Abstract
PspA is an important pneumococcal vaccine candidate that is capable of inducing protection in different animal models. Because of its structural diversity, a PspA-based vaccine should contain at least one fragment from each of the two major families (1 and 2) in order to elicit broader protection. In the present work, we have tested the potential of PspA hybrids containing fused portions of family 1 and 2 (PspA1ABC-4B and PspA1ABC-3AB) PspA fragments to induce protection against pneumococci bearing distinct PspA fragments. Sera from mice immunized with these hybrid PspA fragments were able to increase C3 deposition on pneumococci bearing PspA fragments from both families, in contrast with sera made against the PspA family 1 (PspA1ABC) and PspA family 2 (PspA3ABC) fragments, which were effective only within the same family. Although PspA hybrids were able to extend protection against pneumococcal infection with strains bearing diverse PspA fragments, the immunity elicited by family 2 was clade dependent, suggesting that PspA fragments from family 2 clades 3 and 4 should both be included in a comprehensive PspA vaccine. These results indicate that PspA fusion proteins constitute an efficient immunization strategy for future PspA-based antipneumococcal vaccines since they are able to extend protection provided by a protein derived from a single transcript.
Streptococcus pneumoniae is a major human pathogen that causes a number of life-threatening diseases, such as pneumonia, meningitis, and bacteremia, in addition to otitis media and sinusitis. Altogether, pneumococcal diseases account for at least 1 million deaths worldwide every year among children under 5 years of age, most of them in developing countries (6). The rapid increase in antibiotic resistance, high cost, and limited coverage of the currently available conjugate vaccine further aggravate the problem and reinforce the need for more affordable and more broadly protective strategies for immunization against pneumococcal infection.
Several proteins have been investigated as vaccine candidates against infection with S. pneumoniae, including pneumococcal surface protein A (PspA), an exposed virulence factor found in all pneumococcal strains. PspA is highly immunogenic and protective in different animal models (reviewed by Tai [23]), and a recent human trial showed an increase in specific antibody levels after immunization with a recombinant PspA fragment (18). Furthermore, sera from humans immunized with PspA were able to passively protect mice against challenge with various pneumococcal strains (3), reinforcing the importance of humoral immunity in this model.
PspA is composed of five domains, (i) a signal peptide, (ii) an α-helical and charged N-terminal domain with a heptad pattern typical of coiled-coil proteins, (iii) a proline-rich region that acts as a flexible tether linking the N and C termini, (iv) a choline-binding domain with a stretch of 10 repeats of 20 amino acids that anchors PspA to the pneumococcal cell wall and membrane, and (v) a short hydrophobic tail at the C terminus (26, 28). The N-terminal domain of PspA is considered to be the most exposed part of the molecule and binds protective monoclonal antibodies (9, 15). It also exhibits serological variability, especially its most C-terminal 100 amino acids (15). This region, also referred to as the B window or clade-defining region (CDR), was the basis for the classification of PspA into six clades, which were grouped into three families (10). Surveillance studies show that PspA from families 1 and 2 are present in 94 to 99% of the isolates in different countries in the Americas (1, 2, 17, 25), with prevalence rates of about 50% each (10, 19, 25). Previous studies have shown that the level of cross-reactivity among different PspA fragments roughly follows the degree of similarity among the amino acid sequences within the CDR, with a tendency for a higher degree of cross-reactivity among PspA fragments within the same family, rather than between families (16). On the basis of these data, it has been suggested that a PspA-based vaccine should contain at least one fragment from each of the two major families in order to broaden protection (10). We have previously demonstrated that DNA vaccines that encode PspA family 1 and 2 hybrids induce protection against the homologous family. Immunization with family 1 PspA induced detectable protection against family 2, but family 2 DNA induced no protection against family 1 (16).
An important defense mechanism against pneumococcal infections is opsonin-dependent phagocytosis, with activation of the classic and alternative complement pathways and deposition of C3b on the pneumococcal surface (7, 11, 12). It has been demonstrated that PspA inhibits complement activation (24), and anti-PspA antibodies can overcome this effect (20), leading to increased C3 deposition on the bacterial surface and enhancing clearance. Ren et al. showed that both family 1 and 2 PspA proteins can interfere with complement activation and subsequent deposition on pneumococci (21).
In the present study, we constructed PspA chimeric proteins containing the N-terminal domain of a family 1 PspA fused to the CDR or the whole N-terminal domain of family 2 molecules. Since PspA exerts its effect on virulence by interfering with complement, we analyzed the ability of antibodies generated against the hybrids to increase complement deposition on bacteria bearing PspAs from both families. Finally, the hybrids were tested for the ability to induce protection against pneumococcal strains bearing diverse PspA fragments. There was a strong correlation between complement deposition in the presence of sera against the hybrids and protection elicited by immunization with these molecules, thus providing an insight into the mechanisms by which PspA hybrids might confer cross-protection.
MATERIALS AND METHODS
Construction of PspA fragments and hybrids.
All cloning procedures were performed with Escherichia coli DH5α grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml). DNA fragments encoding portions of the N-terminal regions of PspA clades 1, 3, and 4 were amplified by PCR from pTG-pspA1-4 or pTG-pspA3-2 (16). The primers used in this procedure are listed in Table1. The gene products were ligated to the pGEMT-easy vector (Promega), and the sequences were confirmed by DNA sequencing. The pGEMT-easy-pspA constructs were digested with the appropriate restriction endonucleases, and the resulting fragments were ligated to the linearized pAE-6xHis vector (14). The pspA1ABC-4B hybrid was obtained with primers that allowed the removal of the signal sequence present in pTG-pspA1-4 and then ligated to previously digested pAE-6xHis. The pspA1ABC-3AB hybrid was constructed by fusing the 3′ terminus of pspA1ABC with the 5′ terminus of pspA3AB through complementary cohesive ends added to the primers and then ligated to pAE-6xHis.
