Abstract
The environmental pathogen Legionella pneumophila possesses five proteins with Sel1 repeats (SLRs) from the tetratricopeptide repeat protein family. Three of these proteins, LpnE, EnhC, and LidL, have been implicated in the ability of L. pneumophila to efficiently establish infection and/or manipulate host cell trafficking events. Previously, we showed that LpnE is important for L. pneumophila entry into macrophages and epithelial cells. In further virulence studies here, we show that LpnE is also required for efficient infection of Acanthamoeba castellanii by L. pneumophila and for replication of L. pneumophila in the lungs of A/J mice. In addition, we found that the role of LpnE in host cell invasion is dependent on the eight SLR regions of the protein. A truncated form of LpnE lacking the two C-terminal SLR domains was unable to complement the invasion defect of an lpnE mutant of L. pneumophila 130b in both the A549 and THP-1 cell lines. The lpnE mutant displayed impaired avoidance of LAMP-1 association, suggesting that LpnE influenced trafficking of the L. pneumophila vacuole, similar to the case for EnhC and LidL. We also found that LpnE was present in L. pneumophila culture supernatants and that its export was independent of both the Lsp type II secretion system and the Dot/Icm type IV secretion system. The fact that LpnE was exported suggested that the protein may interact with a eukaryotic protein. Using LpnE as bait, we screened a HeLa cell cDNA library for interacting partners, using the yeast two-hybrid system. Examination of the protein-protein interaction between LpnE and a eukaryotic protein, obscurin-like protein 1, suggested that LpnE can interact with eukaryotic proteins containing immunoglobulin-like folds via the SLR regions. This investigation has further characterized the contribution of LpnE to L. pneumophila virulence and, more specifically, the importance of the SLR regions to LpnE function.
Legionella pneumophila is an intracellular pathogen and the causative agent of Legionnaires' disease. The bacterium infects alveolar macrophages and establishes a replicative vacuole derived from the endoplasmic reticulum by utilizing the Dot/Icm type IV secretion system (48, 53). The ability of L. pneumophila to propagate itself within macrophages appears to have developed from its parasitic relationship with protozoa in aquatic environments. The genome sequences of three L. pneumophila strains have highlighted the large number of eukaryotic protein-like proteins that this pathogen has acquired (11, 13). These proteins are predicted to allow L. pneumophila to manipulate host cell processes, and several bacterial proteins with similarity to eukaryotic proteins have been identified as substrates of the Dot/Icm system (12, 19, 39, 41). Others, such as the ecto-nucleoside triphosphate diphosphohydrotase Lpg1905, have distinct roles in pathogenesis (47). The genome sequences have revealed that L. pneumophila possesses five genes encoding proteins with predicted tetratricopeptide repeat (TPR) motifs. At least two of these genes, lpnE and enhC, contribute to enhanced entry into human tissue culture cell lines (14, 42).
The TPR motif was originally reported for cell division cycle proteins of Saccharomyces cerevisiae (32, 49). Today this motif is known to be ubiquitous in nature, as it is found within functionally unrelated proteins from all genera. A TPR is defined as a degenerate 34-residue motif with a consensus amino acid arrangement of alternate large and small residues and high amino acid conservation observed specifically at positions 8, 20, and 27 (49). These conserved residues allow the TPR to create a pair of antiparallel alpha helices. Multiple motifs, ranging from 3 to 16 in number among TPR-containing proteins, lead to the formation of an alpha superhelical structure (17). This complex and unique structure gives rise to distinct substrate grooves that facilitate specific protein-protein interactions. The ability of TPR proteins to interact with other proteins enables them to play a vital role in eukaryotic cell processes, such as mitosis, transcription repression, and protein import (20, 37, 52). Bacteria also utilize TPR proteins for a range of functions, including gene regulation, flagellar motor function, chaperone activity, and virulence (9, 16, 43, 54). Several chaperones required for type III secretion system-mediated translocation of virulence proteins into host cells contain TPR domains, including PcrH from Pseudomonas aeruginosa, LcrH from Yersinia species, and CesD from enteropathogenic Escherichia coli (8, 9, 54).
The Sel1 repeat (SLR) motif comprises a subtype of TPR, named after the extracellular protein from Caenorhabditis elegans for which it was first described (29). Sel1 and its homologues are involved in cell-to-cell interactions that specify the fate of C. elegans cells during development through binding of the membrane proteins Lin-12 and Glp-1 (29). The SLR motif has a less stringent definition than the TPR motif, with the length of an SLR ranging from 36 to 44 amino acids (40). However, the motif consensus sequences are comparable and, subsequently, the motif folding is considered to be equivalent. As such, SLR proteins are also predicted to mediate important protein-protein interactions, and the motif is found preferentially in eukaryotic proteins (40).
All EnhC, LpnE, and LidL proteins contain SLR-type TPR motifs and have been shown to be important for L. pneumophila -host interactions (14, 42). In this study, we investigated the contribution of LpnE to the infection of amoebae and A/J mice by L. pneumophila. In addition, we examined the involvement of LpnE in trafficking of the Legionella-containing vacuole and the basis of protein-protein interactions mediated by the SLR regions of LpnE to increase our understanding of how this protein is linked to L. pneumophila virulence.
MATERIALS AND METHODS
Bacterial and yeast strains, growth conditions, and plasmids.
Bacterial and yeast strains and plasmids used in this study are listed in Table 1. Bacteria were cultured aerobically at 37°C. L. pneumophila strains were grown in ACES [N-(2-acetamido)-2-aminoethanesulfonic acid] buffered yeast extract broth or on buffered charcoal yeast extract (BCYE) plates (23), and unless otherwise stated, Escherichia coli strains were cultured in Luria-Bertani (LB) broth or agar. Antibiotics were added to media at the following final concentrations for L. pneumophila (or E. coli ): kanamycin, 25 μg/ml (100 μg/ml); chloramphenicol, 6 μg/ml (12.5 μg/ml); and ampicillin, 100 μg/ml (100 μg/ml). Saccharomyces cerevisiae was cultured in YPD (1% yeast extract, 2% peptone, and 2% dextrose) or synthetic dropout medium (Clontech, CA) lacking appropriate nutrients at 30°C.
TABLE 1.
