Abstract
Treponema denticola, a spirochete indigenous to the oral cavity, is associated with host inflammatory responses to anaerobic polymicrobial infections of the root canal, periodontium, and alveolar bone. However, the cellular mechanisms responsible for the recognition of T. denticola by the innate immune system and the underlying cell signaling pathways that regulate the inflammatory response to T. denticola are currently unresolved. In this study, we demonstrate that T. denticola induces innate immune responses via the utilization of Toll-like receptor 2 (TLR2) but not TLR4. Assessment of TLR2/1 and TLR2/6 heterodimers revealed that T. denticola predominantly utilizes TLR2/6 for the induction of cellular responses. Analysis of the mitogen-activated protein kinase (MAPK) signaling pathway in T. denticola-stimulated monocytes identified a prolonged up-regulation of the MAPK extracellular signal-related kinase 1/2 (ERK1/2) and p38, while no discernible increase in phospho-c-Jun N-terminal kinase 1/2 (JNK1/2) levels was observed. With the aid of pharmacological inhibitors selectively targeting ERK1/2 via the mitogen-activated protein kinase/extracellular signal-related kinase 1/2 kinase and p38, we further demonstrate that ERK1/2 and p38 play a major role in T. denticola-mediated pro- and anti-inflammatory cytokine production.
Treponema denticola's increased presence within the periodontium of the host is associated with localized inflammation (8, 9, 10, 18, 22, 30). Moreover, T. denticola is part of the microbial “BANA-positive red complex” (T. denticola, Tannerella forsythia, and Porphyromonas gingivalis) associated with advanced periodontal disease (19, 32). Dental implant failures also have numerous spirochetes in the infected infrabony area between the implant and the inflamed peri-implant tissue (17, 21, 27). Localized alveolar osteitis or “dry socket” is a painful postextraction anaerobic infection where a blood clot fails to form and increased numbers of T. denticola cells have been cultured from these extraction sites (23). Direct smears of pericoronitis exudates indicate these acutely infected areas have increased levels of treponemes (37). In addition, spirochetes have been observed in endodontic infections and associated apical regions of the tooth (4, 31, 33). Clearly, the oral spirochetes are integral components of a polymicrobial anaerobic flora that infect and cause inflammatory diseases within the oral cavity.
The host's immune response to periodontium-associated pathogens has been shown to be an important determinant in the pathology of periodontal disease by regulating the production of inflammatory cytokines involved in gingival inflammation and alveolar bone loss (7, 13, 14, 26). The ability of the host to recognize and initially respond to microbes as well as microbial components has been largely attributed to a family of type I transmembrane Toll-like receptors (TLRs) (20, 34, 38). Previous studies analyzing how T. denticola stimulates innate immune responses have demonstrated that T. denticola can stimulate human gingival epithelial cells (HGECs) through TLR2 (3). However, the functional significance of other TLRs involved in mediating cellular responses to T. denticola and how this recognition mediates qualitative and quantitative aspects of the host inflammatory response are currently not well understood (29). The aim of the current study was to characterize the TLRs involved in the ability of T. denticola to stimulate innate immune cells and to identify the underlying cell signaling pathways that regulate the inflammatory response to T. denticola.
MATERIALS AND METHODS
Strains and culture conditions.
T. denticola strain ATCC 35405 was cultured under anaerobic conditions using a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) at 35°C in an atmosphere of 80% nitrogen, 10% hydrogen, and 10% carbon dioxide. The strain was grown in modified NOS medium (brain heart infusion broth, 12.5 g/liter; trypticase, 10 g/liter; yeast extract, 2.5 g/liter; sodium thioglycolate, 0.5 g/liter; l-cysteine, 1 g/liter; l-asparagine, 0.25 g/liter; sodium bicarbonate, 0.2%; heat-inactivated rabbit serum, 10%; thiamine pyrophosphate, 0.0006%) (15) to late log phase (approximately 109 cells/ml), centrifuged at 4,420 × g, and washed three times in phosphate-buffered saline without calcium or magnesium (PBS) (Cambrix Bio Science, Walkersville, MD). Cells were brought to a final cell density of 1010 cells/ml in PBS, utilizing dark-field microscopy and a Petroff-Hausser counting chamber (Hausser Scientific, Horsham, PA).
Cell culture.
