Abstract
A d-serine deaminase (DsdA) mutant of uropathogenic Escherichia coli strain CFT073 has a hypercolonization phenotype in a murine model of urinary tract infection (UTI) due to increased virulence gene expression by an unknown mechanism (B. J. Haugen et al., Infect. Immun. 75:278-289, 2007). DsdC is a d-serine-dependent activator of dsdXA transcription. DsdC may regulate the virulence genes responsible for hypercolonization. The loss of DsdA leads to increased intracellular accumulation of d-serine. In this study we show that deletion of the genes encoding l-serine deaminases SdaA and SdaB resulted in a mutant that accumulates higher intracellular levels of l-serine than CFT073. CFT073 sdaA sdaB has a mild competitive colonization defect whereas a CFT073 dsdA sdaA sdaB triple mutant shows a greater loss in competitive colonization ability. Thus, the inability to generate serine-specific catabolic products does not result in hypercolonization and the ability to catabolize serine represents a positive physiological trait during murine UTI. CFT073 dsdC and CFT073 dsdC dsdA mutants continue to outcompete the wild type in the UTI model. These results confirm that loss of DsdA activity results in the hypercolonization phenotype and that DsdC does not play a direct role in the elevated-colonization phenotype. Interestingly, a CFT073 dsdA mutant with deletions of d-serine transporter genes dsdX and cycA shows wild-type colonization levels of the bladder but is attenuated for kidney colonization. Thus, d-serine acts as a signal for hypercolonization and virulence gene expression by CFT073 dsdA, whereas overall catabolism of serine represents a positive Escherichia coli fitness trait during UTI.
Urinary tract infections (UTIs) in adult women impose an estimated cost of $2.4 billion per year in the United States (12). Most women will experience at least one UTI in their lifetime, resulting in an estimated 6.8 million physician visits, 1.2 million emergency room visits, and nearly a quarter million hospitalizations each year. Escherichia coli remains by far the primary causative agent of community-acquired UTIs (11).
The urinary tract is a normally sterile environment, and it poses daunting challenges to colonization by E. coli and other microorganisms. In addition to the cleansing flow of urine, numerous innate and acquired immune factors challenge the growth of uropathogenic Escherichia coli (UPEC) in the urinary tract. The host defense involves phagocytic attack, antimicrobial peptides, complement lytic and opsonizing factors, and reactive oxygen and nitrogen species. In addition, the urinary tract offers high-salt and high-osmolarity conditions while limiting E. coli nutrients common to the intestinal tract, especially neutral sugars and iron (4). Thus, we hypothesize that the ability of UPEC to import and metabolize the available carbon and nitrogen sources present in the urinary tract plays a special role in its ability to colonize and cause disease at that site.
From a bacterial nutritional standpoint, urine is a dilute mixture of amino acids and small peptides, quite similar to tryptone broth, with the notable exception of the abundance of urea in urine (4). The growth of E. coli in tryptone broth is well characterized, where growing cells preferentially and sequentially utilize serine and then aspartate while secreting acetate. Once these amino acids are depleted, cells then import and use tryptophan and acetate, followed by alanine, glutamine, and threonine (31). This order of nutrient preference holds true for E. coli grown in broth cultures (21) as well as on semisolid motility agar (1).
However, E. coli growth in the urinary tract may more closely resemble growth inside a chemostat than a closed laboratory system. The production of urine from the kidneys provides a continuous supply of a preferred nutrient, such as serine, and hypothetically leads to dilution of the excreted product, acetate. While studies of amino acid utilization mainly focus on the l-forms of the amino acids, d-serine is among the most abundant amino acids in human urine, where it is present at a range of concentrations from 3 to 115 μg/ml, with d-serine accounting for as much as 97% of the total serine (S. Pellett and R. A. Welch, unpublished observation) (10). We originally hypothesized that d-serine in urine represents a host defense mechanism because d-serine can be bacteriostatic in minimal medium at concentrations of 50 μg/ml (13). d-Serine toxicity is related to its interference with l-serine and pantothenate biosynthesis (20), but this inhibition can be overcome if the bacterium possesses an intact d-serine utilization locus (13). Approximately 85% of E. coli pyelonephritis and urosepsis isolates carry at least one operon for d-serine utilization (24). Strikingly, nearly all isolates of the common extraintestinal E. coli O18 K1 H7 pathotype possess two copies of the d-serine deaminase locus (16). Encoded within these loci are a d-serine-specific transporter (DsdX) (2), a d-serine deaminase (DsdA), and a d-serine-responsive, positive transcription factor (DsdC) that is required for the expression of dsdX and dsdA (9, 20). Expression of the dsdXA operon is also subject to catabolite repression by glucose (16, 24; G. A. Baisa, A. J. Schmidt, and R. A. Welch, unpublished data). Aside from DsdX, d-serine can also be imported through the d-alanine and glycine transporter CycA (6, 30). DsdA degrades d-serine to ammonia and pyruvate (20). This activity allows the utilization of d-serine as a sole carbon and nitrogen source and provides resistance to the bacteriostatic effect of d-serine.