TABLE 1.
Primers used to amplify pspA gene fragments
| Fragment | Primera |
|---|---|
| pspA1ABC | 5′ CTC GAG GAA GAA GCG CCC GTA GC 3′ (forward) |
| 5′ TAG TTA GGT ACC TGG TTG TGG TGC TGA AG 3′ (reverse) | |
| pspA3ABC | 5′ CTC GAG GAA GAA GCG CCC GTA GC 3′ (forward) |
| 5′ TAG TTA GGT ACC TTT TGG TGC AGG AGC TGG 3′ (reverse) | |
| pspA3AB | 5′ GGT ACC GAA GAA GCG CCC GTA GC 3′ (forward) |
| 5′ TAG TTA GGT ACC TTT TGG TGC AGG AGC TGG 3′ (reverse) | |
| pspA4B | 5′ TAG GGT ACC TTA TTC TTC ATC TCC ATC AGG G 3′ (reverse) |
The sequences in boldface type are the restriction sites for XhoI (CTC GAG) and KpnI (GGT ACC).
PspA expression and purification.
Competent E. coli BL21(DE3)pLys or Si cells (Invitrogen) were transformed with pAE6xHis vectors containing the pspA constructs. Protein expression was induced in the mid-log-phase cultures by 1 mM IPTG (Sigma) or 300 mM NaCl, respectively. The recombinant proteins, bearing an N-terminal histidine tag, were purified from the soluble fraction through affinity chromatography with Ni2+ charged chelating Sepharose resin (HiTrap Chelating HP; GE HealthCare) in an Akta Prime apparatus (GE HealthCare). Elution was carried out with 250 mM imidazole. The eluted fractions were further sujected to anion-exchange chromatography (MonoQ Sepharose; GE HealthCare) to eliminate contaminants, and the purified PspA fractions was collected at approximately 200 mM NaCl. The purified fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), dialyzed against 10 mM Tris-HCl (pH = 8)-20 mM NaCl-0.1% glycine, and stored at −20°C.
Pneumococcal strains.
All of the strains used in this study are described in Table 2. Pneumococci were maintained as frozen stocks (−80°C) in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) with 10% glycerol. In each experiment, the isolates were plated on blood agar prior to growth in THY.
TABLE 2.
Pneumococcal strains used in this study
| Strain | Serotype | PspA family | PspA clade(s) | Source |
|---|---|---|---|---|
| 245/00 | 14 | 1 | 1 | Universidade Federal de Goiás, Goiânia, Brazil |
| A66.1 | 3 | 1 | 1, 2 | University of Alabama at Birmingham |
| D39 | 2 | 1 | 2 | University of Alabama at Birmingham |
| 679/99 | 6B | 2 | 3 | Instituto Adolfo Lutz, São Paulo, Brazil |
| 3JYP2670 | 3 | 2 | 4 | University of Alabama at Birmingham |
Animals and immunization.
Female BALB/c mice from the Instituto Butantan (São Paulo, Brazil) were immunized subcutaneously with 5 μg of recombinant PspA derivatives in Ringer's solution with 50 μg of Al(OH)3 as an adjuvant (final volume of 100 μl per mouse). The adjuvant alone was used as a control. The animals were given three doses of protein at 14-day intervals. Sera were collected from mice by retro-orbital bleeding 1 day before each dose and 2 weeks after the third immunization.
Analysis of serum cross-reactivity.
Cross-reactivity of anti-PspA antibodies was analyzed by enzyme-linked immunosorbent assay (ELISA). Polysorp 96-well plates (Nunc, Roskilde, Denmark) were coated with PspA (1 μg/ml) extracted from pneumococcal strains bearing PspA fragments of clades 1 to 4 by choline chloride washing (full-length PspA) as described elsewhere (5). The plates were washed three times with phosphate-buffered saline (PBS) containing 0.1% Tween 20, blocked with 10% nonfat dry milk in PBS overnight at 4°C, and washed again with PBS. The plates were then incubated with serial dilutions of sera from the immunized mice in PBS-1% bovine serum albumin at 37°C for 1 h. The plates were then washed again and incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG; 1:15,000; Sigma) in PBS-1% bovine serum albumin at 37°C for 1 h. Following another round of washes, antibodies were detected by adding OPD substrate (0.04% o-phenylenediamine in citrate-phosphate buffer [pH 5] containing 0.01% H2O2). After color development (10 min), the reaction was interrupted with 1.25 M H2SO4 and the A492 was determined. The reciprocal titer was considered the last dilution of serum that registered an optical density of 0.10.
Antibody binding assay.
S. pneumoniae strains were grown in THY to a concentration of ∼108 CFU/ml (optical density of 0.4 to 0.5) and harvested by centrifugation at 2,000 × g for 2 min. The pellets were washed once with PBS, resuspended in the same buffer, and incubated in the presence of pooled sera from mice immunized with PspA fragments for 30 min at 37°C. After another wash with PBS, the samples were incubated with 100 μl of fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG Fc (MP Biomedicals) diluted 1:1,000. Samples were analyzed with a FACScalibur (BD Biosciences).
Complement deposition assay.