Human peripheral blood mononuclear cells (PBMC) were obtained from three to five healthy donors (University of Louisville, Institutional Review Board, Human Subjects Protection Program, study number 503.05) and isolated from citrated venous blood by collecting the buffy coat and eliminating red blood cell contamination with Histopaque 1077 (Sigma-Aldrich, St. Louis, MO) density gradients. Human monocytes were purified from PBMC samples by negative selection with a monocyte isolation system (Miltenyi Biotec, Auburn, CA). This procedure routinely resulted in samples that were more than 95% pure CD14+ cells, as shown by flow cytometry. C57BL/6 wild-type, TLR1−/−, TLR2−/−, TLR4−/−, and TLR6−/− female mice, 6 to 8 weeks of age, were used as the source of peritoneal macrophages. Mice were injected by the intraperitoneal route with 3 ml of 3% thioglycolate broth. After 4 days, peritoneal cells were harvested by flushing the peritoneal cavity with 10 ml of ice-cold PBS. After being washed in PBS containing 1% fetal calf serum and 2 mM EDTA, cells were resuspended at a concentration of 2.5 × 106 cells/ml in RPMI 1640 supplemented with 10% fetal bovine serum, 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate, 2 mM l-glutamine, 20 mM HEPES, 50 U of penicillin/ml, and 50 μg of streptomycin/ml (RPMI 1640 complete medium). HEK293 (human embryonic kidney) cell lines stably expressing TLR2 or TLR4 in conjunction with an NF-κB-dependent secreted alkaline phosphatase were grown in RPMI 1640 complete medium according to the supplier's protocol (Invivogen, San Diego, CA). To assess the functional role of extracellular signal-related kinase 1/2 (ERK1/2) and p38 in T. denticola-induced cytokine production by PBMCs, cells were pretreated for 2 h with the mitogen-activated protein kinase (MAPK) kinase (MEK) inhibitor U0126 (50 μM), the p38 inhibitor SB203580 (10 μM), or a dimethyl sulfoxide (DMSO) control (0.01%). After a 20-h stimulation in RPMI 1640 complete medium, cytokine levels in cell supernatants were measured by enzyme-linked immunosorbent assay (ELISA) according to the protocol suggested by the supplier (eBioscience; San Diego, CA).
Reagents.
All cell culture reagents were purchased from Gibco Technologies (Grand Island, NY). HEK293 cell lines stably expressing TLR2 or TLR4 along with an NF-κB-dependent secreted alkaline phosphatase reporter gene were purchased from Invivogen (San Diego, CA). TLR2-specific (FSL1) and TLR4-specific (Escherichia coli lipopolysaccharide) agonists were used as appropriate controls (Invivogen, San Diego, CA). Antibodies used for the detection of total and phosphorylated, extracellular signal-regulated kinases (ERK1/2) and p38 MAPK were obtained from Cell Signaling (Beverly, MA). MAPK inhibitors U0126 (MEK1/2 inhibitor) and SB203580 (p38) were obtained from Calbiochem (San Diego, CA).
Western blot.
Macrophages (5 × 106/ml; 1-ml volume) in 24-well plates were pretreated with RPMI 1640 complete medium, U0126 (MEK1/2 inhibitor; MEK1/2 is the direct upstream kinase responsible for phosphorylating ERK1/2), SB203580 (p38 inhibitor), or 0.01% DMSO for 2 h before the addition of T. denticola. At various time points, cells were washed with ice-cold PBS and then lysed on ice for 10 min in RIPA buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1 mM sodium orthovanadate, 1 mM sodium fluoride, 1 mM phenyl methylsulfonyl fluoride, and 1 mg/ml aprotonin). The resulting whole-cell lysates were passed through a 20-gauge needle three times and then incubated on ice for an additional 30 min. Cell debris were pelleted by centrifugation, and the supernatants were collected and stored at −20°C until assayed. Twenty micrograms of total cellular protein was suspended in lithium dodecyl sulfate buffer, heated for 10 min at 70°C, resolved by lithium dodecyl sulfate-polyacrylamide gel electrophoresis, and then transferred to polyvinylidene difluoride membranes using Novex (Invitrogen, Carlsbad, CA). Probing and visualization of immunoreactive bands were performed using ECL Plus (Amersham Pharmacia Biotech, Denver, CO), following the supplier's protocol. Levels of phosphorylated and total proteins were determined using densitometer scans of the blots using a Kodak Image Station 4000MM system (Eastman Kodak, New Haven, CT).
