Abstract
Evaluation of the protective efficacy of recombinant T-cell-reactive proteins of Coccidioides posadasii in a murine model of coccidioidomycosis has led to the discovery of potential vaccines against this respiratory disease. A recombinant proline-rich antigen (rAg2/Pra) has been reported to be a leading vaccine candidate. However, contradictory results exist on the protection afforded by this antigen. Subcutaneous vaccination of either C57BL/6 or BALB/c mice with rAg2/Pra plus adjuvant followed by intraperitoneal challenge with C. posadasii resulted in a significant reduction of the fungal burden at 12 to 14 days postchallenge compared to that in nonvaccinated animals. Use of the same vaccination protocol followed by intranasal (i.n.) challenge of C57BL/6 mice with an equal number of organisms culminated in chronic pulmonary infection or death over a 90-day period. Early studies of Ag2/Pra suggested that it is a component of an immunogenic complex. We reveal in this study that C. posadasii produces a homolog of the reported proline-rich antigen, designated Prp2, which shows 69% protein sequence identity and 86% similarity to Ag2/Pra. Protection against i.n. challenge of C57BL/6 mice was evaluated by vaccination with the single bacterially expressed homolog, rAg2/Pra, or rPrp2 in combination with rAg2/Pra, each in the presence of the same adjuvant. The combined vaccine provided significantly better protection than either of the single recombinant protein vaccines. Results of enzyme-linked immunospot assays of the immunized mice revealed that the two proline-rich homologs contain unique T-cell epitopes. In combination, the recombinant proteins stimulate a more heterogeneous and protective T-cell repertoire than the monovalent vaccines.
Coccidioides posadasii and C. immitis are the etiologic agents of coccidioidomycosis (San Joaquin Valley fever), a human respiratory disease which is typically contracted by inhalation of spores produced by soilborne mold (6). The two species are phenotypically indistinguishable but were separated on the basis of phylogenetic analysis of single-nucleotide polymorphisms, genes, and microsatellites (17, 35). The two species of Coccidioides exhibit epidemiological differences; while C. immitis appears to be limited to the San Joaquin Valley of California, C. posadasii is found throughout the Southwestern United States, Central Mexico, and certain arid regions of Central and South America (8). Coccidioides spp. are formidable pathogens of humans. The course of disease presentation ranges from a primary respiratory infection that often resolves spontaneously to extrapulmonary dissemination, which can be acute, chronic, or progressive and life-threatening (10). Fossil evidence indicates that coccidioidal infections of homeotherms in North America has occurred during at least the past 8,500 years (36), suggesting that Coccidioides has evolved over a relatively long time span as a pathogen of Homo sapiens. Coccidioides is considered to be the most virulent of the primary fungal pathogens of humans (13). For example, infection of laboratory monkeys via the pulmonary route with as few as 10 spores has been shown to result in rapidly progressive disease (5). Coccidioides is an environmental pathogen. Climatic factors influence the seasonality of fungal blooms in the soil (7), which are at least partly responsible for recurrent epidemics of coccidioidomycosis in human populations. Approximately 20 million persons live in counties within West Texas, New Mexico, Arizona, and Southern California which are major regions of endemicity for this respiratory disease. It is estimated that up to 100,000 new cases of human primary coccidioidal infection occur each year in the United States (20), accounting for millions of dollars in health care costs. Vaccination against coccidioidomycosis has been argued to be a cost-effective intervention and a feasible endeavor, since natural infection almost always confers lifelong immunity to reinfection (3, 6, 46).