Complement was previously inactivated by incubation of sera at 56°C for 30 min. Bacteria were grown and centrifuged as previously described. The pellets were washed once, centrifuged, and resuspended in PBS. Samples (80 μl) were incubated in the presence of anti-PspA sera at a final concentration of 20% for 30 min at 37°C. Bacteria were then washed once with PBS, resuspended in 90 μl of gelatin Veronal buffer (Sigma), and incubated in the presence of fresh-frozen normal mouse serum (from BALB/c mice) at 37°C for 30 min. After another wash with PBS, the samples were incubated with 100 μl of FITC-conjugated goat antiserum to mouse complement C3 (MP Biomedicals) at a dilution of 1:1,000 on ice for 30 min in the dark. Finally, the bacteria were washed two more times with PBS, resuspended in 1% formaldehyde, and stored at 4°C in the dark until analysis with a FACScalibur (BD Biosciences). This is a modified version of the method described by Ren et al. (20).
Challenge.
Two weeks after the last immunization, mice were challenged intraperitoneally with strain St 679/99 (400 CFU) or intravenously with fresh cultured strain A66.1 (1,000 CFU) or 3JYP2670 (106 CFU) and monitored for 10 days. Differences between the overall survival rates of the groups were analyzed by the Fisher exact test.
RESULTS
PspA expression and purification.
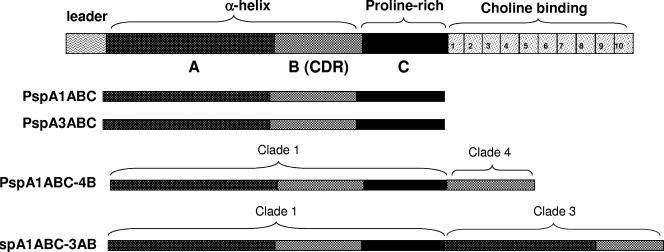
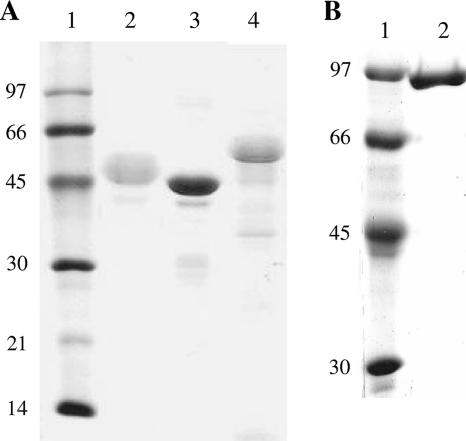
PspA fragments and fusion proteins containing different portions of the N-terminal region from clade 1, 3, or 4 are shown in Fig. 1. These were expressed in fusion with a His tag in transformed competent E. coli strains and purified through Ni2+ affinity chromatography and anion-exchange chromatography to ensure the high purity necessary for the immunization experiments. The SDS-PAGE of the purified recombinant proteins is shown in Fig. 2. There was a difference in the predicted masses and the “apparent sizes” of the fragments estimated by SDS-PAGE, an effect that has been previously reported by other groups (27) and is thought to be partially due to the high percentage of prolines in these molecules.
FIG. 1.
Schematic diagram of PspA fragments. At the top is the whole PspA molecule, containing the N-terminal α-helical domain (including regions A and B), the proline-rich region (C), and the choline-binding domain. Each recombinant fragment is shown with its distinct domains.
FIG. 2.
SDS-PAGE of recombinant PspA fragments purified from E. coli. (A) Lanes: 1, standard molecular weight marker; 2, PspA1ABC; 3, PspA3ABC; 4, PspA1ABC-4B. (B) Lanes: 1, standard molecular weight marker; 2, PspA1ABC-3AB. The values on the left are molecular sizes in kilodaltons.
Serum reactivity to native PspA fragments.
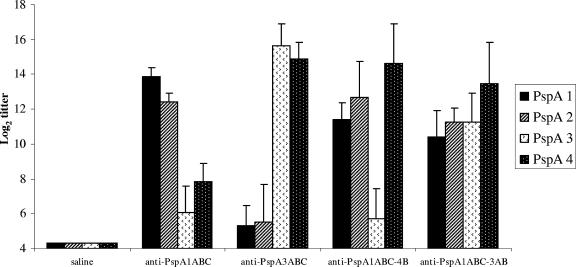
BALB/c mice were immunized with three doses of PspA1ABC, PspA3ABC, PspA1ABC-4B, or PspA1ABC-3AB in alum at 14-days intervals. Two weeks after the second immunization, sera were collected and tested for reactivity with full-length PspAs of clades 1 to 4. The results are shown in Fig. 3. Anti-PspA1ABC antibodies showed a strong reaction with family 1 PspAs (clades 1 and 2), while their reactivity with family 2 (clade 3 and 4) was much weaker (P < 0.001). Anti-PspA3ABC antibodies, on the other hand, showed higher titers against family 2 than against family 1 PspAs (P < 0.001), while their reactivity with family 1 PspAs was not significantly higher than that of control sera. In both cases (anti-PspA1 and anti-PspA3), no significant differences in reactivity between sera within each family were observed.
FIG. 3.
Serum reactivity against full-length PspA fragments from different families. Sera from mice immunized with recombinant PspA1ABC, PspA3ABC, PspA1ABC-4B, or PspA1ABC-3AB were analyzed by ELISA against PspA fragments from clades 1 to 4 extracted from pneumococci. Each result shown is the log2 of the titer.
