Abstract
Serine repeat antigens (SERAs) are a family of secreted “cysteine-like” proteases of Plasmodium parasites. Several SERAs possess an atypical active-site serine residue in place of the canonical cysteine. The human malaria parasite Plasmodium falciparum possesses six “serine-type” (SERA1 to SERA5 and SERA9) and three “cysteine-type” (SERA6 to SERA8) SERAs. Here, we investigate the importance of the serine-type SERAs to blood-stage parasite development and examine the extent of functional redundancy among this group. We attempted to knock out the four P. falciparum serine-type SERA genes that have not been disrupted previously. SERA1, SERA4, and SERA9 knockout lines were generated, while only SERA5, the most strongly expressed member of the SERA family, remained refractory to genetic deletion. Interestingly, we discovered that while SERA4-null parasites completed the blood-stage cycle normally, they exhibited a twofold increase in the level of SERA5 mRNA. The inability to disrupt SERA5 and the apparent compensatory increase in SERA5 expression in response to the deletion of SERA4 provides evidence for an important blood-stage function for the serine-type SERAs and supports the notion of functional redundancy among this group. Such redundancy is consistent with our phylogenetic analysis, which reveals a monophyletic grouping of the serine-type SERAs across the genus Plasmodium and a predominance of postspeciation expansion. While SERA5 is to some extent further validated as a target for vaccine and drug development, our data suggest that the expression level of other serine-type SERAs is the only barrier to escape from anti-SERA5-specific interventions.
New drugs and a vaccine are urgently needed to facilitate effective and sustained control of the protozoan parasite Plasmodium falciparum, the most significant cause of malaria. Novel enzymes that are both accessible and essential to parasite development in the blood are attractive therapeutic targets. Proteases that function in the erythrocytic cycle are promising in this regard, as many of them appear to have unique properties and/or function in parasite-specific processes such as hemoglobin digestion and parasite invasion of host erythrocytes (5, 21, 25, 29).
Accordingly, the P. falciparum serine repeat antigens (SERAs) also have potential as drug targets. These proteins are expressed in the parasitophorous vacuole of mature, erythrocytic-stage parasites (8, 13, 20), and all of them possess a papain protease-like central domain flanked by two novel and relatively cysteine-rich domains. Several SERA proteins, termed the “serine-type” SERAs, possess an atypical serine residue in the active-site position of the enzyme domain usually occupied by a canonical cysteine. The enzyme domain from at least one serine-type SERA, SERA5, retains a degree of catalytic function; however, it is uncertain if this proteolytic activity has biological relevance (12). The P. falciparum genome contains a total of nine SERA genes, six of which encode serine-type SERA proteins and three of which encode more classical cysteine-type SERAs (2, 11, 20). These two types of SERA proteins form distinct phylogenetic groups not just within P. falciparum but also across the Plasmodium genus, suggesting different biological roles for members of each type (3, 12). The precise nature and relevance of these roles remain to be determined.
While it has been established that all members of the P. falciparum SERA gene family, eight of which are tandemly arrayed genes on chromosome 2 (SERA1 to SERA8), and a ninth gene on chromosome 9 (SERA9), are expressed in a coregulated manner late in the blood-stage cycle, it is clear that each member is expressed at very different levels (2, 4, 18-20). This expression pattern is similar across all parasite lines examined to date, and multiple SERAs are expressed in the one parasite, suggesting that family members are not differentially expressed. Proteomic data, although not strictly quantitative, suggest a strong concordance between mRNA and protein levels, with SERA5 peptides detected with extraordinary frequency (16). SERA5 ranked fourth (159 peptides) in a list of 712 trophozoite/schizont proteins detected. SERA4 peptides were also well represented in this analysis, ranking 79th (22 peptides), together with SERA6 (208th; nine peptides), SERA3 (209th; nine peptides), SERA7 (380th; three peptides), and SERA9 (457th; two peptides). A separate proteomic analysis of P. falciparum schizonts yielded almost identical results (17). It is important that some SERA family members that are expressed weakly in the erythrocytic cycle may be predominantly expressed at other stages.
Several lines of evidence suggest that a possible role for some members of this family is to promote merozoite egress from infected erythrocytes. Apart from their appropriate location in the parasitophorous vacuole, it has been shown that antibodies specific for SERA5 appear to impede schizont rupture (23). Related to this, the targeted deletion of a P. falciparum cysteine-type SERA homologue (SERA8) in P. berghei, which is predominantly expressed in sporozoites, results in the arrest of sporozoite egress from oocysts in the mosquito (1). While it is attractive to contemplate a generic role in parasite egress for this family of proteins at different life stages, it remains to be demonstrated if this is the case. Moreover, the reason for so many SERA genes, and for a variable number between species (3), remains a mystery.
In order to probe the biological role of the SERAs during the blood stage, we previously attempted to disrupt each of the SERA genes clustered on chromosome 2 using a single-crossover recombination approach (7). Using this technology, we disrupted the peripherally located genes (SERA2, SERA3, SERA7, and SERA8) but were unsuccessful in targeting the peripherally located gene SERA1 or any of the more strongly expressed genes, SERA4, SERA5, and SERA6 (20). Since then, another more powerful gene-targeting technology has been developed for P. falciparum. This approach involves selecting double-crossover integrants using a combination of both positive and negative selectable markers (9). In this study, we used this method as well as repeating the single-crossover approach in an attempt to disrupt the two most strongly expressed members of the SERA family, SERA4 and SERA5, and the remaining two serine-type SERAs, SERA1 and SERA9. While the SERA5 gene remained refractory to deletion, we were able to disrupt SERA1, SERA4, and SERA9. SERA4-null parasites displayed a twofold increase in the expression of SERA5 mRNA. We also performed a comprehensive analysis of genome sequences of Plasmodium species parasites and demonstrate that, in contrast to the cysteine-type members, the serine-type SERAs form a monophyletic group that have expanded postspeciation. Together, these data provide evidence for a key, probably single, role for serine-type SERAs in the erythrocytic cycle.
