Abstract
Streptococcus gordonii colonization of damaged heart surfaces in infective endocarditis is dependent upon the recognition of host receptors by specific bacterial surface proteins. However, despite several attempts to identify the mechanisms involved in this interaction, the nature of the bacterial proteins required remains poorly understood. This study provides clear evidence that several S. gordonii surface proteins participate in the interaction with platelets to support platelet adhesion and induce platelet aggregation. S. gordonii strains were found to support strong (DL1-Challis, SK12, SK184, and Blackburn) or moderate (UB1545 Δhsa and CH1-Challis) adhesion or failed to support platelet adhesion (M5, M99, and Channon). In addition, under flow conditions, platelets rolled and subsequently adhered to immobilized S. gordonii at low shear (50 s−1) in an Hsa-dependent manner but did not interact with S. gordonii DL1 at any shear rate of >50 s−1. S. gordonii strains either induced (DL1-Challis, SK12, SK184, UB1545 Δhsa, and M99) or failed to induce (M5, CH1-Challis, Channon, and Blackburn) platelet aggregation. Using a proteomic approach to identify differential cell wall protein expression between aggregating (DL1) and nonaggregating (Blackburn) strains, we identified antigen I/antigen II family proteins SspA and SspB. The overexpression of SspA or SspB in platelet-nonreactive Lactococcus lactis induced GPIIb/GPIIIa-dependent platelet aggregation similar to that seen with S. gordonii DL1. However, they failed to support platelet adhesion. Thus, S. gordonii has distinct mechanisms for supporting platelet adhesion and inducing platelet aggregation. Differential protein expression between strains may be important for the pathogenesis of invasive diseases such as infective endocarditis.
Infective endocarditis (IE) describes a family of persistent microbial infections that typically target previously damaged or diseased heart valves (24). Most often, IE occurs after infection by viridans streptococci or Staphylococcus aureus (25). IE is characterized by the development of an infected platelet thrombus on a heart valve resulting in valve failure and almost certain death in the absence of treatment. Even with treatment, mortality can be as a high as 20%, as early signs are nonspecific and diagnosis is often delayed or uncertain (23).
Mammalian platelets are small (2 to 4 μm), discoid, short-lived fragments derived from megakaryocyte precursors (15). Platelets play a crucial role not only in the formation of a normal hemostatic plug but also in the formation of a pathological thrombus, particularly within arteries subjected to high shear stress (26). Platelets circulate in a resting state through blood vessels lined by an endothelial layer. Following an insult to the endothelial layer, platelets undergo a series of changes. Upon adhesion, platelets become activated, change shape, secrete granule contents, and aggregate to prevent blood loss (2). Thus, platelets rely on specific adhesive interactions with extracellular matrices (e.g., collagen, von Willebrand factor [vWf], and fibronectin). In order to achieve firm adhesion, platelets possess many receptors specific for extracellular matrix proteins.
Streptococcus gordonii is a prominent member of the viridans group of oral bacteria that occur primarily on the tooth surface (18). S. gordonii, among others, is well known for its ability to colonize damaged heart valves and is among the bacteria most frequently identified as being the primary etiological agents of subacute bacterial endocarditis (8). Thus, the pathogenic potential of these bacteria at nonoral sites may well depend on specific surface proteins that normally function primarily in the colonization of the tooth surface. Many attempts at identifying the bacterial proteins responsible for binding to platelets have been made (10). Early studies suggested that S. gordonii strains could not interact with or stimulate platelet activation (7). However, more recent studies have shown that S. gordonii can support platelet adhesion and induce platelet aggregation (16, 34, 35, 39), interactions that might contribute to the establishment of thrombotic vegetations found in IE patients. A key step leading to the development of a thrombotic vegetation is the interaction of proteins expressed on the surface of S. gordonii with specific platelet membrane receptors.
Recent studies have demonstrated that S. gordonii GspB or its close homologue, Hsa, a highly glycosylated surface-anchored protein, interacts with platelets through the recognition of specific sialic acid residues found on the major platelet adhesion receptor GPIbα (4, 34, 35). The primary function of GspB/Hsa is to support bacterial adhesion to the tooth pellicle (36); therefore, the ability to interact with platelets through GPIbα is a novel function for these proteins. Most species of oral streptococci express high-molecular-mass cell wall-associated antigen I (AgI)/AgII family polypeptides, designated SspA (172 kDa) and SspB (164 kDa), in S. gordonii (27). These polypeptide adhesins recognize multiple ligands including salivary agglutinin glycoprotein (gp-340) (30), collagen type I (13), β1 integrins (27), and other oral microorganisms such as Porphyromonas gingivalis, Candida albicans, and Actinomyces naeslundii (6, 9, 17, 21). Even though SspA and SspB are 96% identical, there is supporting evidence that there are different ligand binding properties and differential expression between AgI and AgII protein family members (14).
Recently, we demonstrated that the deletion of Hsa from S. gordonii reduced platelet adhesion under static conditions (∼40% inhibition) and had little or no effect on platelet aggregation (16). Furthermore, the deletion of SspA/SspB alone from S. gordonii failed to affect the ability to support platelet adhesion or induce platelet aggregation. These results suggest that other surface proteins expressed on S. gordonii may be responsible for supporting platelet adhesion and inducing platelet aggregation. In this paper, we used a proteomic approach to identify surface-expressed proteins of S. gordonii that may be responsible for interacting with human blood platelets.
MATERIALS AND METHODS
Materials.
The platelet agonist thrombin receptor-activating peptide (TRAP) was purchased from Bio-Data (Horsham, PA). Monoclonal antibody Abciximab was obtained from Eli Lilly (Leiden, The Netherlands). The peptide RGDS (arginine, glycine, aspartic acid, and serine) was purchased from Calbiochem (Nottingham, United Kingdom). Brain heart infusion (BHI) and M17 media were purchased from Oxoid (Basingstoke, United Kingdom). All other laboratory reagents were purchased from Sigma-Aldrich.
Bacterial strains and growth conditions.