Aside from our work, the possible significance of d-serine catabolism by UPEC during human UTI was suggested by Roos et al., who found that dsdA expression for E. coli asymptomatic bacteriuria strain 83972 is consistently up-regulated 2.5- to 8-fold in chronically infected humans compared to its expression during growth in a glucose minimal medium (25). UPEC strain CFT073 dsdA displays an extended lag phase compared to the wild type during growth in human urine but eventually grows to cell densities similar to those of the wild type after 24 h (24). This suggests that the accumulated inhibitory d-serine is catabolized, secreted, or incorporated into a nontoxic product. We previously demonstrated that the CFT073 dsdA strain counterintuitively outcompetes wild-type CFT073 in the murine urinary tract and is hyperflagellated and hypermotile in vitro (8, 24). In vivo transcriptome analysis that compared the genes expressed by the CFT073 dsdA mutant during murine infection to those of the wild type revealed that a set of virulence factors including P and F1C fimbriae and hemolysin are up-regulated in the mutant strain (24). dsdX is also up-regulated in vivo, and its expression depends upon the intracellular presence of d-serine (9), which suggests that d-serine accumulates to higher levels in CFT073 mutants unable to degrade d-serine. Our current model proposes that the inability to degrade d-serine in a CFT073 dsdA mutant results in elevated intracellular d-serine concentrations during murine UTI, which lead to either direct hypermodulation of a d-serine-dependent regulon or an indirect alteration of catabolic potential. Thus, we posit that d-serine accumulation represents a signal to UPEC that it has entered the urinary tract, a site relatively low in glucose, making catabolite repression unlikely. We recently found that a fimB-like recombinase gene, ipuA, linked to the CFT073 dsdCXA genes mediates bidirectional phase switching of the type 1 fimbria fimS site and reciprocal changes in motility (5). We also observe that dsdA undergoes reversible on-and-off expression with a reduced switching frequency in a CFT073 ipuA mutant background (S. Pellett, P. Roesch, H. Hamilton, and R. A. Welch, unpublished data). Therefore, our model must also take into account the fact that the expression of dsdA undergoes a reversible phase-switching control. Phase variation of dsdA expression implies that there are alternative levels of intracellular d-serine in populations of CFT073 cells, and our model examines both the wild type and dsdA mutants.
In this study, we set out to determine if intracellular accumulation of serine is important for colonization of the urinary tract by CFT073. First we examined the effects of l- versus d-serine accumulation on colonization of the murine urinary tract. l-Serine is degraded by one of two deaminases, SdaA and SdaB (28). We demonstrate that a CFT073 sdaA sdaB mutant accumulates l-serine through loss of l-serine deaminase activity, but unlike CFT073 dsdA, this mutant colonizes the mouse urinary tract at levels only slightly lower than those of the wild-type strain. However, a CFT073 dsdA sdaA sdaB strain is attenuated in the mouse model, suggesting that serine catabolism is important for colonizing the urinary tract. Next we sought to define if a UTI colonization response to d-serine depends on the d-serine-responsive transcription factor DsdC. We provide evidence that the colonization effect in CFT073 dsdA is not fully mediated through DsdC, because a CFT073 dsdC dsdA mutant continues to exhibit an increased-colonization phenotype. Lastly we show that d-serine transport is required for the elevated CFT073 dsdA colonization phenotype and loss of d-serine transport negatively affects kidney infection. Our results support a role for d-serine but not l-serine in signaling and virulence gene regulation of UPEC.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Urosepsis E. coli strain CFT073 was originally isolated from the blood and urine of a woman admitted to the University of Maryland Medical System (15). WAM2615 is a nalidixic acid-resistant derivative of CFT073 in which 445 bp of dsdA is deleted (24). WAM2880 is a derivative of CFT073 that has lacZYA deleted (22). WAM3147 is a derivative of CFT073 that has dsdX and cycA deleted; WAM3169 is a derivative of CFT073 that has cycA, dsdX, and dsdA deleted (2). E. coli AAEC185 was used as a host for construction of recombinant plasmids (3). Other strains constructed for this study are described below. L broth and LB agar (Fisher) were used for the propagation of all strains except where noted below. Carbenicillin, kanamycin, and chloramphenicol were used at concentrations of 250, 50, and 20 μg/ml, respectively.
Construction of CFT073 mutants by λ-Red recombination.
The construction of CFT073 dsdC dsdA, CFT073 sdaA sdaB, and CFT073 dsdA sdaA sdaB was done using the λ-Red recombination system designed by Datsenko and Wanner (7). Wild-type CFT073 harboring pKD46 (WAM 2811) was used for construction of CFT073 sdaA sdaB; all other λ-Red deletions were generated in WAM2615 harboring pKD46 (WAM2850). Oligonucleotides 1132 and 1133 were designed to result in precise deletion of dsdC, oligonucleotides 1122 and 1123 were designed to result in precise deletion of sdaA, and oligonucleotides 1124 and 1125 were designed to result in precise deletion of sdaB. All oligonucleotide sequences are listed in Table 1. After replacement of each gene with an antibiotic resistance cassette, the cassettes were excised by FLP recombinase, encoded by pCP20, leaving a single FLP recombination target site, as described elsewhere (7). Excision of the antibiotic resistance cassettes was confirmed by PCR and loss of the antibiotic resistance on the appropriate media.
TABLE 1.
