Abstract
Chlamydiae are intracellular bacteria that develop within a membrane-bound vacuole called an inclusion. To ensure that the inclusion is a safe niche for chlamydial replication, chlamydiae exploit a number of host cell processes, including membrane-trafficking pathways. Recently, several Rab GTPases were found to associate with the inclusions of various chlamydial species. Here we report that Cpn0585, a Chlamydia pneumoniae inclusion membrane protein (Inc), interacts with multiple Rab GTPases. The results from yeast two-hybrid experiments revealed that an amino-terminally truncated form of Cpn0585 (Cpn0585102-651) interacts with Rab1, Rab10, and Rab11 but not with Rab4 or Rab6. Cpn0585-Rab GTPase interactions are direct and GTP dependent as shown in glutathione S-transferase pull-down assays using native and recombinant Cpn0585. In C. pneumoniae-infected HEp-2 cells transfected with enhanced green fluorescent protein (EGFP)-tagged Rab GTPases, the colocalization with Cpn0585 at the inclusion membrane was partial for EGFP-Rab1 and EGFP-Rab10, but extensive for wild-type EGFP-Rab11A and the constitutively active GTPase-deficient EGFP-Rab11AQ70L. Moreover, Cpn0585 colocalized with EGFP-Rab11AQ70L as early as 2 h postinfection. Upon delivery into live C. pneumoniae-infected cells, Cpn0585628-651-specific antibodies bound to the inclusion membrane, demonstrating that the Rab GTPase-interacting domain of Cpn0585 faces the host cell cytosol. Finally, ectopic expression of Cpn0585102-651 partially inhibited the development of C. pneumoniae inclusions in EGFP. but not in EGFP-Rab11AQ70L-expressing HEp-2 cells. Collectively, these data suggest that Cpn0585 is involved in the recruitment of Rab GTPases to the inclusion membrane and that interfering with this function may adversely impact the fitness of the C. pneumoniae inclusion for chlamydial replication.
Members of the Chlamydiaceae family are obligate intracellular gram-negative bacteria that include the human pathogens Chlamydia trachomatis and Chlamydia pneumoniae. While C. trachomatis is responsible for ocular and sexually transmitted diseases that can result in blindness and infertility, C. pneumoniae is a common cause of upper respiratory infections and pneumonia and has been associated with chronic inflammatory conditions such as atherosclerosis, chronic obstructive pulmonary disease, and asthma (5, 9, 22).
Like all chlamydiae, C. pneumoniae and C. trachomatis have a unique biphasic developmental cycle that alternates between the infectious metabolically inactive elementary body (EB) and the noninfectious, replicating, and metabolically active reticulate body (RB). This developmental cycle occurs within the chlamydial inclusion, a membrane-bound vacuole that is actively modified through processes that require early chlamydial protein synthesis (17). During such remodeling, the inclusion is trafficked to the peri-Golgi region, where it avoids fusion with lysosomes and instead fuses with Golgi-derived exocytic vesicles that are delivered to the inclusion via a multivesicular body (MVB)-dependent pathway (4, 24, 49). These Chlamydia-mediated vesicular trafficking events transform the inclusion into a compartment from which chlamydiae can acquire nutrients and interfere with multiple host cell functions that collectively promote a safe environment for chlamydial replication (4, 19, 34). Despite their importance as potential targets for therapeutic intervention, the chlamydial and corresponding host cell factors that control the intracellular trafficking and fusogenicity of the inclusion are only beginning to be elucidated. Because the inclusion membrane represents the interface between Chlamydia and the host cell, bacterium-derived products translocated through or associated with this membrane are candidate mediators of these processes (44). Elucidating the interactions between chlamydial and host cell proteins is critical for the creation of a safe vacuolar environment for bacterial replication and therefore may reveal potential targets for the development of novel antichlamydial therapies.
Rab GTPases, the largest family of Ras-like small GTPases, are involved in the generation, transport, docking, and fusion of vesicles, as well as in multiple other membrane trafficking functions, mainly through the specific recruitment of effector molecules (38, 64). To execute their functions, Rab GTPases cycle between a cytoplasmic, GDP-bound, inactive state and a membrane-associated, GTP-bound, active state (38, 50, 64). Evidence indicates that a number of intracellular bacterial pathogens, including Mycobacterium tuberculosis, Salmonella enterica serovar Typhimurium, and Legionella pneumophila exploit Rab GTPases to regulate the biogenesis of the phagosomes they inhabit and to ensure their intracellular survival (13, 35, 58).
Analysis of the intracellular localization of several enhanced green fluorescent protein (EGFP)-tagged Rab GTPases in Chlamydia-infected cells revealed that a subset of Rabs is selectively recruited to the chlamydial inclusion in both a species-dependent and a species-independent fashion (46). Specific associations with C. pneumoniae, C. trachomatis, and C. muridarum inclusions were detected for Rab1 (46), which functions in endoplasmic reticulum-to-Golgi and intra-Golgi trafficking (56), and for Rab4 and Rab11 (46), which participate in endosome-to-trans-Golgi network and endosomal trafficking, as well as in transferrin receptor recycling (33, 42, 57, 62). In contrast, association of Rab6, which is involved in Golgi-to-endoplasmic reticulum retrograde trafficking (31, 61), was observed only with C. trachomatis inclusions, while an association of Rab10, another Golgi-associated Rab (10), was detected only with C. pneumoniae and C. muridarum inclusions (46). These findings suggested that chlamydial proteins with access to the host cell cytosol might be involved in the recruitment of Rab GTPases to the inclusion.
Because of their topology at the interface of the chlamydial vacuole and host cell cytosol, members of a family of inclusion membrane proteins termed Incs represent potential bacterial molecules that interact with and recruit Rab GTPases to the inclusion. As a group, Incs from the same or different chlamydial species share minimal primary sequence identity with each other or with proteins in public databases, but they bear a conserved secondary structure consisting of a unique bilobed hydrophobic region of 50 to 80 amino acids and domains that are exposed at the cytosolic face of the inclusion (43, 44). Incs had been suggested to participate in the recruitment of Rab GTPases to the inclusion membrane (46). However, it was not until recently that an Inc from C. trachomatis, namely, CT229, was shown to interact with Rab4A (45).
Recently, we reported that Cpn0585, a C. pneumoniae-specific protein that localizes to the inclusion membrane (30), is targeted by the anti-C. pneumoniae CD8+ T-cell response in infected mice (63). The two defined CD8+ T-cell epitopes (63; C. Cortes and B. Wizel, unpublished data) map to a region of Cpn0585 that is predicted to localize to the cytoplasmic face of the inclusion. Interestingly, certain segments of the putative cytosolic domain of Cpn0585 share sequence similarity with a number of Rab GTPase-binding proteins. Moreover, this domain of Cpn0585 is predicted to include several coiled-coil segments, a secondary structure commonly found among proteins that interact with Rab GTPases (51). Thus, we hypothesized that Cpn0585 is a Rab GTPase-interacting protein.
Here we report that Cpn0585 interacts with several Rab GTPases in a guanine nucleotide-dependent manner and that these interactions occur through a domain of this Inc that faces the cytosol of the host cell. The data obtained in the present study suggest that Cpn0585 functions as an effector molecule that recruits Rabs to the C. pneumoniae inclusion membrane and reveal a biologically relevant link between Cpn0585 and Rab GTPases in the development of C. pneumoniae.
MATERIALS AND METHODS
Chlamydia and cell culture.
EBs of C. pneumoniae isolate Kajaani 6 were propagated within HL cells (University of Washington) (27) in Chlamydia medium as described previously (63). EBs were divided into aliquots in a sucrose-phosphate-glutamate buffer and stored at −70°C. The infectivity, as measured by inclusion-forming units (IFU) of purified organisms, was titrated in cycloheximide-treated (1 μg/ml) HL cell monolayers. HEp-2 cells (ATCC CCL 23; American Type Culture Collection, Manassas, VA) were grown in complete RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT), 20 mM HEPES, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 20 μg of gentamicin/ml (all from Invitrogen, Carlsbad, CA) at 37°C and 5% CO2. HEp-2 cell monolayers were infected with C. pneumoniae at the indicated multiplicity of infection (MOI) in Chlamydia medium, which consists of complete Iscove modified Dulbecco medium (Invitrogen) with 0.5 mg of glucose/ml and 0.26 mg of sodium bicarbonate/ml.
Antibodies.
