salmonella and eukaryotic cell markers (Materials and Methods). Bacteria were predominantly located in the lamina propria 12 h postinoculation and were rarely associated with the enterocyte layer (Fig. 1E and F). No bacteria were found within the lymphoid follicles associated with the Peyer's patch tissue, the muscularis mucosa, or the submucosa. Bacteria of both serovars were exclusively located within major histocompatibility complex class II (MHC-II)-positive cells with features of phagocytic antigen-presenting cells (Fig. 1E and F). Sequential 0.25-μm optical Z-plane sections confirmed the bacteria to be intracellular (data not shown). Even though large amounts of inoculum were used, relatively few bacteria could be detected in tissue sections, precluding a statistical analysis of the data.
Analysis of the role of virulence-associated loci in lymphatic translocation of serovar Dublin and establishment of systemic infections.
We next investigated the genetic basis of translocation of serovar Dublin to and through MLNs in calves. A library of defined signature-tagged mutants of serovar Dublin 3246 was constructed (Materials and Methods). The library contained a total of 36 mutants (each marked with a unique oligonucleotide tag) lacking T3SS-1 and T3SS-2 components and cognate secreted effectors, SPI-3, SPI-4, and SPI-5 genes, plasmid virulence genes, and other putative virulence loci. The library also included three calf-virulent tagged serovar Dublin 3246 strains as positive controls and three chicken-virulent tagged serovar Gallinarum SG9 strains as negative controls.
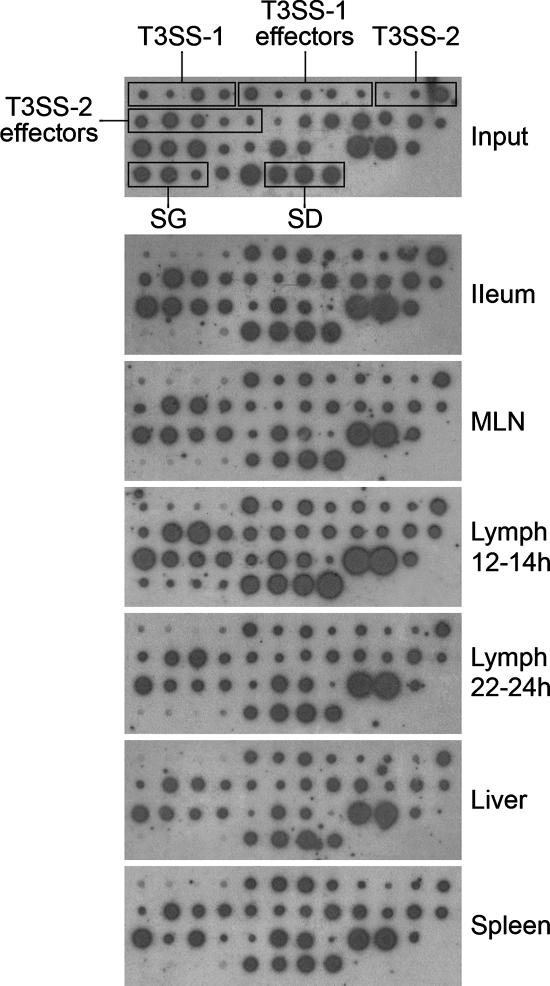
The fate of each mutant was followed after oral inoculation by amplification and hybridization of the unique tags from the input and output pools (Materials and Methods). Three calves (A to C) were anesthetized ca. 11 h after oral inoculation, and a cannula was inserted in an efferent lymph vessel draining a MLN(s) in the distal ileal loop. Efferent lymph was collected in 2-h intervals at 12 to 24 h postinoculation. Efferent lymph vessels of two other calves (D and E) were cannulated 4 days postinoculation. Biopsies from ileal mucosa, MLNs, liver, and spleen were collected at the end of each experiment. Duplicate dot blot hybridizations were performed with 32P[dCTP]-labeled tags amplified from bacteria in the input and output pools from each site in the five calves. Representative dot blots from one calf showing the fate of mutants 12 to 24 h after oral inoculation are shown in Fig. 2. To aid the assignment of attenuation scores for each of the mutants across duplicate blots and replicate animals, hybridization signals were compared to those from the corresponding input pool and scored 0 to 3 (where 0 indicates no signal, 1 indicates ca. one-third input signal, 2 indicates ca. two-thirds input signal, and 3 indicates a signal equivalent to the input signal). The mean scores for each mutant in the lymph and tissues are shown in Tables 1 and 2, respectively. Differences in signal intensity for each mutant in the input reflect variations in cytosine content and the efficiency of hybridization of tags under the conditions used, not the size of the mutant population.