Oligonucleotides used in this study
| IDa | Purpose | Sequenceb |
|---|---|---|
| 557 | dsdC cloning | CTGCCAGCGTATCAGCGAGACTGC |
| 571 | dsdC cloning | CATTTCACCGTCATATCGCCAAACG |
| 579 | dsdC mutagenesis | CAGCGACAATTaCTCTGCCGCGAGAGCGAAGGAC |
| 580 | dsdC mutagenesis | GTCCTTCGCTCTCGCGGCAGAGtAATTGTCGCTG |
| 593 | dsdC mutagenesis; PstI | ggctgcagCTGCCAGCGTATCAGCGAGACTGC |
| 594 | dsdC mutagenesis; KpnI | ggggtaccCATTTCACCGTCATATCGCCAAACG |
| 1122 | sdaA deletion | AAATTCGCCCATCCGTTGCAGATGGGCGAGTAAGAGTAcatatgaatatcctccttag |
| 1123 | sdaA deletion | CGTTACTGGAAGTCCAGTCACCTTGTCAGGAGTATT-ATCtgtgtaggctggagctgcttc |
| 1124 | sdaB deletion | CGGGAAGAGGCCTCGCAAAAAGAGGCCTCTGGAG-AGCGAcatatgaatatcctccttag |
| 1125 | sdaB deletion | CGGGCGGCGCTTCCTCCGTTTTAACGCGATGTATTT-CCTtgtgtaggctggagctgcttc |
| 1132 | dsdC deletion | CATAATAAGGTTATTTAGGAACCAGATGAT-TTAATGTATgtgtaggctggagctgcttcg |
| 1133 | dsdC deletion | TTTTGCCGCTATTTTTTTACACTTAAGAGAA-AAATGAGcatatgaatatcctccttag |
ID, identification number.
Sequences are displayed 5′ to 3′. Lowercase bases indicate bases that do not match the genomic sequence of CFT073 and are thus mutagenic. Sequences matching useful restriction enzyme recognition sites are in bold, and the identity of the enzyme is noted in the “Purpose” column.
Murine model of UTI.
Urinary tract colonization was assayed using the competitive murine model of UTI as previously described (23, 24). CFT073 lacZYA (WAM2880) was used as the wild type to allow discrimination of mutant strains as pink colonies and WAM2880 as white colonies on MacConkey lactose agar medium. This strain colonizes mouse bladders and kidneys indistinguishably from the wild type (22). Strains were grown independently in L broth without shaking at 37°C for 7 days, with passage to fresh medium on the second, fourth, and sixth days. Cultures of wild-type or mutant bacteria were concentrated by centrifugation at 7,500 rpm in an Eppendorf 5215D centrifuge for 2 min, resuspended in phosphate-buffered saline, and then mixed in a 1:1 ratio. Inocula of 50 μl, carrying between 1 × 108 and 5 × 108 CFU, were delivered via a catheter to the bladders of isoflurane-anesthetized Swiss Webster or CBA/J mice (Harlan). Unless otherwise noted, mice used were Swiss Webster mice; competitive colonization phenotypes of a given mutant in coinfections with wild-type CFT073 produce similar results in Swiss Webster and CBA/J mice (B. J. Haugen and P. Redford, personal communication). After 2 days, mice were euthanized by CO2 asphyxiation, and the bladders and kidneys were excised, homogenized, serially diluted in phosphate-buffered saline, and plated to MacConkey lactose agar medium. When tested strains carried plasmids for complementation studies, the medium contained antibiotics at the proper concentrations to select for bacteria that maintained the plasmids. After enumeration, the competitive index (ratio of recovered CFU of mutant/recovered CFU of wild type) colonization levels were graphed and analyzed using a paired Wilcoxon signed-rank test and Prism 4.0c (GraphPad). It should be noted that in our original report of the CFT073 dsdA hypercolonization phenotype, we compared means of the CFU in animals, whereas in this report we switched to median values, which permit a fairer comparison of the scattered data seen in this animal model.
Construction of dsdC-STOP mutant strain of CFT073.
Restriction and DNA modification enzymes were purchased from either New England Biolabs or Promega Corporation. dsdC-specific primers for PCR were designed using the CFT073 genome sequence. Two overlapping PCR products were generated by pairing primer 557 with mutagenic primer 579 and primer 571 with mutagenic primer 580 (Table 1). The template was wild-type CFT073 genomic DNA prepared using the Wizard genomic DNA isolation kit (Promega). The PCR amplification used the Expand High Fidelity PCR System (Roche Diagnostics) according to the manufacturer's protocol. The resulting PCR products were gel purified with the QIAQuick gel purification kit (QIAGEN) and then mixed as template for a final PCR using primers 593 and 594 (Table 1). The resulting product was then digested with KpnI and PstI, gel purified as described above, and ligated into KpnI-PstI-restricted pEP185.2 (kind gift of Virginia Miller, Washington University). The resulting construct containing the mutant dsdC allele, a glutamate 37 codon mutated to a stop codon, was cloned into CC118 λpir and sequenced using T3 and T7 primers. The suicide plasmid construct was transformed into a S17λpir background and then mated with a spontaneous nalidixic acid-resistant mutant of CFT073. Single-crossover chromosomal integrates of the recombinant plasmid were selected, and d-cycloserine was used to enrich for cells that had undergone a second crossover event and excised the suicide plasmid backbone from the CFT073 chromosome. Chromosomal DNA was isolated from the putative dsdC mutants and examined via PCR analysis using dsdC-specific primers to confirm and then sequence across the mutation. Phenotypic analysis of the putative mutants was carried out by inoculating colonies onto morpholinepropanesulfonic acid (MOPS) glycerol minimal medium and MOPS minimal d-serine agar medium.
Complementation of CFT073 dsdC.