Furthermore, the antibodies elicited against the PspA1ABC-4B hybrid showed strong reactivity with both PspAs from family 1, as well as with the clade 4 PspA-family 2. Failure of anti-PspA1ABC-4B antibodies to react with the clade 3 PspA fragment from the same family (family 2) may be due to the fact that the fragment of PspA family 2 included in this hybrid comprises only the most variable one-third of the N-terminal domain. Finally, PspA1ABC-3AB elicited antibodies that showed strong reactions with both family 1 and family 2 PspAs (clades 1 to 4), probably because of the presence of the more conserved A region from family 2. On the whole, these results show that although both PspA fusions were able to increase antibody cross-reactivity, the presence of a larger fragment from family 2 in the PspA1ABC-3AB hybrid seems to be important to extend the reactivity of sera to all four PspA clades.
Anti-PspA binding to the pneumococcal surface.
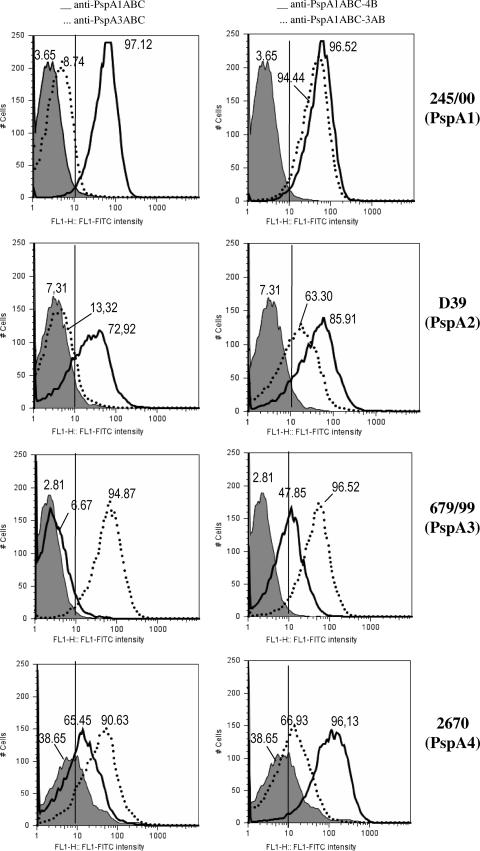
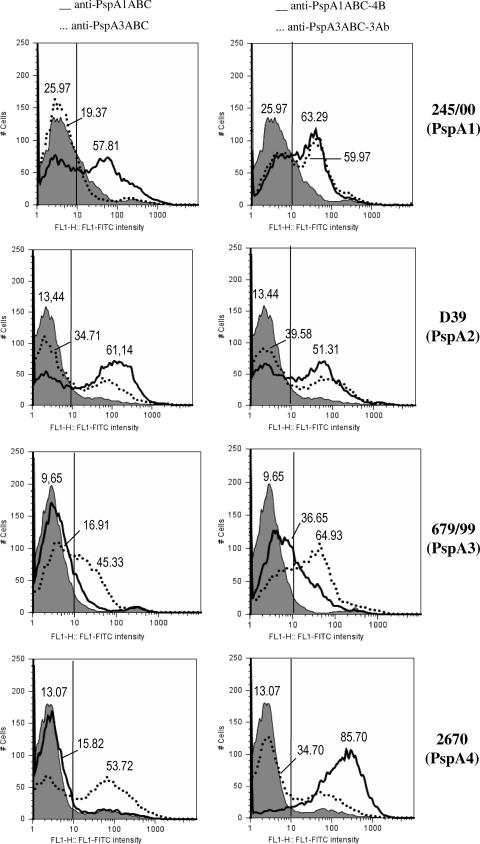
In order to investigate the ability of anti-PspA antibodies to bind intact pneumococci, sera from immunized mice were incubated with pneumococci bearing PspAs from clades 1 to 4, washed, and incubated with FITC-labeled anti-mouse IgG. The percentage of fluorescent bacteria in each group was measured by flow cytometry and is shown in Fig. 4. Anti-PspA1ABC antibodies showed strong binding to both family 1-bearing strains St 245/00 and D39 (solid lines on the left side), while no significant binding to PspA3-containing strain St 679/99 was detected. Interestingly, the percentage of PspA4-containing bacteria (3JYP2670) that became fluorescent after incubation with anti-PspA1ABC antibodies was higher than that of the control (solid line, left side), revealing a significant cross-reaction.
FIG. 4.
Binding of anti-PspA antibodies to the pneumococcal surface. Sera from immunized mice were tested for the ability to bind S. pneumoniae strains bearing PspAs of clades 1 to 4. The percentage of fluorescent bacteria (greater than 10 fluorescence intensity units) is shown for each sample. Left side, anti-PspA1ABC (solid lines) and anti-PspA3ABC (dotted lines); right side, anti-PspA1ABC-4B (solid lines) and anti-PspA1ABC-3AB (dotted lines). Serum from mice immunized with alum was used as a control for each bacterium and is represented by the gray-shaded areas.
The opposite pattern was observed for anti-PspA3ABC antibodies, with strong binding to PspA family 2-containing strains (St 679/99 and 3JYP2670), while binding to PspA family 1-containing strains (St 245/00 and D39) was only about twice that of the control (left side, dotted lines). The levels of binding within the same family were similar for anti-PspA3ABC, while the anti-PspA1ABC binding to D39 strain containing PspA2 was slightly lower than binding to the strain bearing a homologous PspA fragment.