Sources and strains of S. gordonii used in this study are listed in Table 1. Deletion of hsa in S. gordonii DL1 by allelic exchange mutagenesis to generate strain UB1545 Δhsa was described previously (16). S. gordonii strains were maintained on blood agar and subsequently grown in BHI broth in sealed tubes incubated statically overnight (stationary phase) at 37°C for use in experiments. Lactococcus lactis MG1363 was maintained on GM17 agar plates and subsequently grown in GM17 medium containing 0.5% glucose (14) in sealed tubes for 16 h at 30°C. Plasmids derived from pUB1000 were maintained in L. lactis by including 5 μg/ml erythromycin in the growth medium. Bacteria were harvested and washed by centrifugation at 15,000 × g for 5 min, washed three times, and finally resuspended in phosphate-buffered saline (pH 7.5). For all experiments, S. gordonii suspensions were adjusted to an optical density (OD) at 600 nm (OD600) of 1.0 for adhesion studies and an OD600 of 1.6 for aggregation studies.
TABLE 1.
S. gordonii strains used in this study
| Strain | Source |
|---|---|
| DL1-Challis | R. Cole, National Institutes of Health, Bethesda, MD |
| SK12 | M. Kilian, Arhus University, Denmark |
| M5 | B. Rosan, University of Pennsylvania, Philadelphia, PA |
| CH1-Challis | D. Clewell, University of Michigan, Ann Arbor, MI |
| UB1545 Δhsa | H. Jenkinson, University of Bristol, Bristol, United Kingdom |
| M99 | P. Sullam, University of California, San Francisco, CA |
| Channon | R. Cole, National Institutes of Health, Bethesda, MD |
| Blackburn | R. Cole, National Institutes of Health, Bethesda, MD |
Lectin binding assay.
To assess cell surface expression of Hsa or GspB, levels of binding of succinylated wheat germ agglutinin (sWGA; EY Laboratories, San Mateo, CA) to immobilized streptococcal cells were determined. Bacteria were cultured in tryptone-yeast extract-glucose medium (17) at 37°C for 16 h. Cells were harvested, washed in Tris-buffered saline (TBS) (50 mM Tris-HCl [pH 7.5] containing 0.15 M NaCl), and suspended in TBS to approximately 1 × 109 cells/ml. Portions (50 μl) were added to wells of an Immobilon (Nunc) microtiter plate and incubated at 37°C for 2 h. Wells were washed with TBS, and sWGA (2 μg/ml in TBS containing 1% bovine serum albumin [BSA], 1 mM CaCl2, and 1 mM MgCl2) was added. Plates were incubated at 25°C for 1 h with gentle shaking and washed three times with TBS. Streptavidin-horseradish peroxidase (0.1 μg/ml in TBS) was added and incubated at 25°C for 45 min. After three washes with TBS, o-phenylenediamine (0.5 mg/ml in 50 mM phosphate citrate buffer [pH 5.0]) was added, plates were incubated at 25°C for 20 min, and the absorbance at 450 nm was measured.
Platelet preparation.
Whole blood was drawn from the antecubital vein of healthy volunteers who had abstained from taking nonsteroidal anti-inflammatory drugs in the previous 10 days. Ethical approval for collection of blood was obtained from the Royal College of Surgeons in Ireland Ethics Committee. To prevent coagulation, 9 volumes of blood were added to 1 volume of acid-citrate-dextrose or 3.8% sodium citrate. Platelet-rich plasma (PRP) was prepared by centrifugation of anticoagulated whole blood at room temperature at 150 × g for 10 min. Platelets were separated from plasma proteins by gel filtration. PRP was adjusted to pH 6.5 with acid-citrate-dextrose, and apyrase (1 U/ml) and prostaglandin E1 (1 μM) were added prior to centrifugation at room temperature at 630 × g for 10 min. The resultant supernatant of platelet-poor plasma was removed, and the platelet pellet was resuspended in 2 ml modified HEPES-Tyrode's buffer (JNL buffer) (6 mM dextrose, 130 mM NaCl, 9 mM NaHCO3, 10 mM Na citrate, 10 mM Tris base, 3 mM KCl, 0.8 mM KH2PO4, and 0.9 mM MgCl2). The platelet suspension was then applied to a chromatograph column containing 5 ml packed Sepharose-2B, which was previously equilibrated with JNL buffer. The resultant platelet fractions were pooled. The platelet concentration was adjusted to 2 × 108 platelets/ml on a Sysmex-100 particle counter.
Static platelet adhesion assay.
Microtiter plates (96 wells) were coated with 0.1 ml bacteria (OD of 1.0), fibrinogen (20 μg/ml), or BSA (20 μg/ml). The plate was incubated at 37°C for 2 h. Following this, the plate was washed and blocked with 1% BSA for a further 1 h at 37°C. The plate was washed three times in JNL buffer to remove any unbound protein. Fifty microliters of gel-filtered platelets (final concentration of 2 × 108 platelets/ml) was added to each well and allowed to adhere for 30 min at 37°C. Each well was gently washed three times with 0.1 ml JNL buffer to remove any nonadhered platelets. Adherent platelets were then lysed with 0.1 ml lysis buffer containing a substrate for acid phosphatase (0.1 M Na acetate [pH 5.5], 0.1% Triton X-100, 10 mM p-nitrophenol phosphate) and incubated for 2 h at 37°C. The reaction was stopped by the addition of 1 M NaOH to the mixture. The resultant color was read at 405 nm in a microtiter plate reader (Wallac Victor2; Perkin-Elmer).
S. gordonii-induced platelet aggregation.
Platelet aggregation was assessed by monitoring light transmission using a PAP-4 platelet aggregometer (Bio/Data Corp., Horsham, PA). Platelets were tested for normal responses to TRAP (5 μM) and/or ADP (20 μM). Bacteria were prepared as described above for platelet adhesion assays. Fifty microliters of bacterial cells (OD600 of 1.6) was mixed with 450 μl PRP. The light transmission of PRP without added bacteria and the light transmission of platelet-poor plasma were defined as 0% and 100% light transmission, respectively, and platelet aggregation was expressed as a final percentage of light transmission after 25 min. Five independent assays were performed for each strain.
Shear-induced platelet adhesion.
