Abstract
Many Streptococcus pyogenes (group A streptococcus [GAS]) virulence factor- and transcriptional regulator-encoding genes cluster together in discrete genomic regions. Nra is a central regulator of the FCT region. Previous studies exclusively described Nra as a transcriptional repressor of adhesin and toxin genes. Here transcriptome and proteome analysis of a serotype M49 GAS strain and an isogenic Nra mutant of this strain revealed the complete Nra regulon profile. Nra is active in all growth phases tested, with the largest regulon in the transition phase. Almost exclusively, virulence factor-encoding genes are repressed by Nra; these genes include the GAS pilus operon, the capsule synthesis operon, the cytolysin-mediated translocation system genes, all Mga region core virulence genes, and genes encoding other regulators, like the Ihk/Irr system, Rgg, and two additional RofA-like protein family regulators. Surprisingly, our experiments revealed that Nra additionally acts as a positive regulator, mostly for genes encoding proteins and enzymes with metabolic functions. Epidemiological investigations revealed strong genetic linkage of one particular Nra-repressed regulator, Ralp3 (SPy0735), with a gene encoding Epf (extracellular protein factor from Streptococcus suis). In a serotype-specific fashion, this ralp3 epf gene block is integrated, most likely via transposition, into the eno sagA virulence gene block, which is present in all GAS serotypes. In GAS serotypes M1, M4, M12, M28, and M49 this novel discrete genetic region is therefore designated the eno ralp3 epf sagA (ERES) pathogenicity region. Functional experiments showed that Epf is a novel GAS plasminogen-binding protein and revealed that Ralp3 activity counteracts Nra and MsmR regulatory activity. In addition to the Mga and FCT regions, the ERES region is the third discrete chromosomal pathogenicity region. All of these regions are transcriptionally linked, adding another level of complexity to the known GAS growth phase-dependent regulatory network.
Streptococcus pyogenes (group A streptococcus [GAS]) is an exclusively human pathogen. GAS causes diseases ranging from commonly mild superficial infections of the skin and mucous membranes of the nasopharynx to severe but rare toxic and/or invasive diseases (7, 8, 12). GAS is equipped with many virulence factors which allow this successful pathogen to infect and survive within the host. Different subsets of virulence factors are required for the various stages of the infection process (21). Balanced and timely expression of virulence factor genes is therefore crucial for pathogen fitness.
Besides the 13 common two-component signal transduction systems (TCS), about 30 stand-alone transcriptional regulators sense and integrate environmental information in GAS. Specific transcriptional induction and repression processes finally control expression of the appropriate virulence gene subsets. Seven GAS TCS and four transcriptional regulators have been implicated in host-pathogen interactions (34, 51). The longest known transcriptional regulator is Mga, which was shown to act as a direct and indirect positive transcriptional regulator of many virulence genes within the so called “Mga chromosomal region” (44). Mga acts together with transcriptional regulators belonging to the RofA-like protein (Ralp) family in a growth phase-dependent regulatory network (3, 19, 26).
The Ralp family regulators characterized so far occur as two variants (RofA [also designated Ralp1] and Nra [also designated Ralp2]) and are encoded in a serotype-specific fashion within the FCT genomic region, which represents a putative pathogenicity island (5, 16, 25, 43). In silico inspection of the GAS serotype M1 genome sequence (15) allowed identification of two additional Ralp regulators, designated Ralp3 and Ralp4 (19), which were partially characterized during review of this work (29, 45).
The FCT region resembles the Mga region in terms of the high number of encoded putative and confirmed virulence factors, including several microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (40) which mediate GAS adherence to eukaryotic cells and internalization processes via fibronectin and collagen binding (10, 11).
Embedded in the FCT regions of GAS serotypes M4, M6, M12, and M28, the fibronectin-binding MSCRAMM protein F1 is associated with the RofA version of the Ralp regulators (5, 25, 54). Transcription of the protein F1-encoding gene was shown to be under direct positive control of RofA (16, 17). In GAS serotype M3, M5, M18, and M49 strains, a gene encoding a collagen-binding MSCRAMM (Cpa) replaced the protein F1 gene at its position upstream of the Ralp gene. The Cpa-encoding gene is associated with the Nra version of Ralp regulators in the strains belonging to these serotype (25, 27, 43).
Recent work has shown that Cpa is important for GAS adherence to collagen type I, and its expression is apparently linked to cases of arthritis and osteomyelitis (27).
Of note, the FCT regions of GAS serotype M12 and M28 strains encode the highest number of MSCRAMMs. In addition to protein F1, these two serotypes carry a gene encoding Cpa plus a gene encoding a second fibronectin-binding MSCRAMM, designated protein F2 (28). The protein F2-encoding gene also occurs in serotypes M3, M5, M18, and M49 and is exclusively associated with the presence of a second transcriptional regulator of the AraC/XylS family, designated MsmR (35). As demonstrated for a serotype M49 strain, protein F2 is apparently the most important fibronectin-binding MSCRAMM in all protein F1-negative strains, since it almost exclusively mediates epithelial cell adherence and internalization (28).
Of note, the FCT region of GAS and partially homologous genomic structures of S. pneumoniae and S. agalactiae encode all the enzymes and structural proteins essential for pilus formation (47, 54). The GAS pili could be involved as alternative adhesins in epidermal GAS infections (1, 31). Because of their broad occurrence in streptococci and their putative biological function, GAS pilus proteins are currently being discussed as promising vaccine antigens (54).
The Nra version of the Ralp family regulators was first described in a GAS serotype M49 strain by Podbielski et al. (43). The amino acid sequence of Nra shares 62% homology with RofA and is expressed predominantly in the transition growth phase. As a negative transcriptional regulator, Nra suppresses transcription of its own gene and transcription of Cpa- and protein F2-encoding genes (33, 43). The Cpa gene is the first gene of a polycistronic message that includes all potential pilus-encoding genes, suggesting that pilus formation is apparently under negative control of Nra. Phenotypically, Nra is in control of GAS interactions with host cells by basically modulating transcription of virulence genes responsible for host cell adherence, internalization, and cytotoxicity. The latter feature is achieved via transcriptional control of genes encoding SpeA (superantigen), SpeB (cysteine protease), and streptolysin S (33).
Most recently, the activity of the second transcriptional regulator (MsmR) encoded in the FCT region of a serotype M49 GAS strain was characterized (35). Transcriptome analysis revealed that the MsmR regulon includes up to 24 genes under positive MsmR control and 36 genes for which MsmR acts as a transcriptional repressor. Direct binding of MsmR to promoter regions of genes encoding protein F2, Nra/Cpa, and Nga/streptolysin O could be demonstrated (35). In particular, the influence of MsmR on transcription of genes encoding the structural components of a GAS “type III secretion machinery”-like cytolysin-mediated translocation (CMT) system, recently discovered and characterized by Madden et al. (32), plays an important role in the GAS-host cell interaction (35). All genes located in the FCT region, including nra and msmR, are under positive MsmR control. The MsmR transcription level is highest in the transition growth phase. Therefore, MsmR and Nra could mutually influence transcription by responding to signals that are not known yet. Thus, these two factors contribute to balanced control of crucial virulence factor expression in GAS within the framework of the already established Mga-Ralp growth phase-dependent regulatory network (26, 35).
In order to better define the Nra part of this regulatory network, we identified the complete Nra regulon using established GAS whole-genome DNA arrays (6). We confirmed and supplemented the data by investigating the global growth phase-dependent Nra regulon via proteome analysis, with a focus on surface-expressed and secreted GAS subproteomes. Our results demonstrated that Nra activity is connected with another Ralp regulator (SPy0735 or Ralp3), which is in charge of virulence factor expression from a novel pathogenicity island-like genomic region present in a subset of GAS serotype strains. Like the Mga and FCT regions, this novel region, designated the epf ralp3 eno sagA (ERES) region, contains genes encoding important known and novel virulence factors. Apparently, the ERES region is integrated into the Mga-Ralp network, indicating its importance for GAS pathogenesis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
GAS serotype M49 strain 591 was obtained from R. Lütticken (Aachen, Germany). The GAS strains belonging to different serotypes used in this study have been previously described by Podbielski (41). Escherichia coli strain DH5α (Gibco-BRL) was used as the host for plasmids pUC18Erm1, pSF151, and pFW5-luc (2, 43, 53). E. coli XL10-gold (Stratagene) served as the host for plasmid pQE30 (Qiagen). All E. coli strains were cultured in Luria-Bertani medium at 37°C with agitation. The GAS wild-type strain and isogenic derivatives were cultured in Todd-Hewitt broth (Invitrogen) supplemented with 0.5% yeast extract (Invitrogen) (THY medium) at 37°C under a 5%CO2-20%O2 atmosphere, unless otherwise specified. For selection and maintenance of mutants, antibiotics were added to the media at the following concentrations: ampicillin, 100 μg ml−1 for E. coli; kanamycin, 50 μg ml−1 for E. coli and 300 μg ml−1 for GAS; spectinomycin, 100 μg ml−1 for E. coli and 60 μg ml−1 for GAS; and erythromycin, 300 μg ml−1 for E. coli and 5 μg ml−1 for GAS.