Dendritic cells are potent antigen-presenting cells (APCs) and play a key role in the activation of naïve T lymphocytes in response to coccidioidal infection (2, 10). Stimulation of CD4+ T cells in a vaccine-induced host response to Coccidioides is pivotal for protection against coccidioidomycosis (6). It has been shown that CD8+ T cells also contribute to protection, particularly under conditions of depletion or absence of CD4+ T cells (16). Our approach to the development of a vaccine against coccidioidomycosis has been to select and evaluate Coccidioides proteins that stimulate a T helper 1 (Th1) immune response pathway (12, 32, 43, 44). Activation of Th1-associated immunity has been shown in clinical evaluations and animal model studies to be essential for host survival and containment of the fungal pathogen (6, 10). Most of the T-cell-reactive proteins of Coccidioides which have been characterized to date have been isolated from the parasitic cell wall. A leading vaccine candidate and the subject of at least 34 published reports during the past 27 years is a cell wall-associated, proline-rich antigen that has been designated both antigen 2 (Ag2) and Ag2/Pra (10). A significant body of evidence in the literature suggests that both the recombinant Ag2/Pra (rAg2/Pra) protein and a genetic vaccine with AG2/PRA elicit a protective CD4+ T-cell-mediated response to coccidioidal infection. However, conflicting data exist in reported studies of Ag2/Pra-vaccinated mice challenged by the intraperitoneal versus the intranasal (i.n.) route. For example, while the genetic vaccine was effective against intraperitoneal challenge, genetically vaccinated mice infected by the natural i.n. route showed no significant reduction of the fungal burden in the lungs compared to nonvaccinated control animals challenged by the same route (24, 25). Early studies of native, cell wall-associated Pra suggested that it is highly glycosylated (14) and structurally heterogenous (9). Although attempts to isolate the native glycoprotein have failed (10), two-dimensional immunoelectrophoresis (2D-IEP) separations of cell wall extracts which contained Ag2/Pra revealed that the precipitin corresponding to this antigen is a complex consisting of two or more serologically related components (9). The latter appear to be of different molecular sizes, since they partially separated in the 2D-IEP gel. Our intent in this study was to both clarify the apparent limited protective efficacy of Ag2/Pra against murine pulmonary infection with Coccidioides and determine whether additional proline-rich, cell wall-associated antigens are produced, comprising an immunogenic molecular complex.
MATERIALS AND METHODS
Fungal growth conditions.
The C. posadasii isolate (C735) used throughout this study was a clinical strain originally obtained from a patient with disseminated coccidioidomycosis. The saprobic and parasitic phases of the fungus were grown under previously described conditions (22). Parasitic-phase cultures, which have been shown to yield cells in near-synchronous stages of development during the first generation of the parasitic cycle (22), were harvested after different periods of incubation (36 to 132 h) after inoculation with arthroconidia.
Vaccination, animal challenge, and evaluation of protection.
Immunoprotection experiments were conducted with C57BL/6 mice (females; 8 weeks old), using the same immunization protocol as that previously described (43). In the initial experiment, two groups of mice (15 per group) were immunized subcutaneously with bacterially expressed rAg2/Pra (1 μg per dose) or adjuvant alone. The selected dosage of the protein vaccine for C57BL/6 mice was determined to provide optimal survival after i.n. challenge with Coccidioides (41). The procedure used to generate the recombinant protein (rAg2/Pra) is described below. The adjuvant employed was unmethylated CpG dinucleotides in a synthetic oligodeoxynucleotide preparation (32). The adjuvant composition and concentration were the same as those reported previously (43). The mice were immunized by the subcutaneous route, with the same amount of antigen plus adjuvant, 2 weeks after the first immunization. The animals were challenged with 50 viable arthroconidia by the i.n. route 4 weeks after the second immunization. The arthroconidia were isolated from 2-week-old plate cultures of the saprobic phase. Mice were scored for survival over a 90-day period postchallenge. Survival differences between groups of i.n. inoculated mice were analyzed for statistical significance by the Kaplan-Meier method as reported previously (43). The survival experiments were repeated three times. In a separate experiment, six groups of mice (nine mice per group) which had been vaccinated with rAg2/Pra plus adjuvant as described above were examined for fungal burdens in the lungs and spleen upon sacrifice at 15, 20, 30, 40, 60, and 90 days postchallenge. Fungal burdens in the lungs and spleens of survivors were determined as previously described (43). The CFU were expressed on a log scale, and the Mann-Whitney U test was used to compare differences in the median CFU values for statistical significance, as reported previously (43). The detection limit of the CFU assay was 10 colonies per organ homogenate (1og10 CFU = 1).