The ability of platelets to adhere to immobilized S. gordonii DL1 under flowing conditions was investigated using a parallel flow chamber (Glycotech, Rockville, MD). Bacteria were washed and harvested as described above. Glass coverslips were coated with bacteria for 2 h at room temperature. Unbound bacteria were removed by gentle washing with phosphate-buffered saline three times. Coverslips were subsequently blocked with 1% BSA for 60 min at 37°C. Platelets (3 × 109) were perfused over the immobilized bacteria at a shear rate of 50 s−1 or 800 s−1. Platelet interaction was visualized by phase-contrast microscopy (63× LD-Achroplan objective) using a Zeiss Axiovert-200 epifluorescence microscope. Images were captured every second up to 480 s by a liquid-chilled Quantix-57 charge-coupled-device camera (Photometrics Ltd., Tucson, AZ). Platelet rolling was analyzed using MetaMorph (Universal Imaging Corp., Downingtown, PA).
Cell wall fractionation of S. gordonii.
S. gordonii cells were grown to stationary phase (18 h) in BHI broth without shaking at 37°C. Cell wall proteins were isolated as described previously (19). In brief, S. gordonii cells (OD of 1.6) were centrifuged at 15,000 × g for 5 min, and the pellet was incubated in 1 ml Tris-EDTA-lysozyme (TEL) solution (100 mM Tris-HCl [pH 8] containing 5 mM EDTA, 1.0% lysozyme, and protease inhibitor cocktail) for 3 h at 37°C with constant inversion. The supernatant containing released cell wall proteins was collected by centrifugation at 3,000 × g for 10 min at 4°C. AgI/AgII polypeptides in cell wall extracts of different S. gordonii strains were detected by Western immunoblot analysis using polyclonal antibodies (diluted 1:5,000) to Streptococcus mutans AgI/AgII as described previously (16).
Gel electrophoresis liquid chromatography-tandem mass spectrometry.
For tryptic digestion and subsequent mass spectrometry analysis, 50 μg of S. gordonii cell wall proteins was separated on 4 to 20% gradient precast acrylamide gels (Pierce) using a Miniprotean 3 apparatus (Bio-Rad), and the resulting gel was stained with colloidal Coomassie brilliant blue G-250 (Bio-Rad). Visible banding differences in gel lanes from S. gordonii extracts were excised and digested in gel with trypsin according to methods described previously by Shevchenko et al. (31). The resulting peptide mixtures were resuspended in 1% formic acid and analyzed by nanoelectrospray liquid chromatography mass spectrometry. A high-performance liquid chromatography instrument (Dionex) was interfaced with an LTQ ion trap mass spectrometer (ThermoFinnigan). Chromatography buffer solutions (buffer A [5% acetonitrile and 0.1% formic acid] and buffer B [80% acetonitrile and 0.1% formic acid]) were used to deliver a 60-min gradient (35 min to 45% buffer B, 10 min to 90%, hold for 10 min, 3 min to 5%, and hold for 15 min). A flow rate of 2 μl/min was used at the electrospray source. Spectra were searched using the XTANDEM algorithm (5) against the UniProt Knowledgebase (release 7.0). Proteins were considered identified if the XTANDEM score was more negative than a threshold of −5.
Expression of SspA and SspB proteins in L. lactis.
For the expression of SspA and SspB polypeptides on the surface of L. lactis cells, sspA and sspB genes were amplified from S. gordonii DL1 and cloned into plasmid pUB1000 (17). The pUB1000 vector contains the promoter region of a constitutively expressed L. lactis gene, usp45, fused to the leader peptide coding sequence of S. gordonii sspA. Downstream are the target sequence for sortase recognition and anchoring to the cell wall and a Rho-independent transcription termination element. A unique SalI site is immediately downstream of the signal sequence, and a unique BamHI site is upstream of the LPxTG sortase consensus element. In-frame cloning of amplified sspA or sspB into the SalI/BamHI sites results in the constitutive expression of SspA and SspB proteins on the lactococcal cell surface (17).
Statistical analysis.
Statistical analyses were performed using InStat Statistical (GraphPad software) and Excel (Microsoft) software. Data shown are the means plus or minus standard errors of the means (SEM), and comparisons between mean values were performed using the Student paired t test.
RESULTS
S. gordonii-platelet adhesion.
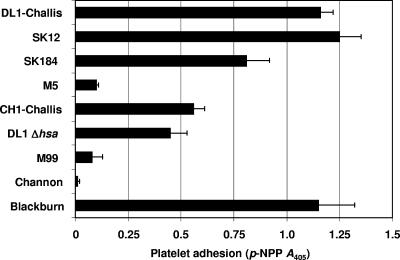
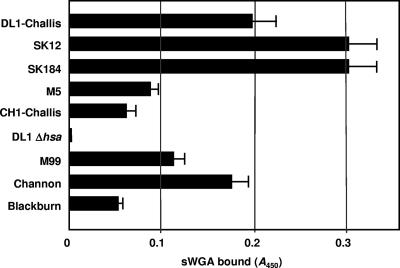
Platelet adhesion to immobilized bacteria is thought to be the initial step leading to thrombosis buildup. Using different strains of S. gordonii, we identified three significantly different phenotypes capable of supporting platelet adhesion (P < 0.05) (Fig. 1). Strains DL1-Challis, SK12, SK184, and Blackburn all supported strong platelet adhesion. Strain CH1-Challis supported moderate platelet adhesion, while strains M5, M99, and Channon failed to support adhesion. To determine cell surface expression levels of Hsa or GspB, we utilized sWGA, a lectin with binding specificity for N-acetyl-d-glucosamine (GlcNAc) present on mature GspB (3, 4). We have previously shown by Western and dot blot analyses that sWGA specifically recognizes Hsa on the surface of S. gordonii DL1 (16). sWGA detects a single diffuse high-molecular-mass band at ∼300 kDa present in cell wall extracts from S. gordonii DL1 but absent from wall extracts from strain UB1545 Δhsa (16). In a lectin binding assay, sWGA did not react with immobilized cells of strain UB1545 Δhsa (Fig. 2). Surface expression levels of Hsa-like proteins were greater on strains SK12 and SK184 than on strain DL1 but much lower on strains M5 and Blackburn (Fig. 2). Significantly, strain CH1-Challis expressed <40% of the level of sWGA-interacting glycoprotein of DL1-Challis, even though these strains were thought to be identical.