Peptide affinity-purified rabbit anti-Cpn0585 was prepared by using the carboxy (C)-terminal 23-mer RLQEENAQLRAEVERLEQEQFQG representing residues 629 to 651 of Cpn0585 (Open Biosystems, Huntsville, AL). Other antibodies used were: rabbit anti-Chlamydia EBs (Fitzgerald Industries International, Concord, MA), mouse anti-major outer membrane protein (anti-MOMP; clone RR402; University of Washington) (41), mouse anti-glutathione S-transferase (anti-GST; Invitrogen), mouse anti-hemagglutinin (anti-HA; Invitrogen), Alexa Fluor 594 F(ab′)2-goat anti-rabbit immunoglobulin G (IgG; Invitrogen), Alexa Fluor 594 F(ab′)2-goat anti-mouse IgG (Invitrogen), horseradish peroxidase-conjugated goat anti-rabbit IgG (MP Biomedicals, Solon, OH), and horseradish peroxidase-conjugated goat anti-mouse IgG (Jackson Immunoresearch Laboratories, West Grove, PA).
Plasmid constructions.
For yeast two-hybrid screening, the amino (N)-terminally truncated coding sequence of Cpn0585 (Cpn0585102-651) was amplified by PCR from C. pneumoniae genomic DNA with a forward primer designed with a 5′ EcoRI site (5′-CGG AAT TCC TTT ATG ATT CTC AGG GCC T) and a reverse primer with a 5′ BamHI site (AGA GGA TCC CAT CTA CGA TGT TTT CTT ATC CTT G). The PCR product was cloned into the EcoRI and BamHI sites of pGADT7 containing the GAL4 activation domain (GAL4AD) or pGBKT7 containing the GAL4-binding domain (GAL4BD) (Clontech Laboratories, Mountain View, CA). Rab GTPases with deletions on the sites needed for C-terminal lipid modification were cloned into pGBKT7 as described previously (45, 46). The Rab11B gene was amplified by PCR from a plasmid containing the human EGFP-tagged Rab11B (pEGFP-Rab11) (46) using a forward primer with a 5′ EcoRI site (GAA TTC ATG GGG ACC CGG GAC GAC GAG) and a reverse primer with a 5′ XhoI site (CTC GAG TCA TGC AGC TTG TTG GGC TTC). The PCR product was cloned into pGEM-T Easy (Promega, Madison, WI) and then subcloned into the EcoRI and SalI sites of pGBKT7. To generate pGBKT7-Rab11BQ70LΔCCQNL, we first constructed pEGFP-Rab11BQ70L by site-directed mutagenesis (QuikChange; Stratagene, San Diego, CA) using 5′ and 3′ mutagenic oligonucleotides (GGA CAC CGC TGG CCT GGA GCG CTA CCG CG and CGC GGT AGC GCT CCA GGC CAG CGG TGT CC) and pEGFP-Rab11B (46) as a template. Subsequently, pGBKT7-Rab11B was constructed by PCR amplification of Rab11Q70L from pEGFP-Rab11BQ70L using a forward primer with a 5′ EcoRI site (GAA TTC ATG GGG ACC CGG GAC GAC GAG) and a reverse primer with a 5′ XhoI site (CTC GAG TCA CAG GTT CTG GCC AGC AGT GC). After cloning the PCR product into pCRII-TOPO (Invitrogen), the EcoRI/XhoI fragment containing the Rab11BQ70L coding region was cloned into the EcoRI/SalI site of pGBKT7. Finally, amino acids 214 to 219 (CCQNL) were deleted from pGBKT7-Rab11BQ70L by site-directed mutagenesis using 5′ and 3′ mutagenic oligonucleotides (CAG AAG CCC AAC AAG CTG CAG CAG AAC CTG TGA CTC GAC CTG and CAG GTC GAG TCA CAG GTT CTG CTG CAG CTT GTT GGG CTT CTG). pGBKT7-Rab11BS25NΔCCQNL was constructed by PCR amplification using pEGFP-Rab11BS25N (52) as a template, the forward Rab11B-specific primer described above, and a reverse primer with a 5′ XhoI site (CTC GAG TCA TGC AGC TTG TTG GGC TTC). After the PCR product was cloned into pGEM-T Easy, the EcoRI/XhoI fragment containing the Rab11BS25N coding region was cloned into the EcoRI/SalI site of pGBKT7. For immunofluorescence studies, mammalian expression plasmids containing human EGFP-tagged Rab1, Rab4A, Rab4AQ67L, Rab6A, Rab10, and Rab11A were constructed as previously described (45, 46, 52). pEGFP-Rab11AQ70L was constructed by PCR mutagenesis using the forward primer with a 5′ EcoRI site (GAA TTC ATG GGC ACC CGC GAC GAC GAG), reverse primer with a 5′ XhoI site (CTC GAG TTA GAT GTT CTG ACA GCA CTG C), and pGreenLatern-Rab11AQ70L (kindly provided by Craig Roy, Yale University, New Haven, CT) as a template. The PCR product was cloned into pGEM-T Easy, and then the EcoRI/XhoI fragment containing the Rab11AQ70L coding region was cloned into the EcoRI/SalI sites of pEGFP-C2 (Clontech). pEGFP-Cpn0585102-651 was constructed by PCR amplification from C. pneumoniae genomic DNA, using the forward primer with a 5′-EcoRI site (CCG GAA TTC ACT TTA TGA TTC TCA GGG CCT) and the 5′ BamHI site-containing reverse primer described above. Cpn0585102-651 was also PCR amplified from C. pneumoniae genomic DNA for cloning into pcDNA3.1/myc-His(−)A (Invitrogen) by using the forward primer with a Kozak sequence and a 5′ XhoI site (GCC GCC ATG CTC GAG CTT TAT GAT TCT CAG GGC) and the reverse primer with a 5′ BamHI site (GGA GGA TCC TTA GGT ACC TCC TTG AAA TTG CTC TTG CTC CAG). For GST pull-down assays, plasmids containing GST-Rab1A, GST-Rab4A and GST-Rab6A were constructed by cloning the coding regions of human Rab1A, Rab4A, and Rab6A, contained within the BamHI/XhoI fragments of the respective cDNA clones (provided by Guthrie cDNA Resource Center, Guthrie Research Institute, Sayre, PA) into the BamHI and XhoI sites of pGEX-4T-1 (GE Healthcare Bio-Sciences, Piscataway, NJ). GST-Rab11A was constructed by PCR amplification of the Rab11A gene from pEGFP-Rab11A (46) by using a forward primer with a 5′ EcoRI site (GAA TTC TAT GGG CAC CCG CGA CGA CGA G) and a reverse primer with a 5′ XhoI site (CTC GAG TTA GAT GTT CTG ACA GCA CTG C). The PCR product was cloned into pGEM-T Easy, and then the EcoRI/XhoI fragment containing the Rab11A coding region was cloned into the EcoRI/XhoI sites of pGEX-4T-1. To generate recombinant Cpn0585 protein (rCpn0585), the full coding sequence of this Inc (residues 1 to 651) was PCR amplified from C. pneumoniae genomic DNA by using a forward primer with a 5′ BamHI site (CCA GGA TCC TCG CAA CAC CCG CTC AAA AAT CC) and a reverse primer with a 5′ SalI site (GGG CGT CGA CTC CTT GAA ATT GCT CTT GCT C). The BamHI/SalI-digested PCR product was cloned into the same sites of pET-21d(+) (Novagen EMD Biosciences, San Diego, CA).
Yeast two-hybrid assays.
The yeast two-hybrid screen was performed by using a Matchmaker GAL4 two-hybrid system 3 according to the manufacturer's protocols (Clontech) as previously described (48). pGBKT7- and pGADT7-based plasmid DNA constructs were, respectively, transformed into Saccharomyces cerevisiae strains AH109 and Y187 (Clontech) by using the S. cerevisiae Easycomp transformation kit (Invitrogen), and transformants were selected on synthetic complete plates lacking appropriate nutrients. After mating of the AH109 with Y187 yeast strains, interacting clones were identified by plating diploids onto synthetic complete plates lacking leucine (L), tryptophan (W), histidine (H), and adenine (A) using a multiprolonged liquid transfer tool (V&P Scientific, Inc., San Diego, CA). The interaction detected when mating yeast-containing pGBKT7-p53 with yeast containing pGADT7-T (simian virus 40 large T-antigen) was used as a positive control. The negative control is the diploid containing pGADT7-Cpn0585102-651 and pGBKT7-p53.
Transfection studies.