Antibodies generated against PspA hybrids PspA1ABC-4B and PspA1ABC-3AB strongly recognized strains from both families. In the case of the PspA family 2-containing strains, however, the strength of binding correlated directly with the homology of the family 2 PspA clade (3 or 4) present in the bacterium and immunizing fusion protein. Anti-PspA1ABC-3AB showed higher binding to the PspA3-containing strain and lower binding to the PspA4-containing strain, while anti-PspA1ABC-4B showed the opposite effect.
It is interesting, however, that antibodies generated against the PspA3ABC fragment showed efficient binding to the PspA4-containing strain, a cross-reactivity effect that was not seen with the PspA1ABC-3AB hybrid. This may be an indication that the antibodies generated against the hybrid are somewhat different from those made against the independent PspA family 2 fragment. This could be explained by conformational differences or the absence of region C from family 2 PspA fragments in the hybrids.
Complement deposition in the presence of anti-PspA antibodies.
We also determined the ability of the elicited antibodies to increase complement deposition on the bacterial surface. Pneumococci were incubated with anti-PspA1ABC, anti-PspA3ABC, anti-PspA1ABC-4B, or anti-PspA1ABC-3AB and 10% fresh-frozen normal mouse serum, washed, and labeled with FITC-conjugated goat anti-mouse C3. The percentage of bacteria coated with C3 was determined by flow cytometry and is shown in Fig. 5.
FIG. 5.
Complement deposition onto the pneumococcal surface in the presence of antibodies to PspA. Bacteria containing PspAs of clades 1 (St 245/00), 2 (D39), 3 (St 679/99), and 4 (3JYP2670) were incubated in the presence of sera from mice immunized with PspA1ABC (left side, solid lines), PspA3ABC (left side, dotted lines), PspA 1ABC-4B (right side, solid lines), or PspA1ABC-3AB (right side, dotted lines) and normal mouse serum, washed, and incubated with FITC-conjugated goat anti-mouse C3. The percentage of fluorescent bacteria is shown for each group. Serum from mice immunized with alum was used as a control (gray-shaded areas on both sides).
Antibodies induced against PspA1ABC increased the amount of C3 deposited onto the pneumococcal strains bearing PspA fragments from clades 1 and 2 by two- to fourfold while showing almost no ability to enhance C3 deposition on the strains expressing clade 3 and clade 4 PspAs (Fig. 5, solid lines, left side).
Anti-PspA3ABC serum, on the other hand, caused a 4-fold enhancement of C3 deposition on PspA3- and PspA4-bearing strains while showing no enhancement of C3 deposition on the strain expressing PspA clade 1 and only a 2.5-fold enhancement of the C3 levels on the surface of the PspA2-containing strain (D39) (Fig. 5, dotted lines, left side).
Finally, antibodies raised against the hybrids PspA1ABC-4B and PspA1ABC-3AB (Fig. 5, right side, solid and dotted lines, respectively) strongly augmented the amount of surface-bound C3 on pneumococci expressing family 1 and 2 PspAs. However, the immune sera to the PspA fusion proteins caused different levels of C3 deposition on the four strains that correlated with sequence similarities between the hybrid and the PspA fragment present in the bacterium. This effect was most evident with PspA family 2-containing strains (679/99 and 2670); anti-PspA1ABC-4B showed a higher percentage of fluorescent bacteria when the PspA clade 4-bearing strain was considered, and anti-PspA1ABC-3AB was more effective at enhancing C3 deposition on the strain expressing PspA clade 3.
On the whole, the antibody binding and complement deposition data show that immune sera reveal the highest binding and complement deposition against strains expressing PspA of the same family used for immunization. Evidence of cross-reactivity between clades within the same family was observed, although the cross-reactions were sometimes less pronounced for clades in PspA family 2 than that for those in PspA family 1. There was a general correlation between the antibody binding data and the complement deposition data, but there were some interesting exceptions. For example, when the 3JYP2670 (PspA clade 4) target strain was used, the antisera to PspA1ABC and PspA1ABC-3AB showed similar binding but the latter serum resulted in three times as much complement deposition as the former.
Protection against a pneumococcal challenge.
Mice immunized with the PspA fragments or hybrids were challenged intravenously 2 weeks after the third dose with S. pneumoniae serotype 3 strains A66.1 (PspA clades 1 and 2) and 3JYP2670 (PspA clade 4) or intraperitoneally with serotype 6B strain 679/99 (PspA clade 3) and monitored for 10 days. This use of different challenge routes was due to differences in the virulence of the strains.
Table 3 shows the final survival rates of the groups. After a challenge with A66.1 (containing PspA clades 1 and 2), the highest and only statistically significantly different survival rates were obtained for groups immunized with recombinant proteins containing the clade 1 PspA fragment (PspA1ABC, PspA1ABC-4B, or PspA1ABC-3AB). The animals immunized with Pspa3ABC showed protection with borderline significance (P = 0.06).
TABLE 3.
Survival of mice immunized with PspA fragments and hybrids after a challenge with pneumococcal strains containing PspA fragments from different families and clades
| Groupd | No. of mice alive/total (P value)
|
||
|---|---|---|---|
| Strain A66.1 (PspA1 and -2, serotype 3)a | Strain St 679/99 (PspA3, serotype 6B)b | Strain 3JYP2670 (PspA4, serotype 3)a | |
| Alum | 0/6 | 0/6 | 0/12 |
| PspA1ABC | 4/6 (0.03)c | 2/6 (0.2) | 1/12 (>0.2) |
| PspA3ABC | 3/5 (0.06) | 5/6 (0.007)c | 0/12 |
| PspA1ABC-4B | 4/6 (0.03)c | 2/6 (0.2) | 5/12 (0.02)c |
| PspA1ABC-3AB | 6/6 (0.001)c | 4/6 (0.03)c | 2/12 (0.2) |
Intravenous challenge.