FIG. 1.
Platelet adhesion to immobilized S. gordonii under static conditions. Strains of S. gordonii were immobilized on to the surface of plastic wells and incubated with plasma-free platelets (2 × 108 platelets/ml). Wells were washed, and adhered platelets were lysed using 0.1% Triton X-100. As a measure of the number of platelets bound, alkaline phosphatase activity was determined by the change in absorbance (A) at 405 nm. The P value was <0.05.
FIG. 2.
Determination of relative levels of cell surface expression of Hsa or GspB using sWGA. S. gordonii cells (2 × 107) were applied to plastic wells, fixed, and reacted with biotin-conjugated sWGA and peroxidase-linked streptavidin, as described in Materials and Methods. Strains M99, Channon, and Blackburn each carried a gspB-like gene, while all the other strains contained hsa-like genes (not shown).
Consistent with previous observations (16, 34), strain UB1545 Δhsa was only about 50% reduced in its ability to bind platelets (Fig. 1). These results suggest that additional factors, other than Hsa, contribute to static platelet adherence. In addition, platelet adhesion to immobilized S. gordonii occurred in the absence of any plasma proteins, therefore suggesting a direct interaction between a specific platelet membrane receptor and a bacterial cell wall-specific protein. Furthermore, platelets adhered to immobilized S. gordonii in a calcium-dependent manner (data not shown). S. gordonii DL1 was selected as a representative of the adherent strains and used for all subsequent studies.
S. gordonii-induced platelet aggregation.
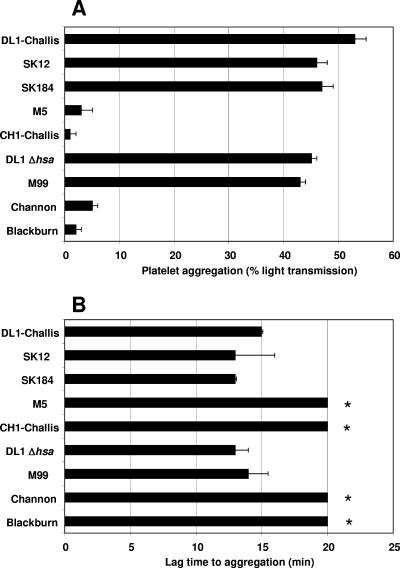
A number of strains of S. gordonii were tested for their abilities to induce platelet aggregation (Fig. 3A). Aggregation occurred in an all-or-nothing response whereby altering the concentration of the bacteria led to a change in lag time but did not affect the maximum aggregation. Lag time can be defined as the time from the addition of bacteria to PRP to the first signs of platelet aggregation. Comparison of lag times (Fig. 3B) to those of wild-type strain DL1, which we previously demonstrated to induce platelet aggregation (16), identified two significantly different groups (P < 0.05). One group had a lag time of 14 ± 1 min, and the other group failed to induce platelet aggregation (no observable signs of platelet aggregation after 20 min). Strains DL1, SK12, SK184, and M99 induced platelet aggregation, whereas strains M5, CH1, Channon, and Blackburn failed to do so. Interestingly, UB1545 Δhsa did induce platelet aggregation.
FIG. 3.
S. gordonii-induced platelet aggregation. (A) Strains of S. gordonii were tested for their abilities to induce platelet aggregation. S. gordonii (OD600 of 1.6) was added to platelets in a stirring cuvette heated to 37°C. Platelet aggregation was assayed by light transmission at 37°C using a PAP-4 aggregometer (Bio-Data). Results are expressed as percent aggregation. (B) Lag time is expressed as the time(s) from the addition of bacteria to the platelets to the first signs of aggregation. Bars represent mean values from at least five independent experiments, and error bars indicate SEM. * indicates that no aggregation was seen after 20 min. The P value was <0.05.
Platelets were incubated with the GPIIb/GPIIIa inhibitor Abciximab or the common integrin recognition motif blocking peptide RGDS. Aggregation experiments were then preformed using either DL1 (as a representative of the proaggregatory strains) or the platelet agonist TRAP. Our results demonstrate that these inhibitors prevented platelet aggregation by S. gordonii DL1 and TRAP (P < 0.01) (Table 2). This indicates that S. gordonii DL1-induced platelet aggregation is indeed true aggregation involving fibrinogen binding to GPIIb/GPIIIa (an essential step in aggregate formation).
TABLE 2.
Inhibition of S. gordonii-induced platelet aggregation by GPIIb/IIIa inhibitorsa
| Agonist | Platelet aggregation (%) ± SEM
|
||
|---|---|---|---|
| Control | Abciximab | RGDS peptide | |
| TRAP | 73 ± 7 | 0* | 0* |
| S. gordonii DL1 | 51 ± 5 | 0* | 0* |
PRP was pretreated with GPIIb/GPIIIa inhibitors Abciximab (10 μg/ml) or RGDS (500 μM) or no inhibitor (control) for 10 min before the addition of the agonists (20 μM TRAP or S. gordonii DL1 [OD of 1.6]). * indicates a P value of <0.01.
Identification of S. gordonii surface receptors responsible for supporting shear-induced platelet adhesion.
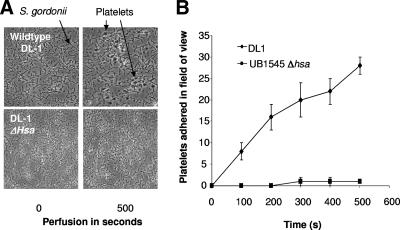
Several previous publications in the last few years suggested the importance of the interactions between S. gordonii Hsa and platelet GPIbα (16, 34, 35). Indeed, we have demonstrated that the deletion of Hsa from the surface of strain DL1 (UB1545 Δhsa) reduces platelet adhesion by 60% (16). In vivo, bacteria would interact with platelets under shear conditions rather than under static conditions to support platelet adhesion. Therefore, we investigated this interaction by perfusing PRP over immobilized S. gordonii DL1. Almost immediately upon commencement of low shear (50 s−1), the platelets began to interact with DL1 cells with a typical “rolling” fashion followed by firm adhesion. In contrast to this, platelet adhesion to the Hsa deletion mutant (UB1545 Δhsa) was ablated when PRP was perfused at low shear (50 s−1) (Fig. 4A and B). Additionally, platelets failed to “roll” or adhere to immobilized DL1 at a higher shear rate of 500 s−1 or above (data not shown). At sites of injury, circulating platelets typically bind to endothelium-bound vWf via the membrane receptor GPIbα. In control experiments, platelets failed to interact with immobilized vWf under low-shear conditions (50 s−1) but “rolled” across immobilized vWf at a shear rate of >1,000 s−1.