The CFT073 dsdC gene, including its Shine-Dalgarno sequence, was PCR amplified with primers 557 and 571. The PCR product was digested with PstI and BamHI enzymes, gel purified, and ligated with pACYC177 previously digested with the same two restriction enzymes. This plasmid was then used for complementation analysis.
Motility plates.
Strains were inoculated onto LB agar medium and grown overnight at 37°C. Isolated colonies were selected and used to inoculate tryptone broth cultures that were grown overnight at 37°C in a shaking incubator. Five milliliters of the overnight culture was inoculated onto the center of a low-agar (0.2%) tryptone plate and was incubated for 12 to 18 h at 37°C.
Growth studies.
Strains were grown overnight in 2 ml of the appropriate liquid medium. Three-milliliter cultures of fresh medium were inoculated with 30 ml of the appropriate overnight. Optical density at 600 nm (OD600) was monitored by spectrophotometry, and the CFU per ml of the culture were determined by hourly dilution plating to LB agar solid plate medium.
Serine accumulation.
All strains were grown in MOPS minimal medium. MOPS minimal medium was made as described previously (19) with the carbon source substitution of 43.4 mM glycerol, 2.66 mM glycine, and 380 mM dl-isoleucine for glucose and the omission of thiamine, giving a final composition of 1.32 mM K2HPO4, 9.52 mM NH4Cl, 0.523 mM MgCl2, 0.276 mM K2SO4, 10 mM FeSO4, 0.5 mM CaCl2, 50 mM NaCl, 40 mM MOPS, 4 mM Tricine, 3 nM (NH4)6(MO7)24, 0.4 mM H2BO3, 30 nM CoCl2, 10 nM CuSO4, 80 nM MnCl2, 10 nM ZnSO4, 43.4 mM glycerol, 2.66 mM glycine, and 380 mM dl-isoleucine. A 2-ml overnight MOPS culture of each strain was inoculated into 25 ml fresh MOPS medium and grown at 37°C with shaking until the culture entered mid-log phase (OD600, ∼0.5). If the accumulation of d-serine was to be examined, 500 mg/ml d-serine was added to the culture when it reached an OD600 of ∼0.45 and allowed to incubate for 15 min but was otherwise prepared for the assay as indicated. A 1-ml aliquot of the culture was removed, pelleted by centrifugation, and stored at −80°C for use in the deaminase activity assay (see below). Ten milliliters of the culture was pelleted by centrifugation and washed twice with MOPS-Tris buffer (0.1 M MOPS, 8 mM MgSO4, and 8 mM Tris base; pH was adjusted to 7.0) at 4°C. Washed cells were resuspended in 2 ml MOPS-Tris buffer plus 0.4% glycerol, giving a total protein concentration of approximately 0.3 mg/ml. The cell suspension was allowed to equilibrate to 37°C for 5 min, at which point chloramphenicol was added to a final concentration of 50 mg/ml and a 50-ml aliquot was removed and stored at −80°C for a subsequent bicinchoninic acid (BCA) assay to measure actual protein concentration. The chloramphenicol-treated cells were incubated for an additional 15 min to ensure cessation of protein synthesis. The cell suspension was split into 50-ml aliquots. Ten milliliters of l- or d-[14C]serine, 5.5 Ci/mol and 0.6 mM, a quantity sufficient to saturate the transporters (2), was added to the suspensions, giving a final concentration of 0.1 mM l- or d-[14C]serine, and allowed to incubate for various times. Five milliliters of MOPS-Tris buffer was added to the cultures to stop transport, and the suspension was filtered through a 0.45-mm-pore-size nitrocellulose membrane filter. The filter was washed with an additional 5 ml of MOPS-Tris buffer. Filters were allowed to dry and then were placed in scintillation vials containing 3 ml Biosafe counting cocktail (Research Products International Corporation). The samples were counted in a Packard Tri-Lab 2100 TR liquid scintillation analyzer. All disintegration-per-minute values determined by sample counting were normalized to the activity of l- or d-[14C]serine, 5.5 Ci/mol, and to the protein concentration of the sample as determined by BCA assay to express counted signal in terms of nmol l- or d-serine/mg total protein.
Serine deaminase activity assay.
Cell pellets harvested during the accumulation experiments were resuspended in 1 ml 1 M KPO4. Fifty milliliters of the suspension was reserved and stored at −80°C for determination of total protein concentration by BCA assay. Fifty milliliters of the suspension was added to a glass test tube containing an additional 250 ml of KPO4 and 10 ml of Pop Culture lysis solution (Novagen), two tubes per cell pellet. Tubes were incubated at 37°C for 15 min to lyse cells. Tubes were then treated with 100 ml of either 9.5 mM d-serine or 100 ml H2O. Tubes were incubated at 37°C for 20 min. Samples were then treated with 0.9 ml of a 3.89 M 2,4-dinitrophenylhydrazine solution (2,4-dinitrophenylhydrazine dissolved in 1.2 N HCl). Samples were incubated at room temperature for 20 min. Reactions were stopped by addition of 1.7 ml 2.5N NaOH. The OD520 of each sample was measured, and the final optical density of the assay mixture correlates in a linear fashion with pyruvate concentration over an optical density range of 0 to 1.5. Because serine deaminase proteins DsdA, SdaA, and SdaB convert d-serine or l-serine to ammonia and pyruvate, the presence of pyruvate is a function of deaminase activity within the assay as described elsewhere (14). Values were normalized to concentration of total protein determined by the BCA assay.