DNA techniques.
Purification of chromosomal DNA from GAS and other streptococcal species was performed as outlined by Kreikemeyer et al. (24). Plasmid DNA manipulation, transformation of E. coli and GAS, and other conventional manipulations were performed essentially as described previously (43). Probes for Southern hybridization were generated by PCR using the following primer sets: RALP3_probeF (5′-CTCTATCAAACCTCGATATTCT-3′)/RALP3_probeR (5′-CCATTTTCCGATGATTTCCTGA-3′) for ralp3, Epf_probeF (5′-GATGCTATTGCAGAATTAGAGA-3′)/Epf_probeR (5′-CAATAGCTTTCTTAAGTTCTGG-3′) for epf, and Eno_For (5′-CGGTGGATCACACTCAGATG-3′)/Eno_Rev (5′-GTCGTCACCAACCAATTGAA-3′) for eno.
Construction of recombinant vectors and GAS strains.
For construction of the ralp3 deletion mutant of serotype M49 strain 591, a 3.3-kbp fragment including the entire SPy0735 gene and the neighboring sequence was amplified from chromosomal DNA of strain 591 with primer spy0735wholeF3/SacI (5′-GGCGGCGAGCTCGCTGCAATAGATTGCTTGTCTA-3′) and primer spy0735wholeR/Hind (5′-GGCGGCAAGCTTGGCATAGTAGCGAACCTCTTCT-3′). The PCR product was cloned into HindIII/SacI sites of pUC18Erm1. The resulting recombinant plasmid was utilized as a template for a second PCR with primer spy0735nikiF/Bam (5′-GGCGGCGGATCCATGAAGGTGTAACATAACTGAA-3′) and primer spy0735nikiR/Sph (5′-GGCGGCGCATGCCACAAGAACCTGTCTTCTCCTT-3′), in which the whole plasmid was amplified but the SPy0735 gene was left out. The generated PCR product was ligated with an aphA3 kanamycin resistance cassette, including the orthologous promoter and terminator regions, which was amplified with primer aphA3For/Bam (5′-GGCGGCGGATCCGCTACCAAGACGAAGAGGAT-3′) and primer aphA3Rev/Sph (5′-GGCGGCGCATGCGGACAGTTGCGGATGTACTT-3′) and with pSF151 as the template. The resultant plasmid, pVK2006, was transformed into serotype M49 strain 591, and then transformants were selected on kanamycin-containing agar plates to recover all strains with crossover events. In the second step, all transformants were plated in duplicate on THY medium containing kanamycin or erythromycin. Deletion of the SPy0735 gene in isolates exhibiting kanamycin resistance and erythromycin sensitivity resulted from double-crossover events and was confirmed by site-specific PCRs. The following primers were used; spy0733R (5′-TGGAATAGAGCGTAGAATCTTGT-3′), aphR3 (5′-GTCCGGTCGATCAGGGAGGATA-3′), aphF3 (5′-CCTTTATACCGGCTGTCCGTCATT-3′), Epf_probeF, Epf_probeR, RALP3_probeF, and RALP3_probeR.
To construct a vector for complementation of ralp3, a fragment including the putative promoter and terminator regions was amplified with primers ralp3F/Eco (5′-GCGAATTCACGTTGTCAGCCAGCAGACACCCA-3′) and ralp3R/Sph (5′-GCGCATGCCTAGAAGAATAAGTCTGATACTGT-3′). The fragment was subsequently cloned into pAT18, which was followed by transformation into the ralp3 mutant.
For construction of an Epf mutant, an internal fragment of the gene was amplified using primers epfKOF (5′-TTGCCGTACCTACTGCATTCA-3′) and epfKOR (5′-CCTTGTTCCTCACCTTCTATT-3′) and cloned into pSF151 via BamHI/EcoRI sites. The resulting plasmid was integrated into the chromosome of strain 591. The correct integration was confirmed by site-specific PCR with primers aphR3 and spy0735nikiF/Bam.
For construction of plasmids expressing recombinant Epf (rEpf), primers rEPF-F/Sph (5′-GCGGAAGCTGAAGAACTGCCAGAT-3′) and rEPF-R/SalI (5′-ACCTGATTTTGGAAGATGGTTCGCT-3′) were used to amplify the epf gene without the region encoding the deduced signal sequence and sorting signal. The fragment was cloned into pQE30 via SphI/SalI sites and transformed into E. coli XL10-gold.
Construction of nra-luc, prtF2-luc, cpa-luc, pel sagA-luc, and msmR-luc plasmids has been described previously (24-28, 35, 43). The plasmids were introduced into the ralp3 deletion mutant as outlined by Podbielski et al. (43).
For construction of ralp3-luc, epf-luc, and eno-luc plasmids, the following primer sets were used: spy0735lucF/Nhe (5′-TGACCTTACGCATTCTCCAAT-3′)/spy0735lucR/Bam (5′-TGCGACAATTGGTCCTGATA-3′) for ralp3, epflucF/Nhe (5′-GCAGCGGTTGACGCAGCTAAGA-3′)/epflucR/Bam (5′-AGCCAGGTTCTTAGTCTTTACT-3′) for epf, and EnolucF/Nhe (5′-CTGTAGGTGACGAAGGTGGAT-3′)/EnolucR/Bam (5′-CGATTTCTGTCTTACCGTCTA-3′) for eno. The luc constructs were introduced into the ralp3 mutant as described above.
RNA preparation, Northern blotting, and Southern blotting.
Total RNA was isolated from cells in the growth phases indicated below by the hot phenol method (6, 48). RNA quality and integrity were assessed by electrophoretic analysis and measurement of the A260/A280 ratios. Denaturing agarose gel electrophoresis and Northern blot hybridization with digoxigenin-dUTP-labeled probes have been described previously (42). Digoxigenin-dUTP-labeled gene probes were generated by PCR.
Quantitative assays for luciferase activity.
The GAS luc reporter strains were cultured in THY medium in ambient air with or without 5% CO2, unless otherwise stated. To measure luminescence, aliquots from bacterial cultures were withdrawn at 1-h intervals and processed as described previously (43). The Luc assay with each individual strain was repeated four or five times on different days. As previously described for this test system (24, 27, 28, 35, 43), the individual data from all days were quite different. However, the overall expression profiles matched. Thus, the data shown here are the data from one representative experiment.
Microarray analysis and real-time reverse transcription (RT)-PCR assay.
For microarray analysis, total RNA was extracted on two independent occasions from serotype M49 wild-type and isogenic nra mutant strains grown to exponential, transition, and stationary phases, and amino allyl-modified cDNA was synthesized (6). The cDNA was labeled with CyDye using a CyScribe postlabeling kit (Amersham) according to the manufacturer's instructions. The DNA microarrays, containing 50-mer oligonucleotides designed from open reading frame sequences of a partial serotype M49 genome sequence (accession no. NZ_AAFV00000000) and previously reported serotype M1, M3, and M18 sequences (NC_002737, NC_004606, and NC_003485), were constructed by MWG (6). Oligonucleotide chip slides were prehybridized for 20 min at 42°C in a solution containing 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.5% sodium dodecyl sulfate (SDS), and 10 mg ml−1 bovine serum albumin (BSA). The prehybridization buffer was removed by washing the slides in distilled water five times and drying them by centrifugation. The denatured cDNA was hybridized to slides for 16 h at 42°C in the dark. Before the image was scanned, the slides were washed for 5 min at 42°C in 2× SSC-0.1% SDS (pH 7.0), for 5 min at 42°C in 1× SSC (pH 7.2), and for 2 min at room temperature in 0.1× SSC prewarmed to 37°C and dried by centrifugation after they were washed in distilled water. Scanning the image and statistical analyses were carried out using an Affymetrix 428Array scanner (Affymetrix) and ImaGene version 5.6 and GeneSight version 3.5 software (BioDiscovery) as previously reported (6).
For real-time RT-PCR assays, three independent extractions of total RNA from the wild type and the nra mutant were performed as described above. Synthesis of cDNA was performed with a Superscript First-Strand synthesis system for RT-PCR (Invitrogen). DNA contamination was assessed by PCR in non-RT samples. Primer sets for selected genes were designed using Primer Express software (version 1.0; PE Applied Biosystems). The primers used have been listed by Beyer-Sehlmeyer et al. (6) and Nakata et al. (35). RT-PCR amplification was done using the SYBR green method with an ABI PRISM 7000 sequence detection system (Applied Biosystems). The data were analyzed using an ABI PRISM 5700 sequence detection system (version 1.6; Applied Biosystems). The level of DNA gyrase subunit A gene (gyrA) transcription was used for normalization.