FIG. 4.
Platelet adhesion to immobilized S. gordonii under low-shear conditions. PRP was perfused over immobilized S. gordonii at 50 s−1. Platelet rolling followed by firm adhesion was visualized using phase-contrast microscopy. Images were captured every second up to 300 s. Images are representative fields taken from one of three independent experiments that yielded similar results.
Identification of S. gordonii surface receptors responsible for inducing platelet aggregation.
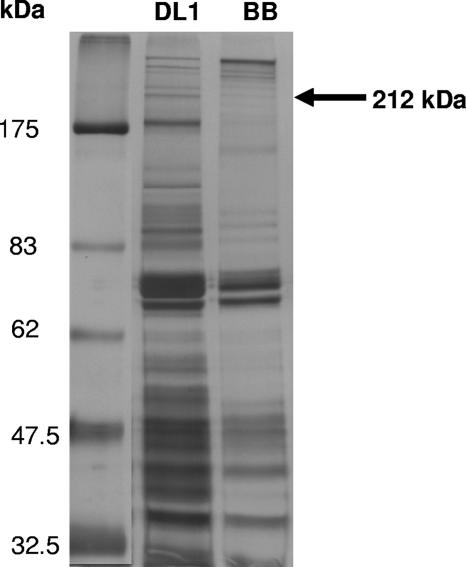
We used a proteomic approach to identify the proteins responsible for inducing platelet aggregation by comparing S. gordonii DL1 (induces platelet aggregation) and S. gordonii Blackburn (representative strain that failed to induce platelet aggregation). S. gordonii cells at the stationary phase of growth were incubated with TEL, and supernatant samples containing cell wall proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A number of differences in the levels of protein expression between S. gordonii strains DL1 and Blackburn were apparent (Fig. 5). Bands present in DL1 and absent in Blackburn were excised, digested with trypsin, and subjected to tandem mass spectrometry analysis. The AgI/AgII family agglutinin receptor precursor SSP-5 (or SspA/SspB) protein was positively identified by 11 unique peptides with an XTandem score of −96.8 in the ∼212-kDa band from S. gordonii DL1, which was absent from Blackburn (Table 3).
FIG. 5.
Profile of proteins expressed on the surface of S. gordonii DL1 or Blackburn (BB). Cell wall proteins were extracted with TEL buffer (see Materials and Methods) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein concentrations in each sample loaded onto the gel were identical as determined by Bio-Rad assay (20 μg/ml). The arrow indicates the band that was excised from DL1 and identified by mass spectrometry as SspA/SspB. A major 180-kDa band present in DL1 and absent in Blackburn identifies as a putative cell surface protein unrelated to SspA/SspB.
TABLE 3.
Identification of SspA/SspB by tandem mass spectrometrya
| Unique peptide no. for SSP5_STRGN protein | Peptide sequence |
|---|---|
| 1 | YEADLAAIKK |
| 2 | LAAYQTELAR |
| 3 | NAAIKAENEEIK |
| 4 | NEDGVDIDR |
| 5 | TEALTAGRPK |
| 6 | TTSFVLVDPLPTGYK |
| 7 | GNQVSGVSVQQYDSLEAAPK |
| 8 | GAFQLFSADNPEEFYK |
| 9 | TFSSLNLTMK |
| 10 | VAYASNTVR |
| 11 | VEDPSAPIPVSVGK |
The agglutinin receptor precursor SSP-5 (SspA/SspB) protein was positively identified with an XTandem score of −96.8 in the 212-kDa band of S. gordonii strain DL1 only. The UniProt accession number is P16952, and the Etest score is 1.5E − 05.
Effect of S. gordonii surface protein SspA/SspB expressed in L. lactis on platelet aggregation.
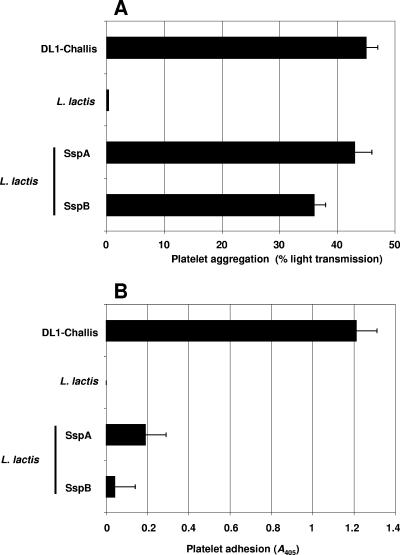
We previously reported that the deletion of SspA and SspB from S. gordonii DL1 did not significantly affect platelet aggregation (16). One explanation for this could be that S. gordonii-induced platelet aggregation is a multifactorial event in which the bacteria have evolved to contain compensatory proteins to induce platelet aggregation. Therefore, we used surrogate host L. lactis for the expression of proteins of interest (17). L. lactis failed to induce platelet aggregation or support platelet adhesion, suggesting that it does not express proteins capable of interacting with platelets. The expression of SspA or SspB in L. lactis led to platelet aggregation with a lag time and a percent aggregation similar to those of S. gordonii DL1 (Fig. 6A). Platelet aggregation induced by L. lactis expressing either SspA or SspB was inhibited by Abciximab (43% ± 3% for L. lactis SspA versus 0% for Abciximab and 36% ± 2% for L. lactis SspB versus 0% for Abciximab), suggesting that this event is indeed due to true platelet aggregation (P < 0.05). Consistent with previous observations (16), SspA and SspB did not play a role in supporting platelet adhesion (P value was not significant) (Fig. 6B).
FIG. 6.