Bacterial and yeast strains and plasmids used in this study
| Strain or plasmid | Serogroup and/or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| L. pneumophila strains | ||
| 130b (ATCC BAA-74) | O1; clinical isolate | 22 |
| 130b lpnE::km | lpnE insertion mutant of 130b (Kmr) | 42 |
| 130b lpnE::km/ (pMIP:lpnE) | lpnE::km mutant carrying pMIP:lpnE (Kmr Cmr) | 42 |
| 130b lspDE::km | lspDE insertion mutant of 130b (Kmr) | 47 |
| 130b dotA::cm | dotA insertion mutant of 130b (Cmr) | 47 |
| E. coli strains | ||
| XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac F′ [proAB+lacIqlacZ ΔM15 Tn10] (Tetr) | Stratagene |
| BL-21 (DE3) | F−omp T hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| KC8 | pyrF::Tn5 hsdR leuB600 trpC9830 lacD74 strA galK hisB436 | Clontech |
| S. cerevisiae strains | ||
| PJ69-4a | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::Gal7-lacZ | 35 |
| AH109 | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1TATA-HIS3 MEL1 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ | 35 |
| Y187 | MATα ura3-52 his3-200 ade2-101 trp1-901 leu2-3,112 gal4Δ met−gal80Δ URA3::GAL1UAS-GAL1TATA-lacZ MEL1 | 30 |
| Plasmids | ||
| pMip | pMMB207 with the promoter region of mip cloned into SacI/XbaI sites | 42 |
| pMip:lpnE | pMip carrying lpnE and the ribosome binding site cloned into XbaI/PstI sites | 42 |
| pMIP:lpnE1-51 | pMIP with an lpnE truncation encoding the first 51 amino acids cloned into XbaI/PstI sites | This study |
| pMIP:lpnE1-122 | pMIP with an lpnE truncation encoding the first 122 amino acids cloned into XbaI/PstI sites | This study |
| pMIP:lpnE1-266 | pMIP with an lpnE truncation encoding the first 266 amino acids cloned into XbaI/PstI sites | This study |
| pMIP:lpnE52-375 | pMIP with an lpnE truncation encoding amino acid 52 to the stop codon cloned into XbaI/PstI sites | This study |
| pGBT9 | GAL4(1-147) DNA binding domain; TRP1 Ampr | 4 |
| pGAD424 | GAL4(768-881) activation domain; LEU2 Ampr | 4 |
| pGADT7 | GAL4(768-881) activation domain; LEU2 Ampr; hemgglutinin epitope tag | Clontech |
| pMAL-c2x | Expression vector to generate translation fusions to malE; Ptac Ampr | New England Biolabs |
DNA, PCR, and cloning techniques.
Plasmid DNA was purified using a QIAprep spin miniprep kit (QIAGEN, Hilden, Germany), and bacterial genomic DNA was isolated as described previously (2). DNA-modifying enzymes were used according to the manufacturer's instructions (Promega, WI). PCR amplification was performed using 200 ng of template DNA and approximately 0.25 μg of each oligonucleotide. Oligonucleotide sequences and annealing conditions used are listed in Table 2. Plasmid constructs were confirmed by DNA sequencing using a PRISM ready reaction dye deoxy terminator cycle sequencing kit and a 3730 DNA analyzer (Applied Biosystems, CA). DNA sequences were assembled using Sequencher 3.1.1 (Gene Codes Corp., MI).
TABLE 2.
Sequences of oligonucleotides used in this study
| Primer | Sequence (5′-3′) | Annealing temp (°C) |
|---|---|---|
| lpnE F | GCT CTA GAG ATA GCT CTT AAA AAT AAG G | 46 |
| lpnE1-51 R | TTC TGC AGT TAA TCT CCT TTA TCC GCT TC | 46 |
| lpnE1-122 R | TTC TGC AGT TAC GCA TTA TTT TGA TCC GC | 46 |
| lpnE1-266 R | TTC TGC AGT TAG GCA TTC CCC AGA TCT G | 46 |
| lpnE52-375 F | GCT CTA GAA AGG AAT TAT AAA TGG CCA AGG CTC AAT ATT TGC TGG GTA AG | 46 |
| lpnE R | AAC TGC AGG AAA ACA GGT AAC AGG C | 46 |
| MBP-lpnE F | GCT CTA GAA TGG ACA TGA AAA AAT ATA TT | 44 |
| lpnE-Y2HF | CGG GAT CCG TAT GGA CAT GAA AAA ATA TAT T | 44 |
| OBLS1 IgF | CGA ATT CAT GGC AGG TCT GGA GGA TGT G | 56 |
| OBLS1 IgR | GG GAT CCC GAC TCA TCG TCA ATT TCA C | 56 |
| OBLS1 FnF | CGA ATT CAT GCC CCG GAA GCT CGA CGT C | 56 |
| OBLS1 FnR | CGG GAT CCA CTA CGG CCA TGT CCG CTG | 56 |
| OBLS1 IgFnR | CGG GAT CCT TAT TTT ACT CTG AGA AGC GTA G | 50 |
Coculture of Acanthamoeba castellanii and L. pneumophila.
A. castellanii ATCC 50739 was grown in 75-mm2 tissue culture flasks (Sarstedt, Leicestershire, United Kingdom) with PYG 712 medium at 20°C without agitation (10). Cultures of A. castellanii that were 72 h old were harvested and seeded into 24-well tissue culture trays (Sarstedt) at a density of 2.5 × 105 cells/well in A.c. buffer (0.1% trisodium citrate, 0.4 mM CaCl2, 2.5 mM KH2PO4, 4 mM MgSO4, 2.5 mM Na2HPO4, 0.05 mM ferric pyrophosphate). Stationary-phase L. pneumophila strains, also resuspended in A.c. buffer, were added at a multiplicity of infection (MOI) of 0.01, and the coculture was incubated at 37°C with 5% CO2. At specific time points, samples from duplicate wells for each strain were plated onto BCYE agar and assessed for replication of L. pneumophila. Results were expressed as mean log10 CFU ± standard deviations for three independent experiments.
Pulmonary infection of A/J mice with L. pneumophila.
To examine the comparative virulence of L. pneumophila 130b and the lpnE::km derivative within an established animal model, mixed infections of mutant and wild-type L. pneumophila were performed as described previously (45, 46). Briefly, 6- to 8-week-old female A/J mice (Jackson Laboratory, ME) were anesthetized and inoculated intratracheally with 106 CFU of wild-type and mutant bacteria at a ratio of 1:1. Twenty-four and 72 h after inoculation, mice were sacrificed and their lung tissue isolated. Tissue was homogenized using a Pro 200 homogenizer (Pro Scientific Inc., CT), and complete host cell lysis was attained by incubation of the homogenized tissue in 0.1% saponin for 10 min at 37°C. Serial dilutions were plated onto both plain and antibiotic-selective BCYE to obtain the total number of viable bacteria and the number of lpnE mutant bacteria colonizing the lung tissue. Mixed infections were also performed with lpnE::km/(pMip:lpnE) and lpnE::km/(pMIP:lpnE52-375) strains in competition with L. pneumophila 130b/(pMIP).
Transcomplementation of L. pneumophila 130b lpnE::km with truncated lpnE.
Truncated forms of lpnE were amplified using the oligonucleotides listed in Table 2 and cloned into the XbaI/PstI sites of pMIP (42). These plasmids were introduced into L. pneumophila 130b lpnE::km via electroporation as described previously (21). Briefly, 50-ml logarithmic-phase cultures were harvested via centrifugation (10,000 × g, 4°C, 15 min) and washed with cold phosphate-buffered saline (PBS) and then cold 10% (vol/vol) glycerol before being resuspended in 10% glycerol with approximately 500 ng of plasmid DNA and subjected to electrophoresis at 2,300 V, 100 Ω, and 25 mF.
Tissue culture conditions and L. pneumophila invasion assays.
The human monocytic cell line THP-1 and the human alveolar epithelial cell line A549 were propagated and prepared for infection as described previously (42). Stationary-phase strains of L. pneumophila were added at an MOI of 5 for THP-1 cells and an MOI of 100 for A549 cells and allowed to infect cells for 2 h in 5% CO2 at 37°C. Cells were then treated with 100 μg/ml gentamicin for 1 h to kill extracellular bacteria and washed with PBS before being lysed with 0.01% digitonin. Serial dilutions of the inoculum and bacteria recovered from lysed cells were plated on BCYE agar, and results are expressed as percentages of the inoculum that resisted killing by gentamicin (means ± standard deviations for at least three independent experiments).