RESULTS
Deletion of enantiomer-specific serine deaminase genes results in accumulation of the corresponding serine enantiomer.
In this study, we set out to define the requirements for hypercolonization by CFT073 dsdA when in competition with the wild type by the examination of several aspects of serine metabolism. We observed that during infection of mice infected with only CFT073 dsdA, the strain highly upexpresses dsdX (8). Transcription of dsdX and dsdA is dependent upon the LysR transcriptional regulator DsdC (9). The up-regulation of dsdX in a dsdA mutant suggests that d-serine cannot be degraded and thus accumulates and drives transcription of dsdX in a DsdC-dependent manner. With the evidence that d-serine accumulation is correlated with hypercolonization, we sought to determine whether the effects of d-serine accumulation are specific to the d-enantiomer.
In order to test the hypothesis that d-serine accumulation results in hypercolonization, we monitored d- and l-14[C]serine uptake in wild-type CFT073, CFT073 dsdA, CFT073 sdaA sdaB, and CFT073 dsdA sdaA sdaB. The regulation of sdaA and sdaB is not very well understood, and it is unclear which deaminase would be of greater importance in vivo during a UTI, so we chose to examine CFT073 sdaA sdaB as our model l-serine-accumulating strain. We had previously seen an up-regulation of dsdX in CFT073 dsdA in vivo, but we had not previously directly monitored d-serine uptake with CFT073 dsdA. While studying d-serine uptake in CFT073 (2), it was observed that when wild-type CFT073 was pretreated with d-serine and then used to monitor d-serine uptake, uptake proceeded normally for the first minute, but the strain did not subsequently accumulate additional d-serine, unlike the dsdA mutant, which continued to accumulate d-serine for 10 min following addition of d-[14C]serine (A. Anfora, unpublished observation). We replicated and expanded these observations, which are presented in Table 2. We show in Table 2 that any strains with a deletion of dsdA accumulated d-serine after 10 min of incubation. Similarly, strains with sdaA sdaB deletions accumulated more l-serine than did strains able to express SdaA and SdaB, as shown in Table 3. An inverse relationship was observed between the accumulation of a serine enantiomer and the enantiomer-specific serine deaminase activity (data not shown).
TABLE 2.
Accumulation of l-[14C]serine
| Strain |
l-Serine accumulation (ng/mg)a
|
|
|---|---|---|
| 1 min | 10 min | |
| Wild type | 3.68 ± 0.44 | 7.44 ± 0.86 |
| dsdA | 4.77 ± 0.05 | 16.84 ± 1.13 |
| sdaA sdaB | 16.73 ± 0.47b | 41.78 ± 2.32b |
| dsdA sdaA sdaB | 23.26 ± 7.54b | 41.93 ± 11.82b |
Values are means ± standard deviations (two replicates per group unless otherwise noted).
Significantly different (P < 0.05) compared to wild-type value at indicated time point as determined by two-way analysis of variance with Bonferroni post-test comparing all columns using Graphpad Prism 4.0c.
TABLE 3.
Accumulation of d-[14C]serine
| Strain |
d-Serine accumulation (ng/mg)a
|
|
|---|---|---|
| 1 min | 10 min | |
| Wild typec | 6.87 ± 7.28 | 7.86 ± 2.29 |
| dsdA | 37.30 ± 10.77b | 57.66 ± 21.23b |
| sdaA sdaB | 2.95 ± 2.46 | 5.14 ± 2.48 |
| dsdA sdaA sdaB | 31.14 ± 5.71b | 70.27 ± 7.68b |
| dsdC | 16.84 ± 1.14 | 69.62 ± 4.37b |
Values are means ± standard deviations (two replicates per group unless otherwise noted).
Significantly different (P < 0.05) compared to wild-type value at indicated time point as determined by two-way analysis of variance with Bonferroni post-test comparing all columns using Graphpad Prism 4.0c.
Three replicates in group.
CFT073 mutants that cannot catabolize l-serine are impaired in their ability to colonize the murine urinary tract.
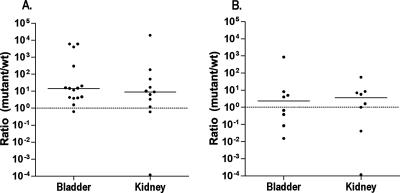
To determine if the enhanced-colonization phenotype occurs with the accumulation of l-serine as it does with d-serine, the CFT073 sdaA sdaB mutant was tested in competition with wild-type CFT073 (Fig. 1A). Compared to the wild type, the CFT073 sdaA sdaB mutant was at a clear disadvantage in the bladder (median ratio = 0.28, P = 0.020) and nearly equal in its ability to colonize the kidney (median ratio = 0.50, P = 0.042). We then tested the CFT073 dsdA sdaA sdaB strain in a coinfection with wild-type CFT073 (Fig. 1B). CFT073 dsdA sdaA sdaB displayed a competitive disadvantage in comparison with wild-type CFT073 sdaA sdaB in the bladder (median ratio = 0.034, P < 0.001) and displayed a strong trend towards being at a disadvantage in the kidney (median ratio = 0.084, P = 0.091). Thus, deletion of the genes for SdaA and SdaB negates the competitive advantage seen for CFT073 dsdA in coinfection with wild-type CFT073.
FIG. 1.