HEp-2 cells (4 × 106) were transiently transfected with plasmid DNA (2 μg) by using the Nucleofector I device (Amaxa, Gaithersburg, MD), a Nucleofector kit V, and the T-027 program, according to the manufacturer's instructions. After nucleofection, the cells were washed, resuspended in complete medium, and seeded into 24-well plates (Costar, Cambridge, MA) containing glass coverslips (3 × 105/2 ml/well) for overnight culture. HEp-2 cell monolayers were then infected with C. pneumoniae at the indicated MOIs and incubated for an additional 48 h in Chlamydia medium supplemented with cycloheximide (1 μg/ml).
Confocal microscopy.
Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 60 min, treated with permeabilization buffer (0.2% bovine serum albumin-0.05% saponin in PBS) for 10 min, and then stained sequentially for 60 min each with anti-Cpn0585 antibody (2 μg/ml) and Alexa Fluor 594 F(ab′)2-goat anti-rabbit IgG (1:200). Coverslips were mounted onto glass slides by using ProLong Antifade (Invitrogen) and viewed by confocal laser-scanning microscopy. An UltraVIEW LCI confocal laser-scanning imaging system equipped with krypton, argon, and He-Ne lasers (Perkin-Elmer, Waltham, MA) on a Nikon ECLIPSE TE2000-S inverted microscope with a PlanAPO 100×/1.4 oil immersion objective lens (Nikon, Melville, NY) was used. When indicated, the numbers of C. pneumoniae inclusions in each coverslip were counted in 25 ×1,000 high-powered fields (HPF). A digital charge-coupled device camera (Ultra Pix; Hamamatsu Photonics, Okayama, Japan) with a resolution of 1344 × 1024 × 12 was used to capture images. Images were acquired by using ImagingSuite Acquisition & Processing Software (v.5.0; Perkin-Elmer) and then processed using Adobe Photoshop CS (v.8.0; Adobe Systems, Mountain View, CA).
Protein production and purification.
Escherichia coli BL21(DE3)pLysS cells (Invitrogen) harboring various GST-Rab GTPase plasmids were grown at 37°C in Luria-Bertani broth to an optical density of 0.8 at 600 nm prior to their induction with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 30°C. Expressed GST-Rab GTPase fusion proteins were purified as previously described (7, 45) using glutathione-Sepharose 4B (GE Healthcare Bio-Sciences). rCpn0585 was expressed in BL21(DE3)pLysS E. coli cells grown at 37°C in Luria-Bertani broth to an optical density of 0.5 at 600 nm and then induced with 1 mM IPTG for 2 h at 37°C. Bacteria were disrupted in lysis buffer by using a French press as described previously (54), and protein was purified by affinity chromatography using HisTrap FF columns (GE Healthcare Bio-Sciences). Eluted rCpn0585 protein was pooled and dialyzed extensively against 50 mM Tris-1 M NaCl (pH 7.5) and then against PBS. The solution of rCpn0585 was centrifuged at 24,000 × g at 4°C for 20 min prior to being used in GST pull-down assays.
GST pull-down assays.
C. pneumoniae- and mock-infected HEp-2 cells were lysed in lysis buffer containing 20 mM Tris-HCl (pH 7.4), 50 mM NaCl, 0.5% NP-40, 2.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, and a cocktail of protease inhibitors (Sigma-Aldrich, St. Louis, MO). Soluble lysate protein extract was obtained by rocking for 45 min at 4°C and centrifugation for 1 h at 24,000 × g at 4°C. Protein concentrations were determined by using the BCA protein assay (Pierce, Rockford, IL). Lysates were precleared by sequential 1-h incubation periods with glutathione-Sepharose 4B and GST-bound glutathione-Sepharose 4B beads. GST-Rab GTPase-charged glutathione-Sepharose 4B beads were loaded with 2 mM GTPγS or GDP (Sigma-Aldrich) as previously described (7, 45). To conduct GST pull-down experiments, beads charged with 10 μg of GTPγS- or GDP-loaded GST Rab GTPase fusion proteins were incubated with 500 μg of precleared cell lysate protein or with 270 ng of rCpn0585 (350 μl) for 16 h at 4°C. Beads were washed six times with buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, and 2.5 mM MgCl2 and then twice with a similar buffer but without detergent. Bound proteins were eluted by boiling beads for 5 min with 40 μl of Laemmli sodium dodecyl sulfate (SDS) loading buffer and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (29) and immunoblotting.
SDS-PAGE and immunoblotting.
Proteins were separated under reducing conditions by SDS-10% PAGE (29) and transferred onto a 0.45-μm-pore-size Trans-Blot nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA). After blocking with PBS containing 5% (wt/vol) nonfat milk and 0.05% Tween 20, membranes were immunoblotted with rabbit anti-Cpn0585 antibody (1:5,000) and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000). Both primary and secondary antibodies were diluted in PBS-0.2% nonfat milk-0.05% Tween 20. Blots were extensively washed after each incubation step and developed by chemiluminescence using the ECL plus reagent (GE Healthcare Bio-Sciences). The same blots were used to detect GST fusion proteins and GST after membrane stripping, blocking, and probing with mouse anti-GST antibody (1:5,000) and horseradish peroxidase-conjugated goat anti-mouse IgG (1:10,000).
Delivery of antibodies into live C. pneumoniae-infected cells.
HEp-2 cells grown in coverslips in 24-well plate (2 × 105 cells/well) were infected with C. pneumoniae (MOI of 5) and cultured for 24 h in Chlamydia medium with 1 μg of cycloheximide/ml. Cells were washed three times with PBS, and 450 μl of Iscove medium without serum was added to each well. A total of 0.5 μg of rabbit anti-Cpn0585 or rabbit anti-Chlamydia EB antibodies diluted in 50 μl of 20 mM HEPES were mixed with 2 μl of PULSin reagent (Polyplus-Tranfection, New York, NY), vortex mixed, and 15 min later added to the appropriate wells. After 4 h of incubation at 37°C and 5% CO2, the cells were washed twice with PBS and cultured for 24 h more in Chlamydia medium with 1 μg of cycloheximide/ml. At 48 h postinfection, cells were fixed and stained with Alexa Fluor 594 F(ab′)2-goat anti-rabbit IgG. After extensive washing with PBS and prior to mounting the coverslips onto glass slides, cells were incubated for 10 min with 1 μM SYTOX Green (Invitrogen), which stains the nucleic acid of C. pneumoniae bacteria and HEp-2 cells.
RESULTS
Regions of Cpn0585 share sequence similarity with multiple Rab GTPase-interacting proteins.
Although Rab GTPases 1, 4, 10, and 11 were demonstrated to associate with the C. pneumoniae inclusion membrane (45), the effector molecules involved in this specific recruitment remain unidentified. Our work on the T-cell response to C. pneumoniae identified Cpn0585 as a target of protective CD8+ T cells in infected mice (39, 63; I. Pinchuk and B. Wizel, unpublished data; Cortes and Wizel, unpublished) and led us to hypothesize that this C. pneumoniae antigen could represent a Rab GTPase-binding protein. First, Cpn0585 is an Inc (30) that shares many of the features that characterize other members of this family of proteins, including an N-terminal region with a bilobed hydrophobic domain that may anchor the protein to the inclusion membrane (residues 52 to 74 and 81 to 101) and a C-terminal region that is thought to face the cytosol of host cells (residues 102 to 651) (Fig. 1) (3). Second, a number of Rab-interacting proteins such as GM130 (36), FIP3 (23), and golgin-84 (14) are listed in the BLAST search (1) output for Cpn0585. Moreover, a BLAST 2 sequence alignment (55) showed that the C-terminal region of Cpn0585 encompasses the 40 to 45% similarity that this Inc shares with GM130, FIP3, and golgin-84 (Fig. 1). Of note, the segments of these Rab-binding proteins that are similar to Cpn0585 also include the Rab-binding domains. Third, the C-terminal region of Cpn0585 is predicted to include six coiled-coil domains (6) (Fig. 1), a structural motif involved in protein-protein interactions and commonly found among Rab-binding proteins (14, 59, 60). Finally, Cpn0585 contains two CD8+ T-cell epitopes, one represented by residues 459 to 466 (63) and the other that maps within residues 101 to 115 (Cortes and Wizel, unpublished) (Fig. 1). Although it is unknown how these Cpn0585 epitopes are generated, their location within the C-terminal region strongly suggest accessibility to the cytosol of the host cell and, hence, to the major histocompatibility complex class I antigen processing machinery. Altogether, these data led us to formulate the hypothesis that Cpn0585 is a Rab GTPase-binding protein and that the interacting domain of this Inc faces the cytosol of host cells.