Proteome analysis and protein identification.
The S. pyogenes serotype M49 wild-type strain and the corresponding Nra mutant were cultured in THY medium at 37°C in a 5% CO2-20% O2 atmosphere. The bacteria were grown as batch cultures until the desired growth phase was reached. The cultures were harvested by centrifugation (4,000 rpm, 10 min, 25°C). The supernatant containing the secreted GAS proteome was collected, sterile filtered (0.22 μm; Neolab filter device), and precipitated with 12% (vol/vol) trichloroacetic acid.
In order to access the surface-associated proteome, bacterial pellets were washed twice in phosphate-buffered saline (PBS)-30% sucrose (pH 7.4) (isotonic osmoprotective buffer) and resuspended to an optical density at 600 nm (OD600) of 20 in this buffer. Subsequently, bacterial suspensions were digested under ambient conditions with phage lysin C (100 U/ml) (23, 36). This enzymatic treatment proved to be superior to the mutanolysin digestion described in a recent study (23). The digestion was terminated after 20 min, and phage lysin C-released surface proteins found in the supernatant of the digest were subsequently dialyzed (Serva dialysis tubes; molecular mass cutoff, 3,000 Da).
Aliquots of both the secreted and surface-associated proteomes were ultrafiltered with Centricon ultrafiltration tubes (500 μl to 4 ml; molecular mass cutoff, 5,000 Da; Amicon), aliquoted (100 μg total protein), and subsequently vacuum dried. For proteomic analysis, the aliquots were separated by isoelectric focusing (pH 3 to 10 nonlinear gradient) and SDS-polyacrylamide gel electrophoresis (12.5% acrylamide) (18). Gels were stained with Coomassie brilliant blue and digitalized using a Umax Mirage II scanner and Corel Photo Paint 12.0 software. Spot analysis was performed with Progenesis gel analysis software. For the spots which occurred in multiple isoforms, the quantities of all isoforms were summed for comparison of the wild type and Nra mutant. Background subtraction and normalization procedures were performed for all gels analyzed. Detected spots were isolated by proteolytic “in-gel” trypsin digestion (20) with methylated trypsin (Promega, Mannheim, Germany) and were identified by matrix-assisted laser desorption ionization-time of flight analysis (Reflex III; Bruker Daltonics, Bremen, Germany), followed by peak analysis with the Flex Analysis software and the Mascot database utilizing Biotools 2.3 database search software (Bruker Daltonics).
GAS adherence to matrix proteins, rEpf-binding enzyme-linked immunosorbent assay, and biofilm formation.
To quantify the binding of GAS wild-type and mutant strains to immobilized matrix proteins, 96-well enzyme-linked immunosorbent assay plates (Greiner Bio-One, Solingen, Germany) were coated overnight at 4°C with 5 μg/well human matrix proteins (Sigma) in PBS. Plates were blocked with 1% BSA in PBS for 1 h at 37°C, and after three washes in 0.05% Tween 20-PBS, early-stationary-phase GAS strains diluted to an A600 nm of 0.4 in PBS were incubated in the coated wells for 30 min at 37°C. As a control, GAS strains were incubated in noncoated and BSA-coated wells. After four washes with PBS, a goat anti-GAS-horseradish peroxidase (HRP) conjugate (Dunn Labortechnik GmbH, Asbach, Germany) diluted 1:5,000 in PBS was added, and the plates were incubated for 1 h at room temperature. After four washes with PBS, bound antibodies were visualized using a 3,3′,5,5′-tetramethylbenzidine peroxidase enzyme immunoassay substrate kit (Bio-Rad). After 10 min, the reactions were stopped with 0.5% H2SO4, and the OD450 was monitored.
In order to test rEpf binding to human matrix proteins, plates were coated, blocked, and washed as described above. Then 5 μg/well rEpf was added and allowed to bind for 30 min at 37°C. After subsequent washing steps, bound rEpf was detected by incubation with 100 μl/well Ni-nitrilotriacetic acid-HRP conjugate (1:2,000 in PBS; Qiagen, Germany) and detection with 3,3,5′,5-tetramethylbenzidine as outlined above.
The GAS serotype M49 wild-type strain, the Nra mutant, the Ralp3 mutant, and the Epf mutant were tested to determine their biofilm-building capacities as described by Lembke et al. (30).
Eukaryotic cell adherence and internalization.
Human epithelial HEp-2 (= ATCC CCL23) cells were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (Gibco). Adherence to and internalization in HEp-2 cells was assessed by using the antibiotic protection assay (38). Briefly, HEp-2 cells were infected with GAS strains at a multiplicity of infection of 1:10 for 2 h. For quantification of bacterial adhesion, GAS-infected cells were washed with PBS and lysed with distilled water. Serial dilutions of the lysate were plated on THY agar plates to count CFU. For assessment of bacterial internalization, the cells were washed with PBS and incubated with Dulbecco modified Eagle medium supplemented with penicillin (50 U/ml) for an additional 2 h. Then the cells were washed and lysed, and the bacterial numbers were counted.
Eukaryotic cell viability after infection with GAS was determined by fluorescent staining using calcein AM and ethidium homodimer-1 (LIVE/DEAD viability/cytotoxicity kit for animal cells; catalog no. L-3224; Molecular Probes). Images were observed with a fluorescence microscope (Olympus BX60) at a magnification of ×600 and were quantitatively assessed by counting the number of cells stained with ethidium homodimer-1 in three independent microscopic fields.
Preparation of His-tagged rEpf.
Hyperexpression of the rEpf protein was induced by adding 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to mid-exponential-phase E. coli cells, and then the cells were grown at 30°C for 5 h. Recombinant proteins were purified by using a QIAexpress protein purification system (Qiagen) as outlined in the manufacturer's instructions. Eluted protein was concentrated by ultrafiltration with a Centricon-100 centrifugal concentrator (Amicon) and diluted with PBS. The concentration of recombinant proteins was determined using a bicinchoninic acid protein assay kit (Pierce). The integrity of the recombinant proteins was analyzed by SDS-polyacrylamide gel electrophoresis, gel staining, and Western blotting using the HRP-Ni-nitrilotriacetic acid conjugate (Qiagen) as reported previously (28).
Hemolysis assay.
The wild-type, mutant, and complemented strains were grown overnight as described above. Only cultures with the same final OD600 after incubation were used for the hemolysis assays. GAS culture supernatants recovered from stationary phase were collected after centrifugation, and then an equivalent volume of 2% defibrinized sheep erythrocytes in PBS was added. After 30 min of incubation at 37°C, the samples were centrifuged and the absorbance at 540 nm was measured to determine the release of hemoglobin. After subtraction of the background and streptolysin O-dependent hemolysis values, values were compared to a standard curve for erythrocytes lysed in water.
Statistical analysis.
A statistical analysis for all functional tests was performed with the Mann-Whitney U test or by one-way analysis of variance and Scheffe's test. A P value of <0.01 was considered significant.
RESULTS
Growth phase-dependent Nra transcriptome.
To extend our knowledge concerning Nra target genes identified in our previous investigations (33, 43), we characterized the Nra regulatory activity on the whole-genome level with a focus on three different time points during GAS growth. The exponential phase (3 h) and late stationary phase (16 h) are characterized by low-level Nra expression, whereas in transition phase (6 to 8 h) maximal transcription of Nra was noted. The transition phase was also the time when there were the most notable phenotypic differences between a wild-type strain and the corresponding Nra mutant (33, 43).
A summary of differentially transcribed genes identified by DNA array analysis and confirmatory real-time RT-PCR is presented in Tables S1 to S4 in the supplemental material. The results suggested that Nra is an active regulator in all growth phases investigated (see Tables S1 to S4 in the supplemental material). Based on a cutoff level of at least a twofold difference between the wild type and the Nra mutant, the highest number of differentially transcribed genes was found in the transition phase of growth (for details concerning the analysis of array data, see reference 6). At this time point, a total of 112 genes exhibited increased transcript abundance, suggestive of negative Nra control, whereas 84 genes were apparently directly or indirectly under positive Nra control in the wild type. During the exponential growth phase 67 and 53 genes displayed augmented transcript abundance and decreased transcript abundance, respectively, whereas in the stationary phase of growth the corresponding numbers were 84 and 29 genes, respectively.
The transcript amounts of 18 and 8 genes were consistently found to be increased and decreased, respectively, in all three growth phases investigated (see Table S4 in the supplemental material). Among the 18 genes that were obviously transcriptionally repressed in the wild type are the Nra gene itself and all the genes encoded in the Cpa pilus operon located adjacent to Nra in the serotype M49 FCT region. Additionally, the capsule synthesis operon (has) and all virulence factor-encoding genes in the Mga core region are apparently under negative Nra control (see Table S4 in the supplemental material). However, as the central regulator of the virulence genes in the Mga region, Mga alone was found to be repressed by Nra in the exponential phase of growth.