MATERIALS AND METHODS
Plasmid design and construction.
DNA was amplified via PCR from a mixed trophozoite/schizont parasite genomic DNA (gDNA) template. Preparation of genomic DNA was performed as described previously (28). Each reaction mixture contained 200 μM deoxynucleoside triphosphate, 200 nM each primer, 0.5 units of Platinum Taq DNA polymerase high-fidelity enzyme (Gibco BRL), 2.0 mM MgSO4, and 200 ng template. The double-crossover transfection vectors contain 5′- and 3′-targeting sequences for homologous recombination into the genome. These sequences were obtained by amplifying SERA sequences from P. falciparum gDNA using the oligonucleotides specified in Tables S1 and S2 in the supplemental material. The resulting fragments were cloned into the SacII/BglII (5′ target) and AvrII/EcoRI or AvrII/ClaI (3′ target) sites of pHTK (9). Ligation of PCR products into the appropriate plasmid was performed as previously described (26), with the exception that polyethylene glycol 8000 (1%) and ATP (1 mM) were added to each ligation reaction mixture. The ligated DNA was electroporated into Escherichia coli strain PMC103 using the Bio-Rad Gene Pulser II electroporation system and the Pulse Controller unit.
Parasite culture and transfection.
P. falciparum asexual erythrocytic-stage parasites (D10 or 3D7 lines) were cultivated at 37°C, 5% CO2, 1% N2, and 1% O2 (27) in 4% hematocrit using human O-positive erythrocytes and fed every second day with complete culture medium (RPMI-HEPES supplemented with 0.2% NaHCO3, 5% heat inactivated human serum, and 5% Albumax). Enriched trophozoite and schizont preparations used in Western blot analyses were obtained by Percoll purification (29). Ring-stage parasites (∼5% parasitemia) were transfected with 100 μg of purified plasmid DNA (plasmid maxi kit; Qiagen) (6) using modified electroporation conditions (10), and drug cycling commenced according to methods described previously by Crabb et al. and Duraisingh et al. (7, 9). Briefly, parasites were cultured in a 10-cm culture dish for 48 h prior to selection with 2.5 nM WR99210. Transfected parasites were visible after 3 weeks of continuous culture. Parasites containing integrated forms of the plasmid were obtained by drug cycling, where drug was removed for 2 to 3 weeks before being reapplied.
Southern blotting.
Total parasite DNA was isolated from saponin (0.15%)-lysed trophozoite- and schizont-infected erythrocytes at 5% parasitemia. Buffer A (50 mM NaAc [pH 5.2], 100 mM NaCl, 1 mM EDTA) and sodium dodecyl sulfate (SDS) (3% [wt/vol]) were added to the saponin-lysed pellet and mixed by inversion. The resultant mixture was extracted using phenol-chloroform (1:1) (Progen). The aqueous phase was ethanol precipitated as described previously (26), and the DNA pellets were air dried and resuspended in 500 μl of Tris-EDTA. Manipulation of recombinant DNA and analysis of nucleic acids by Southern blot hybridization were carried out using standard procedures (26).
SDS-polyacrylamide gel electrophoresis and Western blotting.
Parasite proteins extracted from trophozoite and schizont preparations that had been purified by Percoll density gradient (64%) were separated by SDS-polyacrylamide gel electrophoresis (15) on 4 to 20% polyacrylamide gradient Tris-HEPES-sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels under reducing conditions and transferred onto polyvinylidene difluoride membranes (Amersham) using the Bio-Rad Mini-Protean II electrophoresis cell and a Mini Tran-Blot electrophoretic transfer cell apparatus according to the manufacturer's instructions. After transfer, polyvinylidene difluoride membranes were blocked in 5% (wt/vol) skim milk in phosphate-buffered saline overnight at 4°C. Membranes were probed with rabbit anti-SERA-specific polyclonal antibodies. The antibodies and methodology used were described in a previous study (20). Horseradish peroxidase-conjugated sheep anti-rabbit or sheep anti-human immunoglobulin G (Silenus Laboratories) diluted 1/3,000 was used for detection.
Growth rate assay.
Ring-stage wild-type and knockout parasites were synchronized by sorbitol lysis twice at 4-h intervals and plated in duplicate at 0.5% parasitemia into complete culture medium containing a 4% hematocrit in the absence of drug selection. Methanol-fixed and Giemsa-stained thin blood smears were made every 48 h for a total of 28 days. Fresh medium was added every 48 h, and cultures were maintained below a parasitemia of 5%.
Microarray.
Microarrays comprised of 6,912 P. falciparum 70-mer oligonucleotides were printed in duplicate on polylysine-coated microscope slides (Operon Technologies) as described previously (4). Highly synchronous P. falciparum D10 parasite cultures were obtained by double-sorbitol treatment (5%; Sigma) and used to measure SERA gene expression levels between mature (trophozoite/schizont) and young (ring) D10 parasites. For this, parasitized erythrocytes were lysed by saponin lysis (0.15%), and the RNA was extracted using RNeasy Minicolumns (Qiagen). Total RNA (20 μg) was reverse transcribed overnight using SuperScript II (Gibco BRL). The reactive amino group of aa-dUTP was used to conjugate 2 μg of the purified cDNA preparations with 100 μg of the fluorescent dye esters N-hydroxysuccinimide-Cy3 (Amersham) and N-hydroxysuccinimide-Cy5 (Amersham). Microarrays were hybridized overnight at 42°C with 500 ng of each fluorescently labeled probe in a solution containing 25% (vol/vol) formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.1% (wt/vol) SDS. Arrays were washed twice at room temperature with 1× SSC-0.2% SDS, followed by two stringent washes (0.1× SSC) prior to scanning on a ScanArray 4000 apparatus (GSI Lumonics). Data were extracted from the raw images and mined using GeneSpring (Silicon Genetics).