Platelet interaction with L. lactis strains expressing SspA and SspB. (A) L. lactis strains (OD600 of 1.6) were individually added to platelets in a stirring cuvette heated to 37°C. Platelet aggregation was assayed by light transmission at 37°C using a PAP-4 aggregometer. L. lactis carrying an empty vector failed to induce platelet aggregation; however, L. lactis expressing SspA or SspB induced platelet aggregation (P < 0.01). Results are expressed as percent aggregation. (B) L. lactis cells expressing SspA or SspB were immobilized onto the surface of plastic wells and incubated with plasma-free platelets (2 × 108 platelets/ml). Wells were washed, and adhered platelets were lysed using 0.1% Triton X-100. As a measure of the number of bound platelets, alkaline phosphatase activity was determined by the change in A405. L. lactis expressing SspA or SspB failed to support platelet adhesion (P value was not significant). Bars represent mean values from at least five independent experiments, and error bars indicate SEM.
DISCUSSION
The ability of viridans streptococci to interact with human blood platelets is thought to be a critical factor in the pathogenesis of infective endocarditis. There have been many attempts to identify the molecular mechanisms underlying these interactions. Recent studies identified a family of cell wall-anchored, serine-rich repeat proteins found on several streptococci including GspB/Hsa of S. gordonii (4, 33), Fap1 of Streptococcus parasanguinis (38), and SrpA of Streptococcus sanguinis (28). All of these glycoproteins recognize sialyl-T antigen, an O-linked saccharide found on platelet GPIbα. These studies provide details on the specific proteins mediating streptococcus-platelet interactions. However, none of these studies addressed the role of these proteins in platelet function, such as their ability to induce platelet aggregation or support platelet adhesion.
Platelet adhesion to immobilized S. gordonii varies between strains. This appears to be a direct protein-protein interaction, as plasma proteins were not required for adhesion. These results are consistent with our previous observations where we demonstrated that S. gordonii binds soluble GPIb (glycocalacin) in a plasma-free environment (16). The extent of platelet adhesion to immobilized S. gordonii may be associated with expression levels of serine-rich glycoproteins. In particular, DL1, SK12, and SK184 express higher levels of surface Hsa, while strain M5 showed a lower expression level (Fig. 2). The correlation between serine-rich glycoprotein expression and platelet adhesion was not so evident for M99, Channon, and Blackburn, which produce GspB-like proteins. It is possible that GspB binding activity may be more sensitive to the conformation of GPIbα than Hsa. GspB recognizes only carbohydrates in the membrane-proximal portion of GPIbα, in contrast to Hsa, which also binds N-linked sugars near the tips of the α and β chains (3). This may offer an explanation as to why M99 does not support adhesion in our experiments. Deletion of Hsa from DL1 (UB1545 Δhsa) reduced platelet adhesion by 60%, most likely due to the inability of platelet GPIbα to bind to S. gordonii. Consistent with previous findings that Hsa binds to GPIbα (3, 33), we have demonstrated that the preincubation of platelets with a monoclonal antibody to GPIbα reduced platelet adhesion by 50% (data not shown). These results suggest that under static conditions, at least two S. gordonii proteins are involved in supporting platelet adhesion, Hsa and an unidentified protein or proteins. Studies are undergoing at present to identify this protein. Strains DL1, SK12, SK184, and Blackburn express both proteins; strains CH1-Challis and UB1545 Δhsa express an unidentified protein (not Hsa); and strains M5, M99, and Channon fail to express either protein.
A major limitation to our current understanding stems from the fact that all of the previous studies have been carried out under static conditions. It has been argued in the literature that data obtained in vitro using static binding assays may not be relevant to the fluid dynamic environment encountered in the vasculature (37). There are a growing number of papers in the literature from the last few years suggesting that the local fluid environment of the circulation critically affects the molecular pathways of cell-cell interactions (1, 22). Under fluid shear conditions, platelets interacted with immobilized S. gordonii with a typical rolling behavior followed by firm adhesion. This rolling behavior followed by firm adhesion is typical of platelet interactions with subendothelial matrix proteins at sites of vessel injury. This interaction typically occurs as a result of platelet GPIbα binding to endothelium-bound vWf. This interaction occurs under high-shear conditions but not under low-shear conditions (2). Deletion of Hsa from DL1 (UB1545 Δhsa) ablated platelet interactions with immobilized S. gordonii under shear conditions. We therefore conclude that Hsa is critical for firm adhesion under shear conditions and that this event is most likely mediated by an interaction with platelet GPIbα. Furthermore, Hsa must exist in a suitable conformation for direct interaction with GPIbα under low-shear conditions.
Platelet aggregation is an artificial in vitro process that is used to study one aspect of thrombus formation. In vivo, thrombus formation usually occurs when platelets bind to a damaged endothelial layer. These platelets become activated and secrete agents such as ADP that activate nearby platelets and recruit them into the growing thrombus. This recruitment phase is the in vivo counterpart to aggregation. It is well established that there is a lag time in bacterially induced platelet aggregation in vitro. Lag time is defined as the time from the addition of bacteria to the platelets to the first signs of aggregation. The lag time is representative of the recognition of binding moieties within membrane receptors and binding of plasma proteins, including specific antibody, fibrinogen, or complement (11, 12, 20, 32). Therefore, bacteria can induce platelet aggregation via several mechanisms: specific antibody binding to the platelet Fc receptor, complement formation on the bacterial surface, or simply bacteria binding directly to a membrane platelet receptor. Unlike platelet adhesion, platelet aggregation is dependent on the presence of plasma proteins. Experiments to identify the nature of this interaction are under way. Platelet aggregation induced by S. gordonii varied between strains. The reason for this variation is not yet clear, except that different strains most likely express a different profile of surface proteins that interact with platelets. In particular, strains DL1, SK12, SK184, and M99 induced platelet aggregation with a similar lag time of 12 to 15 min, while strains M5, CH1-Challis, Channon, and Blackburn all failed to induce platelet aggregation after 20 min. Interestingly, deletion of Hsa (UB1545 Δhsa) failed to prevent platelet aggregation (Fig. 3). Strain CH1, a subculture of the Challis strain isolated over 50 years ago, has been documented to carry a point mutation within the secA2 coding sequence. This generates a translational stop at codon 770 (instead of at codon 793), a truncated polypeptide, and, presumably, a loss of protein function (3). SecA2 facilitates the transport (secretion) of Hsa and possibly other proteins from the cytoplasm. It is likely that alterations in the expression of a protein other than Hsa in strain CH1 results in an inability to prevent platelet aggregation. We have been unable to identify the same secA2 point mutation, or any other nonsense mutation, within the C-terminal coding regions of secA2 from any of the other S. gordonii isolates (data not shown). Therefore, it is possible that expression levels of a proaggregatory protein could be altered through some other mechanism in nonaggregating strains such as M5, Channon, and Blackburn.