Avoidance of LAMP-1 by L. pneumophila-containing vacuoles.
Immunofluorescence was used to investigate the ability of Legionella-containing vacuoles to avoid lysosomal fusion, using the lysosome-associated membrane protein LAMP-1 as a marker for lysosomal membranes, in both A549 and THP-1 cells. Immunofluorescence was performed as described previously (47). Briefly, 105 A549 cells were seeded onto 12-mm glass coverslips (Menzel-Glaser, Braunschweig, Germany) and grown overnight before being infected with stationary-phase L. pneumophila at an MOI of 100. Alternatively, 105 THP-1 cells were allowed to differentiate in the presence of phorbol 12-myristate 13-acetate for 72 h, ensuring differentiation into adherent, elongated cells, before being infected with stationary-phase L. pneumophila at an MOI of 5. Infection proceeded for 5 h or 24 h before cells were washed with PBS and fixed with 4% (wt/vol) paraformaldehyde (pH 7.4). Following fixation, the coverslips were blocked for 1 h with PBS containing 10% fetal bovine serum and then stained with monoclonal mouse anti-L. pneumophila (6026; ViroStat, ME) and, subsequently, anti-mouse immunoglobulin G (IgG)-Alexa Fluor 594 (Invitrogen, CA). Following labeling of the extracellular bacteria, cells were permeabilized for 1 h with PBS containing 10% fetal bovine serum and 0.05% saponin. Cells were washed with PBS and then stained with anti-L. pneumophila and rabbit anti-human LAMP-1 H-288 (Santa Cruz Biotechnology, CA). Anti-rabbit-Alexa Fluor 488 and anti-mouse-Alexa Fluor 594 or -Alexa Fluor 405 were used as secondary antibodies, and all antibodies were diluted in PBS with 10% fetal bovine serum, used at 1:50 (Legionella), 1:100 (LAMP-1), or 1:200 (secondary antibodies), and incubated with the cells for 1 h at 37°C. At the end of this staining procedure, intracellular bacteria appeared green, LAMP-1 appeared red, and extracellular bacteria appeared red or blue. Coverslips were mounted in DAKO fluorescent mounting medium (DAKO Corporation) and stored at 4°C in the dark. Slides were examined under a 100× objective on an Olympus BX51 microscope (Olympus, Tokyo, Japan). Images were acquired using an Olympus DP-70 digital camera and merged using DP controller software, version 1.1.1.71. LAMP-1 avoidance was scored blind for various strains of L. pneumophila, and data for at least 50 intracellular bacteria from three independent coverslips were expressed as mean percentages of LAMP-1 avoidance (± standard deviations). Differences in LAMP-1 avoidance were assessed for significance using an unpaired two-tailed t test.
Production of recombinant LpnE and polyclonal anti-LpnE antibodies.
Full-length lpnE was amplified by PCR, using MBP-lpnE F and lpnE R (Table 2), and cloned into the XbaI/PstI sites of the pMAL-c2x expression vector (New England Biolabs, MA) to generate a maltose binding protein (MBP) fusion with LpnE. Production of this fusion protein was induced in E. coli BL21 with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and subsequently purified using amylose agarose as described by the manufacturer (Scientifix, Australia). Polyclonal antibodies were generated against column-purified MBP-LpnE resuspended in incomplete Freund's adjuvant by inoculation of 500 μg of protein into a rabbit on days 0, 28, and 49 before exsanguination on day 66 (Chemicon International, Inc., CA). Immune serum was adsorbed against L. pneumophila 130b lpnE::km and diluted 1:500 for immunoblotting in 0.05% Tween (vol/vol) in Tris-buffered saline.
TCA precipitation of culture supernatants.
L. pneumophila extracellular proteins were precipitated from bacterial culture supernatants as described previously (38). Fifty-milliliter samples of stationary- or late-logarithmic-phase broth cultures of L. pneumophila were subjected to centrifugation (10,000 × g, 4°C, 15 min), and the supernatants were filtered through 0.22-μm filters (Millipore, MA). A 10% (wt/vol) final concentration of trichloroacetic acid (TCA) was added to precipitate the proteins, which were pelleted by centrifugation (10,000 × g, 4°C, 1 h) and washed with 100% (vol/vol) cold methanol before being dried and resuspended in 2× sodium dodecyl sulfate (SDS) sample buffer. TCA precipitation included the use of previously described lspDE and dotA mutants (47). TCA-precipitated culture supernatants and whole-cell lysate samples of strains of interest were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose for immunoblotting. Rabbit anti-E. coli RpoB antibodies (34) were used at 1:4,000 as a control for cytoplasmic protein contamination of supernatant samples, and rabbit anti-Lpg1905 (1:500) (47) was used to control for loading of TCA-precipitated proteins. When investigating the presence of truncated forms of LpnE in culture supernatants, Novex 4 to 20% acrylamide-Tris-glycine gels were utilized to ensure the recovery of small peptides.
Yeast two-hybrid system.
lpnE was amplified with the lpnE-Y2HF and lpnE R oligonucleotides (Table 2) and cloned into pGBT9 digested with BamHI and PstI. The resulting plasmid was transformed into S. cerevisiae AH109 by the lithium acetate method as described previously (28). This strain was mated with S. cerevisiae Y187 pretransformed with a human HeLa Matchmaker GAL4 library according to the manufacturer's protocol (Clontech). Protein-protein interactions were selected by plating the mating mixture onto synthetic dropout medium plates lacking adenine, histidine, tryptophan, and leucine. Library plasmids were isolated from resulting yeast colonies and rescued in E. coli KC8 on M9 minimal medium containing the required nutrients, except for tryptophan. Library plasmids and pGBT9:lpnE were transformed into S. cerevisiae PJ69-4A, and β-galactosidase liquid culture assays (o-nitrophenyl-β-d-galactoside) were performed in triplicate according to the Clontech manual for the Matchmaker yeast two-hybrid system. The interaction between LpnE and specific regions of obscurin-like protein 1 (OBSL1) was examined in the same manner. A region of OBSL1 encompassing multiple immunoglobulin (Ig)-like domains and a second region encoding an Ig-like domain and a fibronectin (Fn) domain were cloned into pGAD424 digested with EcoRI and BamHI. The single Ig domain of the Fn fragment (IgFn) was also cloned into the EcoRI/BamHI fragment of pGAD424. The resulting plasmids were introduced into S. cerevisiae PJ69-4A, and the yeast strains were assessed for β-galactosidase activity.
RESULTS
Contribution of lpnE to infection of A. castellanii and A/J mice.