FIG. 1.
Schematic representation of Cpn0585: predicted secondary structures, similarity with Rab GTPase-interacting proteins, and location of CD8+ T-cell epitopes. As determined with the Kyte-Doolittle and Berger algorithms (6, 28), Cpn0585 contains a bilobed hydrophobic region located near its N terminus (black boxes) and six segments with high predicted probability (55 to 93%) to form coiled-coil structures (gray boxes; CC1 to CC6). Two CD8+ T-cell epitopes map to a region of Cpn0585 downstream the bilobed hydrophobic domain (striped boxes; residues 101 to 115 and residues 459 to 466) (60; Cortes and Wizel, unpublished). Using BLAST 2 sequence alignment (55), the C-terminal region of Cpn0585 (residues 102 to 651) shares 40 to 45% sequence similarity and 20 to 25% sequence identity with the indicated Rab-binding proteins. The diagram depicts the regions of Rab-binding proteins that are similar to Cpn0585. The Rab binding domains of FIP3 (residues 695 to 756) (15), GM130 (residues 536 to 616) (60), and golgin-84 (residues 220 to 615) (14) are within the regions that bear similarity with Cpn0585 (black segments).
The C-terminal region of Cpn0585 interacts with Rab1, Rab10, and Rab11 in a GTP-dependent manner.
To determine whether Cpn0585 is a Rab GTPase-binding protein, we examined the ability of this Inc to interact with a collection of Rab GTPases by using a yeast two-hybrid assay. The C-terminal region of Cpn0585 (Cpn0585102-651) was cloned into the GAL4 DNA activation domain (GAL4AD) plasmid pGADT7 and expressed in a MATα yeast strain, while Rab GTPases 1, 4, 6, 10 and 11 were cloned into the GAL4 DNA-binding domain (GAL4BD) plasmid pGBKT7 and expressed in a MATa yeast strain. Constitutively active Rab GTPases lacking the C-terminal domain involved in membrane localization were used in the present study (16, 45).
Pairwise crosses were performed between the Cpn0585102-651- and Rab GTPase-expressing strains. Of the Rab GTPases that were tested, Rab1, Rab4A, Rab4B, Rab10, Rab11A, and Rab11B associate with the C. pneumoniae inclusion membrane, whereas Rab6A and Rab6B do so with the C. trachomatis inclusion membrane (46). As shown by the induction of the GAL4-dependent reporter genes and growth on plates lacking His (H) and Ade (A), Cpn0585102-651 was found to interact with Rab1, Rab10, Rab11A, and Rab11B but not with Rab4A or Rab4B (Fig. 2A). As expected, GAL4AD-Cpn0585102-651 did not interact with GAL4BD-Rab6A or GAL4BD-Rab6B (Fig. 2A). Cpn0585 did not activate transcription of the GAL4-dependent reporter genes by itself because a yeast diploid resulting from mating the strains containing pGADT7-Cpn0585102-651 and pGBKT7-p53 failed to grow on H- and A-deficient media. In contrast, growth was observed for the positive control diploid yeast strain containing pGBKT7-p53 and pGADT7-T (Fig. 2A). Yeast transformed with Cpn0585102-651 cloned in pGBKT7 and mated with Rab GTPases cloned into pGADT7 confirmed that the interaction of Rab1, Rab10, and Rab11 with Cpn0585 is specific (data not shown). The specificity of this interaction is also supported by results from previous yeast two-hybrid assays in which CT229 was identified as the only Rab GTPase-interacting protein from a collection of 27 tested Incs that included CT119 and Cpn0186, the C. trachomatis and C. pneumoniae versions of IncA (45; M. Scidmore, unpublished data).
FIG. 2.
Screening Cpn0585-Rab GTPase interactions with the yeast two-hybrid assay. (A) S. cerevisiae strain AH109 containing the indicated pGBKT7-Rab GTPase constructs were mated with S. cerevisiae strain Y187 expressing pGAD-Cpn0585102-651. To identify possible interacting clones, diploids selected on plates lacking Leu and Trp (LW) were diluted in sterile water and transferred to plates lacking Leu, Trp, His, and Ade (LWHA). The diploid containing pGADT7-Cpn0585102-651 and pGBKT7-53 was used as a negative control. A positive interaction was detected by using the diploid containing pGADT7-T (simian virus 40 large T-antigen) and pGBKT7-53. (B) S. cerevisiae strain Y187 expressing pGADT7-Cpn0585102-651 was mated with S. cerevisiae strain AH109 expressing wild-type pGBKT7-Rab11B, GTP-constitutively active pGBKT7-Rab11BQ70L, or the GDP-dominant-negative pGBKT7-Rab11BS25N. Diploids were selected and screened as described for panel A. (C) Expression of GAL4AD-Cpn0585102-651 (arrow) was determined by immunoblotting of yeast cells transformed with pGADT7-Cpn0585102-651 (lane 1) or pGADT7 alone (lane 2) using anti-HA antibody.
If Cpn0585 is a Rab GTPase effector protein, it should interact with Rabs only in the GTP-bound active but not in the GDP-bound inactive state. Indeed, GAL4AD-Cpn0585102-651 was found to interact with either the wild-type GAL4BD-Rab11B or the constitutively active GTP-bound GAL4BD-Rab11BQ70L but not with the dominant-negative GDP-bound GAL4BD-Rab11BS25N (Fig. 2B).
Expression of Cpn0585102-651 in yeast was verified by immunoblotting studies. A band of 78 kDa that corresponds to the fusion protein GAL4AD-Cpn0585102-651 was detected in yeast cells transformed with pGADT7-Cpn0585102-651 but not in yeast cells transformed with the empty vector (Fig. 2). Overall, these data demonstrate that Cpn0585 is capable of interacting with multiple Rabs in a GTP-dependent manner and that the Rab-binding domain(s) are located between amino acids 102 and 651.
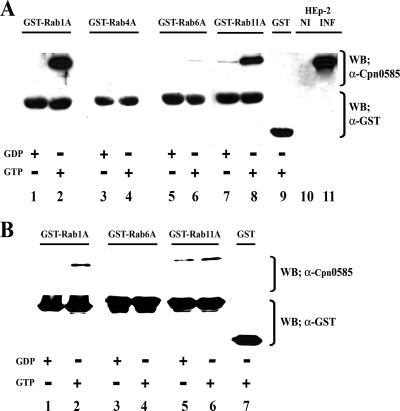
To confirm the yeast two-hybrid results, we examined the interaction of Cpn0585 with multiple Rab GTPases in GST pull-down assays. Detergent extracts from C. pneumoniae-infected HEp-2 cells were prepared 48 h postinfection and incubated with GTPγS- or GDP-loaded GST-Rab1A, -Rab4A, -Rab6A, and -Rab11A fusion proteins. The interactions between C. pneumoniae-derived Cpn0585 and Rab GTPases were detected by immunoblotting with a highly specific anti-Cpn0585 antibody. Native Cpn0585 was found to interact in a GTP-dependent manner with GST-Rab1A and GST-Rab11A but not with GST-Rab4A, GST-Rab6A, or GST (Fig. 3A). All detected Cpn0585-Rab GTPase interactions were most likely direct since pure full-length rCpn0585 was also pulled-down using GTPγS-loaded GST-Rab1A and -Rab11A but not with GST-Rab6A or GST (Fig. 3B). We did not test GST-Rab10 in these assays, since this fusion protein was insoluble when expressed in bacteria. These data confirm the yeast two-hybrid studies and strongly suggest a direct and GTP-dependent association between Cpn0585 and the Rab GTPases 1 and 11.
FIG. 3.
Cpn0585 interacts with GST-Rab1A and GST-Rab11A in a GTP-dependent manner. GST and GST-Rab GTPase fusion proteins were expressed in E. coli and purified by affinity chromatography using glutathione-Sepharose 4B beads as described in Materials and Methods. (A) Glutathione-bound GST-Rab1A, Rab4A, Rab6A, and Rab11A were loaded with GTPγS or GDP and incubated overnight with 500 μg of precleared cell lysate protein from C. pneumoniae-infected HEp-2 cells. Beads were washed, and bound protein was eluted and separated by SDS-PAGE. Lanes loaded with 40 μg of protein extracts prepared from C. pneumoniae-infected (INF) and uninfected (NI) HEp-2 cells were used as controls. Immunoblot analysis was performed using anti-Cpn0585 antibodies and, after the membrane was stripped, with anti-GST antibodies. (B) GTPγS- or GDP-loaded GST-Rab1A, -Rab6A, and -Rab11A fusion proteins were incubated with 270 ng of rCpn0585 and processed for GST pull-down assays as described in panel A. The data shown are representative of two experiments.