Quantitative reverse transcription (RT)-PCR.
Parasitized erythrocytes at 36 to 40 h postsynchronization were lysed by saponin lysis, and RNA was extracted using RNeasy Minicolumns (Qiagen). Total RNA (5 μg) was reverse transcribed using SuperScript II (Gibco BRL), and 500 ng of the resultant cDNA was added to the QuantiTect SYBR green PCR mixture (Qiagen) containing the gene-specific oligonucleotides specified in Table S1 in the supplemental material. PCR was performed using a LightCycler apparatus (Roche), and reaction conditions were as follows: 95°C (15 min), followed by 40 cycles of 95°C (15 s), 45°C (30 s), and 67°C (20 s). Serial dilutions of P. falciparum D10 genomic DNA were used as a standard reference control. The same preparation procedure was followed when performing quantitative PCR on parasite preparations isolated following a time course for wild-type D10 and D10-ΔSERA4 parasite lines. Parasites in each line were collected at 30 h postsynchronization and subsequent 4-h intervals for 20 h and prepared as described above.
Database searches and phylogenetic analysis.
HMMER (http://hmmer.janelia.org) profiles were generated for the conserved protease and C-terminal domains of aligned P. falciparum 3D7 SERA proteins. These profiles were used to search a database of six-frame translations of genomic sequences as well as protein sequences of apicomplexan species. From the results, matches were extracted for the following species: P. falciparum (three strains), Plasmodium reichenowi, Plasmodium vivax, Plasmodium knowlesi, Plasmodium yoelii, Plasmodium berghei, Plasmodium chabaudi, Plasmodium gallinaceum, and Theileria annulata. In the case of T. annulata, only the protease domain was matched by the hidden Markov model (HMM) profiles generated. Alignment of the corresponding T. annulata protein sequence with P. falciparum cysteine-type SERA sequences indicated the presence of a divergent (and therefore unrecognized by the C-terminal HMM profile) but homologous C-terminal domain in which all seven cysteine residues were conserved. The C-terminal portion of the T. annulata sequence was extracted from the multiple alignment and used for further analyses.
A multiple alignment was constructed from concatenated protease and C-terminal domains using Hmmalign. This alignment consisted of 354 characters over 71 sequences. P. reichenowi SERA8 was removed because the obtained sequence was partial. For cysteine-type SERAs lacking a C-terminal domain (e.g., P. falciparum SERA8, P. yoelii SERA5, and P. vivax SERA12), gaps were inserted to pad the alignment. Phylogenetic analyses were carried out using MrBayes (24) with fixed Blosum62 transition probabilities and substitution rates of Γ(4) + I. In parallel, topologies were inferred using neighbor-joining and maximum likelihood methods as implemented by Phylip (Phylogeny Inference Package version 3.6; J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle). Internal branch splits congruent between Markov chain Monte Carlo (MCMC), maximum likelihood, and neighbor-joining methods were recorded. In order to examine support for the Cys-Ser split in the inferred phylogeny, we removed all trees from the MCMC sampling that contained this branch partition and reanalyzed the remainder of the data.
RESULTS
SERA5 is the only serine-type SERA gene that is refractory to genetic deletion.
Two of the six serine-type SERA genes, SERA2 and SERA3, have previously been disrupted in P. falciparum blood stages using a single-crossover approach; however, this same methodology was not successful in deleting three other genes in this class, SERA1, SERA4, and SERA5 (20). The sixth member of this group, SERA9, has not been targeted previously. Here, we attempted to disrupt the genes encoding four serine-type SERA genes, SERA1, SERA4, SERA5, and SERA9, for which knockout lines had not previously been generated. We employed the single-crossover recombination approach to target SERA1 because although this approach was unsuccessful in our previous study (20), unlike the other genes in our previous work, it was not repeated on multiple occasions. A double-crossover recombination approach was used for SERA4, SERA5, and SERA9 (Fig. 1). For these genes, 5′ (FL1) and 3′ (FL2) regions of the target genes were cloned into the transfection plasmid pHTK to facilitate gene targeting (9). Each plasmid was transfected into D10 and/or 3D7 parasite lines, and stable parasite populations were obtained using the positive selection agent WR99210. Negative selection was then applied by culturing the parasite populations in the presence of ganciclovir. In some cases, populations were routinely subjected to drug cycling on and off with WR99210 and ganciclovir (cycle 1 [C1] and C2, etc.) to facilitate selection for integrant lines.
FIG. 1.
Targeted disruption of four serine-type SERA genes. (A) Schematic representation detailing the outcome of expected integration events in SERA knockout parasite lines. Solid lines under the exons represent the location of the gene targeting regions used to clone fragments into pHH1 (SERA1), pTKΔSERA4, pTKΔSERA5, and pTKΔSERA9. The targeting regions of the coding sequence are indicated by the gray shaded regions. (B) Southern blots summarizing the result of the attempted targeted disruption of each SERA gene using restriction-digested genomic DNA from each transfected line following drug cycling and parental parasites. Genomic DNA was digested BstBI (B)/SphI (S) for SERA1, AflIII (A) for SERA4, XmnI (X) for SERA5, and BclI (Bc)/NotI/SwaI (Sw) for SERA9. The membranes were probed with each respective targeting sequence shown above (A). The positions of endogenous (E), plasmid (P), and integration-specific fragment (In) events that result from digestion with the respective enzymes and the probing of blots with the specific targeting region (gray) are indicated, and their molecular weights (in thousands) are given below each relevant blot. RE, restriction enzyme.