Statistics.
The significance of differences between groups was evaluated by analysis of variance and the Tukey multiple comparison test using the Instat program (GraphPad, San Diego, CA). Differences between groups were considered significant if P values were <0.05.
RESULTS
T. denticola utilizes TLR2 and not TLR4.
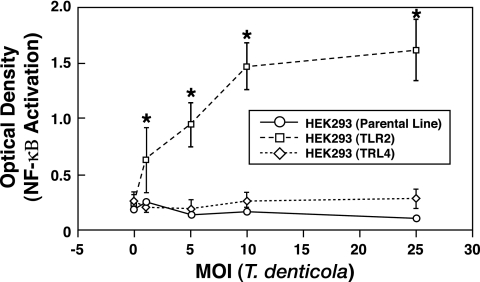
To initially identify the role of TLR2 and TLR4 usage by T. denticola, HEK293 cells expressing TLR2 or TLR4 were stimulated with T. denticola. Compared to either nonstimulated HEK293 cells expressing TLR2 or TLR4 or the parental line, HEK293 cells expressing TLR2 showed a significant increase (P < 0.05) in NF-κB activation at all multiplicities of infection (MOIs) tested when stimulated with T. denticola (Fig. 1). In sharp contrast, HEK293 cells expressing TLR4 did not exhibit any discernible (P > 0.05) increase in NF-κB activity above nonstimulated levels when stimulated with T. denticola (Fig. 1). These results demonstrate that T. denticola can stimulate NF-κB activation via TLR2 but not TLR4.
FIG. 1.
T. denticola utilizes TLR2 and not TLR4 to activate NF-κB. HEK293 cells expressing TLR2 or TLR4 were stimulated with various MOIs of T. denticola for 24 h and then analyzed for activation of NF-κB by the secreted alkaline phosphatase assay. An asterisk indicates statistical significance with a P value of <0.05 compared to results for parental HEK293 cells. The data represent the arithmetic means ± standard deviations for five separate experiments.
T. denticola does not induce innate immune responses in the absence of TLR2.
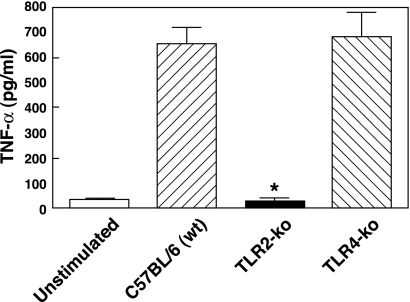
Although the data shown in Fig. 1 demonstrate that T. denticola can utilize TLR2, they do not definitively rule out the possibility that additional TLRs may be involved in the ability of T. denticola to stimulate innate immune responses. Therefore, we next employed the use of TLR2 and TLR4 knockout mice in order to determine if T. denticola was capable of mediating cellular responses in the absence of TLR2. Peritoneal macrophages obtained from wild-type, TLR2-deficient, or TLR4-deficient mice were stimulated with T. denticola and assessed for tumor necrosis factor alpha (TNF-α) production. As shown in Fig. 2, no significant (P > 0.05) differences were observed in the levels of TNF-α between wild-type and TLR4-deficient macrophages stimulated with T. denticola. These results are in agreement with those shown in Fig. 1, demonstrating that TLR4-expressing HEK293 cells did not exhibit activation of NF-κB when stimulated with T. denticola. However, in contrast to either wild-type or TLR4-deficient cells, TLR2-deficient macrophages stimulated with T. denticola did not induce any significant increase in TNF-α compared to levels for nonstimulated control (Fig. 2). Taken together, these data demonstrate that T. denticola requires TLR2 to stimulate innate immune responses.
FIG. 2.
T. denticola requires TLR2 for the activation of innate immune responses. Peritoneal macrophages obtained from wild-type (wt), TLR2-deficient (TLR2-ko), or TLR4-deficient (TLR4-ko) mice were stimulated with T. denticola and assessed for TNF-α production 20 h poststimulation. The asterisk indicates statistical significance at P values of <0.05 compared to results for wild-type cells. The data represent the arithmetic means ± standard deviations for three separate experiments.
TLR2 coreceptor utilization.