Several other important virulence factor- and regulator-encoding genes were exclusively affected during one or two growth phases. Of these, virulence genes encoding the fibronectin-binding adhesin protein F2, the factors of the CMT machinery (28, 32, 46), a putative hemolysin (HylX), two hyaluronidases (HylP2 and HlyP3), streptococcal pyrogenic exotoxin A (SpeA), a putative extracellular matrix-binding protein (Epf), the Mac1/IdeS protease, streptokinase (Ska), and the laminin-binding protein (Lmb) exhibited increased transcript abundance in the Nra mutant (see Tables S1 to S4 in the supplemental material). Judging from the augmented transcript amounts, Nra is also a direct or indirect negative transcriptional regulator for two additional Ralp family transcriptional regulators, Ralp4 (SPy0216) and Ralp3 (SPy0735), the global regulator Rgg, and at least one of the TCS, Ihk/Irr.
It is also apparent from the transcriptome data that a high number of almost exclusively virulence genes were under negative Nra control, whereas only a few virulence genes were found to be under positive regulatory control. A disproportionately high number of positively controlled genes were genes encoding proteins required for metabolic functions and metabolite transport, genes encoding proteins with unknown functions, and many genes encoding phage-related proteins (see Tables S1 to S4 in the supplemental material). Particularly in the transition phase of growth, many genes encoding metabolic enzymes, proteases, ABC transporters, chaperones, heat shock proteins, carboxylases, peptidases, and proton-translocating ATPases belong to the Nra regulon. For example, the complete maltose/maltodextrin binding, translocation, and utilization complex is apparently induced if Nra is expressed (see Table S2 in the supplemental material).
Transition phase Nra secretome and surfome.
In order to determine if the observed transcriptional changes lead to concomitant differential protein expression, a comparative proteome analysis of surface-associated (surfome) and secreted (secretome) proteins from the wild type and Nra mutant during the transition growth phase was performed. From a summary of the proteome analysis (see Table S5 in the supplemental material), it is apparent that a total of 28 differentially expressed proteins were found to be under negative control, whereas Nra apparently acts as positive regulator for 39 proteins. An overrepresented portion of proteins which either were expressed exclusively in the Nra mutant (spots missing in the wild-type gels) or exhibited strong up-regulation in the Nra mutant are proteins in the virulence factor category. This result matches the results of the transcriptional analysis. Nearly all Mga regulon-encoded proteins, including Emm, C5a-peptidase, SclA, Lsp, and SfbX, showed higher expression levels in the Nra mutant background. The reason why the Sof49 gene, which is transcribed with the gene encoding SfbX in a bicistronic operon, was not found to be differentially expressed could not be determined in this study.
Also, the expression of proteins constituting the CMT system (NAD glycohydroloase and streptolysin O) is under negative control of Nra. Moreover, the expression levels of enolase (Eno) and an extracellular matrix-binding protein (Epf) (see transcriptome data and below) were found to be higher in the Nra mutant background. With the exception of streptokinase, SclB, maltose-binding protein, pullulanase, and SofM49, all the down-regulated proteins in the Nra mutant belong to metabolism and housekeeping categories, suggesting that Nra is a positive regulator for expression of the genes encoding these proteins.
Not unexpectedly, no correlation of transcript abundance and protein amount was noted for several proteins. These discrepancies could be explained by changes in mRNA stability and other posttranscriptional and posttranslational regulation processes. These effects cannot be measured by transcriptome and proteome methods, which shows the limitations of these global analyses.
Of note, many surface-localized proteins were also found in the secreted compartment, suggesting that there is turnover and/or cleavage from the surface during the transition growth phase. The cysteine protease SpeB, which was also expressed at higher levels in the Nra mutant background, is a putative enzyme with such cleavage activity. Altogether, most of the changes in mRNA abundance due to direct or indirect control of Nra during the transition phase uncovered by transcriptome analysis matched results of proteome investigations.
Ralp3, a GAS transcriptional regulator located in the novel ERES pathogenicity region.
The most interesting aspect of the Nra transcriptome and proteome was the connection of Nra with other Ralp regulators. Our decision to further explore the Nra-Ralp3 connection was based on the following facts: (i) at the time of study setup until the first submission, the Ralp4 (RivR) (45) and Ralp3 (29) regulators had not been investigated at all and (ii) in silico inspection of the Ralp3 region suggested features of pathogenicity islands.
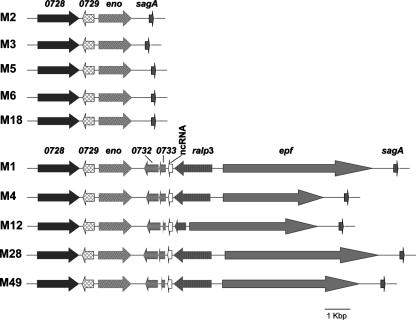
In silico inspection of the publicly available GAS genome sequences for the presence of ralp3 genes revealed that the distribution was serotype dependent (Fig. 1). ralp3 homologous genes were found exclusively in serotypes M1, M4, M12, M28, and M49 and were absent from sequenced serotype M2, M3, M5, M6, and M18 strains (Fig. 1). Ralp3 is apparently a putative transcriptional regulator surrounded by at least three GAS virulence factors. This fact particularly justified closer inspection of this region. Eno (enolase, an enzyme and plasminogen-binding protein) and SagA (streptolysin S, the β-hemolysin of GAS) represent the eno sagA (ES) gene block, which is present in all GAS serotypes. In contrast, Epf (a putative extracellular matrix-binding protein) is encoded adjacent to ralp3 exclusively in a subset of GAS serotype strains. This novel pathogenicity island-like region was termed the ERES region in the selected serotypes in which the RE (ralp3 epf) gene block was found in addition to the ES gene block.
FIG. 1.
Presence and organization of the S. pyogenes ES and ERES gene blocks in GAS strains belonging to various serotypes. The organization of both regions is shown for different S. pyogenes strains whose genomes have been sequenced. The data were extracted from the genome sequences at NCBI. The SPy numbering is the numbering for the serotype M1 genome sequence (15). eno encodes GAS enolase; sagA encodes GAS streptolysin S; ralp3 encodes GAS RofA-like protein 3; epf encodes a novel GAS plasminogen-binding protein; and ncRNA is a putative untranslated small RNA species. The accession number and strain of each serotype used for the comparative analysis are as follows: serotype M1, accession number NC_002737 and strain SF370; serotype M2, NC_008022 and MGAS10270; serotype M3, NC_004606 and SSI-1; serotype M4, NC_008024 and MGAS10750; serotype M5, NC_009332 and Manfredo; serotype M6, NC_006086 and MGAS10394; serotype M12, NC_008023 and MGAS2096; serotype M18, NC_003485 and MGAS8232; serotype M28, NC_007296 and MGAS6180; and serotype M49, AAFV00000000 and 591.
The deduced primary Ralp3 amino acid sequence predicted a typical helix-turn-helix motif in the N-terminal protein domain, and mean levels of amino acid sequence similarity and identity of 52 and 29%, respectively, were calculated for Ralp3 compared to Nra and RofA protein sequences.
The fact that the ERES region contains remnants of two open reading frames (SPy0732 and SPy0733) which display homology to transposase genes could explain the presence or absence of the RE gene block in selected GAS serotype strains (Fig. 1).
Of note, GAS serotype M1, M4, M12, and M28 strains all encode the RofA version of Ralp in their FCT regions; therefore, so far M49 is the only serotype for which Nra has been shown to exert a regulatory influence on ralp3 transcription. Whether RofA is functionally linked with the ERES region in the serotypes listed above needs to be determined.
In order to extend the in silico analysis, the distribution of the ralp3, epf, and eno genes was investigated further by Southern blot hybridization and site-specific PCR using strains that are considered laboratory strains. Whereas eno was detected in each of the GAS laboratory strains tested (serotypes M1, M2, M3, M4, M5, M6, M12, M18, M28, and M49), ralp3 and epf were found only in GAS serotypes M1, M4, M12, M28, and M49 (data not shown). To further extend the investigation to clinical isolates and to test if the presence or absence of the genes is strain specific, we examined additional GAS clinical isolates. In serotype M2, M3, and M6 clinical isolates no epf and ralp3 genes could be detected (see Table S6 in the supplemental material). In contrast, both genes were detected in several serotype M1, M4, M12, and M28 clinical isolates, suggesting that the presence of both genes is generally linked with certain serotypes and is not a strain-specific feature (see Table S6 in the supplemental material).
Moreover, the eno, ralp3, and epf gene probes did not hybridize with chromosomal DNA from one strain each of S. intermedius, S. gordonii, S. anginosus, S. mitis, S. oralis, S. sanguis, S. parasanguis, S. equi, S. mutans, S. agalactiae, S. pneumoniae, S. aureus, S. epidermidis, S. lugdunensis, Enterococcus faecalis, and Enterococcus faecium (data not shown). These results were confirmed by PCR (data not shown).