Bladder and kidney colonization of mice with wild-type (wt) CFT073 and CFT073 sdaA sdaB. Mice were transurethrally inoculated with a 1:1 ratio of CFT073 lacZYA and CFT073 sdaA sdaB (A) or CFT073 lacZYA and CFT073 dsdA sdaA sdaB (B). Each data point represents the competitive index that is the ratio of recovered mutant CFU/g tissue to recovered wild-type CFU/g tissue for the designated organ tissue from one mouse 48 h postinoculation. Horizontal bars represent the median values of ratios for the organ type, which were equal to 0.28 (P = 0.020) and 0.502 (P = 0.042) (A) and 0.034 (P < 0.001) and 0.084 (P = 0.091) (B) for the bladder and kidney, respectively.
DsdC is not required for CFT073 dsdA elevated colonization of the murine urinary tract.
In this study, we set out to define the requirements for hypercolonization by CFT073 dsdA when in competition with the wild type. We observed that during infection of mice without the presence of the wild type, CFT073 dsdA upexpresses dsdX (8). Transcription of dsdX and dsdA is dependent upon the LysR transcriptional regulator DsdC (9). The up-regulation of dsdX in a dsdA mutant suggests that d-serine cannot be degraded and thus accumulates and drives transcription of dsdX in a DsdC-dependent manner. With the evidence that d-serine accumulation is correlated with hypercolonization, we sought to determine whether the effects of d-serine accumulation are mediated through the transcription factor DsdC. We deleted the entire coding sequence of dsdC from CFT073 dsdA using the λ-Red-mediated recombination system and tested the resulting strain in competition with the wild type in the murine model of UTI. This strain continued to colonize the bladders and kidneys of mice at statistically significant levels higher than those of the wild type (Fig. 2). Thus, the CFT073 dsdA elevated-colonization phenotype does not depend on DsdC-regulated genes.
FIG. 2.

Bladder and kidney colonization of mice with wild-type (wt) and CFT073 dsdA dsdC. Mice were transurethrally inoculated with a 1:1 ratio of CFT073 lacZYA and CFT073 dsdA dsdC. Each data point represents the competitive index that is the ratio of recovered mutant CFU/g tissue to recovered wild-type CFU/g tissue for the designated organ tissue from one mouse 48 h postinoculation. Horizontal bars represent the median values of ratios for the organ type, which are 4.0 and 4.4 for the bladder and kidney, respectively (P < 0.004 and P < 0.005, respectively).
CFT073 dsdC hypercolonizes during competitive murine UTI.
As stated above, dsdC is necessary for the transcription of dsdX and dsdA, so we hypothesized that a CFT073 dsdC mutant would also accumulate d-serine via CycA and thus continue to colonize at levels greater than those of the wild-type strain. To test this hypothesis, we replaced the wild-type allele of dsdC with dsdC-STOP, an allele with an introduced stop codon early in the coding sequence. This strain does not grow in the presence of 500 μg/ml d-serine. The CFT073 dsdC strain was tested in competition with the wild type in CBA/J mice, and we observed a hypercolonization phenotype similar to that of CFT073 dsdA dsdC, with the dsdC mutant outcompeting the wild type 15- and 9-fold in the bladder and kidney, respectively (Fig. 3A). To confirm this observation, the dsdC-STOP mutation was complemented with a plasmid expressing dsdC. The resulting strain grows in the presence of 500 μg/ml d-serine. This complemented strain was tested in the mouse model of UTI in competition with a wild-type strain carrying the same complementing plasmid (Fig. 3B). Complementation of the CFT073 dsdC strain reduced the colonization of the bladder and kidney to levels that were not statistically different from those of the wild type.
FIG. 3.
Bladder and kidney colonization of mice with wild-type (wt) and CFT073 dsdC. Mice were transurethrally inoculated with a 1:1 ratio of CFT073 lacZYA and CFT073 dsdC-STOP (A) or the same strains carrying a plasmid carrying dsdC (B). Each data point represents the competitive index that is the ratio of recovered mutant CFU/g tissue to recovered wild-type CFU/g tissue for the designated organ tissue from one mouse 48 h postinoculation. Horizontal bars represent the median values of ratios for the organ type, which were equal to 15 (P < 0.002) and 9.0 (P < 0.032) (A) and 2.3 (P = 0.54) and 3.6 (P = 0.68) (B) for the bladder and kidney, respectively.
CFT073 requires intracellular accumulation of d-serine to hypercolonize.
The above results confirmed that the inability to degrade d-serine results in hypercolonization due to accumulation of d-serine. The source of d-serine either may be from the host or may be endogenously produced by CFT073 during infection. If the source of the d-serine were the host, a CFT073 dsdA derivative strain unable to import d-serine would colonize similarly to the wild type. CFT073 encodes two d-serine transporters, CycA and DsdX (2). CFT073 cycA dsdXA is unable to transport radiolabeled d-serine or use d-serine as a sole carbon source (2). We tested CFT073 cycA dsdXA in competition with wild-type CFT073 in the murine model of UTI (Fig. 4). We observed no competitive difference in bladder colonization by this strain (median ratio = 0.69, P = 0.56), but interestingly, in the kidney, this mutant colonized at levels approximately 10-fold less than those of the wild type (P = 0.015).
FIG. 4.