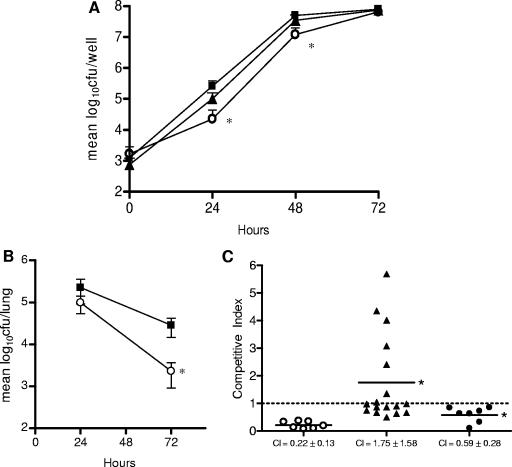
Previously, we reported that lpnE was required for efficient uptake of L. pneumophila into A549 epithelial cells and THP-1 macrophages (42). In this work, we extended our virulence studies to examine the contribution of lpnE to replication of L. pneumophila in A. castellanii and in the lungs of A/J mice. Coculture of L. pneumophila lpnE::km and A. castellanii demonstrated that lpnE was necessary for efficient infection of the amoeba (Fig. 1A). For up to 48 h postinfection, the lpnE mutant was recovered in significantly smaller numbers than wild-type L. pneumophila 130b. Bacterial numbers 72 h after infection were comparable between 130b and the lpnE mutant. The small reduction in CFU observed for the lpnE mutant during the first 48 h of A. castellanii infection was restored to wild-type levels upon transcomplementation of the lpnE mutant with full-length lpnE (Fig. 1A).
FIG. 1.
(A) L. pneumophila infection of A. castellanii. Amoebae were infected at an MOI of 0.01 with wild-type L. pneumophila 130b (▪) or the lpnE::km (○) or lpnE::km/(pMIP:lpnE) (▴) mutant. Bacterial CFU were determined at 24, 48, and 72 h postinfection and are represented as mean log10 CFU ± standard deviations for three independent experiments. *, significantly different from 130b (P = 0.034 at 24 h and P = 0.003 at 48 h [unpaired, two-tailed t test]). (B) Lung colonization of A/J mice by L. pneumophila 130b (▪) and an lpnE::km mutant (○). Strain 130b and the lpnE::km mutant were introduced into the lungs of A/J mice via intratracheal inoculation at a ratio of 1:1. Twenty-four and 72 h following infection, the numbers of CFU for 130b and the lpnE::km mutant were determined. Data are expressed as means ± standard deviations of the log10 CFU recovered per lung (n = 7 or 8). *, significantly different from 130b at 72 h (P = 0.018; unpaired, two-tailed t test). (C) CIs of derivatives of L. pneumophila 130b in mixed infections with the wild-type parent strain. The pairs tested were as follows: lpnE::km mutant versus 130b (○), lpnE::km/(pMIP:lpnE) mutant versus 130b/(pMIP) (▴), and lpnE::km/(pMIP:lpnE52-375) mutant versus 130b/(pMIP) (•). *, CI was significantly different from that for the lpnE::km mutant versus 130b (P < 0.05; unpaired, two-tailed t test).
The reduced ability of the lpnE mutant to infect macrophages, alveolar epithelial cells, and amoebae highlighted the importance of LpnE for host-pathogen interactions in both mammalian and protozoan hosts. The contribution of lpnE to infection of A/J mice was also examined by evaluating the ability of the lpnE mutant strain to compete with wild-type L. pneumophila 130b following intratracheal infection of A/J mice. Lung colonization was examined 24 and 72 h after infection (Fig. 1B). The mutant strain was recovered in similar numbers to wild-type L. pneumophila 130b 24 h after inoculation, but at 72 h significantly fewer mutant bacteria than L. pneumophila 130b bacteria were recovered. We also calculated the competitive index (CI) of the lpnE mutant by comparing the ratio of mutant to wild-type CFU in the lungs of mice at 72 h to the ratio of mutant to wild-type CFU in the inoculum. In general, a mutant with a CI of <0.5 is considered attenuated (5). At 72 h, the mean CI of the lpnE mutant was 0.22 ± 0.13 (Fig. 1C). Complementation of the lpnE mutant with full-length lpnE restored the CI of the mutant strain to 1.75 ± 1.58. These values were significantly different (P = 0.019; unpaired two-tailed t test), indicating that lpnE was needed for efficient colonization of the lungs of A/J mice (Fig. 1C).
Trafficking of the lpnE mutant in tissue culture cells.
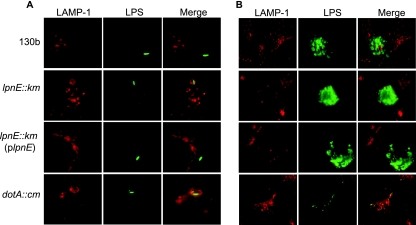
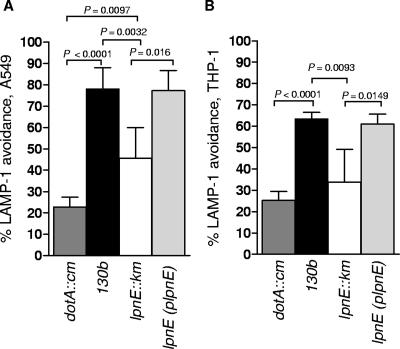
Although we showed previously that despite an initial invasion defect the lpnE mutant can replicate in THP-1 and A549 cells to wild-type levels (42), we did not examine the ability of the mutant to avoid trafficking to late endosomes and lysosomes. Here we investigated the capacity of the lpnE mutant vacuole to avoid fusion with late endosomes by determining the percentage of lpnE vacuoles that avoided the late endosomal marker LAMP-1 5 h after infection. We performed these experiments with A549 cells and THP-1 cells, with the latter undergoing extended treatment with phorbol 12-myristate 13-acetate to ensure differentiation into elongated cells, which are easier to view using immunofluorescence microscopy. We found that 5 h after infection, the majority of vacuoles containing a dotA mutant were positive for LAMP-1 in both A549 and THP-1 cells (Fig. 2 and 3). In contrast, the majority of L. pneumophila 130b vacuoles avoided LAMP-1 (Fig. 2 and 3). Interestingly, the lpnE mutant exhibited an intermediate trafficking phenotype. The percentage of lpnE mutant vacuoles that avoided LAMP-1 was significantly different from those observed for both the wild-type and the dotA mutant in A549 cells and for the wild-type in THP-1 cells (Fig. 3). This indicated that the inactivation of lpnE altered vacuole trafficking in tissue culture cells. Complementation of the lpnE mutant with full-length lpnE restored the ability of L. pneumophila to evade fusion with LAMP-1 (Fig. 3). Interestingly, the percentage of LAMP-1 avoidance for wild-type L. pneumophila was notably lower for THP-1 cells than for A549 cells 5 h after infection. This suggested that THP-1 cells had a greater capacity to drive endosome fusion, which may relate to their monocytic lineage and/or the extended differentiation time used to produce flat cells for microscopy. Since the lpnE mutant showed decreased avoidance of LAMP-1, we also examined the ability of the lpnE mutant to form vacuoles containing multiple (>10) bacteria. Similar to wild-type L. pneumophila, the lpnE mutant was still able to form vacuoles containing multiple bacteria, and these did not associate with LAMP-1 (Fig. 2B).
FIG. 2.