FIG. 2.
Analysis of the role of virulence loci in invasion, spread to MLNs, lymphatic translocation, and infection of organs. Representative dot blots from calf A show the prevalence of Salmonella mutants in tissues at 24 h and in efferent lymph at 12 to 14 h and 22 to 24 h postinoculation relative to the input. SD, serovar Dublin controls; SG, serovar Gallinarum controls.
The three tagged serovar Dublin controls were present in efferent lymph and all enteric and systemic sites examined at both 1 and 4 days after oral inoculation. In contrast, the serovar Gallinarum controls were recovered in smaller quantities than the input amounts from the ileal mucosa, MLNs, and efferent lymph at early time intervals and were not detected in efferent lymph after 22 h. The serovar Gallinarum controls were cleared from the ileal mucosa and MLNs by 4 days postinoculation and were not detected in the liver or spleen. Gentamicin treatment of lymph samples collected 12 to 24 h or 4 days after oral inoculation eliminated >98% of bacteria. These data are consistent with our findings in surgically manipulated animals (above) and bacterial recoveries following oral inoculation of calves with the parent strains (29) and indicate that translocation occurs in a predominantly cell-free niche following exposure by the natural route.
A significant finding from these experiments was that the tagged mutants represented in the liver and spleen at 24 h postinoculation were the same ones, in the same proportions, as found in MLNs draining the distal ileal loop and efferent lymph (Fig. 2 and Tables 1 and 2). Thus, all the serovar Dublin mutants that persisted in large numbers at 4 days postinfection in the spleen (19 mutants with attenuation scores of 2.5 or more) were detected in similar proportions in the MLNs and efferent lymph, implying that dissemination via draining MLNs and efferent lymph is relevant in the seeding of distal organs.
Mutants lacking T3SS-1 structural or translocon components were significantly impaired in their ability to translocate from the intestine to the MLNs, efferent lymph, liver, or spleen. Since multiple T3SS-1 mutants exhibited the same phenotype, it is unlikely that the attenuation observed is due to polar or second-site effects. Mutants lacking individual T3SS-1 effectors were detected in the lymph and reached the liver and spleen at 1 and 4 days postinoculation, albeit in reduced numbers in the case of sptP and sopB (Tables 1 and 2). This implies redundancy in the effector repertoire and/or the need for effectors to act in concert. In marked contrast, all T3SS-2 structural and effector mutants were present at all sites 1 day postinoculation, indicating that early dissemination of serovar Dublin from the intestines does not require SPI-2. By day 4, many T3SS-2 structural mutants were cleared from the liver and spleen and were less abundant in the ileal mucosa, MLNs, and efferent lymph, confirming that T3SS-2 plays a role in persistence at enteric sites (8, 26). The abundance of the sifA mutant was reduced in the spleen by day 4, suggesting a possible role in systemic persistence.