When S. suis EF (strain D282 [52]) and EF* (strains 2840 and 1890 [52]) reference isolates were tested for presence of the three genes, we detected hybridizing bands for eno and epf gene probes only under low-stringency conditions. The presence of these genes was also confirmed by PCR (data not shown). A ralp3 gene was not detected in the S. suis isolates investigated by using Southern blot experiments or PCR (data not shown).
Together, the findings demonstrated that the complete ERES region is obviously GAS specific and that the RE gene block was not horizontally acquired from the other gram-positive species mentioned above.
Transcription of ralp3 and influence of ralp3 on FCT region and ERES region genes.
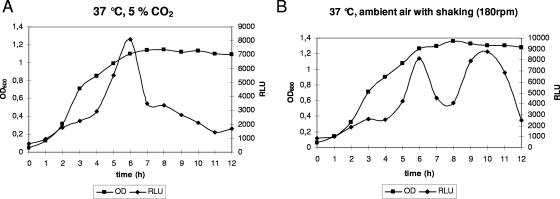
In advance of a functional characterization of Ralp3, we investigated the ralp3 transcriptional profile in the different GAS growth phases in liquid medium. Northern blot analysis indicated that ralp3 was transcribed as a monocistronic message (data not shown). This finding allowed genetic manipulation of the Ralp3-encoding gene without obvious polar effects. First, we constructed a luciferase reporter gene recombinant in the wild-type background, which carried the luc gene fused to the putative ralp3 promoter region. As shown in Fig. 2A, transcription of the Ralp3-encoding gene increased in a linear fashion during the first few hours of exponential growth. In late exponential phase the transcription rate suddenly exceeded the multiplication rate of the bacteria, leading to a sharp peak of activity at the transition phase of growth. Throughout the stationary phase, ralp3 transcription gradually decreased if bacteria were grown under a 5% CO2-20% O2 atmosphere. When bacteria were grown in a shaking culture in ambient air, a second prominent ralp3 transcription peak was noted in the stationary growth phase (Fig. 2B).
FIG. 2.
Temporal transcriptional activity of ralp3. Growth phase-dependent luciferase activity was measured in the ralp3 luc strain. The strain was grown at 37°C in THY medium under a 5% CO2-20% O2 atmosphere (A) or with ambient air with shaking (180 rpm) (B). Growth was monitored over time by measuring the culture OD600. Luciferase activity is expressed in relative light units (RLU). The results of one representative experiment out of five independent experiments are shown.
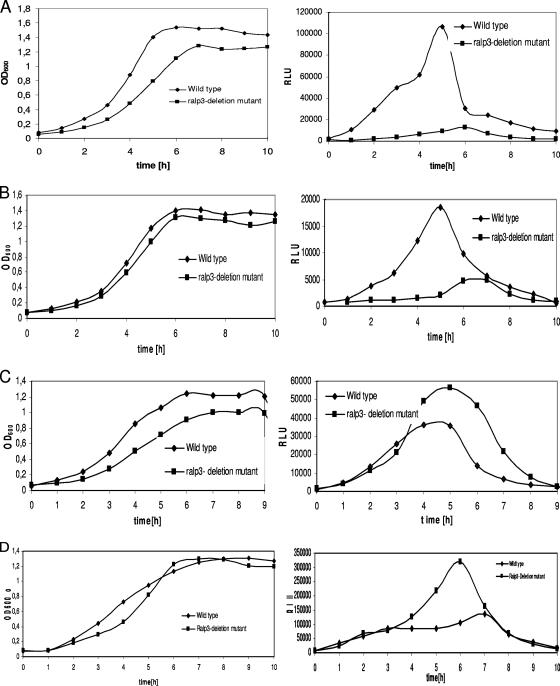
For further transcriptional and functional analysis we next constructed a Ralp3 deletion mutant in which an aphA3 resistance cassette replaced the ralp3 gene. When the ralp3 gene mutant was tested to determine its growth kinetics profile, it consistently displayed a slightly decreased multiplication rate and final optical density under all atmospheric conditions tested (Fig. 3 and 4). The reason for this phenotype could not be determined in this study.
FIG. 3.
Temporal transcriptional activity of FCT region genes in the wild-type and Ralp3 mutant backgrounds. Growth phase-dependent luciferase activity was measured in THY medium under a 5% CO2-20% O2 atmosphere at 37°C. Growth was monitored over time by measuring the culture OD600 (left panels). Luciferase activity is expressed in relative light units (RLU) (right panels). The results of one representative experiment out of five independent experiments are shown. (A) Activity of nra (transcriptional profile of nra-luc); (B) activity of cpa (transcriptional profile of cpa-luc); (C) activity of msmR (transcriptional profile of msmR-luc); (D) activity of prtF2 (transcriptional profile of prtF2-luc).
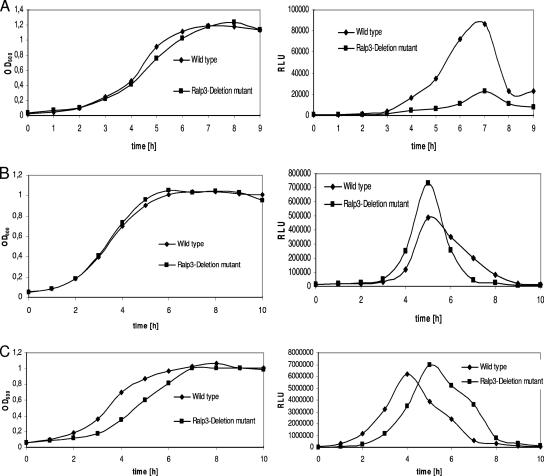
FIG. 4.
Temporal transcriptional activity of ERES region genes in the wild-type and Ralp3 mutant backgrounds. Growth phase-dependent luciferase activity was measured in THY medium under a 5% CO2-20% O2 atmosphere at 37°C. Growth was monitored over time by measuring the culture OD600 (left panels). Luciferase activity is expressed in relative light units (RLU) (right panels). The results of one representative experiment out of five independent experiments are shown. (A) Activity of epf (transcriptional profile of epf-luc); (B) activity of sagA (transcriptional profile of sagA-luc); (C) activity of eno (transcriptional profile of eno-luc).
Next, using the genetic background of the Ralp3 mutant, we introduced luciferase reporter gene fusions of the FCT region genes nra, msmR, prtF2, and cpa in order to investigate the potential reverse influence of Ralp3 on the FCT region genes. As shown in Fig. 3A, transcription of the Nra-encoding gene was repressed throughout the growth curve, suggesting that Ralp3 had a positive direct or indirect effect on Nra transcription and giving the first indication of a bidirectional regulatory axis. Additionally, transcription of the gene encoding the collagen-binding adhesin Cpa was found to be repressed in the Ralp3 mutant background (Fig. 3B). In contrast, the corresponding reporter gene Ralp3 mutants showed an increased rate of transcription of the genes encoding the FCT region regulator MsmR (Fig. 3C) and the fibronectin-binding MSCRAMM protein F2 (Fig. 3D), especially in the transition and stationary growth phases. The measurements did not allow discrimination between a direct Ralp3 effect on the three genes and an indirect effect via the Ralp3-Nra regulatory axis.
To investigate the putative Ralp3 transcriptional influence on the adjacent ERES region genes, eno, epf, and sagA luciferase fusions were introduced into the Ralp3 mutant background. As shown in Fig. 4A, Ralp3 had a positive transcriptional influence on epf transcription, since notably reduced epf transcription in the Ralp3 mutant background was detectable. In contrast, transcription of sagA encoding streptolysin S was found to be increased in the Ralp3 mutant background, indicative of direct or indirect negative Ralp3 control of this virulence gene (Fig. 4B). No significant changes in eno transcription were noted in the Ralp3 mutant (Fig. 4C), although the eno-luc fusion mutant consistently showed delayed growth, resulting in delayed peak transcription of eno. It was also apparent that there were transcription maxima for eno, sagA, and epf in the mid-exponential, transition, and early stationary growth phases, respectively (Fig. 4A to C). In addition to elucidation of Ralp3 dependence, these profiles indicated the specific times during growth at which the virulence factors are potentially important.
Phenotypic consequences of Ralp3 activity.