Intraperitoneal challenge.
Survival rates were significantly different from those of controls (P < 0.05, Fisher's exact test). Survival was measured 10 days after a challenge.
Strains are listed with their PspA clades and capsular types.
For the mice challenged with St679/99 (PspA clade 3), only the groups immunized with PspA3ABC and the hybrid PspA1ABC-3AB showed a significant rise in survival compared to controls. PspA1ABC and the hybrid PspA1ABC-4B—which do not contain fragments from clade 3—did not protect against this challenge strain. As for a challenge with highly virulent strain 3JYP2670, containing PspA clade 4, only the animals immunized with a fusion protein containing the clade 4 fragment (PspA1ABC-4B) had a statistically significant increase in survival rates. In the mice immunized with the PspA1ABC-3AB hybrid and challenged with clade 4 strain 3JYP2670, although a few mice survived to the end of the experiment, survival was not different from that of controls.
Altogether, these results suggest that protection against a fatal pneumococcal challenge is dependent on the level of similarity between the immunizing recombinant protein and the PspA fragment present on the bacterium.
DISCUSSION
The present work investigated the potential of PspA chimeric proteins—containing fused fragments from families 1 and 2—to be used as a broader-coverage antipneumococcal vaccine. First, we examined the level of cross-reactivity elicited by immunization with the hybrids in comparison with fragments of each family alone. Sera against the single PspA fragments—PspA1ABC and PspA3ABC—revealed a strong reactivity within the same family and much weaker reactions with the other family. Antibodies induced against the hybrids, on the other hand, were reactive with both PspA families, but there were differences in the level of recognition of family 2 PspA fragments. While for PspA1ABC-4B, cross-reactivity with family 2 was restricted to PspA clade 4, anti-PspA1ABC-3AB antibodies were strongly reactive with all four PspA clades. This suggests that the inclusion of longer fragments of each family, containing more conserved regions, is important to broaden cross-protection against strains of different PspA clades. Kolberg et al. (13) have shown that a combination of only two anti-PspA monoclonal antibodies generated by immunization of mice with several heat-killed pneumococci was able to detect virtually all of the clinical isolates tested in the study; one of them was reactive with a PspA family 1 strain, and the other was reactive with a family 2 strain. On the basis of our results presented here and earlier results obtained with monoclonal antibodies to PspA (8), it could be inferred that the monoclonal antibodies used by Kolberg et al. must be reactive to the more conserved region of the family 2 PspA fragments. This reinforces the hypothesis that a PspA-based vaccine containing a single fragment from each major family could be effective against virtually all pneumococci.
Once the cross-reactivity pattern of sera generated against PspA fragments of different clades and the respective hybrids was determined, we investigated the ability of such antibodies to interact with PspA molecules on intact pneumococcal strains, an important requirement for functionality. Flow cytometry analysis revealed that antibodies generated against PspA fragments—PspA1ABC and PspA3ABC—bound strongly to pneumococci containing PspA fragments of the same family while only weak or negligible binding to the heterologous family was observed. The only exception was that the percentage of PspA4-positive bacteria after incubation with anti-PspA1ABC was twice that of the control. This may be due to the exposure, in this strain, of more conserved epitopes, allowing for the interaction with anti-PspA1 sera.
In contrast to these general observations with the PspA fragments, antibodies to the fusion proteins containing sequences from both PspA family 1 and family 2 molecules showed significant binding to PspA fragments of both families. Even so, a higher percentage of PspA3-containing bacteria were positive after incubation with anti-PspA1ABC-3AB, and anti-PspA1ABC-4B bound more efficiently to the PspA4-bearing strain.
Overall, the binding of antibody to the bacteria was in accordance with the ELISA findings, which also showed a strong cross-reactivity within the same family as the immunogen and lower recognition of PspA fragments of the heterologous family.
This study demonstrated that the recombinant proteins maintained enough of the original structure that antibodies elicited in response to them also bound cell surface PspA on intact bacteria. Even more important was our observation that fusion proteins containing PspAs of the two families were able to elicit antibodies reactive with both PspA families and that they protected against strains expressing PspA of either family.
Despite the evidence that sera against the recombinant proteins could interact with live pneumococci, it was necessary to investigate whether such antibodies could enhance complement deposition on the bacterial surface, a property thought to be critical for protection (20). Since all PspA fragments and hybrids induced antibodies primarily of the IgG1 isotype (not shown), we can assume that the differences observed in our experiments were due to differences in epitope recognition by these antibodies.