These results suggest that S. gordonii-induced platelet aggregation is a more complex process than platelet adhesion. To identify the proteins responsible for inducing platelet aggregation, we used a proteomic approach to determine differences in protein expression between DL1 (representative of the strains that induce platelet aggregation) and Blackburn (representative of the strains that failed to induce platelet aggregation). A major differential protein band with an apparent molecular mass of 212 kDa was identified, SspA/SspB. Previously, we demonstrated that the deletion of SspA/SspB from DL1 had no effect on platelet aggregation or platelet adhesion (16). However, the deletion of Hsa and SspA/SspB from DL1 ablated platelet aggregation, suggesting that these proteins work in concert or may play a compensatory role in inducing platelet aggregation, whereby the deletion of one protein allows the other protein to interact with platelets and induce aggregation (16). To confirm the primary role of SspA and SspB in inducing platelet aggregation, each protein was overexpressed in the surrogate host L. lactis. The results presented here suggest that SspA and SspB are both individually capable of inducing platelet aggregation. By contrast, and consistent with previous results, neither SspA nor SspB supported platelet adhesion, further suggesting specific roles for these proteins in inducing platelet aggregation.
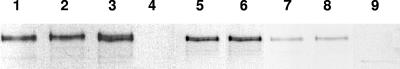
In summary, S. gordonii has the ability to support platelet adhesion and induce platelet aggregation, critical steps leading to the development of a thrombus. In this paper, we describe two S. gordonii proteins that play an essential role in each of these events. Under static conditions, two proteins mediate strong platelet adhesion, Hsa binding to platelet GPIbα and a second interaction that remains uncharacterized. Under low-shear conditions, platelets interact with immobilized S. gordonii in a typical “rolling” fashion, an interaction that is mediated by Hsa. S. gordonii-induced platelet aggregation is mediated by three proteins, Hsa, SspA, and SspB. Platelet aggregation still occurs in the absence of Hsa or in the absence of SspA/SspB; however, deletion of the genes encoding all three proteins effectively ablates aggregation (16). On the basis of our studies, strains of S. gordonii may be characterized as platelet aggregation and adhesion positive (Agg+ Adh+) (DL1-Challis, SK12, and SK184), aggregation positive and adhesion negative (Agg+ Adh−) (M99), Agg− Adh+ (CH1-Challis and Blackburn), or Agg− Adh− (Channon and M5). Strains M5 and Blackburn do not produce detectable levels of AgI/AgII in cell wall extracts (Fig. 7), while strains M99 and Channon show much lower levels of AgI/AgII than does DL1 (Fig. 7). Reduced AgI/AgII expression thus tends to correlate with the Agg− phenotype, supporting the evidence that AgI/AgII proteins play a major role in platelet aggregation. Taken collectively, these results suggest that S. gordonii-induced platelet aggregation is a multifactorial event involving at least three individual proteins. The next step is to identify the fourth adhesin responsible for supporting static platelet adhesion and identify the platelet receptors involved in inducing platelet aggregation.
FIG. 7.
Western immunoblot analysis of SspA/SspB (AgI/AgII) expression in cell wall extracts of S. gordonii strains. Lanes: 1, Challis-DL1; 2, SK12; 3, SK184; 4, M5; 5, Challis-CH1; 6, DL1 Δhsa; 7, M99; 8, Channon; 9, Blackburn. Equal amounts of protein (5 μg) were loaded into each lane, and nitrocellulose blots were reacted with polyclonal AgI/AgII antiserum (diluted 1:5,000), as described in Materials and Methods, to detect a doublet band of ∼200 kDa.
Acknowledgments
We thank Gerard Cagney for his useful discussions and help with the proteomics.
This work was funded by a postdoctoral research fellowship to S.W.K. (PD2005/06), a program grant to D.C. (RP2002/09) from the Health Research Board of Ireland, and a Wellcome Trust project grant (064832) to H.F.J.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 24 September 2007.