Screening of the library further showed that mutation of the SPI-3 genes mgtC and rmbA had no effect on recovery from tissues 24 h postinoculation; however, by day 4 the mgtC mutant was either present at reduced levels or was cleared from the MLNs, liver, and spleen (Table 2). Disruption of the SPI-4-encoded T1SS and its secreted substrate SiiE impaired persistence in the ileal mucosa, MLNs, and efferent lymph from 1 to 4 days. However, the mutants were well represented in the liver and spleen, consistent with the role of SPI-4 in enteric but not systemic disease in calves (25, 26). Mutations in SPI-5 enteritis-associated genes (pipB, pipC, and pipD) had no apparent effect on the persistence or translocation of serovar Dublin. Plasmid virulence genes influenced systemic persistence as expected, since spvR and spvB mutants were present at reduced levels or were cleared from efferent lymph, liver, and spleen by 4 days postinoculation. Mutation of somA, mig-14, and slyA led to reduced numbers of bacteria in the liver and spleen by 4 days postinoculation. The abundance of the mig-14 mutant was also reduced in the MLNs, and the mutant was absent in efferent lymph at this time. In contrast, no significant effect on virulence was observed by mutation of virK, pmrAB, sodC1 or sodC2. Mutants lacking spv, mgtC, mig-14, somA, and slyA, like T3SS-2 mutants, exhibited reduced persistence in the ileal mucosa, suggesting that they may play a role in sustaining a replicating pool of bacteria required to seed the distal organs at later time points.
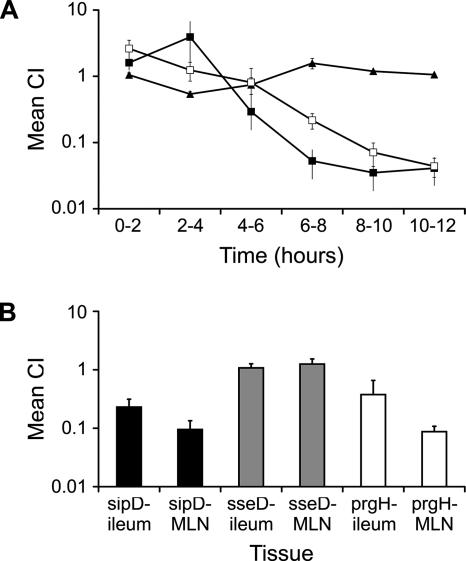
Competition experiments confirm that lymphatic translocation does not require T3SS-2.
To verify these findings and to validate the attenuation scores assigned by detection of signature tags, selected serovar Dublin T3SS-1 and T3SS-2 mutants were analyzed in competition experiments. Ligated ileal loops were coinfected with the parent strain and isogenic sipD (T3SS-1), prgH (T3SS-1), or sseD (T3SS-2) mutants (three calves per mutant in a 1:1 ratio with the parent) (see Materials and Methods). Efferent lymph was collected continuously in 2-h intervals for 12 h, and the CIs were derived at each interval. The serovar Dublin sseD mutant, which was previously found to be avirulent when injected intravenously in calves (2), translocated via efferent lymph in comparable numbers to the parent throughout (Fig. 3A). In contrast, the sipD and prgH mutants were found in significantly lower numbers than the wt during the period in which translocation correlates with host specificity (CIs at 8 to 12 h less than 0.1; P values of <0.05). Analysis of ileal mucosa and MLN at postmortem examination revealed that the sseD mutant colonized both sites as well as the parent, whereas the sipD and prgH mutants colonized the ileal mucosa and MLN in significantly lower numbers than the wt (Fig. 3B) (P values of <0.05). These results confirmed that early lymphatic translocation is dependent on T3SS-1 but independent of T3SS-2.
FIG. 3.
T3SS-1, but not T3SS-2, is required for early lymphatic translocation of serovar Dublin. (A) Coinfection experiments comparing the ability of serovar Dublin T3SS-1 and T3SS-2 mutants to compete with wt during lymphatic translocation. The graph shows the CIs for the prgH (□), sipD (▪), and sseD (▴) mutants relative to wt in efferent lymph up to 12 h after inoculation. Each point represents the mean ± standard error of the mean from three calves. (B) CIs for serovar Dublin sipD, sseD, and prgH mutants relative to wt in ileal mucosa and MLNs collected 12 h after loop inoculation. Bars represent the means ± standard errors of the means of triplicate tissue samples from three calves per mutant.