Bladder and kidney colonization of mice with wild-type (wt) and CFT073 cycA dsdXA. Mice were transurethrally inoculated with a 1:1 ratio of CFT073 lacZYA and CFT073 cycA dsdXA. Each data point represents the competitive index that is the ratio of recovered mutant CFU/g tissue to recovered wild-type CFU/g tissue for the designated organ tissue from one mouse 48 h postinoculation. Horizontal bars represent the median values of ratios for the organ type, which were equal to 0.69 (P = 0.56) and 0.12 (P = 0.015) in the bladder and kidney, respectively.
DISCUSSION
Why is a CFT073 mutant unable to produce DsdA at a competitive advantage in the murine urinary tract compared to the wild type? Our interest in d-serine metabolism is prompted by the linkage of the dsdCXA genes to FimB- and FimE-like recombinase genes and preliminary evidence that dsdA expression undergoes reversible phase switching. It is not clear if the competitive advantage of CFT073 dsdA is due to intracellular accumulation of d-serine or an inability to generate metabolic by-products of d-serine catabolism. Since d- and l-serine are degraded to pyruvate and ammonia by the appropriate deaminase, we first examined the effects of mutations in the l-serine deaminase genes sdaA and sdaB on colonization of the murine urinary tract. The CFT073 sdaA sdaB mutant is able to accumulate elevated amounts of l-serine but not d-serine, is attenuated for colonization of the murine bladder, and colonizes the kidney similarly to the wild type. This suggests that hypercolonization does not result from general serine accumulation, only d-serine accumulation. Intracellular d-serine is more likely to serve as an in vivo signal for expression of virulence determinants such as Pap fimbriae and hemolysin than is l-serine, as evidenced by loss of colonization following l-serine accumulation. Additionally, CFT073 dsdA sdaA sdaB is able to accumulate both the l- and d-serine enantiomers but is reduced in its ability to colonize both the bladder and the kidney of mice compared to the wild type. These results suggest that d-serine is able to serve as a nutrient for CFT073 in the urinary tract. The dsdA sdaA sdaB strain UTI results suggest that loss of l-serine deaminase genes negates the colonization advantage that the dsdA mutation provides to CFT073. The deleterious effect of the loss of l-serine deaminase may be tied to decreased energy production in the tricarboxylic acid cycle. l-Serine is preferentially consumed in a mixed pool of amino acids in tryptone broth (31). Consumption of l-serine provides pyruvate for the tricarboxylic acid cycle and the acetate switch (21, 31).
CFT073 mutants able to accumulate d-serine independently of a DsdA deletion were then examined to determine if the colonization advantage of CFT073 dsdA is dependent upon accumulation of d-serine. We turned our attention to the transcriptional regulator DsdC. Here we hypothesized that accumulation of d-serine leads to hypermodulation of DsdC activity with expression effects on genes other than at the dsdCXA locus. The mouse colonization results reveal that dsdC is not necessary for hypercolonization by CFT073 dsdA, as CFT073 dsdA dsdC maintains an elevated-colonization phenotype. Further, CFT073 dsdC also displayed a hypercolonization phenotype. Since both DsdC and d-serine are required to transcribe dsdA, it appears that the inability to produce d-serine deaminase results in increased colonization ability. This correlates with our previous observations that d-serine can enter the cell through CycA as well as DsdX and that during murine UTI by CFT073 dsdA, the d-serine-responsive gene dsdX is up-regulated relative to wild-type expression levels (8). These observations further suggest that the increased colonization observed with CFT073 dsdA may not be solely dependent upon a loss of ability to degrade d-serine but may also require intracellular accumulation of d-serine to provide the competitive advantage.
d-Serine accumulation requires expression of one or both of the two d-serine transporters, DsdX and CycA (2). Interestingly a CFT073 cycA dsdXA mutant unable to transport d-serine or to produce DsdA (2) colonized mouse bladders at wild-type levels but was attenuated 10-fold in the kidney compared to the wild type. This suggests that modulation of the colonization phenotype in strains deficient in d-serine deaminase activity results from the intracellular accumulation of exogenous d-serine. In addition we surmise that expression of traits important for CFT073 kidney colonization is aided by uptake of d-serine. We cannot rule out the possibility that there is another substance present during murine UTI that DsdA normally modifies which is accumulated in the dsdA mutant backgrounds, although all indications are that d-serine is the prime substrate.
How d-serine accumulation results in hypercolonization is not clear, but we can envision several possibilities. Examination of the transcriptome of CFT073 dsdA in the context of murine UTI revealed increased expression of P and F1C fimbriae and the hemolysin, along with OmpF (8). The mechanism of these transcriptional changes is unknown but may result from the influence of d-serine on other transcription factors. A similarly broad up-regulation of virulence genes was observed in UPEC strain 536 deleted for H-NS (18). Thus, one hypothesis is that d-serine accumulation in CFT073 dsdA antagonizes H-NS repression of virulence genes, which results in a hypercolonization phenotype in the murine model of UTI.
Another possibility is that d-serine is incorporated directly into a cellular product, such as peptidoglycan, altering the properties of the cell envelope. In vitro, d-serine serves as a substrate for d-alanine-d-alanine ligase, suggesting a potential for d-serine to inhibit the formation of, or even be incorporated into, the terminal positions of peptidoglycan (26). The peptidoglycan of seven different species of bacteria becomes less cross-linked and contains d-serine instead of d-alanine at the fourth and fifth peptide positions when d-serine is added to different complex media (29). Alterations in peptidoglycan structure alter the virulence phenotype of Salmonella enterica serovar Typhimurium (17). A Serratia marcescens dapA strain, unable to synthesize the peptidoglycan components lysine and mesodiaminopimelate, has elevated hemolysin-related cytotoxicity and an increased ability to attach to surfaces (27). These alterations in peptidoglycan when paired with our previous observations of genes up-regulated in CFT073 dsdA in vivo (26) suggest that remodeling of peptidoglycan may occur following intracellular accumulation of d-serine. We are currently investigating these different hypotheses.