(A) Immunofluorescence of LAMP-1 in A549 cells infected for 5 h with wild-type L. pneumophila 130b or the dotA::cm, lpnE::km, or lpnE::km/(pMIP:lpnE) mutant. LAMP-1 was detected with an anti-LAMP-1 mouse monoclonal antibody diluted 1/100 followed by the secondary antibody, anti-mouse-Alexa Fluor 488, diluted 1/200. Bacteria were detected with anti-lipopolysaccharide (anti-LPS) antibodies raised in rabbits and diluted 1/50, followed by the secondary antibody Alexa Fluor 594 diluted 1/200. (B) Immunofluorescence of LAMP-1 in A549 cells infected for 24 h with wild-type L. pneumophila 130b or the dotA::cm, lpnE::km, or lpnE::km/(pMIP:lpnE) mutant. Primary and secondary antibodies were the same as for panel A.
FIG. 3.
Percentage of Legionella-containing vacuoles that avoided LAMP-1 after infection of A549 cells (A) and THP-1 cells (B) for 5 h. P values of <0.05 (unpaired two-tailed t test) are indicated. LAMP-1 avoidance was scored blind according to the staining patterns indicated in Fig. 4A.
Transcomplementation of the lpnE mutant with truncated forms of lpnE.
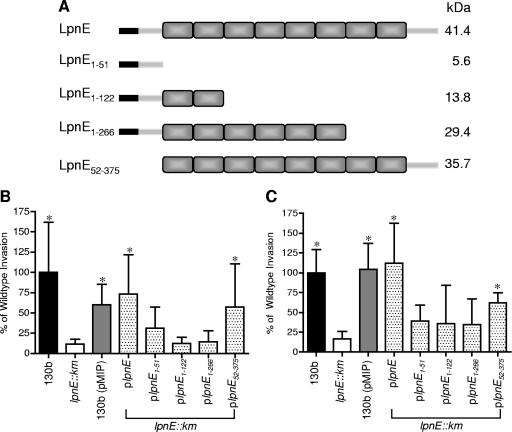
To investigate the putative role of the eight SLR regions of LpnE in host-pathogen interactions, four truncated versions of lpnE were designed to encompass the N terminus (amino acids 1 to 51), the N terminus plus two SLR domains (amino acids 1 to 122), the N terminus and the first six SLR domains (amino acids 1 to 266), and the eight SLR domains plus the C terminus (amino acids 52 to 375) (Fig. 4A). These lpnE truncations were cloned into the pMIP complementation vector, which utilizes the constitutively active mip promoter region (56), and introduced into the lpnE mutant. The ability of these derivatives of L. pneumophila to establish infection of both THP-1 macrophages and A549 alveolar epithelial cells was investigated (Fig. 4B and C). As reported previously, disruption of lpnE significantly reduced the ability of L. pneumophila to infect both cell lines, which was restored upon transcomplementation with full-length lpnE (Fig. 4B and C) (42). In contrast, transcomplementation of the mutant strain with truncated forms of lpnE did not restore wild-type levels of invasion. The introduction of lpnE1-51, lpnE1-122, or lpnE1-266 had no influence on the compromised ability of the lpnE mutant to infect THP-1 or A549 cells. However, lpnE52-375, encoding all of the SLR regions of LpnE but lacking the N-terminal 51 amino acids, was able to partially complement the lpnE mutant defect in both cell types (Fig. 4B and C). We also examined the ability of the lpnE mutant complemented with lpnE52-375 to colonize the lungs of A/J mice in mixed infections with wild-type L. pneumophila. The CI of the lpnE::km strain carrying pMIP:lpnE52-375 in competition with wild-type L. pneumophila/(pMIP) at 72 h was 0.59 ± 0.11, indicating partial complementation of the virulence defect (Fig. 1C).
FIG. 4.
(A) Schematic representation of L. pneumophila SLR protein LpnE and truncated variants created for this study. Shaded rectangles represent the SLR regions, and black rectangles signify the predicted N-terminal 22-amino-acid signal peptide. The truncations were used to complement L. pneumophila lpnE::km, and resulting strains were examined for uptake by THP-1 macrophages (B) and A549 epithelial cells (C). Data are expressed as percentages of the amount of inoculum that was intracellular following a 2-h infection and 1-h gentamicin treatment and are means ± standard deviations for at least three independent experiments. *, significantly different from the lpnE::km mutant (P < 0.05; unpaired two-tailed t test).
Presence of LpnE in culture supernatants.
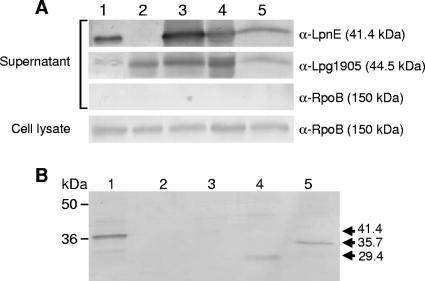
The reduced invasion phenotype of the lpnE mutant in THP-1 and A549 cells and the replication defect of the mutant in A. castellanii and A/J mice suggested that LpnE may play a direct role in host-pathogen interaction. To determine the localization of LpnE in L. pneumophila, rabbit polyclonal antibodies were raised against purified MBP-LpnE and used to detect LpnE within stationary-phase L. pneumophila cultures. Immunoblot analysis showed that LpnE was present in the culture supernatant of L. pneumophila 130b (Fig. 5A), indicating that the protein was extracellular. Interestingly, the presence of LpnE in culture supernatants appeared to be independent of the virulence-related Lsp type II secretion system (46) and also independent of the Dot/Icm type IV secretion system, as LpnE was found in culture supernatants derived from both lspDE and dotA mutant strains of L. pneumophila 130b (Fig. 5A). To confirm that the presence of LpnE in the culture supernatant was not due to cell breakdown and contamination with cytoplasmic proteins, the same samples were incubated with anti-RpoB to detect a known cytoplasmic protein. In addition, we used antibodies to the secreted protein Lpg1905 (47) to control for protein loading (Fig. 5A). Apart from a lack of LpnE expression in the lpnE mutant and overexpression of LpnE from the complementing pMIP plasmid, LpnE was present in equivalent amounts in all other strains tested, including wild-type L. pneumophila 130b and the secretion system mutants (Fig. 5A).
FIG. 5.
Immunoblot analysis of culture supernatants precipitated with TCA and detected with anti-LpnE, anti-Lpg1905, and anti-RpoB antibodies. (A) Stationary-phase TCA-precipitated culture supernatants and whole-cell lysate samples from derivatives of L. pneumophila 130b were separated by SDS-polyacrylamide gel electrophoresis. Lane 1, L. pneumophila 130b; lane 2, lpnE::km mutant; lane 3, lpnE::km/(pMIP:lpnE) mutant; lane 4, ΔdotA mutant; lane 5, ΔlspDE mutant. (B) TCA-precipitated stationary-phase culture supernatants. Lane 1, L. pneumophila lpnE::km/(pMIP:lpnE) mutant; lane 2, lpnE::km/(pMIP:lpnE1-51) mutant; lane 3, lpnE::km/(pMIP:lpnE1-122) mutant; lane 4, lpnE::km/(pMIP:lpnE1-266) mutant; lane 5, lpnE::km/(pMIP:lpnE52-375) mutant. The presence of both LpnE1-266 and LpnE52-375 in the culture supernatant is indicated by arrows and the predicted molecular size of each protein.