In accordance with the binding results, the ability of anti-PspA antibodies to increase complement deposition was dependent on the similarity between the PspA present in the bacterium and that used in the generation of sera. Thus, anti-PspA1ABC antibodies were effective at enhancing the levels of C3 bound only to bacteria containing PspA family 1, while anti-PspA3ABC increased C3 deposition on bacteria expressing PspA family 2. Once again, antibodies to the hybrids broadened the effect, increasing C3 deposition on pneumococci containing PspA fragments of both families. In a study using isogenic pneumococcal strains bearing PspA fragments of clade 2 or 4, Ren et al. (21) showed that the presence of either PspA was able to inhibit complement deposition on the pneumococcal surface compared to the PspA-negative strain. Our data support these results by demonstrating that antibodies raised against PspA family 1 or 2 can increase C3 deposition on strains containing homologous PspA fragments and that cross-reactivity with antibodies to PspA hybrids occurs. After incubation with antibodies to the hybrids, the percentage of fluorescent bacteria containing PspAs of each clade revealed a pattern very similar to that observed with binding; anti-PspA1ABC-3AB was more effective at enhancing C3 deposition on the PspA3-bearing strain, while anti-PspA1ABC-4B interacted better with the PspA4-containing strain. Previous studies investigating complement deposition on pneumococci in the presence of PspA focused on serotype 3 strains (20, 21, 24). In the present work, we demonstrate that anti-PspA antibodies can increase C3 deposition on bacteria of diverse serotypes, including neutral (not charged) capsule 14 strain St 245/00. We have also examined C3 deposition on other pneumococcal isolates bearing different combinations of PspA and capsular types, with results similar to those presented in Fig. 5. This reinforces the role of anti-PspA antibodies in enhancing complement deposition, which can overcome the inhibitory effects of different capsule types.
The presence of anti-PspA IgG antibodies in the bloodstream would greatly improve opsonization and phagocytosis by the immune system. Yuste et al. (29) showed that activation of the complement system can prevent the spread of pneumococcus from the lungs to the bloodstream. Thus, a PspA-based vaccine able to induce high antibody levels could interfere with pneumococcal infection at the early pulmonary phase, as well as in the bloodstream, in case of invasion. This would lead to complement-mediated opsonization and bacterial clearance. On the basis of these data, we challenged the immunized mice with pneumococcal strains bearing diverse PspA fragments in order to verify if there is a correlation between the ability of antibodies induced against the hybrids to increase complement deposition on a wider range of bacteria and the level of protection against sepsis. Immunization with PspA1ABC or PspA3ABC led to significant protection only against the strain containing a PspA of the same family, while the hybrids induced protection across families. Nevertheless, cross-protection against family 2 strains was also dependent on the PspA clade; PspA1ABC-3AB was protective against the PspA3-containing strain, and anti-PspA1ABC-4B was protective against the PspA4-containing strain. Previous studies in our laboratory have shown that cross-reactivity of antibodies induced by DNA vaccination is restricted to the same family, while cross-protection against intraperitoneal challenge is restricted to the same clade (16). These similarities suggest that PspA hybrids are able to broaden protection when presented as either recombinant proteins or as DNA. Our findings are supported by the fact that, in the immunization of humans with PspA, antibody reactivity is stronger against the immunizing clade, while significant cross-reactivity exists within clades of the same family (3, 18). In other studies using intravenous challenge with capsular type 3 and 6B pneumococci, it has been observed that cross-protection can be extended across PspA families, even though antibody cross-reactivity is weak (3, 4). However, when a capsular type 4 pneumococcus was used as the challenge strain, the results were similar to those reported here; the best protection was obtained when the challenge strain and the immunizing antigen were in the same clade (22).
We observed a strong correlation between the levels of complement deposition induced by anti-PspA antibodies and survival against a challenge with different pneumococcal strains, reinforcing the notion that this mechanism may be responsible for protection against a pneumococcal challenge. Nevertheless, there were a few exceptions, which demonstrate the complexity of antibody interaction with pneumococci.
Most PspA fragments, even within clades and families, differ somewhat from each other. It should be especially noted that two of the challenge strains we used expressed PspA fragments that were not identical to any of the PspA fragments used for immunization. Thus, the protection we achieved against these challenge strains provides strong evidence that a PspA vaccine like the ones used here could provide protection against pneumococcal strains in general.
Although it may be necessary to include PspA fragments from two clades of family 2 to prepare a broadly protective PspA vaccine, we believe that the use of PspA hybrids can provide an efficient tool to produce a broadly protective PspA-based antipneumococcal vaccine.
Acknowledgments
We thank Jorge Mário da Costa Ferreira, Jr., for help with the fluorescence-activated cell sorter analysis.
This work was supported by FAPESP, CAPES, Fundação Butantan, and NIH grant AI21548.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 8 October 2007.