Acknowledgments
We thank Laura Walters, Holly Hamilton, Shahaireen Pellett, Rose Szabady, and Gary Baisa for help in experiments and manuscript preparation.
This research was supported by NIH grant R01DK063250. B.J.H. was also supported by NIH National Research Service Award T32 GM07215.
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. [DOI] [PubMed] [Google Scholar]
- 2.Anfora, A. T., and R. A. Welch. 2006. DsdX is the second d-serine transporter in uropathogenic Escherichia coli clinical isolate CFT073. J. Bacteriol. 188:6622-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomfield, I. C., M. S. McClain, and B. I. Eisenstein. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439-1445. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, T., and C. W. Keevil. 1997. A simple artificial urine for the growth of urinary pathogens. Lett. Appl. Microbiol. 24:203-206. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, A., P. Roesch, L. Davis, R. Moritz, S. Pellett, and R. A. Welch. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 74:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosloy, S. D. 1973. d-Serine transport system in Escherichia coli K-12. J. Bacteriol. 114:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haugen, B. J., S. Pellett, P. Redford, H. L. Hamilton, P. L. Roesch, and R. A. Welch. 2007. In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect. Immun. 75:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heincz, M. C., S. M. Bornstein, and E. McFall. 1984. Purification and characterization of d-serine deaminase activator protein. J. Bacteriol. 160:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, Y., T. Nishikawa, K. Satoh, T. Iwata, T. Fukushima, T. Santa, H. Homma, and K. Imai. 1998. Urinary excretion of d-serine in human: comparison of different ages and species. Biol. Pharm. Bull. 21:156-162. [DOI] [PubMed] [Google Scholar]
- 11.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 12.Litwin, M. S., C. S. Saigal, E. M. Yano, C. Avila, S. A. Geschwind, J. M. Hanley, G. F. Joyce, R. Madison, J. Pace, S. M. Polich, M. Wang, and Urologic Diseases in America Project. 2005. Urologic Diseases in America Project: analytical methods and principal findings. J. Urol. 173:933-937. [DOI] [PubMed] [Google Scholar]
- 13.Maas, W. K., R. Maas, and E. McFall. 1995. d-Serine deaminase is a stringent selective marker in genetic crosses. J. Bacteriol. 177:459-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFall, E. 1975. Escherichia coli K-12 mutant forming a temperature-sensitive d-serine deaminase. J. Bacteriol. 121:1074-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moritz, R. L., and R. A. Welch. 2006. The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA. J. Clin. Microbiol. 44:4038-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouslim, C., F. Hilbert, H. Huang, and E. A. Groisman. 2002. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45:1019-1027. [DOI] [PubMed] [Google Scholar]
- 18.Müller, C. M., U. Dobrindt, G. Nagy, L. Emödy, B. E. Uhlin, and J. Hacker. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nørregaard-Madsen, M., E. McFall, and P. Valentin-Hansen. 1995. Organization and transcriptional regulation of the Escherichia coli K-12 d-serine tolerance locus. J. Bacteriol. 177:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prüss, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redford, P., and R. A. Welch. 2006. Role of sigma E-regulated genes in Escherichia coli uropathogenesis. Infect. Immun. 74:4030-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redford, P., P. L. Roesch, and R. A. Welch. 2003. DegS is necessary for virulence and is among extraintestinal Escherichia coli genes induced in murine peritonitis. Infect. Immun. 71:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roesch, P. L., P. Redford, S. Batchelet, R. L. Moritz, S. Pellett, B. J. Haugen, F. R. Blattner, and R. A. Welch. 2003. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 49:55-67. [DOI] [PubMed] [Google Scholar]
- 25.Roos, V., M. A. Schembri, G. C. Ulett, and P. Klemm. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799-1806. [DOI] [PubMed] [Google Scholar]
- 26.Sato, M., K. Kirimura, and K. Kino. 2005. d-Amino acid dipeptide production utilizing d-alanine-d-alanine ligases with novel substrate specificity. J. Biosci. Bioeng. 99:623-628. [DOI] [PubMed] [Google Scholar]
- 27.Soo, P. C., J. R. Wei, Y. T. Horng, S. C. Hsieh, S. W. Ho, and H. C. Lai. 2005. Characterization of the dapA-nlpB genetic locus involved in regulation of swarming motility, cell envelope architecture, hemolysin production, and cell attachment ability in Serratia marcescens. Infect. Immun. 73:6075-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su, H., and E. B. Newman. 1991. A novel l-serine deaminase activity in Escherichia coli K-12. J. Bacteriol. 173:2473-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trippen, B., W. P. Hammes, K. H. Schleifer, and O. Kandler. 1976. Mode of action of d-amino acids on the biosynthesis of peptidoglycan. Arch. Microbiol. 109:247-261. (In German.) [DOI] [PubMed] [Google Scholar]
- 30.Wargel, R. J., C. A. Shadur, and F. C. Neuhaus. 1970. Mechanism of d-cycloserine action: transport systems for d-alanine, d-cycloserine, l-alanine, and glycine. J. Bacteriol. 103:778-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]