In order to test whether the transfected plasmids had integrated into the correct locus, Southern blot analysis was used to probe restriction-digested genomic DNA from both parental parasites and one of the selected parasite clones. Filters were probed with either the 5′ or 3′ DNA fragments representing the unique targeting sequence in each plasmid (Fig. 1). Restriction enzymes were chosen to reveal a distinct size difference in fragments representing the wild-type locus, the integrated locus, and the episomal plasmid when the blots were hybridized to a targeting sequence probe.
In the case of SERA1, Southern blot analysis identified a band of 4.6 kb corresponding to the endogenous SERA1 locus in wild-type 3D7. The disappearance of this fragment together with the appearance of fragments representing the targeted locus confirms successful single-crossover homologous recombination (Fig. 1). It should be noted that plasmids that integrate by single-crossover recombination often insert more than one copy into the locus, and if this has occurred, a band corresponding to that expected for the episomal plasmid would be observed. This was the case for SERA1, where, based on band intensity, one additional copy was inserted (Fig. 1).
With respect to the targeting of the SERA4 gene, hybridization with the FL1 and FL2 targeting sequences of SERA4 identified single bands of 5.4 kb and 1.8 kb in D10 corresponding to the endogenous gene. In the transfected D10-SERA4/C0 line, which had not undergone ganciclovir selection, the bands at 2.6 kb and 4.0 kb indicated the presence of plasmid pTKΔSERA4. However, in D10-ΔSERA4 parasites, which had undergone negative selection pressure, single bands at 6.0 kb and 2.5 kb indicated that the plasmid had integrated into the SERA4 gene via the expected double-crossover event (Fig. 1). Therefore, although we were previously unable to disrupt SERA4 using single-crossover technology, a SERA4 “knockout” was obtained by using the negative selection procedure.
In an attempt to delete SERA5, plasmid pHTKΔSERA5 was transfected into D10 parasites on four separate occasions, and parasites were cycled as described above. Parasites were analyzed by pulsed-field gel electrophoresis, and in all rounds of drug cycling hybridizations of both the hDHFR probe and the pGEM probe to D10-pHTKΔSERA5, either parasites produced a smeared multiple banding pattern consistent with episomal maintenance of the plasmid or the hDHFR probe appeared to hybridize to a chromosome other than chromosome 2, suggesting that the plasmid integrated elsewhere into the genome, possibly by nonhomologous recombination (data not shown). Southern blot analysis revealed that all attempts to integrate the plasmid into the genome were unsuccessful (Fig. 1). Hence, our attempts to disrupt SERA5 in the D10 line made in this study, together with the multiple attempts in both D10 and 3D7 lines described in our previous study (21), suggest an important role for this protein in the erythrocytic cycle.
SERA9, which is located on chromosome 9 and is the only SERA gene not contained in the chromosome 2 locus, has not been previously targeted for deletion. Plasmid pHTKΔSERA9 was transfected into D10 parasites and cycled in the same way as described above. Hybridization with the targeting sequence of SERA9 identified a single band of 2.0 kb in D10 corresponding to the endogenous gene. In the transfected D10-SERA9/C0 line that had not undergone negative selection, the bands at 2.0 kb and 9.6 kb indicate the presence of plasmid pTKΔSERA9. However, in the D10-ΔSERA9 parasites, which had undergone negative selection pressure with ganciclovir, a single band at 5.4 kb indicated that the plasmid had integrated into the SERA9 gene via the expected event (Fig. 1).
In summary, we have successfully disrupted SERA1, SERA4, and SERA9, while SERA5 remained refractory to deletion. As we previously deleted SERA2 and SERA3, it is apparent that five of the six members of the serine-type SERA family are not essential to in vitro blood-stage growth, while SERA5 appears to be important for the maintenance of this life cycle stage.
Deletion of SERA4 leads to a transcriptional up-regulation of SERA5.
The phenotype of SERA4 knockout parasites was analyzed in detail, considering that it is the second-highest-expressing member of the SERA family (after SERA5) in the erythrocytic cycle (2, 20). Late-stage parental D10 and D10-ΔSERA4 parasite proteins were separated on a 10% polyacrylamide gel and subjected to Western blot analysis to confirm that the disruption of the SERA4 gene had specifically affected production of the SERA4 protein (Fig. 2A). Duplicate membranes were probed with either rabbit anti-SERA4 polyclonal antibodies or rabbit anti-Hsp70 polyclonal antibodies as a protein loading control. Full-length SERA4 was detected at approximately 100 kDa in D10 and D10-ΔSERA4/C0 parasites, a line containing an episomal form of the plasmid, but not in the two cloned D10-ΔSERA4 lines. Integration of plasmid pHTKΔSERA4 into the SERA4 gene was expected to result in a truncated SERA4 gene predicted to encode the first 157 amino acids of the 963-amino-acid wild-type SERA4 protein. A truncated form of SERA4 was not detected by Western blot analysis of D10-ΔSERA4 parasites (data not shown).
FIG. 2.
Western blot analysis of SERA4-null parasites. (A) Western blots of total material from synchronized late-blood-stage parasites comparing the parental D10 wild type with two clones of D10-ΔSERA4 were probed with either rabbit anti-Hsp70 polyclonal or rabbit anti-SERA4 polyclonal antibody. The antisera used to probe each membrane are indicated at the bottom of the gel. Molecular mass markers (kDa) are shown to the left of the membrane. (B and C) Western blots of total material from synchronized late-blood-stage parasites from wild-type D10 (B) or D10-ΔSERA4 (clone 1) (C) probed with rabbit anti-SERA1 to anti-SERA9 polyclonal antibodies specific to the N-terminal region of each SERA protein (lanes 1 to 9, respectively). The antiserum used to probe each membrane strip is indicated at the top of the lane. The strip probed with the prebleed pool is indicated by P. Molecular mass markers (kDa) are shown to the left of each membrane.