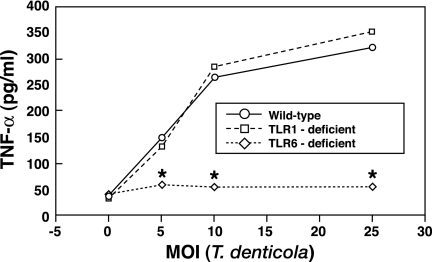
TLR2 can heterodimerize with TLR1 or TLR6, thus altering its interactions and ability to respond to microbial ligands (25, 35, 36). Since our data obtained with HEK293 cells transfected with TLR2 (TLR2 positive) as well as data obtained from TLR2-deficient macrophages demonstrated that cellular responses to T. denticola were mediated by TLR2, we next employed the use of TLR1- and TLR6-deficient macrophages in order to identify the role of TLR2/1 and TLR2/6 in mediating innate immune responses to T. denticola. Compared to wild-type cells, TLR6-deficient macrophages stimulated with T. denticola produced significantly (P < 0.05) less TNF-α at all MOIs tested (Fig. 3). Moreover, a direct comparison of the relative contributions mediated by TLR1 and TLR6 indicated that TLR6-deficient macrophages stimulated with T. denticola produced significantly less (P < 0.05) TNF-α than T. denticola-stimulated TLR1-deficient macrophages, whereas no significant differences were observed in the levels of TNF-α produced by wild-type or TLR1-deficient macrophages (Fig. 3). These data show that T. denticola preferentially utilizes TLR2/6 complexes to induce innate immune responses.
FIG. 3.
Assessment of which TLR2 (TLR2/1 or TLR2/6) coreceptor complex is utilized by T. denticola demonstrates that TLR6 but not TLR1 is involved in mediating cellular responses. TLR6 knockout macrophages produced significantly (P < 0.05) less TNF-α than wild-type macrophages stimulated with T. denticola. An asterisk indicates statistical significance at P values of <0.05 compared to results for wild-type cells. The data represent the arithmetic means ± standard deviations for three separate experiments.
Activation of MAPKs in T. denticola-stimulated macrophages.
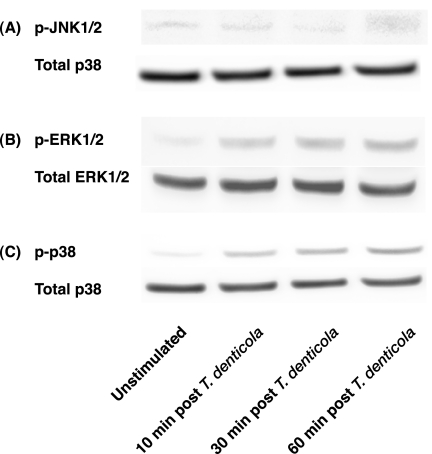
The MAPKs, including ERK1/2, c-Jun N-terminal kinase 1/2 (JNK1/2), and p38, have been shown to play a major role in regulating qualitative and quantitative effects on the host inflammatory response (1, 6). Therefore, to initially investigate whether T. denticola could induce the activation of ERK1/2, JNK1/2, and p38, macrophages were stimulated with T. denticola for 10 to 60 min and analyzed for MAPK activation by Western blotting (Fig. 4). No significant differences in the levels of phospho-JNK1/2 were observed between the nonstimulated control (time = 0) and T. denticola-stimulated cells at 10 to 60 min poststimulation (Fig. 4A). In contrast, increased levels of phospho-JNK1/2 could be detected in cells stimulated with lipopolysaccharide (data not shown). Analysis of the levels of phospho-ERK1/2 and phospho-p38 in T. denticola-stimulated cells revealed increases in the levels of phospho-ERK1/2 and phospho-p38 at 10 to 60 min poststimulation compared to nonstimulated control levels (Fig. 4B and C). Moreover, phospho-p38 and phospho-ERK1/2 levels remained elevated and sustained at both the 30- and 60-min-poststimulation time points (Fig. 4B and C).
FIG. 4.
T. denticola-stimulated monocytes exhibit enhanced levels of phospho-ERK1/2 and phospho-p38 levels. Monocytes were stimulated with T. denticola (MOI = 10) for 0 to 60 min and assayed for phosphorylated levels of JNK1/2 (A), ERK1/2 (B), and p38 (C) by Western blotting using 20 μg of whole-cell lysates. Total levels of ERK1/2 and p38 were monitored to ensure equivalent loading.
Functional significance of MAPK activation in T. denticola-mediated cytokine production.