We also examined export of the truncated forms of LpnE. The predicted products of lpnE1-51 and lpnE1-122 were not detected with anti-LpnE, even in whole-cell lysate samples (data not shown). This indicated that these small truncations of LpnE were either not recognized by our anti-LpnE sera or, alternatively, that lpnE1-51 and lpnE1-122 were not expressed in vitro. However, both LpnE1-266 and LpnE52-375 were expressed and present in culture supernatants (Fig. 5B). This demonstrated that neither the C terminus, absent in LpnE1-266, nor the predicted N-terminal signal sequence, absent in LpnE52-375, was essential for LpnE export. At this stage, the mechanism by which LpnE reaches the culture supernatant is unknown.
LpnE interacts with Ig-like domains of eukaryotic proteins via the SLR regions.
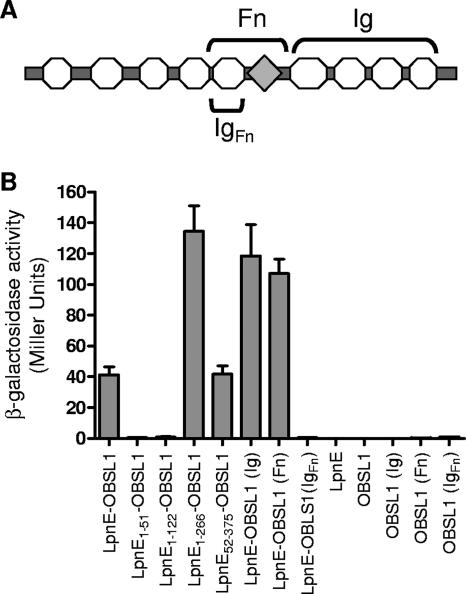
The presence of LpnE in culture supernatants provided further evidence that during infection, the protein may interact with host proteins to stimulate the uptake of L. pneumophila. Since TPR regions in general are known to mediate protein-protein interactions, the ability of LpnE to bind to eukaryotic proteins was examined using the yeast two-hybrid system. Screening of full-length LpnE against a HeLa cell cDNA library identified several putative binding partners of LpnE (Table 3). Recovery of the interacting clones revealed that the most prevalent binding partner, accounting for 28.6% of rescued clones, was OBSL1. This eukaryotic protein, ranging in molecular mass from 130 to 230 kDa, is a member of the Unc-89/obscurin gene family, exhibiting homology to the N-terminal region of the giant muscle protein obscurin, which is known to interact with titin, small ankyrin 1, and myosin within vertebrate skeletal muscle to mediate muscle contraction (3, 27, 57). The function of OBSL1 is unknown, although, similar to obscurin, the protein possesses a number of putative Ig-like domains and an Fn domain (Fig. 6A). While other putative eukaryotic binding partners identified here may warrant further investigation, particularly the epsilon subunit of COPI involved in eukaryotic endosome trafficking and phagosome maturation (6), we initially examined the interaction between LpnE and OBSL1 as the most frequently recovered target. The specificity of the interaction between OBSL1 and LpnE was confirmed and quantified within S. cerevisiae PJ69-4a by using liquid culture β-galactosidase assays (Fig. 6B). The truncated forms of LpnE were also tested for the ability to bind to OBSL1. LpnE1-51 and LpnE1-122 could not bind OBSL1, but LpnE1-266 and LpnE52-375, with six and eight SLR regions, respectively, were able to interact with OBSL1. Curiously, LpnE1-266 exhibited the strongest interaction with OBSL1, but full-length LpnE and LpnE52-375 showed an equivalent strength of interaction with OBSL1, demonstrating that the full repertoire of SLR domains is needed for wild-type-strength protein-protein interaction (Fig. 6B). To investigate the basis of the interaction of LpnE with OBSL1 in more detail, two sections of OSBL1, one comprising the Fn type 3 domain and an Ig domain (Fn) and a second comprising the C-terminal Ig domains of OBSL1 (Ig) (Fig. 6A), were cloned into pGAD424 and examined for the ability to interact with LpnE (Fig. 6B). Both fragments of OBSL1 exhibited strong binding to LpnE, suggesting that the protein-protein interaction occurred through the Ig-like domains common to both fragments. In an attempt to confirm that LpnE interacted specifically with Ig-like domains, we cloned the 8.3-kDa predicted IgFn domain into pGAD424 and examined its interaction with LpnE. β-Galactosidase assays for S. cerevisiae carrying pGBT9:LpnE and pGAD424:IgFn demonstrated no interaction between the IgFn peptide and LpnE. Although this lack of interaction may indicate that the Ig domains do not mediate an interaction between OBSL1 and LpnE, it is more likely that such a small peptide is unable to establish the tertiary structure required for protein-protein interactions.
TABLE 3.
Eukaryotic proteins identified as possible binding partners of LpnE from a yeast-two hybrid HeLa cell cDNA library screen
| Protein | Function | % of recovered clones | Accession no. |
|---|---|---|---|
| OBSL1 | Putative cytoskeletal adaptor protein | 28.6 | KIAA0657 |
| N-Acetylneuraminic acid phosphate synthase | Sialic acid synthesis | 14.3 | AAF75261 |
| Map kinase kinase (kinase 7) | Role in cell signaling | 10.7 | AAV38460 |
| Phosphoserine aminotransferase | l-Serine biosynthesis | 10.7 | AAD42052 |
| Coatomer protein I (COPI) epsilon subunit | Membrane trafficking, possesses TPR domain | 7 | CAA10316 |
| Ras-related GTPase | Small GTPase | 7 | NP_005393 |
| Neuronal protein | Unknown | 7 | AAP97253 |
| Thyroid receptor-interacting protein 6 (TRIP6) | Organization of actin filaments and control of cell migration | 3.6 | AAK21007.1 |
| Solute carrier family 3, isoform A | Transmembrane protein that can mediate integrin-dependent signaling | 3.6 | NP_001012679 |
| ATPase 13A1 | Putative role in cation transport | 3.6 | AAH69211 |
| Na+/K+ ATPase beta-3 chain (ATPB-3) | Maintenance of electrochemical gradient across the plasma membrane | 3.6 | NP_001670 |
FIG. 6.
(A) Domain organization of OBSL1, comprising several Ig-like domains (circles) and one Fn domain (diamond). The regions of OBSL1 subcloned for protein-protein interaction studies, namely, Ig, Fn, and IgFn, are indicated. (B) Protein-protein interactions were assessed using the yeast two-hybrid system β-galactosidase reporter assay. Full-length LpnE, LpnE1-266, and LpnE52-375 showed strong interactions with OBSL1, as did full-length LpnE with two of the truncated forms of OBSL1 but not with IgFn.