REFERENCES
- 1.Beall, B., G. Gherardi, R. R. Facklam, and S. K. Hollingshead. 2000. Pneumococcal pspA sequence types of prevalent multiresistant pneumococcal strains in the United States and of internationally disseminated clones. J. Clin. Microbiol. 38:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandileone, M. C., A. L. Andrade, E. M. Teles, R. C. Zanella, T. I. Yara, J. L. Di Fabio, and S. K. Hollingshead. 2004. Typing of pneumococcal surface protein A (PspA) in Streptococcus pneumoniae isolated during epidemiological surveillance in Brazil: towards novel pneumococcal protein vaccines. Vaccine 22:3890-3896. [DOI] [PubMed] [Google Scholar]
- 3.Briles, D. E., S. K. Hollingshead, J. King, A. Swift, P. A. Braun, M. K. Park, L. M. Ferguson, M. H. Nahm, and G. S. Nabors. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis. 182:1694-1701. [DOI] [PubMed] [Google Scholar]
- 4.Briles, D. E., S. K. Hollingshead, G. S. Nabors, J. C. Paton, and A. Brooks-Walter. 2000. The potential for using protein vaccines to protect against otitis media caused by Streptococcus pneumoniae. Vaccine 19(Suppl. 1):S87-S95. [DOI] [PubMed] [Google Scholar]
- 5.Briles, D. E., J. D. King, M. A. Gray, L. S. McDaniel, E. Swiatlo, and K. A. Benton. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858-867. [DOI] [PubMed] [Google Scholar]
- 6.Briles, D. E., R. M. Perlmutter, D. Hansburg, J. R. Little, and J. M. Davie. 1979. Immune response deficiency of BSVS mice. II. Generalized deficiency to thymus-dependent antigens. Eur. J. Immunol. 9:255-261. [DOI] [PubMed] [Google Scholar]
- 7.Brown, J. S., T. Hussell, S. M. Gilliland, D. W. Holden, J. C. Paton, M. J. Ehrenstein, M. J. Walport, and M. Botto. 2002. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc. Natl. Acad. Sci. USA 99:16969-16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crain, M. J., W. D. Waltman, J. S. Turner, J. Yother, D. E. Talkington, L. M. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels, C. C., T. C. Briles, S. Mirza, A. P. Hakansson, and D. E. Briles. 2006. Capsule does not block antibody binding to PspA, a surface virulence protein of Streptococcus pneumoniae. Microb. Pathog. 40:228-233. [DOI] [PubMed] [Google Scholar]
- 10.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janoff, E. N., C. Fasching, J. M. Orenstein, J. B. Rubins, N. L. Opstad, and A. P. Dalmasso. 1999. Killing of Streptococcus pneumoniae by capsular polysaccharide-specific polymeric IgA, complement, and phagocytes. J. Clin. Investig. 104:1139-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarva, H., T. S. Jokiranta, R. Wurzner, and S. Meri. 2003. Complement resistance mechanisms of streptococci. Mol. Immunol. 40:95-107. [DOI] [PubMed] [Google Scholar]
- 13.Kolberg, J., A. Aase, T. E. Michaelsen, and G. Rodal. 2001. Epitope analyses of pneumococcal surface protein A: a combination of two monoclonal antibodies detects 94% of clinical isolates. FEMS Immunol. Med. Microbiol. 31:175-180. [DOI] [PubMed] [Google Scholar]
- 14.Mastroeni, P., B. Villarreal-Ramos, and C. E. Hormaeche. 1993. Adoptive transfer of immunity to oral challenge with virulent salmonellae in innately susceptible BALB/c mice requires both immune serum and immune T cells. Infect. Immun. 61:3981-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDaniel, L. S., B. A. Ralph, D. O. McDaniel, and D. E. Briles. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323-337. [DOI] [PubMed] [Google Scholar]
- 16.Miyaji, E. N., D. M. Ferreira, A. P. Lopes, M. C. Brandileone, W. O. Dias, and L. C. Leite. 2002. Analysis of serum cross-reactivity and cross-protection elicited by immunization with DNA vaccines against Streptococcus pneumoniae expressing PspA fragments from different clades. Infect. Immun. 70:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mollerach, M., M. Regueira, L. Bonofiglio, R. Callejo, J. Pace, J. L. Di Fabio, S. Hollingshead, and D. Briles. 2004. Invasive Streptococcus pneumoniae isolates from Argentinian children: serotypes, families of pneumococcal surface protein A (PspA) and genetic diversity. Epidemiol. Infect. 132:177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nabors, G. S., P. A. Braun, D. J. Herrmann, M. L. Heise, D. J. Pyle, S. Gravenstein, M. Schilling, L. M. Ferguson, S. K. Hollingshead, D. E. Briles, and R. S. Becker. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 18:1743-1754. [DOI] [PubMed] [Google Scholar]
- 19.Pimenta, F. C., F. Ribeiro-Dias, M. C. Brandileone, E. N. Miyaji, L. C. Leite, and A. L. Sgambatti de Andrade. 2006. Genetic diversity of PspA types among nasopharyngeal isolates collected during an ongoing surveillance study of children in Brazil. J. Clin. Microbiol. 44:2838-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren, B., A. J. Szalai, S. K. Hollingshead, and D. E. Briles. 2004. Effects of PspA and antibodies to PspA on activation and deposition of complement on the pneumococcal surface. Infect. Immun. 72:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren, B., A. J. Szalai, O. Thomas, S. K. Hollingshead, and D. E. Briles. 2003. Both family 1 and family 2 PspA proteins can inhibit complement deposition and confer virulence to a capsular serotype 3 strain of Streptococcus pneumoniae. Infect. Immun. 71:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roche, H., B. Ren, L. S. McDaniel, A. Hakansson, and D. E. Briles. 2003. Relative roles of genetic background and variation in PspA in the ability of antibodies to PspA to protect against capsular type 3 and 4 strains of Streptococcus pneumoniae. Infect. Immun. 71:4498-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai, S. S. 2006. Streptococcus pneumoniae protein vaccine candidates: properties, activities and animal studies. Crit. Rev. Microbiol. 32:139-153. [DOI] [PubMed] [Google Scholar]
- 24.Tu, A. H., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vela Coral, M. C., N. Fonseca, E. Castaneda, J. L. Di Fabio, S. K. Hollingshead, and D. E. Briles. 2001. Pneumococcal surface protein A of invasive Streptococcus pneumoniae isolates from Colombian children. Emerg. Infect. Dis. 7:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yother, J., G. L. Handsome, and D. E. Briles. 1992. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J. Bacteriol. 174:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuste, J., M. Botto, J. C. Paton, D. W. Holden, and J. S. Brown. 2005. Additive inhibition of complement deposition by pneumolysin and PspA facilitates Streptococcus pneumoniae septicemia. J. Immunol. 175:1813-1819. [DOI] [PubMed] [Google Scholar]