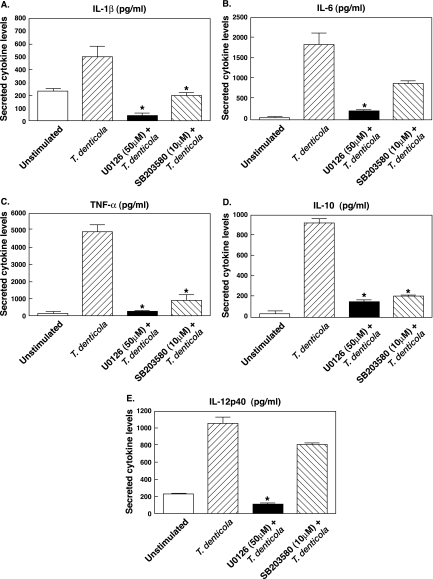
In order to assess and compare the functional role of T. denticola-mediated activation of ERK1/2 and p38, cells were stimulated with T. denticola in the presence or absence of specific inhibitors for ERK1/2 (U0126; MEK1/2 inhibitor) or p38 (SB203580) and assayed for the levels of interleukin 1 beta (IL-1β), IL-6, TNF-α, IL-10, and IL-12 p40 by ELISA. As demonstrated in Fig. 5, T. denticola-stimulated macrophages pretreated with the p38 inhibitor (SB203580) exhibited a significant (P < 0.05) reduction in the levels of IL-1β, TNF-α, and IL-10, whereas the levels of IL-6 and IL-12 p40 were not significantly different from those for cells stimulated with T. denticola alone (Fig. 5). Furthermore, T. denticola-stimulated macrophages pretreated with the ERK1/2 inhibitor showed a significant reduction in all cytokines analyzed compared to results for cells stimulated with T. denticola alone (Fig. 5). These data demonstrate that both p38 and ERK1/2 play a major role in regulating both pro- and anti-inflammatory cytokine production by T. denticola-stimulated immune cells.
FIG. 5.
Functional role of p38 and ERK1/2 in regulating IL-1β (A), IL-6 (B), TNF-α (C), IL-10 (D), and IL-12 p40 (E) production by T. denticola-stimulated (MOI = 10) monocytes. Cells were pretreated for 2 h with the MAPK kinase (MEK) inhibitor U0126 (50 μM), the p38 inhibitor SB203580 (10 μM), or the DMSO control (0.01%). After a 20-h stimulation, cytokine levels in cell supernatants were measured by ELISA. An asterisk indicates statistical significance at a P value of <0.05 compared to results for T. denticola-stimulated cells. The data represent the arithmetic means ± standard deviations for five separate experiments.
DISCUSSION
The host immune response to oral treponemes has been shown to be a key determinant of periodontal inflammation by regulating the production of pro- and anti-inflammatory cytokines that in turn modulate pathogenesis (5, 10, 11, 24, 29). Previous studies by Asai et al. demonstrated that the ability of T. denticola to stimulate IL-8 mRNA from HGECs involved TLR2, since the use of an anti-TLR2 antibody suppressed T. denticola-induced IL-8 production (3); however, the anti-TLR2 antibody used in these studies did not completely inhibit the levels of IL-8 produced by T. denticola-stimulated HGEC. Moreover, the possibility that T. denticola utilized other TLRs in conjunction with TLR2 was not addressed. In this regard, the findings from the present study showed that T. denticola requires TLR2 to stimulate innate immune responses, since TLR2-transfected but not TLR4-transfected HEK293 cells exhibited increases in NF-κB activation. Furthermore, with the aid of TLR2-deficient mouse macrophages, our findings definitively demonstrated that TLR2 is the major TLR utilized by T. denticola for the induction of cellular responses. Moreover, the lack of TNF-α production by T. denticola-stimulated macrophages deficient in TLR2 further suggests that other TLRs are not involved in mediating innate immune responses to T. denticola.
Past studies have shown that TLR2 can mediate cellular immune responses via heterodimerization with TLR1 or TLR6 and that the TLR2/1 or TLR2/6 complex can impart differential recognition of microbial products (25, 35, 36). The current data obtained from HEK293 cells transfected with TLR2 (TLR2 positive) as well as TLR2-deficient macrophages demonstrated that cellular responses to T. denticola were mediated by TLR2. Using TLR1- and TLR6-deficient mouse macrophages, we were able to directly assess the roles of TLR1 and TLR6 in mediating innate immune responses to T. denticola. In this regard, the data obtained from the present study showed that macrophages lacking TLR6 but not TLR1 exhibited reduced cellular responses to T. denticola. Taken together, our studies show that T. denticola requires both TLR2 and TLR6 to induce innate immune responses.