DISCUSSION
Bacterial attachment and subsequent entry into host cells are required for L. pneumophila infection. Furthermore, the ability of L. pneumophila to subvert host cell trafficking pathways occurs immediately upon uptake, indicating that this process is likely linked to mechanisms of entry (36). L. pneumophila uptake by host cells occurs via coiling and conventional phagocytosis. Coiling phagocytosis, which has been observed during L. pneumophila entry into human monocytes, alveolar macrophages, and polymorphonuclear leukocytes and into amoebae, involves the formation of a long, asymmetrical pseudopod that coils around and engulfs the bacterium (1, 7, 33). This is an unusual mechanism of phagocytosis observed only in the uptake of L. pneumophila and some spirochetes, trypanosomatids, and yeasts (44, 51). Recently, it was shown, using Dictyostelium discoideum, that L. pneumophila utilizes specific phagocytic processes that abrogate the use of host cell phosphoinositol 3-kinase, which is traditionally important for host cell membrane dynamics during phagocytosis (55). A range of L. pneumophila virulence factors have been identified as being important for entering host cells. These include pili, the heat shock protein Hsp60, RtxA, the Dot/Icm type IV secretion system, and other novel factors, including the SLR proteins EnhC and LpnE (14, 24, 26, 31, 42, 50). In this study, we extended our previous observations that LpnE contributes to host cell invasion by L. pneumophila and compared the growth characteristics of wild-type L. pneumophila and the lpnE mutant in A. castellanii and in the lungs of A/J mice. In both virulence models, the lpnE mutant was significantly attenuated compared to the wild-type strain. In particular, the difference observed between wild-type L. pneumophila and the lpnE mutant in bacterial replication in the lungs of A/J mice indicated that lpnE plays a significant role in the interaction of L. pneumophila with a mammalian host. Interestingly, a significant difference in lung colonization between the two strains was evident only at 72 h, which did not appear to correlate with the early defect seen in tissue culture cells. However, it is possible that the early infection defect of the lpnE mutant in mammalian cells in vitro did not result in an observable colonization defect until later times in infection, after more than one round of bacterial entry and egress had occurred. Therefore, the virulence defect of the lpnE mutant may reflect a reduced capacity to invade eukaryotic cells and establish a replicative vacuole that, over time, results in reduced mutant numbers compared to wild-type growth.
The presence of five genes encoding SLR proteins in the L. pneumophila genome may point to functional redundancy (11, 13). This is a common feature of L. pneumophila, with functional redundancy clearly present among substrates of the Dot/Icm system (39). However, possession of variant forms of virulence determinants may also point to the ability of L. pneumophila to promote growth within a divergent range of environmental hosts. Further evidence that all of the SLR-containing proteins may be important for L. pneumophila propagation is that their expression is upregulated during the transmissive phase within A. castellanii (10). The transmissive phase is induced late during intracellular replication and is characterized by the upregulation of most virulence determinants, including flagellar components and substrates of the Dot/Icm secretion system. Induction of these factors facilitates bacterial egress from host cells and efficient spread of the infection. Both EnhC (21 SLRs) and LpnE (8 SLRs) contribute to host cell entry, and it is assumed that LidL (12 SLRs), Lpg1356 (8 SLRs), and Lpg1062 (8 SLRs) may also contribute to L. pneumophila virulence. Both EnhC and LidL play a role in early signaling events that regulate trafficking of the Legionella -containing vacuole. Vacuoles containing mutants of these proteins show a reduced ability to evade acquisition of LAMP-1, providing further evidence that the L. pneumophila SLR proteins are important for the initial establishment of infection (15). In this study, we also observed reduced LAMP-1 avoidance by vacuoles containing the lpnE mutant compared to those containing wild-type L. pneumophila.
Complementation with full-length lpnE restored vacuole trafficking to the wild-type phenotype. The shared phenotype of the enhC, lidL, and lpnE mutants may indicate that the SLR proteins act together or in combination to inhibit vacuolar acquisition of LAMP-1, although the mechanism behind this is currently unknown. These data may also indicate that the SLR proteins are not specifically involved in the invasion of host cells, but rather in the mechanisms that immediately follow, namely, establishment of the intracellular replicative niche. The reduced recovery of SLR protein mutants at the initial stages of infection may indicate increased lysosomal degradation of these strains.
Using antibodies generated to recombinant LpnE, we found that LpnE was present in the culture supernatant of L. pneumophila. The presence of LpnE in the culture supernatant appeared to be independent of both the Lsp and Dot/Icm secretion systems and supports the finding that LpnE was not present in the type II secretome, despite the presence of a putative N-terminal signal peptide (18). Of further curiosity is the fact that both LpnE1-266, which lacks the C-terminal region of LpnE, including the last two SLRs, and LpnE52-375, which retains only the SLR regions of LpnE, omitting the putative N-terminal signal peptide, were observed in culture supernatants. This suggested that LpnE uses a unique mechanism of export, independent of N- or C-terminal secretion signals, or that LpnE export may occur through more than one mechanism. At this stage, the mechanism by which LpnE reaches the extracellular environment is unknown.
While the expression of LpnE1-51 and LpnE1-122 could not be detected by Western blotting, it was clear that LpnE1-266 and LpnE52-375 were both localized similarly to full-length LpnE. Despite both of these truncations being present in the culture supernatant, lpnE52-375, encoding eight SLRs, was the only truncation able to partially complement the invasion defect of the lpnE mutant. lpnE52-375 was also able to partially complement the virulence defect of the lpnE mutant in A/J mice. While lpnE52-375 could not completely restore wild-type levels of invasion or lung colonization, this investigation nevertheless highlighted the importance of the full SLR repertoire for function.
Given that SLR domains mediate protein-protein interactions, we examined the ability of LpnE to bind with eukaryotic proteins by using the yeast two-hybrid system. A range of putative eukaryotic binding partners were isolated, the most frequent of which was OBSL1. LpnE and two truncated forms, LpnE1-266 and LpnE52-375, were shown to interact strongly with OBSL1. More specifically, LpnE bound fragments of OBSL1 containing multiple Ig-like domains or an Fn and an Ig-like domain. However, using this technique, we were unable to demonstrate an interaction between LpnE and a single Ig domain. Recently, OBSL1 expression was observed in a range of tissues, and localization of the protein to intercalated discs and the perinuclear region of rat cardiac myocytes indicated its likely function as a cytoskeletal adaptor protein (27). While the interaction between LpnE and OBSL1 may not occur in vivo, it allows speculation that LpnE may interact with other proteins that possess Ig-like folds. Ig-like domains are present in a range of proteins of eukaryotic organisms with roles in cell-cell recognition and in cell surface receptors. Many eukaryotic surface-exposed cell adhesion molecules possess Ig domains to which LpnE may bind, leading to the initiation of host cell signaling events that enhance L. pneumophila internalization and/or establishment of the L. pneumophila replicative vacuole.
There is evidence within eukaryotic systems that proteins containing SLRs are involved in multiprotein complexes with specific and distinct biological functions. For example, the SLR protein Hrd3 is a key component of the endoplasmic reticulum-associated degradation complex that identifies and processes misfolded proteins for ubiquitination (25). Furthermore, it is clear that SLR regions are crucial to the formation of such complexes, and they are considered to act as scaffolding or receptor domains linking components via multiple substrate binding sites. Hence, it is attractive to propose that SLR-containing proteins of L. pneumophila, particularly LpnE, EnhC, and LidL, may form a multiprotein complex linking the bacterium to the host cell via cell adhesion molecules and that they may act in concert to achieve this. However, this study also identified a range of other eukaryotic proteins that may interact with LpnE in vivo. To establish which of these putative binding partners is important during L. pneumophila infection, the in vivo localization of LpnE and the other SLR proteins of L. pneumophila must be investigated. In addition, further elucidation of the interactions between L. pneumophila SLR proteins and both host and bacterial proteins is therefore important to discern the functions of these virulence determinants in L. pneumophila infection.
Acknowledgments
We are indebted to Carmen Buchrieser, Institute Pasteur, for the gift of A. castellanii and to Dane Parker, Department of Microbiology, Monash University, for the RpoB antibodies.