DISCUSSION
The global burden of human typhoidal salmonellosis is estimated at 21.6 million cases and 216,510 deaths per annum (10), and systemic S. enterica infections are a significant economic and welfare issue in animals (39). The mechanisms by which host-specific and -restricted serovars translocate from the intestines and persist at distal sites are poorly understood. Studies of this process have mostly relied on oral inoculation of Ity-susceptible inbred mice with serovar Typhimurium. In the murine model, serovar Typhimurium appears to translocate by at least two routes: in addition to its ability to invade M cells and gain access to gut-associated lymphoid tissue and regional lymph nodes, it can also translocate to the bloodstream within CD18+ phagocytes in a manner that does not require a functional T3SS-1 (38). It has long been assumed that intracellular dissemination to distal sites is facilitated by T3SS-2. By using a bovine cannulation model, we have established that serovar Dublin, a natural systemic pathogen of cattle, translocates from the distal ileum mainly via draining lymphatics in a cell-free niche in a manner that requires T3SS-1 but, remarkably, not T3SS-2. Serovar Gallinarum, which causes fowl typhoid, translocated significantly less well via the lymphatics at 6 to 12 h after loop inoculation and did not reach the liver and spleen, despite the fact that it is known to invade the bovine intestinal mucosa and induce intestinal secretory and inflammatory responses at levels comparable to serovar Dublin (29). Thus, an ability to persist in the intestinal mucosa and translocate to and through MLNs correlates with host specificity in calves.
Following inoculation of distal ileal loops, serovar Dublin was found in draining venules and jugular blood infrequently and only by enrichment. Thus, early translocation from the distal ileum appears to occur mostly via the lymphatic system, consistent with early observations in mice (7) and the large numbers of serovar Dublin organisms detected in MLNs after oral inoculation of calves (29). Samples of jugular blood were intermittently positive by enrichment in some calves as early as 2 h after inoculation, though this may result from deposition of lymph into the bloodstream via the thoracic duct. Although Salmonella organisms are found inside cells in the intestinal mucosa (12, 42; also this study) and liver and spleen (32), serovar Dublin was predominantly extracellular in efferent lymph. Consistent with our findings, using an ovine pseudo-afferent lymph duct cannulation model, Bonneau et al. (4) recently found that an attenuated S. enterica serovar Abortusovis vaccine strain injected subcutaneously in the tongue, cheek, and lips traveled largely free in submaxillary lymph vessels during the first 90 min (88% cell free) but was later found associated with some cell types, including monocytes, granulocytes, and DCs (4).
The data herein provide the first description of the genetic basis of translocation to and through intestinal MLNs by a host-restricted serovar in its natural host exposed via the natural route. The finding that mutation of T3SS-1 significantly attenuated the ability of serovar Dublin to translocate via efferent lymphatics implies that passive sampling of bacteria from the intestinal lumen by DCs or M cells is unlikely to be a major mode of delivery to the lymph. While we cannot preclude the possibility that bacterial entry via such cells may occur, the data indicate that the functions of T3SS-1 (forced phagocytosis and induction of enteritis) are necessary for delivery of serovar Dublin to and subsequently through MLNs. It is noteworthy, however, that following instillation of the prgH or sipD mutants in a 1:1 ratio with the parent strain into distal ileal loops, the wt and T3SS-1 mutant strains were detected in equivalent and relatively high numbers in the first 2 h after loop inoculation. Thus, an initial wave of translocation may occur that is independent of T3SS-1; however, the relevance of this event in establishment of systemic infection should be approached with caution since in the loop model extremely high numbers of bacteria are held statically over the mucosa, and serovar Gallinarum also displayed an early burst of translocation (Fig. 1B), even though it is known not to colonize the distal organs 24 h postinfection (29; also this study). One cannot simply explain the role of T3SS-1 in lymphatic translocation as being due to its role in intestinal invasion, since gentamicin protection assays in a bovine ligated loop model of infection have indicated that serovars Dublin 3246 and Gallinarum SG9 invade the ileal mucosa at comparable initial rates (29). It is possible, however, that T3SS-1 is required to establish a replicating pool in the intestinal mucosa, as T3SS-1 has been reported to influence intracellular proliferation and vacuole biogenesis in cultured epithelial cells (34), and some effectors remain active long after bacterial entry. It is therefore conceivable that differences in the repertoire, sequence, or expression of T3SS-1 and cognate effectors in vivo may dictate the differential virulence of serovars Dublin and Gallinarum in cattle. It remains unclear how serovar Dublin arrives at draining MLNs or if T3SS-1-mediated lysis of phagocytic cells is a relevant mode of escape of Salmonella into a cell-free niche, as proposed (23).