TLR-initiated signaling activates a diverse array of downstream signal transduction pathways that can play critical roles in regulating host inflammatory responses (1, 6). One of the most highly conserved signaling cascades activated in both the innate and adaptive immune systems involves a family of MAPKs including ERK1/2, p38, and JNK1/2. Studies examining the utilization of various MAPK pathways by T. denticola have shown that the MAPKs ERK1/2, JNK1/2, and p38 are strongly but transiently activated in periodontal ligament epithelial cells (16). Purified major surface protein from T. denticola also activated MAPK pathways in human fibroblasts; upon incorporating the use of inhibitors against ERK 1/2 and p38, the authors could inhibit T. denticola-mediated cell proliferation and an increase in cell numbers surviving apoptosis (12). Our current findings demonstrated that T. denticola stimulates the prolonged activation of both ERK1/2 and p38 in monocytes, whereas JNK1/2 activation was not observed. This is in contrast to previous observations that T. denticola cells activate JNK1/2 in periodontal ligament epithelial cells (16) and T. denticola major surface protein activates JNK1/2 in fibroblasts (12). Moreover, lipoteichoic acid from Treponema spp., Treponema maltophilum and Treponema brennaborense, phosphorylates JNK1/2 in myelomonocytic cell lines (28).
Using pharmacological inhibitors for ERK1/2 and p38, we were able to demonstrate that both of these MAPKs play major roles in regulating both pro- and anti-inflammatory cytokine production by T. denticola-stimulated monocytes. Thus, our present findings in conjunction with those of Leung et al. (16) and Jobin et al. (12) suggest that MAPKs play critical roles in regulating innate immune responses to T. denticola. Furthermore, it appears that the activation of different MAPK members, their functional role in regulating innate immune responses, and their kinetics of activation in T. denticola-stimulated cells will vary depending upon the cell type.
The elevated levels of proinflammatory cytokines, including IL-1β, IL-6 and TNF-α, observed in T. denticola-stimulated cultures have also been found in the gingival crevicular fluid and periodontal tissues of patients with inflammatory periodontal disease. Furthermore, IL-1β, IL-6, and TNF-α have been shown to exhibit potent bone resorptive effects due to their pro-osteoclastic properties (14). Clearly, T. denticola has pathogen-associated molecular patterns that are capable of stimulating proinflammatory cytokines essential for initiating and maintaining periodontitis. However, T. denticola-stimulated cultures also exhibited significantly elevated levels of the anti-inflammatory cytokine IL-10, which can potently attenuate host inflammatory responses from both innate and adaptive immune cells, as well as inhibiting osteoclastogenesis. Interestingly, recent studies directly comparing the host-protective effects of IL-10 and IL-12, which are known to be antagonistic to one another, demonstrated that IL-12 plays a more protective role than IL-10 with a Porphyromonas gingivalis murine abscess model (2). Thus, although the balance of pro- and anti-inflammatory cytokine production will likely contribute to the severity of disease under a chronic state of inflammation, other factors, including cytokines that augment cell-mediated immunity, also are likely to be critical for the control and clearance of periodontal pathogens. In toto, net cytokine expression will determine the level of symbiotic détente between the subgingival microflora and the periodontium of the host (10, 13).
In summary, we demonstrate that T. denticola induces innate immune responses via the utilization of TLR2/6 and that the MAPKs ERK1/2 and p38 play major roles in T. denticola-mediated pro- and anti-inflammatory cytokine production.
Acknowledgments
This work was supported by U.S. Public Health Service grant R03 DE016040-02 from the National Institute of Dental and Craniofacial Research and in part by The University of Alabama at Birmingham Digestive Diseases Research Development Center grant #P30 DK064400.
We thank David Fisher, Medical Education and Design Services, The University of Alabama at Birmingham, for his valuable assistance in the technical production of the figures for this article. We also thank Julia Schmitz and Robin Lorenz, Departments of Microbiology and Pathology, The University of Alabama at Birmingham, for their expertise and support in dark-field microscopy through the DDRDC Cell and Molecular Pathology Core.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 8 October 2007.