Cpn0585 colocalizes with Rab1, Rab10, and Rab11 at the C. pneumoniae inclusion membrane.
To further characterize the interaction of Cpn0585 with Rab GTPases, we examined whether EGFP-Rabs colocalized with endogenous Cpn0585 in C. pneumoniae-infected cells. HEp-2 cells transiently transfected with wild-type EGFP-Rab1A, -Rab4A, -Rab6A, -Rab10, or -Rab11A or with EGFP alone were infected with C. pneumoniae for 48 h and, after fixation, the cells were stained with anti-Cpn0585. The reactivity with anti-Cpn0585 was detected at all or nearly all of the periphery of the C. pneumoniae inclusion with both a finely punctuate and patchy staining pattern (Fig. 4). Cpn0585 colocalized mainly with EGFP-Rab11A (Fig. 4R) and partially with Rab1A (Fig. 4F) and Rab10 (Fig. 4O). No colocalization was observed with Rab4A (Fig. 4I), Rab6A (Fig. 4L), or EGFP alone (Fig. 4C). Also, Cpn0585 colocalized with the constitutively active GTP-bound EGFP-Rab11AQ70L but not with the dominant-negative GDP-bound EGFP-Rab11AS25N (data not shown). Collectively, these data provide evidence that Cpn0585 interacts with multiple Rab GTPases and suggest that it may play a role in the recruitment of these Rabs to the chlamydial inclusion.
FIG. 4.
Colocalization of EGFP-Rab GTPases with Cpn0585 in C. pneumoniae-infected HEp-2 cells. HEp-2 cells transiently expressing the indicated EGFP-tagged Rab GTPases (D to R) or EGFP (A to C) were infected with C. pneumoniae at an MOI of 5, and at 48 h postinfection the cells were fixed, stained with anti-Cpn0585 antibodies, and viewed under a confocal laser-scanning microscope. The EGFP (green) (A, D, G, J, M, and P) and anti-Cpn0585 stain (red) (B, E, H, K, N, and Q) signals were merged (C, F, I, L, O, and R). EGFP-Rab1, Rab10, and Rab11A colocalize with Cpn0585 at various regions along the inclusion membrane (F, O, and R). EGFP-Rab4A associates with the inclusion but does not colocalize with Cpn0585 (I). EGFP-Rab6A does not associate with the C. pneumoniae inclusion and is included as a negative control (J to L). In the panels with merged images, asterisks indicate cells that were not transfected with the EGFP constructs but that were infected with C. pneumoniae. In the insets, the arrows and arrowheads indicate areas with or without colocalization between Cpn0585 and EGFP-Rab GTPases, respectively. Bar, 10 μm. The data are representative of three experiments.
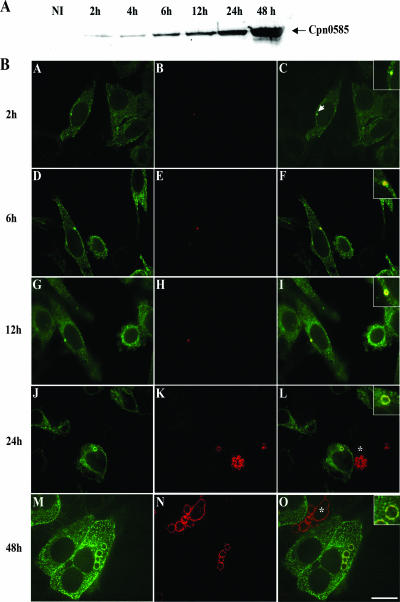
Cpn0585 is expressed at the inclusion membrane of C. pneumoniae-infected cells early after infection, and its C-terminal region faces the host cell cytosol.
We have previously shown that EGFP-Rab11A associates with the C. trachomatis inclusion as early as 1 h postinfection (46). Therefore, Rab GTPase-interacting proteins may be required during the early developmental events of the chlamydial inclusion. Because Cpn0585 is a Rab11-interacting protein, we first examined the expression kinetics of this Inc in C. pneumoniae-infected cells at various time points postinfection and sought to determine whether Cpn0585 associates with Rab11 at an early time point of the C. pneumoniae developmental cycle. Immunoblotting of C. pneumoniae-infected HEp-2 cell detergent-solubilized extracts revealed that Cpn0585 is expressed as early as 2 h postinfection and that the expressed amount of this protein increased as the infection proceeded (Fig. 5A). To determine the point during the chlamydia developmental cycle that Cpn0585 colocalizes with Rab11, the intracellular localization of EGFP-Rab11AQ70L in HEp-2 cells infected with C. pneumoniae at an MOI of 20 was examined at various times postinfection. These studies revealed that Cpn0585 associates with EGFP-Rab11A as early as 2 h postinfection (Fig. 5B, subpanel C) and that this association remains evident at all time points analyzed. A rim-like fluorescence delimiting the inclusion is clearly observed by 12 h postinfection (Fig. 5B, subpanel I) and continues throughout the developmental cycle (Fig. 5B, subpanels J to O). Cpn0585 expression is detected at early time points of the infection on individual RB-containing vesicles (Fig. 5B, subpanel C). In addition, Rab1 and Rab10 were also associated with the C. pneumoniae inclusion membrane at 12 and 6 h postinfection, respectively, suggesting a dynamic association of Rab GTPases during the formation of the C. pneumoniae inclusion (data not shown).
FIG. 5.
Cpn0585: kinetics of expression and colocalization with Rab11A in C. pneumoniae-infected cells. (A) Expression of Cpn0585 at various time points after C. pneumoniae infection. A total of 40 μg of protein from C. pneumoniae-infected HEp-2 cell detergent-solubilized extracts prepared at each of the indicated time points after C. pneumoniae infection was separated by SDS-PAGE and then transferred to a nitrocellulose membrane. Immunoblotting with rabbit anti-Cpn0585 antibodies detected a protein of an estimated molecular mass of 77 kDa as early as 2 h postinfection. Extracts from uninfected HEp-2 cells (NI) were used as a negative control. (B) HEp-2 cells transiently expressing EGFP-Rab11AQ70L were infected with C. pneumoniae at an MOI of 20, and at the indicated time points after infection the cells were fixed, stained with anti-Cpn0585 antibodies, and analyzed by laser scanning confocal microscopy. The signals from EGFP-Rab11AQ70L (green; A, D, G, J, and M) and anti-Cpn0585 stain (red; B, E, H, K, and N) were merged (C, F, I, L, and O). In the insets of panels with the merged images, the colocalization of Cpn0585 and EGFP-Rab11AQ70L is clearly noted as early as 2 h postinfection (arrow in panel C). Asterisks indicate C. pneumoniae-infected cells without EGFP-Rab11AQ70L expression. Bar, 10 μm. The data are representative of three experiments.
The C-terminal domains of a number of Incs from C. trachomatis and C. psittaci are exposed to the host cell cytosol, which may permit interaction with host cytoplasmic proteins (21, 43). To determine whether the C-terminal domain of Cpn0585 faces the cytoplasm of the host cell, an antibody against the last 23 residues of Cpn0585 (residues 629 to 651) was introduced into live HEp-2 cells infected with C. pneumoniae for 24 h using PULSin reagent. The intracellular antibody was visualized at 48 h postinfection in cells that were fixed and then stained with a fluorophore-conjugated secondary antibody. The positive signal detected at the periphery of the inclusion indicated that the anti-Cpn0585 antibodies react with the C. pneumoniae inclusion membrane and that the recognized epitope is exposed to the host cell cytosol (Fig. 6A and C). To demonstrate that this cationic amphiphile liposome delivery reagent does not compromise the integrity of the inclusion membrane, anti-EB antibodies were tested. The intracellular delivery of these antibodies did not stain the intravacuolar EBs (Fig. 6F and H). That both of the antibodies used were immunoreactive with their respective targets was demonstrated in a conventional indirect immunofluorescence assay using cells infected with C. pneumoniae for 48 h. After fixation and permeabilization, anti-Cpn0585 and anti-EB antibodies reacted, respectively, with the inclusion membrane and EBs (Fig. 6E and J). Altogether, these results suggest that the Rab GTPase-interacting region of Cpn0585 faces the cytosol of C. pneumoniae-infected cells. Moreover, the cationic-liposome delivery of antibodies into the cytosol of live C. pneumoniae-infected cells offers a new and less technically demanding approach than microinjection that can be used to study various aspects of chlamydial inclusion biology.