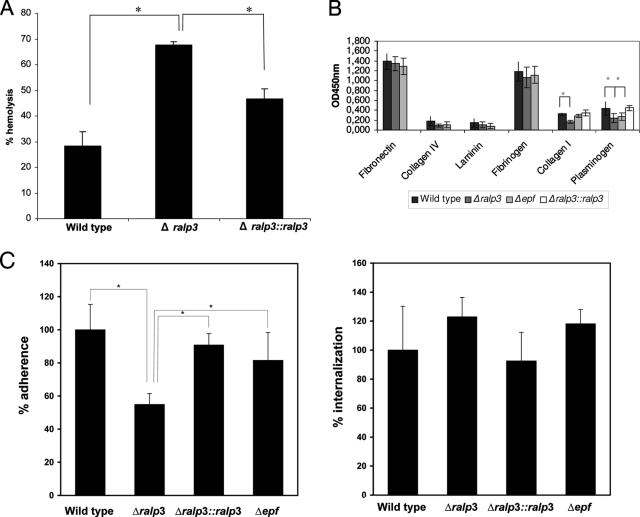
The next set of experiments was performed to investigate the link between transcriptional changes and measurable consequences for the phenotype. As reported above, Ralp3-dependent increased transcription of the streptolysin S-encoding gene sagA was noted (Fig. 4A). This hemolysin is very important for the GAS-host cell interaction, particular for GAS cytotoxicity (22, 35, 37). Thus, the activity of secreted streptolysin S was measured in wild-type, mutant, and complemented strains. A significant increase in streptolysin S activity was found in stationary-phase supernatants from the Ralp3 mutant (P < 0.05) (Fig. 5A), and almost normal wild-type levels were reached if the ralp3 deletion was complemented in trans. This illustrates that the transcriptional change was in fact followed by phenotypic changes.
FIG. 5.
Phenotypic characterization of wild-type and Ralp3 and Epf mutant strains. (A) Streptolysin S hemolytic activity of the wild type, the ralp3 deletion mutant, and the complemented strain. Culture supernatants recovered from stationary phase were analyzed for the ability to lyse sheep erythrocytes. Cholesterol was used as an inhibitor of streptolysin O activity. The data are the means and standard deviations of three independent assays. Asterisk, P < 0.05. (B) Binding of the wild type, the ralp3 deletion mutant, the complemented strain, and the epf insertion mutant to immobilized human matrix proteins, as determined by an adherence enzyme-linked immunosorbent assay. Microtiter plates were coated with the indicated matrix proteins and subsequently incubated with the GAS strains. Binding was detected with anti-GAS polyclonal antibody and is expressed as the OD450. The data are the means ± standard deviations of three independent experiments. Asterisk, P < 0.01. (C) Adhesion to (left panel) and internalization in (right panel) human epithelial cells. HEp-2 cells were infected with GAS strains at a multiplicity of infection of 1:10. The data are the means and standard deviations from three independent experiments. The value for the wild type was defined as 100% for each independent test. Asterisk, significant difference (P < 0.01).
Adherence to host extracellular matrix proteins and adherence to and internalization in host epithelial cells are crucial steps in the GAS infectious process. Comparison of the matrix protein adherence of the wild type and Ralp3 mutant revealed no statistically significant differences in adherence behavior toward fibronectin, fibrinogen, laminin, and collagen IV; however, deletion of the Ralp3 transcriptional regulator significantly decreased GAS adherence to collagen I and plasminogen (Fig. 5B). The wild-type phenotype for adherence to collagen I and plasminogen could be restored by complementation experiments (Fig. 5B). Since fibronectin is the major bridging molecule that allows GAS to adhere to and internalize in host epithelial cells (10) and since adherence to immobilized fibronectin remained the same in the mutant, we did not expect to see major differences in HEp-2 cell adherence or internalization. In contrast to this assumption, adherence of the Ralp3 mutant to HEp-2 cells was significantly reduced by up to 45%. This phenotype could be complemented by ralp3 expression in trans in the Ralp3 mutant background (Fig. 5C). The Ralp3 mutation and complementation had no significant effect on GAS HEp-2 cell internalization (Fig. 5C).
We have recently shown that several GAS serotype strains are able to form biofilms in static conditions and under flow conditions (30). However, mutation of either Nra or Ralp3 did not increase the low biofilm-forming capacity of the serotype M49 strain tested (data not shown).
Functional characterization of Epf.
Mutation of Ralp3 has multiple consequences for important GAS pathogenicity mechanisms. Next, we tried to identify some of the corresponding Ralp3-controlled GAS genes that lead to the observed changes. A potential candidate gene with an unknown function is epf. The deduced serotype M49 Epf amino acid sequence shares 13% overall identity with a protein from S. suis which was described as a putative extracellular protein factor (52). No specific function was assigned to this protein; however, it could be used as a marker for S. suis colonization or infections (50). The serotype M49 epf sequence (NCBI accession no. NZ_AAFV00000000 [6]) comprises 5,637 bp corresponding to a protein with 1,879 amino acid residues (accession no. ZP_00366506) and a deduced molecular mass of 207 kDa. In silico comparisons of Epf sequences from serotype M1, M4, M12, M28, and M49 strains revealed significant size differences for the deduced mature proteins (see Table S7 in the supplemental material). All Epf proteins from these serotype strains share classical signal sequences at the N terminus (amino acids 1 to 34) and LPXTG-like sortase anchoring motifs at the C terminus (for serotype M49 Epf, amino acids 1837 to 1876, Pfam accession no. PF00746), suggesting that they are all surface-located proteins (see Fig. S1 in the supplemental material). Using ClustalW analysis for Epf sequence comparison, the levels of identity between the Epf sequences from five GAS serotype strains ranged from 42 to 84% (http://xylian.igh.cnrs.fr) (see Table S7 in the supplemental material). The S. suis Epf protein is only distantly related to the corresponding proteins from GAS, as indicated by only 12 to 19% identical amino acid residues (see Table S7 in the supplemental material).
Profile searches (http://smart.embl-heidelberg.de/) based on the serotype M49 Epf amino acid sequence revealed several partially or wholly unstructured short peptide motifs (regions with intrinsic disorder) (http://dis.embl.de/) located at the N terminus. The complete central part of the GAS serotype M49 Epf amino acid sequence (amino acids 387 to 1841) is composed of 16 repeat units (see Fig. S1 in the supplemental material). These repeats represent a domain with an unknown function (DUF1542) shared by many surface proteins from gram-positive species. Varying repeat numbers for the different Epf proteins from GAS and S. suis contribute to the deduced size differences (see Fig. S1 in the supplemental material). It should be noted that the level of identity of the Epf sequence from GAS serotype M49 with the S. suis Epf sequence is much higher if single DUF repeats are compared, reaching values of up to 22% identical amino acids. On average, the level of identity of DUF repeats within the GAS M49 Epf sequence is 27% (data not shown).
In order to assign a function to the Epf protein from GAS serotype M49, we expressed this protein in its mature form (excluding the signal sequence and membrane anchor sequences) as a His-tagged fusion protein in E. coli, which allowed us to use the purified protein for functional studies (see Fig. S2A in the supplemental material). Additionally, we generated an Epf insertion mutant strain in the serotype M49 GAS background. Since epf is a monocistronic transcript and the downstream sag operon is transcribed from its own promoter (37), we did not detect any downstream effect on sagA transcription (by Northern blotting or real-time RT-PCR) (data not shown).
The recombinant purified Epf protein was able to bind immobilized plasminogen, whereas no binding of this protein with fibronectin, fibrinogen, laminin, and collagen was detected (see Fig. S2B in the supplemental material). In line with these results, the Epf insertion mutant was significantly attenuated in the capacity to adhere to immobilized plasminogen, suggesting that Epf could act as plasminogen-binding protein (Fig. 5B). Moreover, the Epf insertion mutant was not attenuated in the capacity to adhere to and internalize in HEp-2 epithelial cells (Fig. 5C), suggesting that plasminogen binding is not crucial for serotype M49 GAS host cell adherence and internalization mechanisms.
DISCUSSION
Expression of virulence factor subsets and their associated pathogenesis mechanisms are under control of regulatory networks in GAS (26). Nra, designated a global negative transcriptional regulator of the Ralp family and acting together in a growth phase-dependent network with Mga (Fig. 6), was examined in this study. We used DNA microarray analysis with wild-type and Nra mutant strains to investigate the entire Nra transcriptome in a serotype M49 GAS strain. To further extend data collection from transcription to the protein level, we performed a surface-associated and secreted proteome analysis with material from the transition phase of the growth curve, a time of maximum Nra transcription.
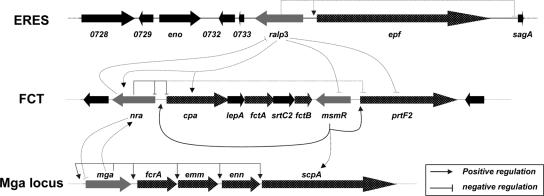
FIG. 6.
Schematic drawing of the ERES-FCT-Mga regulatory network in a GAS serotype M49 strain: transcriptional control of the ERES, FCT, and Mga locus genes in a serotype M49 strain. The dotted lines indicate either potentially direct or indirect regulation. The numbers below the arrows are SPy tag numbers for serotype M1 strain SF370. Gray arrows, genes encoding transcriptional regulators, including ralp3, nra, msmR, and mga; dotted arrows, genes encoding surface-displayed proteins; black arrows, other genes.