REFERENCES
- 1.Akira, A., and K. Takeda. 2004. Toll-like receptor signaling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Alayan, J., E. Gemmell, P. Ford, S. Hamlet, P. Bird, S. Ivanovski, and C. Farah. 2007. The role of cytokines in a Porphyromonas gingivalis murine abscess model. Oral Microbiol. Immunol. 22:304-322. [DOI] [PubMed] [Google Scholar]
- 3.Asai, Y., T. Jinno, and T. Ogawa. 2003. Oral treponemes and their outer membrane extracts activate human gingival epithelial cells through Toll-like receptor 2. Infect. Immun. 71:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgartner, J. C., S. U. Khemaleelakul, and T. Xia. 2003. Identification of spirochetes (treponemes) in endodontic infections. J. Endod. 29:794-797. [DOI] [PubMed] [Google Scholar]
- 5.Beausejour, A., N. Deslauriers, and D. Grenier. 1997. Activation of the interleukin-1β precursor by Treponema denticola: a potential role in chronic inflammatory periodontal disease. Infect. Immun. 65:3199-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutler, B. 2004. Innate immunity: an overview. Mol. Immunol. 40:845-859. [DOI] [PubMed] [Google Scholar]
- 7.Darveau, R. P. 2000. Oral innate host defense responses: interactions with microbial communities and their role in the development of disease, p. 169-218. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology. Horizon Scientific Press, Norfolk, England.
- 8.Edwards, A., D. Dymock, and H. Jenkinson. 2003. From tooth to hoof: treponemes in tissue-destructive diseases. J. Appl. Microbiol. 94:767-780. [DOI] [PubMed] [Google Scholar]
- 9.Ellen, R., and V. Galimanas. 2005. Spirochetes at the forefront of periodontal infections. Periodontol. 2000 38:13-32. [DOI] [PubMed] [Google Scholar]
- 10.Holt, S. C., and J. L. Ebersole. 2006. The oral spirochetes: their ecology and role in the pathogenesis of periodontal disease, p. 323-356. In J. D. Radolf and S. A Lukehart (ed.), Pathogenic Treponema molecular and cellular biology. Caister Academic Press, Norfolk, England.
- 11.Ishihara, K., and K. Okuda. 1999. Molecular pathogenesis of the cell surface proteins and lipids from Treponema denticola. FEMS Microbiol. Lett. 181:199-204. [DOI] [PubMed] [Google Scholar]
- 12.Jobin, M., I. Virdee, C. McCulloch, and R. Ellen. 2007. Activation of MAPK in fibroblasts by Treponema denticola major outer sheath protein. Biochem. Biophys. Res. Commun. 356:213-218. [DOI] [PubMed] [Google Scholar]
- 13.Kinane, D., D. Demuth, S. Gorr, G. Hajishengallis, and M. Martin. 2007. Human variability in innate immunity. Periodontol. 2000. 45:14-34. [DOI] [PubMed] [Google Scholar]
- 14.Kirkwood, K., M. Taba, Jr., C. Rossa, Jr., P. Preshaw, and W. Giannobile. 2006. Molecular biology of the host-microbe interaction in periodontal diseases. Selected topics: molecular signaling aspects of pathogen-mediated bone destruction in periodontal diseases, p. 259-274. In M. G. Newman, H. H. Takei, P. R. Klokkevold, and F. A. Carranza (ed.), Clinical periodontology, 10th ed. Saunders Elsevier, St. Louis, MO.