FIG. 6.
The C-terminal region of Cpn0585 containing the Rab binding domain faces the cytosol of C. pneumoniae-infected cells. Rabbit polyclonal antibodies anti-Cpn0585 (A to D) or anti-EBs (F to I) were introduced into live HEp-2 cells infected for 24 h with C. pneumoniae by using PULSin reagent under the conditions described in Materials and Methods. After 24 h, cells were fixed and stained with an Alexa Fluor 594-conjugated F(ab′)2-goat anti-rabbit IgG. Prior to mounting coverslips onto glass slides, cells were incubated for 10 min with 1 μM SYTOX Green, which stains the nucleic acid of C. pneumoniae bacteria and HEp-2 cells. The signals from SYTOX Green (green; B and G) and Alexa Fluor 594 (red; A and F) were merged (C and H). C. pneumoniae-infected cells that incorporated (arrow) or not (arrowheads) anti-Cpn0585antibodies after PULSin treatment were evident in all experiments. To show that the anti-Cpn0585 and anti-EB antibodies were immunoreactive, a conventional indirect immunofluorescence assay was carried out in parallel using HEp-2 cells that had been infected for 48 h with C. pneumoniae. After fixation and permeabilization, the cells were stained with the anti-Cpn0585 (E) or anti-EB (J) antibodies, followed by the Alexa Fluor 594 F(ab′)2-goat anti-rabbit IgG secondary antibody. Merged signals from antibody (red) and SYTOX Green (green) stainings are shown (E and J). Note the peripheral staining of the inclusion membrane with anti-Cpn0585 (red) and the merged staining of EBs with both anti-EB and SYTOX Green (yellow). The data are from a representative of three experiments.
Ectopic expression of Cpn0585102-651 inhibits C. pneumoniae development, which is partially reversed with Rab11.
Available evidence indicates that ectopic expression of C. caviae IncA or microinjection of neutralizing antibodies to the C. trachomatis Inc CT229 can inhibit chlamydial development (2, 20). To test whether the ectopic expression of the Rab GTPase binding domain-bearing C-terminal segment of Cpn0585 inhibits the development of C. pneumoniae, EGFP-Cpn0585102-651- and EGFP-expressing HEp-2 cells were infected, and the numbers of C. pneumoniae inclusions were determined. Expression of EGFP-Cpn0585102-651 in HEp-2 cells inhibited 45% the number of C. pneumoniae inclusions that developed in EGFP-expressing cells (P < 0.01) (Fig. 7A). In repeat experiments, a 50 to 65% reduction in C. pneumoniae inclusion development was observed (P < 0.01). Interestingly, 90% of the inclusions that developed in EGFP-Cpn0585102-651-expressing cells were aberrant or atypical compared to only 15% in EGFP-expressing cells (Fig. 7A). Atypical inclusions appeared smaller in size and contained large aberrant RBs (Fig. 7B, lower panel). In contrast, the majority of C. pneumoniae inclusions in EGFP- (Fig. 7B, upper panel) and EGFP-Rab GTPase-expressing (data not shown) cells were normal, since these were larger, contained numerous EBs, and stained densely with the anti-MOMP antibody.
FIG. 7.
Modulation of C. pneumoniae inclusion development by the ectopic expression of Cpn0585102-651 and the overexpression of Rab11. (A) Triplicate cultures of HEp-2 cells transiently expressing EGFP or EGFP- Cpn0585102-651 were infected with C. pneumoniae at an MOI of 1, and 48 h later the cells were fixed and stained with anti-C. pneumoniae MOMP (monoclonal antibody RR402). The numbers of normal and aberrant C. pneumoniae inclusions in 25 HPF/well were counted. (B) Representative normal (arrowheads) and aberrant (arrows) inclusions in HEp-2 cells infected for 48 h with C. pneumoniae expressing EGFP (upper panel) or EGFP-Cpn0585102-651 (lower panel). The signals from EGFP (green) and anti-MOMP (red) were merged. (C) Triplicate cultures of HEp-2 cells transiently cotransfected with the indicated pcDNA3.1- and pEGFP-based plasmids were infected with C. pneumoniae at an MOI of 1, and 48 h postinfection the cells were fixed and stained with anti-C. pneumoniae MOMP. The numbers of C. pneumoniae inclusions in 25 HPF/well were counted. Values represent the mean ± the standard deviation of three experiments. *, Differences versus numbers of C. pneumoniae inclusions counted in EGFP-expressing cells (A) or in cells cotransfected with empty pcDNA3.1 and the indicated EGFP-based construct (B) are significant (P < 0.01 [Student t test]).
Because Cpn0585 interacts with multiple Rab GTPases, the ectopic expression of Cpn0585102-651 may compete with native Cpn0585 for binding to Rabs, therefore disrupting chlamydial development. Since Cpn0585 is expressed early in C. pneumoniae-infected cells (Fig. 5A) and this Inc associates predominantly with Rab11 (Fig. 4R), we examined whether overexpression of Rab11 could reverse the inhibition mediated by the ectopic expression of Cpn0585102-651. HEp-2 cells transiently expressing Cpn0585102-651 and either EGFP-Rab11AQ70L, EGFP-Rab4AQ67L, or EGFP were infected, and the numbers of C. pneumoniae inclusions were counted. Both Rab GTPases interact with the C. pneumoniae inclusion membrane (45), but only Rab11 associates with Cpn0585 (Fig. 2, 3, and 4). Coexpression of either EGFP or EGFP-Rab4AQ67L and Cpn0585102-651 inhibited C. pneumoniae inclusion development by more than 50% compared to the expression of EGFP or EGFP-Rab4AQ67L alone (P < 0.01) (Fig. 7C). In contrast, no significant reduction in the number of C. pneumoniae inclusions was observed in cells coexpressing EGFP-Rab11AQ70L and Cpn0585102-651 compared to cells expressing EGFP, EGFP-Rab4AQ67L, or EGFP-Rab11AQ70L alone (Fig. 7C). However, aberrant inclusions were also observed, albeit at lower numbers than in cells expressing EGFP-Cpn0585102-651 alone. These results suggest that although the overexpression of Rab11 can reestablish the biogenesis of C. pneumoniae inclusions, it cannot fully restore the proper environment for normal C. pneumoniae development.
DISCUSSION
Chlamydiae complete a productive infection in susceptible cells by exploiting a number of host cellular processes, which involve interactions between host cell molecules and chlamydial products that are totally or partially exposed to the host cell cytosol. Here, we identified novel GTP-dependent interactions between the C. pneumoniae inclusion membrane protein Cpn0585 and the host cell Rab GTPases 1, 10, and 11. Our results show that, as in C. trachomatis, Inc proteins are key elements in recruiting Rabs to the chlamydial inclusion and reveal for the first time a biologically relevant link between the interaction of Cpn0585 and a Rab GTPase in the development of C. pneumoniae.
Although the exact topology of Cpn0585 in the inclusion membrane is not known, its similarity to IncA suggests that Cpn0585 has two transmembrane domains with the C terminus of the protein facing the cytosol of host cells (3, 43, 44). The results from yeast two-hybrid studies and delivery of cationic liposome-antibody complexes into C. pneumoniae-infected cells strongly suggest that the Rab GTPase-binding domain is accessible to the cytosol of infected cells. Moreover, the C-terminal segment of Cpn0585 includes two CD8+ T-cell epitopes, indicating that this region of the protein may be directly accessible to the cytosol of infected host cells and thus to the major histocompatibility complex class I antigen-processing machinery (63; Cortes and Wizel, unpublished).
Previous reports showed that Cpn0585 mRNA is detected at 8 h postinfection and that the expression of this Inc is induced after infected cells are treated with gamma interferon, suggesting an association of Cpn0585 with chlamydial persistence (25, 32). Since the earliest time point tested for Cpn0585 mRNA expression was 8 h postinfection (25) and our data show that the protein is expressed as early as 2 h postinfection, we postulate that Cpn0585 mRNA synthesis occurs right after C. pneumoniae invasion. Because exposure of C. pneumoniae-infected cells to low concentrations of gamma interferon induces the formation of persistent forms (53), it is possible that an increase of Cpn0585 protein level may participate in the survival of C. pneumoniae by recruiting Rab GTPases to the inclusion.