Western blot analysis was used to determine if the expression profile of other SERA proteins had changed in the parasite line lacking the SERA4 protein. Rabbit antibodies specific for recombinant forms of each of the nine SERA proteins were tested for reactivity with D10 (Fig. 2B) and D10-ΔSERA4 (Fig. 2C) parasite proteins extracted from late-stage parasites. Anti-SERA4, anti-SERA5, and anti-SERA6 antibodies reacted strongly with the expected species in the parental D10 extracts. However, as expected, only SERA5 and SERA6 were detected in the D10-ΔSERA4 knockout line. SERA proteins 1, 2, 3, 7, 8, and 9 were not obvious in either D10 or D10-ΔSERA4 parasites, suggesting that they remain at low levels of expression in both circumstances.
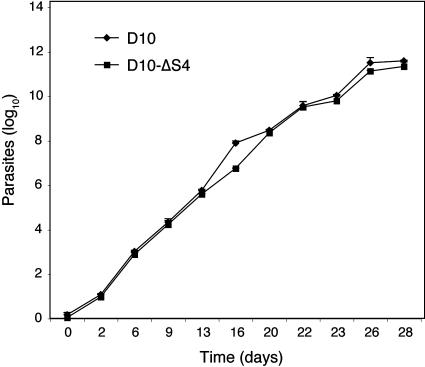
In vitro growth of the D10 parental line was compared with the D10-ΔSERA4 parasite line to determine whether the deletion of SERA4 had any deleterious effect on the ability of these parasites to propagate in blood-stage cultures (Fig. 3). Parasites were synchronized at ring stages, and growth and stage distribution were followed thereafter over a 28-day time period by examination of thin blood smears every 2 days. No detectable difference in growth rates was observed between the parental line and the mutant parasites at any stage over this period.
FIG. 3.
Disruption of SERA4 does not affect in vitro parasite growth. Shown are data from an extended growth rate assay comparing D10 wild-type parasites with the D10-ΔSERA4 (D10-ΔS4) parasite line over a 28-day period. Parasites were maintained below 5%. Error bars represent the standard deviations of mean parasitemias from three individual counts.
In order to investigate whether any members of the SERA multigene family were affected at a transcriptional level in response to the disruption of the SERA4 gene, schizont-stage transcripts were analyzed in both the parental D10 line and the D10-ΔSERA4 line using a combination of microarray and quantitative PCR analyses. Competitive microarray analysis was performed on two occasions using 6,912 70-mer oligonucleotides printed in duplicate on polylysine-coated microscope slides (as described in reference 4). Interestingly, in both arrays, SERA5 was up-regulated by approximately threefold in the D10-ΔSERA4 knockout line in comparison to the parental D10 line (see Fig. S1 in the supplemental material). This was the only gene that showed consistent and obvious transcriptional changes using this method. In addition to this, the coregulated schizont-stage control genes MSP-2, MSP-4, and MSP-5 remained unchanged in the D10-ΔSERA4 parasite line.
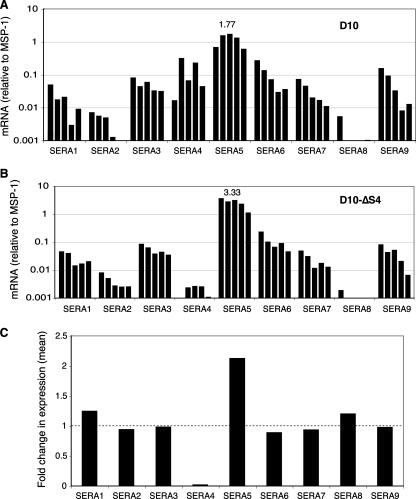
To independently confirm an up-regulation of SERA5 mRNA in SERA4 knockout parasites, we used quantitative RT-PCR. To account for any differences in the synchrony of the parasite lines, we tested RNA extracted from parasites isolated from time courses at 4-h intervals. Expression levels were normalized to the expression of MSP-1, a gene that has a very similar transcriptional profile (Fig. 4). As expected, the level of SERA4 mRNA transcripts was dramatically reduced in the D10-ΔSERA4 parasite line. We consistently observed a twofold increase in SERA5 transcript in SERA4 knockout parasites, while mRNA levels of other SERA genes did not change. These data suggest that a compensatory up-regulation of SERA5 is required in the absence of SERA4. As mRNA and protein levels of SERA appear to be roughly concordant, we expect that such a transcriptional change leads to increased SERA5 protein in SERA4 knockout parasites. However, any change in protein levels was not sufficiently large to be detected by semiquantitative Western blotting (data not shown).
FIG. 4.
Higher levels of SERA5 mRNA are present in SERA4-null parasites. (A and B) SERA gene transcript expression levels in late-blood-stage D10 wild-type (A) andD10-ΔSERA4 (B) parasites measured by quantitative RT-PCR. Parasites in each parasite line were collected at 30 h postsynchronization and subsequent 4-h intervals for 20 h. All expression levels (mRNA) were normalized to the coregulated gene MSP-1. The peak mRNA expression level observed for SERA5 is indicated on the panels to illustrate the difference between D10 and D10ΔS4. (C) Change (n-fold) in gene expression levels between D10 and D10-ΔSERA4 parasite lines. This was calculated by dividing the mean expression levels of the individual values shown in B by those shown in A.
Analysis of complete apicomplexan genome sequences demonstrates that serine-type SERAs form a single monophyletic group.