It is widely assumed that dissemination of Salmonella to distal sites is facilitated by T3SS-2, since it is required for persistence at systemic sites in mice (33), is induced in the intestinal lumen (5), and exhibits contextual regulation to promote systemic virulence (9). The finding that T3SS-2 is dispensable for early systemic translocation was surprising, given that the bacteria were readily detected inside lamina propria cells 12 to 24 h after infection by confocal microscopy. Although T3SS-2 plays a key role in intracellular proliferation in epithelial cells and macrophages, it is not required for replication in murine DCs (19). In this respect, association of serovar Dublin with cells with features of DCs early after invasion of the bovine intestinal epithelium may be highly significant in explaining the apparent absence of a role for SPI-2 genes in early systemic translocation. Consistent with the role of T3SS-2 in bovine intestinal pathogenesis at 5 days after ileal loop inoculation (8) and following oral inoculation of calves (2, 26), T3SS-2 mutants translocated via lymph to the organs less well than the parent strains by 4 days postinoculation. Thus, T3SS-2 may facilitate systemic spread at later times by maintaining a pool of viable bacteria at enteric sites, but it does not appear to be required for lymphatic translocation per se.
Our recent data indicate that systemic virulence of the host-restricted S. enterica serovar Choleraesuis, following oral inoculation of pigs, is associated with reduced net replication and weaker induction of inflammatory responses than serovar Typhimurium (28). These studies suggested that rapid bacterial replication following invasion may trigger responses that confine serovar Typhimurium locally to the intestines, whereas serovar Choleraesuis may disseminate by a strategy of stealth. It remains to be determined if this is a relevant phenomenon in the systemic translocation of serovar Dublin in cattle and, if so, what factors influence net replication of the bacteria following invasion.
The finding that serovar Dublin signature-tagged clones in distal organs following oral exposure are the same ones, in the same proportions, as found in the MLNs and lymph implies that lymphatic translocation may be highly relevant in the systemic virulence of serovar Dublin in cattle. This finding is in marked contrast to recent studies of the dissemination of the enteropathogen Yersinia pseudotuberculosis that showed that at 3 days after intragastric inoculation of mice with a pool of 33 tagged clones, only 3% of those present in the liver and spleen were detected in the MLNs (1). This clonal analysis and other experiments demonstrated that systemic translocation of Y. pseudotuberculosis occurred by a route that bypasses Peyer's patches and replication in intestinal lymph nodes.
In conclusion, by using a bovine cannulation model, we have established that serovar Dublin, a natural systemic pathogen of cattle, translocates from the ileum mainly via draining lymphatics in a cell-free niche in a manner that requires T3SS-1 but not T3SS-2 or other factors established to play a role in systemic pathogenesis following intravenous exposure. The role of T3SS-1 in this process likely reflects its importance in intestinal invasion and implies that passive mechanisms of uptake via lymphoid-associated tissues and DCs alone do not suffice as “portals of entry” to distal sites. The data herein provide the first description of the genetic basis of translocation to and through intestinal MLNs by a host-restricted serovar in its natural host exposed via the natural route. While we have defined the role of key SPIs and other virulence-associated loci in systemic translocation of serovar Dublin, further studies will be needed to define the genetic basis of host specificity.