How does Cpn0585 interact with multiple Rab GTPases and what are the consequences of these interactions? It is likely that Cpn0585 binds different Rabs at various time points during infection and that these interactions depend on the temporal recruitment of particular Rabs to the inclusion. For instance, one of the first events after an EB enters a susceptible cell is the recruitment of Rab11 to the nascent vacuole, which occurs as early as 1 h postinfection (11, 46). At this time point, the primordial inclusion is still located in the periphery of the cell, and it has not been trafficked to the microtubule-organizing center in the perinuclear region (11, 18). During this early stage of infection, Cpn0585 is also synthesized. Hence, this Inc may initiate the recruitment of Rab11 to the inclusion within the first few hours after infection. The continuous association of Cpn0585 with Rab11 is likely important to ensure that the inclusion is trafficked to the peri-Golgi region of the infected cell (45, 46). Once an EB-containing vacuole establishes its niche at the microtubule-organizing center, Cpn0585 may recruit Rab1 and Rab10 that may provide additional sources for membrane lipids that further support the growth of the inclusion. Although Rab1 and Rab10 associate with the C. pneumoniae inclusion membrane at 6 to 12 h postinfection (data not shown), we cannot exclude the possibility that transient interactions may also occur earlier during the infection, since Rab1 has been shown to interact with the Legionella pneumophila-containing vacuole within minutes of bacterial uptake (26). Thus, because active chlamydial protein synthesis is necessary for delivering the inclusion to a safe niche within the cell (49), and Rab GTPases are key elements in vesicle trafficking (38, 64), we propose that Cpn0585 as well as other Incs may direct the inclusion to the microtubule-organizing center as a mechanism to avoid fusion with lysosomes.
Chlamydial inclusions also interact with MVBs, which are complex, dynamic, and heterogeneous late endocytic organelles that serve as a source for biosynthetic precursors essential for intracellular chlamydial growth and inclusion biogenesis (4, 40). Interestingly, Rab11 participates in the formation of MVBs (47). Because our results showed that overexpression of Cpn0585 blocks C. pneumoniae development, it its tempting to speculate that the ectopic expression of Cpn0585 may compete with native Cpn0585 for binding to Rab11, which in turn may interfere in the formation of MVB or in the delivery of MVB to the inclusion.
Cpn0585 may interact with multiple Rab GTPases simultaneously through the existence of more than one Rab binding domain. In fact, our most recent results suggest that Cpn0585 has two overlapping Rab binding domains, one that binds Rab1 and Rab10 (amino acids 387 to 518) and another that binds Rab11 (amino acids 387 to 651). These data are in agreement with the sequence comparison analysis between Cpn0585 and a number of Rab-interacting proteins, which define putative Rab-binding regions within the predicted coiled-coil domains 3, 4, 5, and 6 of Cpn0585. Moreover, the Cpn0585-interacting Rab GTPases are classified within two of four subsets of Rab GTPases organized according to primary sequence (8, 37). While Rab1 and Rab10 belong to the same subfamily of Rab GTPases, Rab11 is classified in another subset. This further supports the notion that Cpn0585 may include two Rab binding domains, a feature shown for other Rab-interacting proteins (59). Finally, we have also found that the fragment Cpn0585102-651 interacts with itself (Cortes and Wizel, unpublished). However, we have not yet determined whether this oligomerization is important for the interaction with Rabs or other processes involved in chlamydial development, as has been described for IncA from C. trachomatis and C. caviae (12). Therefore, studies to determine the affinity of Cpn0585 for Rab GTPases and further define the existence of one or two Rab binding domains are warranted.
The mechanism by which ectopic expression of Cpn0585 blocks chlamydial development is still unclear. Since overexpression of Rab11 reverses the decrease in inclusion number caused by the ectopic overexpression of Cpn0585102-651, we infer that the interaction between native Cpn0585 and Rab11 is crucial during the early stages of C. pneumoniae inclusion formation. Because atypical inclusions still persist in cells cotransfected with Cpn0585 and Rab11, other Rabs, or possibly other Cpn0585 interacting proteins, must be required for normal C. pneumoniae development. Studies to further define the relative contribution of the interactions between Cpn0585 and Rabs 1 and 10 to the formation of C. pneumoniae inclusions are currently under way in our laboratory.
Because no effector molecules have been identified in C. trachomatis that bind Rab1 and Rab11 and no protein homologous to Cpn0585 is encoded in the C. trachomatis genome, sequence comparison studies restricted to the functional regions of Cpn0585 may help to identify the Rab1- and Rab11-interacting proteins in C. trachomatis and reveal other functions associated with molecules located at the inclusion membrane. Moreover, without the ability to genetically manipulate the chlamydial genome, studies that directly target the expression of mammalian factors are a key element to understand the mechanisms that govern the intracellular survival of chlamydiae.
Acknowledgments
We thank Peter Barnes and Viviana Ferreira for critical reading of the manuscript.
This study was supported by grant HL70641 from the National Institutes of Health (B.W.).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 1 October 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzhanov, D., J. Barnes, D. E. Hruby, and D. D. Rockey. 2004. Chlamydial development is blocked in host cells transfected with Chlamydophila caviae incA. BMC Microbiol. 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2:35-47. [DOI] [PubMed] [Google Scholar]
- 4.Beatty, W. L. 2006. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell Sci. 119(Pt. 2):350-359. [DOI] [PubMed] [Google Scholar]
- 5.Belland, R., D. M. Ojcius, and G. I. Byrne. 2004. Chlamydia. Nat. Rev. Microbiol. 2:530-531. [DOI] [PubMed] [Google Scholar]
- 6.Berger, B., D. B. Wilson, E. Wolf, T. Tonchev, M. Milla, and P. S. Kim. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brymora, A., M. A. Cousin, B. D. Roufogalis, and P. J. Robinson. 2001. Enhanced protein recovery and reproducibility from pull-down assays and immunoprecipitations using spin columns. Anal. Biochem. 295:119-122. [DOI] [PubMed] [Google Scholar]
- 8.Burton, J. L., M. E. Burns, E. Gatti, G. J. Augustine, and P. De Camilli. 1994. Specific interactions of Mss4 with members of the Rab GTPase subfamily. EMBO J. 13:5547-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, L. A., and C. C. Kuo. 2004. Chlamydia pneumoniae: an infectious risk factor for atherosclerosis? Nat. Rev. Microbiol. 2:23-32. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. T., C. Holcomb, and H. P. Moore. 1993. Expression and localization of two low-molecular-weight GTP-binding proteins, Rab8 and Rab10, by epitope tag. Proc. Natl. Acad. Sci. USA 90:6508-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dautry-Varsat, A., A. Subtil, and T. Hackstadt. 2005. Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 7:1714-1722. [DOI] [PubMed] [Google Scholar]
- 12.Delevoye, C., M. Nilges, A. Dautry-Varsat, and A. Subtil. 2004. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J. Biol. Chem. 279:46896-46906. [DOI] [PubMed] [Google Scholar]
- 13.Derre, I., and R. R. Isberg. 2004. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72:3048-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diao, A., D. Rahman, D. J. Pappin, J. Lucocq, and M. Lowe. 2003. The coiled-coil membrane protein golgin-84 is a novel Rab effector required for Golgi ribbon formation. J. Cell Biol. 160:201-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eathiraj, S., A. Mishra, R. Prekeris, and D. G. Lambright. 2006. Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364:121-135. [DOI] [PubMed] [Google Scholar]
- 16.Farnsworth, C. C., M. C. Seabra, L. H. Ericsson, M. H. Gelb, and J. A. Glomset. 1994. Rab geranylgeranyl transferase catalyzes the geranylgeranylation of adjacent cysteines in the small GTPases Rab1A, Rab3A, and Rab5A. Proc. Natl. Acad. Sci. USA 91:11963-11967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields, K. A., and T. Hackstadt. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221-245. [DOI] [PubMed] [Google Scholar]