The transcriptome data revealed that Nra is active in all growth phases investigated in this study, with the highest number of controlled genes in the transition phase. Almost equal numbers of genes were found to be negatively and positively controlled by Nra, although Nra was designated a global negative regulator based on results from our previous studies (33, 43). A similar unexpected observation was made by Ribardo and McIver (44), who defined the global Mga regulon from several GAS serotypes. Next to the exponential-growth-phase core Mga regulon under positive Mga control, these authors found an equal number of genes under negative Mga control. Among these Mga-repressed genes were the genes encoding mannose, glucose, and maltose transporters, as well as catabolite regulatory proteins (44).
Also, in our transcriptome study in all investigated growth phases many genes belonging to the metabolism and stress response categories were found to have altered transcript abundance in the Nra mutant background. Among these genes, genes in the maltose/maltodextrin transport and utilization cluster were found to have decreased transcript abundance in the transition phase, suggesting that Nra is a direct or indirect positive regulator. In particular, maltodextrin utilization was recently shown to play a key role in GAS colonization of the oropharynx (49).
The transcriptome and proteome data provided here support the results of earlier investigations of the Mga-Nra growth phase-dependent regulatory network (26). To be precise, Nra could be the regulatory element that is activated by Mga in the exponential phase (44). The new discovery is that Mga and Nra, specifically during the most active growth phases (exponential and transition phases of growth, respectively), not only control virulence factor expression but also have a strong influence on metabolic capacities of GAS. At least for Mga, a link between carbohydrate metabolism and pharyngeal infection in cynomolgus macaques was recently explored (55). The exact role of Nra activity in vivo needs to be determined.
It is noteworthy that Nra activity repressed the transcript abundance of two important GAS regulatory systems, the transcriptional regulator Rgg and the TCS Ihk/Irr. Rgg is required for growth phase-dependent expression of many virulence factors and metabolic enzymes (9, 13), and Ihk/Irr is crucial for GAS evasion of the innate immune defense of the host (56). The presence of these important regulators in the Nra regulon suggests that many observed changes in the Nra transcriptome could be indirect effects; however, this needs to be determined in future studies.
Comparison of the differential transition phase transcriptome with corresponding data from secreted and surface-associated proteomes revealed a good correlation, suggesting that most changes in protein appearance are regulated on the transcriptional level. However, for several virulence factors no match between transcript and protein level was found. This indicates that there are active posttranscriptional and/or posttranslational regulation mechanisms and hints at the limitation of global transcriptome and proteome studies.
The expression of a high number of surface-associated and secreted virulence factors is repressed by Nra; these factors include almost all Mga core regulon virulence factors, like SfbX, C5A peptidase, Lsp, Emm, SpeA, and SclA, as well as the proteins constituting the CMT system (Slo and NAD glycohydrolase [32, 46]) and several other surface adhesins (protein F2 [28]) and GAS pilus proteins (Cpa [27]) (Fig. 6). This transition phase-specific Nra-promoted phenotype supports our earlier observation that Nra activity facilitates GAS persistence by significantly reducing surface protein expression, thereby preventing immune system activation and reducing host cell injury, which promotes intracellular survival (33). At the same time Nra induces expression of metabolic enzymes, phosphotransferase systems, and several stress response-associated proteins. This activity could support metabolic homeostasis and survival in the intracellular niche. In contrast, Mga activity in the exponential phase promotes bacterial adhesion and host immune system disturbance (25, 44). This exponential phase-transition phase Mga-Nra interplay is apparently fine-tuned by another transition phase regulator, MsmR, which counterbalances Nra activity upon detection of unknown signals (35) (Fig. 6).
A new piece of the GAS regulatory network was added by our discovery of another serotype-specific pathogenicity gene block, the RE gene block, which is connected with Nra. During review of this work, Kwinn and colleagues (29) described the RE gene block in a serotype M1T1 GAS strain. These authors designated Epf LSA (large surface-anchored protein). In the serotype M1T1 background, Ralp3 acted as a direct or indirect transcriptional repressor for capsule synthesis genes and genes encoding the cysteine protease SpeB (29). LSA was found to be negatively regulated by Ralp3. The work of Kwinn and colleagues (29) and our results presented here revealed exclusive presence of the RE gene block in selected GAS serotype strains integrated into the middle of a gene block encoding two GAS virulence factors (ES gene block). The presence of the RE gene block is apparently not a strain-specific trait but rather is linked to certain serotypes, as shown by investigation of clinical isolates and multiple strains of the same serotype (Fig. 1; see Table S6 in the supplemental material). However, the presence of the complete ERES gene block could not be linked to the source of isolation.
The complete assembled ERES gene block fulfills many pathogenicity island-specific criteria (14) and could therefore represent the first confirmed GAS pathogenicity island. It is absent in several serotypes, and it encodes remnants of transposases, indicative of previous mobilization. Whether the RE gene block was lost in ES gene block-containing GAS serotypes or was selectively acquired by specific GAS serotypes needs to be investigated. Of note, the RE gene block tested in all serotypes where it is present had a G+C content of 45.5% ± 4% (average ± standard deviation). This is significantly higher than the G+C content of the regular GAS chromosome. However, from which species the specific RE region was horizontally transferred could not be determined in this study.
The complete ERES region contains two core genes, one encoding another Ralp regulator and one coding for a putative extracellular matrix-binding protein (Epf or LSA [29]). Like Nra and in contrast to its activity in the serotype M1T1 GAS strain (29), the Ralp regulator (designated Ralp3 or SPy0735) in the serotype M49 GAS strain investigated here exhibited maximum transcription in the transition phase of growth and is under direct or indirect negative Nra control. Using Ralp3 mutants and reporter gene fusions, we demonstrated a bidirectional regulatory axis for the FCT and ERES regions (Fig. 6). Ralp3 has a direct or indirect positive influence on Nra and Cpa gene transcription. Although in theory active Ralp3 increased nra transcription, which in turn could suppress cpa transcription, we found increased abundance of cpa mRNA in the wild type. This implies that Ralp3 most likely directly acts on cpa transcription, thereby bypassing and/or overruling the Nra effect. However, Ralp3 acted as a negative regulator for MsmR and protein F2.
The two RE region core genes are surrounded by genes encoding the virulence factors enolase (plasminogen binding [39]) and streptolysin S (β-hemolysis [37]). These two genes occur in all serotypes, and the gene encoding streptolysin S is under negative Ralp3 control. Taken together, the complete region was therefore designated ERES.
The high-molecular-weight putative extracellular matrix-binding protein Epf exhibited low homology to an extracellular protein factor from S. suis (EF) (52) which is potentially secreted and so far has no designated specific function in this species yet has been used as a suitable tool for S. suis typing (50). In GAS, Epf has a typical membrane anchor, and increased amounts were found in the Nra mutant surface-associated proteome. Thus, Epf is one of the virulence factors identified in this study with increased expression that is discordant with the transcriptional analysis in the Nra mutant background. The reason for this needs to be elucidated in future studies.
Using rEpf protein and a corresponding Epf mutant, we showed that Epf is a novel GAS plasminogen-binding protein. GAS plasminogen binding is important in invasive disease progression (57). So far, three distinct direct plasminogen-binding proteins have been described in GAS: plasminogen binding group A streptococcal M-like protein/M53, a GAS M protein (4); streptococcal surface enolase, a GAS surface enolase (39); and the moonlighting protein glyceraldehyde-3-phosphate dehydrogenase (also called Plr and streptococcal surface dehydrogenase) (57). The general importance of plasminogen binding and in particular the serotype-specific presence of the novel Epf plasminogen-binding protein justify a more extensive future study of this protein. For the serotype M1T1 Epf homologue LSA no matrix protein-binding activity was reported (29); however, this protein was found to be important in a murine model of systemic infection (29).
In summary, in the transition growth phase the ERES region was proven to add another level of regulation to the existing Mga-Nra/MsmR regulatory network in the serotype M49 background (Fig. 6) and could have similar effects in other ERES region-positive serotypes. In a serotype M49 GAS strain, the Ralp3 regulator included in this region counterbalances the FCT region Nra and MsmR regulatory activities. One potential signal for the ERES region Ralp3 activity could be oxidative stress in the advanced stationary phase, since Ralp3 has another transcription maximum at this time point during growth when GAS is exposed to high oxygen levels.
The potential selective advantage that the ERES region confers to a subset of GAS serotypes under in vivo conditions once the bacteria reach the transition phase is presently a point of speculation. Clearly, during exponential phase Mga is the regulator in charge of GAS, circumventing damage from an active innate and adoptive immune system. During the transition phase, Nra is switched on by Mga and is in charge of bacterial adherence and internalization and the host cell persistence phenotype, with some counterbalance from the other FCT region regulator MsmR. In selected serotypes, Ralp3 could be in charge of dissemination by counteracting Nra and MsmR activity and by promoting plasminogen binding. Determination of the precise mechanisms for these effects and correlation of the molecular epidemiology of the ERES region with a defined clinical behavior of this subset of GAS strains are promising goals for future studies.