H.J.N. is the recipient of an Australian Postgraduate Award (APA) and a Victoria Fellowship, and F.M.S. and A.D.M are supported by Monash University graduate scholarships. Part of this work was supported by NIH grant AI43987 awarded to N.P.C. and by a National Health and Medical Research Council (NHMRC) grant awarded to E.L.H.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 24 September 2007.
REFERENCES
- 1.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 3.Bagnato, P., V. Barone, E. Giacomello, D. Rossi, and V. Sorrentino. 2003. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol. 160:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, P. L., C.-T. Chien, R. Sternglanz, and S. R. Fields. 1993. Using the two-hybrid system to detect protein-protein interactions, p. 153-179. In D. A. Hartley (ed.), Cellular interactions in development: a cellular approach. Oxford University Press, Oxford, United Kingdom.
- 5.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 6.Botelho, R. J., D. J. Hackman, A. D. Schreiber, and S. Grinstein. 2000. Role of COPI in phagosome maturation. J. Biol. Chem. 275:15717-15727. [DOI] [PubMed] [Google Scholar]
- 7.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii : uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broms, J. E., P. J. Edqvist, A. Forsberg, and M. S. Francis. 2006. Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol. Lett. 256:57-66. [DOI] [PubMed] [Google Scholar]
- 9.Broms, J. E., A. Forslund, A. Forsberg, and M. S. Francis. 2003. PcrH of Pseudomonas aeruginosa is essential for secretion and assembly of the type III translocon. J. Infect. Dis. 188:1909-1921. [DOI] [PubMed] [Google Scholar]
- 10.Bruggemann, H., A. Hagman, M. Jules, O. Sismeiro, M. A. Dillies, C. Gouyette, F. Kunst, M. Steinert, K. Heuner, J. Y. Coppee, and C. Buchrieser. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 8:1228-1240. [DOI] [PubMed] [Google Scholar]
- 11.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 13.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fisher, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 14.Cirillo, S. L. G., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 15.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 16.Core, L., and M. Peregro. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 49:1509-1522. [DOI] [PubMed] [Google Scholar]
- 17.Das, A. K., P. T. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DebRoy, S., J. Dao, M. A. Soderberg, O. Rossier, and N. P. Cianciotto. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. USA 103:19146-19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187:7716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodt, G., and S. J. Gould. 1996. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle, R. M., T. W. Steele, A. M. McLennan, I. H. Parkinson, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence analysis of the mip gene of the soilborne pathogen Legionella longbeachae. Infect. Immun. 66:1492-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1998. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect. Immun. 66:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardener, R. G., G. M. Swarbrick, N. W. Bays, S. R. Cronin, S. Wilhovsky, L. Seelig, C. Kim, and R. Y. Hampton. 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151:69-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garduno, R. A., E. Garduno, and P. S. Hoffman. 1998. Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66:4602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisler, S. B., D. Robinson, M. Hauringa, M. O. Raeker, A. B. Borisov, M. V. Westfall, and M. W. Russell. 2007. Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin. Genomics 89:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 29.Grant, B., and I. Greenwald. 1996. The Caenorhabditis elegans sel-1 gene, a negative regulator of lin-12 and glp-1, encodes a predicted extracellular protein. Genetics 143:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Ellege. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 31.Hilbi, H., G. Segal, and H. A. Shuman. 2001. Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42:603-617. [DOI] [PubMed] [Google Scholar]
- 32.Hirano, T., M. Kinoshita, K. Morikawa, and M. Yanagida. 1990. Snap helix with knob and hole: essential repeats in Schizosaccharomyces pombe nuclear protein nuc2 +. Cell 60:319-328. [DOI] [PubMed] [Google Scholar]
- 33.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36:27-33. [DOI] [PubMed] [Google Scholar]
- 34.Ishihama, A., N. Fujita, and R. E. Glass. 1987. Subunit assembly and metabolic stability of E. coli RNA polymerase. Proteins 2:42-53. [DOI] [PubMed] [Google Scholar]
- 35.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kagan, J. C., M. P. Stein, M. Pypaert, and C. R. Roy. 2004. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199:1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King, R. W., J. M. Peters, S. Tugendreich, M. Rolfe, P. Hieter, and M. W. Kirschner. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81:279-288. [DOI] [PubMed] [Google Scholar]
- 38.Leyton, D. L., J. Sloan, R. E. Hill, S. Doughty, and E. L. Hartland. 2003. Transfer region of pO113 from enterohemorrhagic Escherichia coli : similarity with R64 and identification of a novel plasmid-encoded autotransporter, EpeA. Infect. Immun. 71:6304-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittl, P. R. E., and W. Schneider-Brachert. 2007. Sel1-like repeat proteins in signal transduction. Cell. Signal. 19:20-31. [DOI] [PubMed] [Google Scholar]
- 41.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 42.Newton, H. J., F. M. Sansom, V. Bennett-Wood, and E. L. Hartland. 2006. Identification of Legionella pneumophila -specific genes by genomic subtractive hybridization with Legionella micdadei and identification of lpnE, a gene required for efficient host cell entry. Infect. Immun. 74:1683-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okabe, M., T. Yakushi, and M. Homma. 2005. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J. Biol. Chem. 280:25659-25664. [DOI] [PubMed] [Google Scholar]
- 44.Rittig, M. G., K. Schroppel, K. H. Seack, U. Sanders, E. N. N′Diaye, I. Maridonneau-Parini, W. Solbach, and C. Bogdan. 1998. Coiling phagocytosis of trypanosomatids and fungal cells. Infect. Immun. 66:4331-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robey, M., W. A. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP -like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossier, O., S. R. Starkenburg, and N. P. Cianciotto. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72:310-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sansom, F. M., H. J. Newton, S. Crikis, N. P. Cianciotto, P. J. Cowan, A. F. d'Apice, and E. L. Hartland. 2007. A bacterial ecto-triphosphate diphosphohydrolase similar to human CD39 is essential for intracellular multiplication of Legionella pneumophila. Cell. Microbiol. 9:1922-1935. [DOI] [PubMed] [Google Scholar]
- 48.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikorski, R. S., M. S. Boguski, M. Goebl, and P. Hieter. 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307-317. [DOI] [PubMed] [Google Scholar]
- 50.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila : identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szczepanski, A., and H. B. Fleit. 1988. Interaction between Borrelia burgdorferi and polymorphonuclear leukocytes: phagocytosis and the induction of the respiratory burst. Ann. N. Y. Acad. Sci. 539:425-428. [Google Scholar]
- 52.Tzamarias, D., and K. Struhl. 1994. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369:758-761. [DOI] [PubMed] [Google Scholar]
- 53.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 54.Wattiau, P., B. Bernier, P. Deslee, T. Michiels, and G. R. Cornelis. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. USA 91:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber, S. S., C. Ragaz, K. Reus, Y. Nyfeler, and H. Hilbi. 2006. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2:418-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wieland, H., M. Faigle, F. Lang, H. Northoff, and B. Neumeister. 2002. Regulation of the Legionella mip- promotor during infection of human monocytes. FEMS Microbiol. Lett. 212:127-132. [DOI] [PubMed] [Google Scholar]
- 57.Young, P., E. Ehler, and M. Gautel. 2001. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 154:123-136. [DOI] [PMC free article] [PubMed] [Google Scholar]