- 15.Leschine, S., and E. Canale-Parola. 1980. Rifampin as a selective agent for isolation of oral spirochetes. J. Clin. Microbiol. 12:792-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung, W., Q. Wu, P. Hannam, B. McBride, and V.-J. Uitto. 2002. Treponema denticola may stimulate both epithelial proliferation and apoptosis through MAP kinase signal pathways. J. Periodont. Res. 37:445-455. [DOI] [PubMed] [Google Scholar]
- 17.Listgarten, M., and C. Lai. 1999. Comparative microbiological characteristics of failing implants and periodontally diseased teeth. J. Periodontol. 70:431-437. [DOI] [PubMed] [Google Scholar]
- 18.Loesche, W. 1988. The role of spirochetes in periodontal disease. Adv. Dent. Res. 2:275-283. [DOI] [PubMed] [Google Scholar]
- 19.Loesche, W., W. Bretz, D. Kerschensteiner, J. Stoll, S. Socransky, P. Hujoel, and D. Lopatin. 1990. Development of a diagnostic test for anaerobic periodontal infections based on plaque hydrolysis of benzoyl-dl-arginine-naphthylamide. J. Clin. Microbiol. 28:1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medzhitov, R., P. Preston-Hurlburt, and C. A. Janeway. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394-397. [DOI] [PubMed] [Google Scholar]
- 21.Mombelli, A., and N. Lang. 1994. Microbial aspects of implant dentistry. Periodontol. 2000 4:74-80. [DOI] [PubMed] [Google Scholar]
- 22.Moore, W. E. C. 1987. Microbiology of periodontal diseases. J. Periodont. Res. 22:335-341. [DOI] [PubMed] [Google Scholar]
- 23.Nitzan, D. 1983. On the genesis of “dry socket.” J. Oral Maxillofacial Surg. 41:706-710. [DOI] [PubMed] [Google Scholar]
- 24.Nixon, C., M. Steffen, and J. Ebersole. 2000. Cytokine responses to Treponema pectinovorum and Treponema denticola in human gingival fibroblasts. Infect. Immun. 68:5284-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozinsky, A., D. Underhill, J. Fontenot, A. Hajjar, K. Smith, C. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. USA 97:13766-13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page, R. C. 1991. The role of inflammatory mediators in the pathogenesis of periodontal disease. J. Periodont. Res. 26:230-242. [DOI] [PubMed] [Google Scholar]
- 27.Rams, T., and C. J. Link. 1983. Microbiology of failing dental implants in humans: electron microscopic observations. Oral Implant. 11:93-100. [PubMed] [Google Scholar]
- 28.Schroder, N., D. Pfeil, B. Opitz, K. Michelsen, J. Amberger, U. Zahringer, U. Gobel, and R. Schumann. 2001. Activation of mitogen-activated protein kinases p42/44, p38, and stress activated protein kinases in myelo-monocytic cells by Treponema lipoteichoic acid. J. Biol. Chem. 276:9713-9719. [DOI] [PubMed] [Google Scholar]
- 29.Schroder, N., R. Schumann, and U. Gobel. 2006. Innate and adaptive immune responses to oral treponemes, p. 387-402. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic Treponema molecular and cellular biology. Caister Academic Press, Norfolk, England.
- 30.Sela, M. 2001. Role of Treponema denticola in periodontal diseases. Crit. Rev. Oral Biol. Med. 12:399-413. [DOI] [PubMed] [Google Scholar]
- 31.Siqueira, Jr., J. F., and I. N. Rocas. 2003. PCR methodology as a valuable tool for identification of endodontic pathogens. J. Dent. 31:333-339. [DOI] [PubMed] [Google Scholar]
- 32.Socransky, S., A. Haffajee, M. Cugini, C. Smith, and R. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:34-44. [DOI] [PubMed] [Google Scholar]
- 33.Sunde, P. T., I. Olsen, U. Gobel, D. Theegarten, S. Winter, G. Debelian, L. Tronstad, and A. Moter. 2003. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology 149:1095-1102. [DOI] [PubMed] [Google Scholar]
- 34.Takeda, K., and S. Akira. 2004. Toll-like receptors: ligands and signaling, p. 257-270. In S. H. Kaufmann, R. Medzhitov, and S. Gordan. (ed.), The innate immune response to infection. ASM Press, Washington, DC.
- 35.Takeuchi, O., T. Kawai, H. Sanjo, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, K. Takeda, and S. Akira. 1999. TLR6: a novel member of an expanding toll-like receptor family. Gene 231:59-65. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi, O., S. Sato, T. Horiuchi, K. Hoshino, K. Takeda, Z. Dong, R. L. Modlin, and S. Akira. 2002. Cutting edge: Role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169:10-14. [DOI] [PubMed] [Google Scholar]
- 37.Weinberg, A., D. Nitzan, A. Shteyer, and M. Sela. 1986. Inflammatory cells and bacteria in pericoronal exudates from acute pericoronitis. Int. J. Oral Maxillofacial Surg. 15:606-613. [DOI] [PubMed] [Google Scholar]
- 38.Yang, R. B., M. R. Mark, A. Gray, A. Huang, M. H. Xie, M. Zhang, A. Goddard, W. I. Wood, A. L. Gurney, and P. J. Godowski. 1998. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature 395:284-288. [DOI] [PubMed] [Google Scholar]