Supplementary Material
Acknowledgments
We thank Jana Normann and Nikolai Siemens for expert technical assistance. We thank Peter Valentin-Weigand for providing S. suis strains used in this study.
This work was supported by grants Po 391/12-1 and Kr1765/2-1 from the DFG and by BMBF grant BE031-03U213B as a part of the German PathoGenomik Competence Network awarded to A.P. and B.K. B.K. further acknowledges funding by the BMBF “ERANet Pathogenomics” program.
Editor: A. Camilli
Footnotes
Published ahead of print on 24 September 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abbot, E. L., W. D. Smith, G. P. S. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 9:1822-1833. [DOI] [PubMed] [Google Scholar]
- 2.Baev, D., R. England, and H. K. Kuramitsu. 1999. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect. Immun. 67:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckert, S., B. Kreikemeyer, and A. Podbielski. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect. Immun. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge, A., and U. Sjobring. 1993. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem. 268:25417-25424. [PubMed] [Google Scholar]
- 5.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyer-Sehlmeyer, G., B. Kreikemeyer, A. Hoerster, and A. Podbielski. 2005. Analysis of the growth phase-associated transcriptome of Streptococcus pyogenes. Int. J. Med. Microbiol. 295:161-177. [DOI] [PubMed] [Google Scholar]
- 7.Bisno, A. L., M. O. Brito, and C. M. Collins. 2003. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 3:191-200. [DOI] [PubMed] [Google Scholar]
- 8.Carapetis, J. R., A. C. Steer, E. K. Mulholland, and M. Weber. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685-694. [DOI] [PubMed] [Google Scholar]
- 9.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courtney, H. S., D. L. Hasty, and J. B. Dale. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann. Med. 34:77-87. [DOI] [PubMed] [Google Scholar]
- 11.Courtney, H. S., and A. Podbielski. 2004. Group A streptococcal invasion of host cells, p. 239-274. In R. J. Lamont (ed.), Bacterial invasion of host cell. Cambridge University Press, Cambridge, United Kingdom.
- 12.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dmitriev, A. V., E. J. McDowell, K. V. Kappeler, M. A. Chaussee, L. D. Rieck, et al. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J. Bacteriol. 188:7230-7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 15.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, et al. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogg, G. C., and M. G. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogg, G. C., C. M. Gibson, and M. G. Caparon. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an m gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 11:671-684. [DOI] [PubMed] [Google Scholar]
- 18.Görg, A., W. Weiss, and M. J. Dunn. 2004. Current two-dimensional electrophoresis technology for proteomics. Proteomics 4:3665-3685. [DOI] [PubMed] [Google Scholar]
- 19.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henzel, W. J., T. M. Billeci, J. T. Stults, S. C. Wong, C. Grimley, et al. 1993. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc. Natl. Acad. Sci. USA 90:5011-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes, W. 2004. Virulence factors of the group A streptococci and genes that regulate their expression. Front. Biosci. 9:3399-3433. [DOI] [PubMed] [Google Scholar]
- 22.Klenk, M., D. Koczan, R. Guthke, M. Nakata, H. J. Thiesen, et al. 2005. Global epithelial cell transcriptional responses reveal Streptococcus pyogenes Fas regulator activity association with bacterial aggressiveness. Cell. Microbiol. 7:1237-1250. [DOI] [PubMed] [Google Scholar]
- 23.Köller, T., D. Nelson, M. Nakata, M. Kreutzer, V. A. Fischetti, M. O. Glocker, A. Podbielski, and B. Kreikemeyer. PlyC, a novel bacteriophage lysin for compartment dependent proteomics of group A streptococci. Proteomics, in press. [DOI] [PubMed]
- 24.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 25.Kreikemeyer, B., M. Klenk, and A. Podbielski. 2004. The intracellular status of Streptococcus pyogenes: role of extracellular matrix-binding proteins and their regulation. Int. J. Med. Microbiol. 294:177-188. [DOI] [PubMed] [Google Scholar]
- 26.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 27.Kreikemeyer, B., M. Nakata, S. Oehmcke, C. Gschwendtner, J. Normann, et al. 2005. Streptococcus pyogenes collagen type I-binding Cpa surface protein: expression profile, binding characteristics, biological functions, and potential clinical impact. J. Biol. Chem. 280:33228-33239. [DOI] [PubMed] [Google Scholar]
- 28.Kreikemeyer, B., S. Oehmcke, M. Nakata, R. Hoffrogge, and A. Podbielski. 2004. Streptococcus pyogenes fibronectin-binding protein F2: expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 279:15850-15859. [DOI] [PubMed] [Google Scholar]
- 29.Kwinn, L. A., A. Khosravi, R. K. Aziz, A. M. Timmer, K. S. Doran, M. Kotb, and V. Nizet. 2007. Genetic characterization and virulence role of the RALP3/LSA locus upstream of the streptolysin S operon in invasive M1T1 group A streptococci. J. Bacteriol. 189:1322-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lembke, C., A. Podbielski, C. Hidalgo-Grass, L. Jonas, E. Hanski, et al. 2006. Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl. Environ. Microbiol. 72:2864-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lizano, S., F. Luo, and D. E. Bessen. 2007. Role of streptococcal T-antigens in superficial skin infection. J. Bacteriol. 189:1426-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madden, J. C., N. Ruiz, and M. G. Caparon. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell 104:143-152. [DOI] [PubMed] [Google Scholar]
- 33.Molinari, G., M. Rohde, S. R. Talay, G. S. Chhatwal, S. Beckert, et al. 2001. The role played by the group A streptococcal negative regulator Nra on bacterial interactions with epithelial cells. Mol. Microbiol. 40:99-114. [DOI] [PubMed] [Google Scholar]
- 34.Musser, J. M., and F. R. DeLeo. 2005. Toward a genome-wide systems biology analysis of host-pathogen interactions in group A Streptococcus. Am. J. Pathol. 167:1461-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakata, M., A. Podbielski, and B. Kreikemeyer. 2005. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol. Microbiol. 57:786-803. [DOI] [PubMed] [Google Scholar]
- 36.Nelson, D., R. Schuch, P. Chahales, S. Zhu, and V. A. Fischetti. 2006. PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. USA 103:10765-10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nizet, V. 2002. Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 10:575-580. [DOI] [PubMed] [Google Scholar]
- 38.Ozeri, V., I. Rosenshine, D. F. Mosher, R. Fässler, and E. Hanski. 1998. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol. 30:625-637. [DOI] [PubMed] [Google Scholar]
- 39.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 40.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 41.Podbielski, A. 1993. Three different types of organization of the vir regulon in group A streptococci. Mol. Gen. Genet. 237:287-300. [DOI] [PubMed] [Google Scholar]
- 42.Podbielski, A., A. Flosdorff, and J. Weber-Heynemann. 1995. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect. Immun. 63:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 44.Ribardo, D. A., and K. S. McIver. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 62:491-508. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, S. A., G. G. Churchward, and J. R. Scott. 2007. Unraveling the regulatory network in Streptococcus pyogenes: the global response regulator CovR represses rivR directly. J. Bacteriol. 189:1459-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosch, J. W., and M. G. Caparon. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol. Microbiol. 58:959-968. [DOI] [PubMed] [Google Scholar]
- 47.Scott, J. R., and D. Zähner. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62:320-330. [DOI] [PubMed] [Google Scholar]
- 48.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shelburne, S. A., III, P. Sumby, I. Sitkiewicz, N. Okorafo, C. Granville, et al. 2006. Maltodextrin utilization plays a key role in the ability of group A streptococcus to colonize the oropharynx. Infect. Immun. 74:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva, L. M., C. G. Baums, T. Rehm, H. J. Wisselink, R. Goethe, et al. 2006. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet. Microbiol. 115:117-127. [DOI] [PubMed] [Google Scholar]
- 51.Sitkiewicz, I., and J. M. Musser. 2006. Expression microarray and mouse virulence analysis of four conserved two-component gene regulatory systems in group A streptococcus. Infect. Immun. 74:1339-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, H. E., F. H. Reek, U. Vecht, A. L. Gielkens, and M. A. Smits. 1993. Repeats in an extracellular protein of weakly pathogenic strains of Streptococcus suis type 2 are absent in pathogenic strains. Infect. Immun. 61:3318-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 54.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 55.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, et al. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voyich, J. M., D. E. Sturdevant, K. R. Braughton, S. D. Kobayashi, B. Lei, K. Virtaneva, et al. 2003. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. USA 100:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker, M. J., J. D. McArthur, F. McKay, and M. Ranson. 2005. Is plasminogen deployed as a Streptococcus pyogenes virulence factor? Trends Microbiol. 13:308-313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.