Although all known SERAs are relatively closely related, previous phylogenetic analyses of this family suggested that serine-type and cysteine-type SERAs form separate phylogenetic groups indicative of an ancient split between these types and the subsequent species-specific expansion of the serine-type SERAs (3, 12, 14). An ancient split and monophyletic grouping support a conserved function for the serine-type SERAs. However, those previous studies involved only a few Plasmodium species and the use of mostly incomplete genomes. In one study, this led to a prediction of “missing” genes that would be identified upon further sequencing (3). Since that time, there has been a vast increase in sequence data available for a wide array of Plasmodium species and other apicomplexan parasites. Here, we reexamine the phylogeny of the SERA genes utilizing complete genome sequence data in order to ascertain if the original premise holds.
Beginning with HMM profiles generated from P. falciparum SERA sequences, we sought to locate and extract conserved domains of SERA genes from the genomes of sequenced apicomplexan species. By using this approach and by restricting ourselves to species for which whole-genome sequencing has been undertaken, we aimed to generate a map of SERA loci in the surveyed genomes that was as complete as possible. This map of the genomic arrangement (Fig. 5) together with the phylogeny (Fig. 6) constructed from the retrieved sequences shed light on the evolution of the SERA family and lend support to the hypothesis of a single function for serine-type SERAs.
FIG. 5.
SERA loci across the Plasmodium and Theileria genera. HMM hits for the protease and C-terminal SERA domains are indicated schematically on the contigs on which they were found. Breaks in species tracks indicate discontinuities at contig ends. Where they exist, gene annotations have been indicated. When HMM matches existed on multiple short contigs in a species, the contigs were ordered according to orthology relationships with related species inferred from the phylogeny. SERA genes that have been successfully knocked out in P. falciparum are marked in red, while those refractory to deletion are in black. Where orthology is clearly discernible from the phylogeny, it is marked by vertical blocks that join species. Orthologies of P. gallinaceum and T. annulata genes are ambiguous due to a postspecies duplication. This ambiguity is represented by a bifurcation in the orthology block. The phylogeny of Cys SERA genes indicates a duplication of SERA8 present only in P. gallinaceum as well as a duplication resulting in SERA6 and SERA7 in Plasmodium spp. (excluding P. gallinaceum). Ambiguities arising from these duplications are represented by bifurcations in the orthology blocks. Where it is clearly present, the orthology of flanking sequence is also noted. Protease domains marked in green do not contain active-site mutations, whereas protease domains marked in blue contain the cysteine-to-serine active-site mutation. The presence of secondary-site mutations has been marked below the corresponding protease domain. RT-PCR expression levels (this study) and protein abundance as a percentages of SERA5 are shown above the P. falciparum (3D7) track (16, 17). Similarly, relative copy number expression measurements for P. vivax as a percentage of P. vivax SERA4 are shown above the P. vivax track (22). KO, knockout.
FIG. 6.
SERA sequences belong to one of two major phyletic groups that separate according to their catalytic residues. Colored segments that make up the inner disc delineate active-site and secondary-site mutations in SERA sequences. Colored segments that are part of the outer ring mark the clade to which sequences belong. Thick lines in the phylogeny indicate branches shared by MCMC, maximum-likelihood, and neighbor-joining topologies. Thin lines indicate branches supported by MCMC and one other method. Dashed lines indicate branches existing only in the MCMC topology. MCMC branch probabilities are given on the appropriate branches where they are less than 1. Members of the genus Theileria appear to have a single-copy SERA, the only observation to date of a SERA ortholog outside the genus Plasmodium. Radiation in SERA sequences containing the serine active-site mutation tends to be species specific, whereas radiation of cysteine SERA sequences has occurred prior to speciation. Branch lengths for SERA6 and SERA7 tend to be shorter than those of serine-type SERAs and those of SERA8.
To date, orthologs of P. falciparum SERA genes have been discovered only in other species of Plasmodium. Sequence similarity searches indicate that the SERA family is specific to this genus, and entries for SERA genes in orthology databases such as OrthoMCL reflect this. Using the profiles generated, however, we discovered a putative SERA ortholog in Theileria annulata. This is the first time that an ortholog has been identified outside plasmodia. The features and the genomic location of the T. annulata SERA both suggest that the SERA locus in the common ancestor of Plasmodium and Theileria species consisted of a single sequence orthologous to P. falciparum SERA6 or SERA7. T. annulata possesses a gene which appears to be orthologous to PFB0320c (immediately 5′ of the SERA locus) but which is inverted relative to Plasmodium spp. (Fig. 5). No orthology with T. annulata is discernible 3′ of the SERA locus. The orthology of flanking genes on both sides is preserved in rodent malaria parasites, but annotated genes are missing due to the shortness of P. yoelii contigs. PFB0320c codes for a protein containing a hesB domain. P. gallinaceum appears to possess two genes orthologous to P. falciparum SERA8, although low coverage means that information regarding the locus organization is not available. It is interesting that this species does not appear to encode a serine-type SERA, suggesting that their function is not obligatory for Plasmodium biology. Whether this relates to differences in avian and mammalian red blood cells remains to be determined. The presence of an intact C-terminal domain in T. annulata suggests that the lack of the domain in P. falciparum SERA8 is the result of a gene truncation after duplication of the ancestral sequence as opposed to a postduplication domain acquisition.
By examining the consensus tree constructed from MCMC samples not containing the branch dividing the cysteine- and serine-type SERA subgroups, we sought to discover whether there was a plausible evolutionary scenario in which the cysteine SERA subgroup was not monophyletic. The consensus tree constructed from these data maintained the broad grouping of sequences but moved the SERA6/SERA7 subtree to a point in the tree that would require a second Ser→Cys mutation to correct the original Cys→Ser mutation and would also imply that SERA6 and SERA7 have arisen relatively recently. Because the Cys-Ser division is supported by multiple methods of tree reconstruction and is more parsimonious than the alternative hypothesis, it is reasonable to assume monophyly of the cysteine-type SERAs.