REFERENCES
- 1.Bahou, W. F., L. Scudder, D. Rubenstein, and J. Jesty. 2004. A shear-restricted pathway of platelet procoagulant activity is regulated by IQGAP1. J. Biol. Chem. 279:22571-22577. [DOI] [PubMed] [Google Scholar]
- 2.Baruch, D. 2006. Platelet-vessel wall interactions. Therapie 61:371-378. (In French.) [DOI] [PubMed] [Google Scholar]
- 3.Bensing, B. A., J. A. Lopez, and P. M. Sullam. 2004. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibα. Infect. Immun. 72:6528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081-1094. [DOI] [PubMed] [Google Scholar]
- 5.Craig, R., and R. C. Beavis. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466-1467. [DOI] [PubMed] [Google Scholar]
- 6.Demuth, D. R., D. C. Irvine, J. W. Costerton, G. S. Cook, and R. J. Lamont. 2001. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69:5736-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas, C. W., P. R. Brown, and F. E. Preston. 1990. Platelet aggregation by oral streptococci. FEMS Microbiol. Lett. 60:63-67. [DOI] [PubMed] [Google Scholar]
- 8.Douglas, C. W., J. Heath, K. K. Hampton, and F. E. Preston. 1993. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39:179-182. [DOI] [PubMed] [Google Scholar]
- 9.Egland, P. G., L. D. Du, and P. E. Kolenbrander. 2001. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 69:7512-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., T. J. Foster, and D. Cox. 2006. The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 4:445-457. [DOI] [PubMed] [Google Scholar]
- 11.Ford, I., C. W. Douglas, D. Cox, D. G. Rees, J. Heath, and F. E. Preston. 1997. The role of immunoglobulin G and fibrinogen in platelet aggregation by Streptococcus sanguis. Br. J. Haematol. 97:737-746. [DOI] [PubMed] [Google Scholar]
- 12.Ford, I., C. W. Douglas, J. Heath, C. Rees, and F. E. Preston. 1996. Evidence for the involvement of complement proteins in platelet aggregation by Streptococcus sanguis NCTC 7863. Br. J. Haematol. 94:729-739. [DOI] [PubMed] [Google Scholar]
- 13.Heddle, C., A. H. Nobbs, N. S. Jakubovics, M. Gal, J. P. Mansell, D. Dymock, and H. F. Jenkinson. 2003. Host collagen signal induces antigen I/II adhesin and invasin gene expression in oral Streptococcus gordonii. Mol. Microbiol. 50:597-607. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, A. R., C. Gilbert, J. M. Wells, and H. F. Jenkinson. 1998. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect. Immun. 66:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Italiano, J. E., Jr., and R. A. Shivdasani. 2003. Megakaryocytes and beyond: the birth of platelets. J. Thromb. Haemost. 1:1174-1182. [DOI] [PubMed] [Google Scholar]
- 16.Jakubovics, N. S., S. W. Kerrigan, A. H. Nobbs, N. Stromberg, C. J. van Dolleweerd, D. M. Cox, C. G. Kelly, and H. F. Jenkinson. 2005. Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect. Immun. 73:6629-6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakubovics, N. S., N. Stromberg, C. J. van Dolleweerd, C. G. Kelly, and H. F. Jenkinson. 2005. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55:1591-1605. [DOI] [PubMed] [Google Scholar]
- 18.Jenkinson, H. F., and R. J. Lamont. 2005. Oral microbial communities in sickness and in health. Trends Microbiol. 13:589-595. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, P., P. B. Maguire, M. Bennett, D. J. Fitzgerald, R. J. Edwards, B. Thiede, A. Treumann, J. K. Collins, G. C. O'Sullivan, F. Shanahan, and C. Dunne. 2005. Correlation of probiotic Lactobacillus salivarius growth phase with its cell wall-associated proteome. FEMS Microbiol. Lett. 252:153-159. [DOI] [PubMed] [Google Scholar]
- 20.Kerrigan, S. W., I. Douglas, A. Wray, J. Heath, M. F. Byrne, D. Fitzgerald, and D. Cox. 2002. A role for glycoprotein Ib in Streptococcus sanguis-induced platelet aggregation. Blood 100:509-516. [DOI] [PubMed] [Google Scholar]
- 21.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 22.Lehoux, S., and A. Tedgui. 2004. Shear and signal transduction in the endothelial cell. Med. Sci. (Paris) 20:551-556. (In French.) [DOI] [PubMed] [Google Scholar]
- 23.Lukes, A. S., D. K. Bright, and D. T. Durack. 1993. Diagnosis of infective endocarditis. Infect. Dis. Clin. N. Am. 7:1-8. [PubMed] [Google Scholar]
- 24.Moreillon, P., and Y. A. Que. 2004. Infective endocarditis. Lancet 363:139-149. [DOI] [PubMed] [Google Scholar]
- 25.Moreillon, P., Y. A. Que, and A. S. Bayer. 2002. Pathogenesis of streptococcal and staphylococcal endocarditis. Infect. Dis. Clin. N. Am. 16:297-318. [DOI] [PubMed] [Google Scholar]
- 26.Ni, H., and J. Freedman. 2003. Platelets in hemostasis and thrombosis: role of integrins and their ligands. Transfus Apher. Sci. 28:257-264. [DOI] [PubMed] [Google Scholar]
- 27.Nobbs, A. H., B. H. Shearer, M. Drobni, M. A. Jepson, and H. F. Jenkinson. 2007. Adherence and internalization of Streptococcus gordonii by epithelial cells involves beta1 integrin recognition by SspA and SspB (antigen I/II family) polypeptides. Cell. Microbiol. 9:65-83. [DOI] [PubMed] [Google Scholar]
- 28.Plummer, C., H. Wu, S. W. Kerrigan, G. Meade, D. Cox, and C. W. Ian Douglas. 2005. A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br. J. Haematol. 129:101-109. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Prakobphol, A., F. Xu, V. M. Hoang, T. Larsson, J. Bergstrom, I. Johansson, L. Frangsmyr, U. Holmskov, H. Leffler, C. Nilsson, T. Boren, J. R. Wright, N. Stromberg, and S. J. Fisher. 2000. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J. Biol. Chem. 275:39860-39866. [DOI] [PubMed] [Google Scholar]
- 31.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 32.Sullam, P. M., G. A. Jarvis, and F. H. Valone. 1988. Role of immunoglobulin G in platelet aggregation by viridans group streptococci. Infect. Immun. 56:2907-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi, Y., A. L. Sandberg, S. Ruhl, J. Muller, and J. O. Cisar. 1997. A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to α2-3-linked sialic acid-containing receptors. Infect. Immun. 65:5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi, Y., A. Yajima, J. O. Cisar, and K. Konishi. 2004. Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect. Immun. 72:3876-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takamatsu, D., B. A. Bensing, H. Cheng, G. A. Jarvis, I. R. Siboo, J. A. Lopez, J. M. Griffiss, and P. M. Sullam. 2005. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol. Microbiol. 58:380-392. [DOI] [PubMed] [Google Scholar]
- 36.Takamatsu, D., B. A. Bensing, A. Prakobphol, S. J. Fisher, and P. M. Sullam. 2006. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect. Immun. 74:1933-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varki, A. 1994. Selectin ligands. Proc. Natl. Acad. Sci. USA 91:7390-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, H., K. P. Mintz, M. Ladha, and P. M. Fives-Taylor. 1998. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol. Microbiol. 28:487-500. [DOI] [PubMed] [Google Scholar]
- 39.Yajima, A., Y. Takahashi, and K. Konishi. 2005. Identification of platelet receptors for the Streptococcus gordonii DL1 sialic acid-binding adhesin. Microbiol. Immunol. 49:795-800. [DOI] [PubMed] [Google Scholar]