Acknowledgments
We gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (grants S10274, S14573, and C50964X) and the Department for the Environment, Food and Rural Affairs (grants OZ0315 and OZ0319). S.M.P. was supported by a CASE-studentship with Intervet International BV.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 27 August 2007.
REFERENCES
- 1.Barnes, P. D., M. A. Bergman, J. Mecsa, and R. R. Isberg. 2006. Yersinia pseudotuberculosis disseminates from a replicating bacterial pool in the intestine. J. Exp. Med. 203:1591-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bispham, J., B. N. Tripathi, P. R. Watson, and T. S. Wallis. 2001. Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect. Immun. 69:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolton, A. J., M. P. Osborne, T. Wallis, and J. Stephen. 1999. Interaction of Salmonella choleraesuis, Salmonella Dublin, and Salmonella typhimurium with porcine and bovine terminal ileum in vivo. Microbiology 145:2431-2441. [DOI] [PubMed] [Google Scholar]
- 4.Bonneau, M., M. Epardaud, F. Payot, V. Niborski, M.-I. Thoulouze, F. Bernex, B. Charley, S. Riffault, L. A. Guilloteau, and I. Schwartz-Cornil. 2006. Migratory monocytes and granulocytes are major lymphatic carriers of Salmonella from tissue to draining lymph node. J. Leukoc. Biol. 79:268-276. [DOI] [PubMed] [Google Scholar]
- 5.Brown, N. F., B. A. Vallance, B. K. Coombes, Y. Valdez, B. A. Coburn, and B. B. Finlay. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLOS Pathogens 1:252-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carnell, S. C., A. Bowen, E. Morgan, D. J. Maskell, T. S. Wallis, and M. P. Stevens. 2007. Role in virulence and protective efficacy in pigs of Salmonella enterica serovar Typhimurium-secreted components identified by signature-tagged mutagenesis. Microbiology 153:1940-1952. [DOI] [PubMed] [Google Scholar]
- 7.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombes, B. K., B. A. Coburn, A. A. Potter, S. Gomis, K. Mirakhur, Y. Li, and B. B. Finlay. 2005. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect. Immun. 73:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coombes, B. K., M. E. Wickham, M. J. Lowden, N. F. Brown, and B. B. Finlay. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. USA 102:17460-17465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crump, J. A., S. P. Luby, and E. D. Mintz. 2004. The global burden of typhoid fever. Bull. W. H. O. 82:346-353. [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost, A. J., A. P. Bland, and T. S. Wallis. 1997. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet. Pathol. 34:369-386. [DOI] [PubMed] [Google Scholar]
- 13.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567-573. [DOI] [PubMed] [Google Scholar]
- 15.Halle, S., D. Bumann, H. Herbrand, Y. Willer, S. Dahne, R. Forster, and O. Pabst. 2007. Solitary intestinal lymphoid tissue provides a productive port of entry for Salmonella enterica serovar Typhimurium. Infect. Immun. 75:1577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 17.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous detection of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 18.Jang, M. H., M.-N. Kweon, K. Iwatani, M. Yamamoto, K. Terahara, C. Sasakawa, T. Suzuki, T. Nochi, Y. Yokota, P. D. Rennert, T. Hiroi, H. Tamagawa, H. Iijima, J. Kunisawa, Y. Yuki, and H. Kiyono. 2004. Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proc. Natl. Acad. Sci. USA 101:6110-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jantsch, J., C. Cheminay, D. Chakravortty, T. Lindig, J. Hein, and M. Hensel. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5:933-945. [DOI] [PubMed] [Google Scholar]