Various numbers of serine-type SERA genes are observed in different species, ranging from none in P. gallinaceum to nine in P. vivax. The serine-type subgroup of the phylogeny divides neatly into three clades: P. falciparum/P. reichenowi, P. vivax/P. knowlesi, and P.yoelii/P.berghei/P.chabaudi (Fig. 6), and it therefore appears that the duplications that gave rise to multiple copies of the serine-type SERAs have occurred after the speciation events that gave rise to these clades.
The pattern of clade- and species-specific duplication among serine-type SERAs suggests that the expansion of the serine-type SERAs has been relatively recent and followed an ancient cysteine-type-to-serine-type SERA split in an ancestral Plasmodium species. The species-specific expansion of the subfamily, especially in the case of P. vivax and P. falciparum, which infect the same host, supports the hypothesis that a single function for the subfamily exists and that duplication is being driven by host selective pressure.
DISCUSSION
The function of the SERAs expressed during the erythrocytic cycle remains largely unknown, and the role of the serine-type SERAs is particularly uncertain given the noncanonical nature of their active sites. However, from the data presented here, it now appears highly probable that the serine-type SERAs have a crucial function(s) at this life stage. This is supported by the notion that SERA5 appears to be refractory to genetic deletion, while the genes encoding the rest of the members of this group can be disrupted. While such an experiment is not proof of essentiality, the numerous unsuccessful attempts to delete this gene and the knowledge that the SERA locus itself is clearly amenable to gene targeting suggest that the deletion of SERA5 has a deleterious effect on erythrocytic-stage growth. This is consistent with the extraordinarily high level of expression of SERA5, estimated by proteomic studies to be one of the most abundant proteins expressed at the schizont stage. Further evidence of a likely blood-stage function for serine-type SERAs is the possible compensatory increase in SERA5 mRNA expression detected in the SERA4-null parasites. After SERA5, SERA4 is the strongest-expressing member of the SERA family, although it is present only at between 10 and 20% of the levels of SERA5. While the increase at the transcript level was clear, we were not able to detect an obvious increase in SERA5 protein levels in SERA4-null parasites by Western blotting. It is not known how much SERA5 protein is required to compensate for the loss of SERA4 (presumably very little); however, it is below the sensitivity of the serial dilution Western blot technique that we employed here.
The presence of SERA5 in the parasitophorous vacuole and demonstrated (relatively weak) in vitro catalytic activity are suggestive of a role in schizont rupture or cleavage of invasion ligands. However, there remains no proof of this, and indeed, other possible roles at this stage can be envisaged, including possible noncatalytic regulatory functions.
While an important blood-stage role for the serine-type SERAs appears to be likely, the number of different functions that these proteins perform appears to be far fewer than the number of genes present. The comprehensive gene identification and phylogenetic analysis performed in this study confirm that the serine-type SERAs form a single monophyletic group arising from a single ancient duplication and active-site mutation of a cysteine-type SERA. This supports the notion that redundancy is possible between the serine-type SERAs, and indeed, it is possible that there is only one blood-stage role for all members of this group. In addition, the ability to disrupt all except the most strongly expressed of the serine-type SERAs indicates that these more weakly expressed members do not perform independent and distinct functions within the erythrocytic cycle. Moreover, the apparent compensatory increase in SERA5 expression in response to the disruption of SERA4, already discussed above in relation to a blood-stage function, is also consistent with functional redundancy among this group.
If all serine-type SERAs perform the same function, why is SERA5 refractory to genetic deletion? The most likely explanation for this is that the required compensatory change in the expression of another of the serine-type SERAs is too great to be readily achieved in culture. Formal proof of this and proof of the hypothesis of redundant roles for the serine-type SERAs require functional complementation experiments where the SERA5 coding sequence is replaced with that of another serine-type SERA. As SERA genes are large, this remains technically challenging but is theoretically achievable.
Given that we have now seen that the serine-type SERAs form a single monophyletic group, we propose that only one function is likely for members of the serine-type SERAs. This role need not be restricted to the blood stages, and indeed, mRNA profiling suggests that some members of the serine-type SERAs are expressed at other stages (19). With respect to the cysteine-type SERAs, phylogenetic analysis suggests that as many as three orthologous groups represented by P. falciparum members SERA6, SERA7, and SERA8 (Fig. 5 and 6). Some functional data to support this separation exist. The rodent malaria version of the “SERA8-like” SERAs functions in oocyst rupture (1), while “SERA6-like” members are most likely to function in the erythrocytic stage, given their strong expression and apparently essential role at this stage (20).
SERAs that function in the blood stage remain attractive drug targets, and indeed, SERA5 has even progressed considerably as a vaccine candidate. There is now a strong likelihood that interference with serine-type SERA function will interrupt normal erythrocytic development and promote parasite clearance from infected individuals. SERA5 may indeed be obligatory for blood-stage growth. However, if other family members such as SERA4 can compensate if expressed highly enough, it is important that interventions targeting SERA5 should be designed to also block the other serine-type SERAs.
Supplementary Material
Acknowledgments
The genome sequence data for the Ghanian P. falciparum isolate, P. reichenowi, P. knowlesi, P. berghei, P. chabaudi, and P. gallinaceum were produced by the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute. Sequence data for P. vivax, P. yoelii, and Theileria parva were obtained from The Institute for Genomic Research, and sequence data for P. falciparum (HB3) were obtained from the Broad Institute. We thank the Australian Red Cross blood bank for the provision of human blood and serum.