- 18.Grieshaber, S. S., N. A. Grieshaber, and T. Hackstadt. 2003. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell Sci. 116(Pt. 18):3793-3802. [DOI] [PubMed] [Google Scholar]
- 19.Hackstadt, T. 2000. Redirection of host vesicle trafficking pathways by intracellular parasites. Traffic 1:93-99. [DOI] [PubMed] [Google Scholar]
- 20.Hackstadt, T., M. A. Scidmore-Carlson, and C. A. Dooley. 1999. Chlamydia trachomatis inclusion membrane protein required for intracellular development. Mol. Biol. Cell 10(Suppl. S):182A. [Google Scholar]
- 21.Hackstadt, T., M. A. Scidmore-Carlson, E. I. Shaw, and E. R. Fischer. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1:119-130. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, D. L. 1999. Chlamydia pneumoniae, asthma, and COPD: what is the evidence? Ann. Allergy Asthma Immunol. 83:271-292. [DOI] [PubMed] [Google Scholar]
- 23.Hales, C. M., R. Griner, K. C. Hobdy-Henderson, M. C. Dorn, D. Hardy, R. Kumar, J. Navarre, E. K. Chan, L. A. Lapierre, and J. R. Goldenring. 2001. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276:39067-39075. [DOI] [PubMed] [Google Scholar]
- 24.Heinzen, R. A., M. A. Scidmore, D. D. Rockey, and T. Hackstadt. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64:796-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan, R. J., S. A. Mathews, A. Kutlin, M. R. Hammerschlag, and P. Timms. 2003. Differential expression of genes encoding membrane proteins between acute and continuous Chlamydia pneumoniae infections. Microb. Pathog. 34:11-16. [DOI] [PubMed] [Google Scholar]
- 26.Kagan, J. C., M. P. Stein, M. Pypaert, and C. R. Roy. 2004. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199:1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo, C. C., and J. T. Grayston. 1990. A sensitive cell line, HL cells, for isolation and propagation of Chlamydia pneumoniae strain TWAR. J. Infect. Dis. 162:755-758. [DOI] [PubMed] [Google Scholar]
- 28.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Luo, J., T. Jia, Y. Zhong, D. Chen, R. Flores, and G. Zhong. 2007. Localization of the hypothetical protein Cpn0585 in the inclusion membrane of Chlamydia pneumoniae-infected cells. Microb. Pathog. 42:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallard, F., B. L. Tang, T. Galli, D. Tenza, A. Saint-Pol, X. Yue, C. Antony, W. Hong, B. Goud, and L. Johannes. 2002. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews, S., C. George, C. Flegg, D. Stenzel, and P. Timms. 2001. Differential expression of ompA, ompB, pyk, nlpD and Cpn0585 genes between normal and interferon-γ treated cultures of Chlamydia pneumoniae. Microb. Pathog. 30:337-345. [DOI] [PubMed] [Google Scholar]
- 33.McCaffrey, M. W., A. Bielli, G. Cantalupo, S. Mora, V. Roberti, M. Santillo, F. Drummond, and C. Bucci. 2001. Rab4 affects both recycling and degradative endosomal trafficking. FEBS Lett. 495:21-30. [DOI] [PubMed] [Google Scholar]
- 34.McClarty, G. 1994. Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 2:157-164. [DOI] [PubMed] [Google Scholar]
- 35.Meresse, S., O. Steele-Mortimer, B. B. Finlay, and J. P. Gorvel. 1999. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J. 18:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moyer, B. D., B. B. Allan, and W. E. Balch. 2001. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic 2:268-276. [DOI] [PubMed] [Google Scholar]
- 37.Pereira-Leal, J. B., and M. C. Seabra. 2000. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301:1077-1087. [DOI] [PubMed] [Google Scholar]
- 38.Pfeffer, S. R. 2001. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11:487-491. [DOI] [PubMed] [Google Scholar]
- 39.Pinchuk, I., B. C. Starcher, B. Livingston, A. Tvninnereim, S. Wu, E. Appella, J. Sidney, A. Sette, and B. Wizel. 2005. A CD8+ T-cell heptaepitope minigene vaccine induces protective immunity against Chlamydia pneumoniae. J. Immunol. 174:5729-5739. [DOI] [PubMed] [Google Scholar]
- 40.Piper, R. C., and J. P. Luzio. 2001. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic 2:612-621. [DOI] [PubMed] [Google Scholar]
- 41.Puolakkainen, M., J. Parker, C. C. Kuo, J. T. Grayston, and L. A. Campbell. 1995. Further characterization of Chlamydia pneumoniae specific monoclonal antibodies. Microbiol. Immunol. 39:551-554. [DOI] [PubMed] [Google Scholar]
- 42.Ren, M., G. Xu, J. Zeng, C. De Lemos-Chiarandini, M. Adesnik, and D. D. Sabatini. 1998. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95:6187-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rockey, D. D., D. Grosenbach, D. E. Hruby, M. G. Peacock, R. A. Heinzen, and T. Hackstadt. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol. Microbiol. 24:217-228. [DOI] [PubMed] [Google Scholar]
- 44.Rockey, D. D., M. A. Scidmore, J. P. Bannantine, and W. J. Brown. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333-340. [DOI] [PubMed] [Google Scholar]
- 45.Rzomp, K. A., A. R. Moorhead, and M. A. Scidmore. 2006. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 74:5362-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rzomp, K. A., L. D. Scholtes, B. J. Briggs, G. R. Whittaker, and M. A. Scidmore. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savina, A., C. M. Fader, M. T. Damiani, and M. I. Colombo. 2005. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 6:131-143. [DOI] [PubMed] [Google Scholar]
- 48.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638-1650. [DOI] [PubMed] [Google Scholar]
- 49.Scidmore, M. A., D. D. Rockey, E. R. Fischer, R. A. Heinzen, and T. Hackstadt. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect. Immun. 64:5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seabra, M. C., and C. Wasmeier. 2004. Controlling the location and activation of Rab GTPases. Curr. Opin. Cell Biol. 16:451-457. [DOI] [PubMed] [Google Scholar]
- 51.Short, B., A. Haas, and F. A. Barr. 2005. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim. Biophys. Acta 1744:383-395. [DOI] [PubMed] [Google Scholar]
- 52.Smith, A. C., J. T. Cirulis, J. E. Casanova, M. A. Scidmore, and J. H. Brumell. 2005. Interaction of the Salmonella-containing vacuole with the endocytic recycling system. J. Biol. Chem. 280:24634-24641. [DOI] [PubMed] [Google Scholar]
- 53.Summersgill, J. T., N. N. Sahney, C. A. Gaydos, T. C. Quinn, and J. A. Ramirez. 1995. Inhibition of Chlamydia pneumoniae growth in HEp-2 cells pretreated with gamma interferon and tumor necrosis factor alpha. Infect. Immun. 63:2801-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szeliga, J., J. Jordan, C. H. Yang, Z. Sever-Chroneos, and Z. C. Chroneos. 2005. Bacterial expression of recombinant MyoXVIIIA domains. Anal. Biochem. 346:179-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 56.Tisdale, E. J., J. R. Bourne, R. Khosravi-Far, C. J. Der, and W. E. Balch. 1992. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Der Sluijs, P., M. Hull, A. Zahraoui, A. Tavitian, B. Goud, and I. Mellman. 1991. The small GTP-binding protein Rab4 is associated with early endosomes. Proc. Natl. Acad. Sci. USA 88:6313-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Via, L. E., D. Deretic, R. J. Ulmer, N. S. Hibler, L. A. Huber, and V. Deretic. 1997. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272:13326-13331. [DOI] [PubMed] [Google Scholar]
- 59.Vitale, G., V. Rybin, S. Christoforidis, P. Thornqvist, M. McCaffrey, H. Stenmark, and M. Zerial. 1998. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 17:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weide, T., M. Bayer, M. Koster, J. P. Siebrasse, R. Peters, and A. Barnekow. 2001. The Golgi matrix protein GM130: a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2:336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White, J., L. Johannes, F. Mallard, A. Girod, S. Grill, S. Reinsch, P. Keller, B. Tzschaschel, A. Echard, B. Goud, and E. H. Stelzer. 1999. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol. 147:743-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilcke, M., L. Johannes, T. Galli, V. Mayau, B. Goud, and J. Salamero. 2000. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 151:1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wizel, B., B. C. Starcher, B. Samten, Z. Chroneos, P. F. Barnes, J. Dzuris, Y. Higashimoto, E. Appella, and A. Sette. 2002. Multiple Chlamydia pneumoniae antigens prime CD8+ Tc1 responses that inhibit intracellular growth of this vacuolar pathogen. J. Immunol. 169:2524-2535. [DOI] [PubMed] [Google Scholar]
- 64.Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2:107-117. [DOI] [PubMed] [Google Scholar]