- 20.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, M. A., M. W. Wood, P. B. Mullan, P. R. Watson, T. S. Wallis, and E. E. Galyov. 1998. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect. Immun. 66:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLOS Pathogens 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monack, D. M., D. Hersh, N. Ghori, D. Bouley, A. Zychlinsky, and S. Falkow. 2000. Salmonella exploits caspase-1 to colonize Peyer's patches in a murine typhoid model. J. Exp. Med. 192:249-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monaghan, P., P. R. Watson, H. Cook, L. Scott, T. S. Wallis, and D. Robertson. 2001. An improved method for preparing thick sections for immuno/histochemistry and confocal microscopy and its use to identify rare events. J. Microscopy 203:223-226. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, E., A. J. Bowen, S. C. Carnell, T. S. Wallis, and M. P. Stevens. 2007. SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization of cattle. Infect. Immun. 75:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 27.Niess, J. H., S. Brand, X. Gu, L. Landsman, S. Jung, B. A. McCormick, J. M. Vyas, M. Boes, H. L. Ploegh, J. G. Fox, D. R. Littman, and H.-C. Reinecker. 2005. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307:254-258. [DOI] [PubMed] [Google Scholar]
- 28.Paulin, S. M., A. Jagannathan, J. Campbell, T. S. Wallis, and M. P. Stevens. 2007. Net replication of Salmonella enterica serovars Typhimurium and Choleraesuis in porcine intestinal mucosa and nodes is associated with their differential virulence. Infect. Immun. 75:3950-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulin, S. M., P. R. Watson, A. R. Benmore, M. P. Stevens, P. W. Jones, B. Villarreal-Ramos, and T. S. Wallis. 2002. Analysis of Salmonella enterica serotype-host specificity in calves: avirulence of S. enterica serotype Gallinarum correlates with bacterial dissemination from mesenteric lymph nodes and persistence in vivo. Infect. Immun. 70:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penheiter, K. L., N. Mathur, D. Giles, T. Fahlen, and B. D. Jones. 1997. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol. Microbiol. 24:697-709. [DOI] [PubMed] [Google Scholar]
- 31.Rescigno, M., M. Urbano, B. Valzasina, M. Francolini, G. Rotta, R. Bonasio, F. Granucci, J.-P. Kraehenbuhl, and P. Ricciardi-Castagnoli. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2:361-367. [DOI] [PubMed] [Google Scholar]
- 32.Richter-Dahlfors, A., A. M. J. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shea, J. E., C. R. Beuzon, C. Gleeson, R. Mundy, and D. W. Holden. 1999. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect. Immun. 67:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele-Mortimer, O., J. H. Brumell, L. A. Knodler, S. Meresse, A. Lopez, and B. B. Finlay. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 4:43-54. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi, A. 1966. Electron microscope studies of experimental Salmonella infection. Am. J. Pathol. 50:109-136. [PMC free article] [PubMed] [Google Scholar]
- 36.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uzzau, S., D. J. Brown, T. Wallis, S. Rubino, G. Leori, S. Bernard, J. Casadesus, D. J. Platt, and J. E. Olsen. 2000. Host-adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez-Torres, A., J. Jones-Carson, A. J. Baumler, S. Falkow, R. Valdivia, W. Brown, M. Le, R. Berggren, W. T. Parks, and F. C. Fang. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804-808. [DOI] [PubMed] [Google Scholar]
- 39.Wallis, T. S., and P. A. Barrow. July 2005, posting date. Chapter 8.6.2.1, Salmonella epidemiology and pathogenesis in food-producing animals. In R. Curtiss III et al. (ed.), EcoSal-Escherichia coli and Salmonella: cellular and molecular biology, ASM Press, Washington, DC. http://www.ecosal.org. [DOI] [PubMed]
- 40.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 41.Watson, P. R., E. E. Galyov, S. M. Paulin, P. W. Jones, and T. S. Wallis. 1998. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect. Immun. 66:1432-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson, P. R., S. M. Paulin, A. P. Bland, P. W. Jones, and T. S. Wallis. 1995. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect. Immun. 63:2743-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Worley, M. J., G. S. Nieman, K. Geddes, and F. Heffron. 2006. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc. Natl. Acad. Sci. USA 103:17915-17920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, S., R. L. Santos, R. M. Tsolis, S. Stender, W. D. Hardt, A. J. Bäumler, and L. G. Adams. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]