This work was supported by the National Health and Medical Research Council of Australia and the Wellcome Trust, United Kingdom. J.E.M. and T.S. are the recipients of NHMRC Dora Lush postgraduate scholarships, S.K.M. was the recipient of a Melbourne Research scholarship (University of Melbourne), and B.S.C. is an International Research Scholar of the Howard Hughes Medical Institute.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 24 September 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aly, A. S., and K. Matuschewski. 2005. A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 202:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, S., J. Li, S. Itagaki, B. A. Okech, T. G. Egwang, H. Matsuoka, N. M. Q. Palacpa, T. Mitamura, and T. Horii. 2002. Serine repeat antigen (SERA5) is predominantly expressed among the SERA multigene family of Plasmodium falciparum, and the acquired antibody titers correlate with serum inhibition of the parasite growth. J. Biol. Chem. 277:47533-47540. [DOI] [PubMed] [Google Scholar]
- 3.Bourgon, R., M. Delorenzi, T. Sargeant, A. N. Hodder, B. S. Crabb, and T. P. Speed. 2004. The serine repeat antigen (SERA) gene family phylogeny in Plasmodium: the impact of GC content, and reconciliation of gene and species trees. Mol. Biol. Evol. 21:2161-2171. [DOI] [PubMed] [Google Scholar]
- 4.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowman, A. F., and B. S. Crabb. 2006. Invasion of red blood cells by malaria parasites. Cell 124:755-766. [DOI] [PubMed] [Google Scholar]
- 6.Crabb, B. S., and A. F. Cowman. 1996. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 93:7289-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crabb, B. S., M. Rug, T. W. Gilberger, J. K. Thompson, T. Triglia, A. G. Maier, and A. F. Cowman. 2004. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol. Biol. 270:263-276. [DOI] [PubMed] [Google Scholar]
- 8.Delplace, P., B. Fortier, G. Tronchin, J. F. Dubremetz, and A. Vernes. 1987. Localization, biosynthesis, processing and isolation of a major 126 kDa antigen of the parasitophorous vacuole of Plasmodium falciparum. Mol. Biochem. Parasitol. 23:193-201. [DOI] [PubMed] [Google Scholar]
- 9.Duraisingh, M. T., T. Triglia, and A. F. Cowman. 2002. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int. J. Parasitol. 32:81-89. [DOI] [PubMed] [Google Scholar]
- 10.Fidock, D. A., and T. E. Wellems. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. USA 94:10931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodder, A. N., D. R. Drew, V. C. Epa, M. Delorenzi, R. Bourgon, S. K. Miller, R. L. Moritz, D. F. Frecklington, R. J. Simpson, T. P. Speed, R. N. Pike, and B. S. Crabb. 2003. Enzymic, phylogenetic, and structural characterization of the unusual papain-like protease domain of Plasmodium falciparum SERA5. J. Biol. Chem. 278:48169-48177. [DOI] [PubMed] [Google Scholar]
- 13.Knapp, B., E. Hundt, U. Nau, and H. A. Kupper. 1989. Molecular cloning, genomic structure and localization in a blood stage antigen of Plasmodium falciparum characterized by a serine stretch. Mol. Biochem. Parasitol. 32:73-83. [DOI] [PubMed] [Google Scholar]
- 14.Kooij, T. W., J. M. Carlton, S. L. Bidwell, N. Hall, J. Ramesar, C. J. Janse, and A. P. Waters. 2005. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 1:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lasonder, E., Y. Ishihama, J. S. Andersen, A. M. Vermunt, A. Pain, R. W. Sauerwein, W. M. Eling, N. Hall, A. P. Waters, H. G. Stunnenberg, and M. Mann. 2002. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature 419:537-542. [DOI] [PubMed] [Google Scholar]
- 17.Le Roch, K. G., J. R. Johnson, L. Florens, Y. Zhou, A. Santrosyan, M. Grainger, S. F. Yan, K. C. Williamson, A. A. Holder, D. J. Carucci, J. R. Yates III, and E. A. Winzeler. 2004. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 14:2308-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Roch, K. G., Y. Zhou, S. Batalov, and E. A. Winzeler. 2002. Monitoring the chromosome 2 intraerythrocytic transcriptome of Plasmodium falciparum using oligonucleotide arrays. Am. J. Trop. Med. Hyg. 67:233-243. [DOI] [PubMed] [Google Scholar]
- 19.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503-1508. [DOI] [PubMed] [Google Scholar]
- 20.Miller, S. K., R. T. Good, D. R. Drew, M. Delorenzi, P. R. Sanders, A. N. Hodder, T. P. Speed, A. F. Cowman, T. F. de Koning-Ward, and B. S. Crabb. 2002. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J. Biol. Chem. 277:47524-47532. [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell, R. A., and M. J. Blackman. 2005. The role of malaria merozoite proteases in red blood cell invasion. Curr. Opin. Microbiol. 8:422-427. [DOI] [PubMed] [Google Scholar]
- 22.Palacpac, N. M., B. W. Leung, N. Arisue, K. Tanabe, J. Sattabongkot, T. Tsuboi, M. Torii, R. Udomsangpetch, and T. Horii. 2006. Plasmodium vivax serine repeat antigen (SERA) multigene family exhibits similar expression patterns in independent infections. Mol. Biochem. Parasitol. 150:353-358. [DOI] [PubMed] [Google Scholar]
- 23.Pang, X. L., T. Mitamura, and T. Horii. 1999. Antibodies reactive with the N-terminal domain of Plasmodium falciparum serine repeat antigen inhibit cell proliferation by agglutinating merozoites and schizonts. Infect. Immun. 67:1821-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronquist, F., and J. P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 25.Rosenthal, P. J. 2004. Cysteine proteases of malaria parasites. Int. J. Parasitol. 34:1489-1499. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193:673-675. [DOI] [PubMed] [Google Scholar]
- 28.Triglia, T., and A. F. Cowman. 1994. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 91:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, Y., X. Wang, X. Liu, and Y. Wang. 2003. Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 13:601-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.