The same vaccination and challenge protocols were used to compare the survival and fungal burdens of mice immunized with rAg2/Pra versus a recombinant protein homolog of this proline-rich antigen, whose identification and isolation are described below. These comparative studies were conducted by immunization of mice (15 per group) with either the single recombinant protein vaccines (1 μg per dose) plus the CpG adjuvant preparation or the combined pair of recombinant proteins (1 μg each per dose) plus the same adjuvant. The immunization and challenge protocols were the same as those described above. Statistical analyses of the survival and fungal burden data were conducted as described above.
Genome database analysis.
Computational analysis of the C. posadasii (C735) genome database (www.tigr.org) was performed by application of the basic local alignment search tool (BLAST) (1) as previously described (12). The translated sequence of the AG2/PRA gene (15, 49) was used to query the translated genome database. A 1.0-kb fragment of a single contig was selected on the basis of its BLASTX match (expect [E] value, <10−30). Sense and antisense primers were synthesized and employed in a PCR with genomic DNA of C. posadasii to generate a 933-bp amplicon. The nucleotide sequences of the sense and antisense primers were 5′-TGGCCGTTGACAATTCTTTG-3′ and 5′-TTTCTGCGCGAGTCTCCTAAG-3′, respectively. The amplification conditions included an initial denaturation step at 95°C for 2 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min. The 933-bp PCR amplicon was ligated into the pCR2.1 TOPO cloning vector (Invitrogen, Carlsbad, CA), and nucleotide sequence analysis of the insert was performed as reported previously (22). The gene homolog of AG2/PRA identified by this procedure is referred to as PRP2.
PRP2 sequence analysis.
The T-COFFEE algorithm (37) and the CLUSTAL W algorithm (45) were used to align the translated gene sequences. Gaps were treated as noninformative for calculations of sequence identity and similarity. The PROSITE database was used to identify conserved motifs in Prp2 with homology to reported proteins, and the PSORT database was used for prediction of cellular localization of Prp2 as previously described (12).
Bacterial expression of AG2/PRA and PRP2 and purification of the respective recombinant proteins.
Oligonucleotide primers were designed to amplify the full-length cDNAs of AG2/PRA and PRP2. Total RNA was obtained from parasitic-phase cultures of C. posadasii which had been incubated for 132 h and contained spherules in the endosporulation stage of development. Template cDNA was produced from the RNA by reverse transcription as reported previously (19). The nucleotide sequences of the sense and antisense primers used to amplify the AG2/PRA cDNA were 5′-ATAGGCAGCCATATGCAGTTCTC TCACGCTCT-3′ and 5′-GAATTCAAGCTTCTCGAGTGCTGATAGTCTAAATTTAC-3′, which contained engineered NdeI and XhoI restriction sites, respectively (underlined nucleotides). The primer pair yielded a 625-bp AG2/PRA amplicon from a single-stranded template cDNA. The amplification conditions were as follows: 95°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. The nucleotide sequences of the PRP2-specific sense and antisense primers were 5′-ATAGGCAGCCATATGAAGTTCTCTCA CACCCTC-3′ and 5′-GAATTCAAGCTTCTCGAGTCAACTCCTTCTTCCTCGT-3′, respectively. The primers contained the same engineered restriction sites as those described above. This primer pair generated a 427-bp cDNA amplicon of PRP2. The two amplicons were cloned separately into the pCR2.1 TOPO plasmid vector (Invitrogen), and the sequences of the respective cDNA inserts were confirmed by nucleotide sequence analysis as reported previously (21). The sequencing plasmids were digested with NdeI and XhoI, and the AG2/PRA and PRP2 cDNAs were ligated into pET32b (Novagen, Madison, WI) and pET28b (Novagen) plasmid expression vectors, respectively. Because of the absence of an NdeI site in pET32b, blunt-end ligation was necessary for insertion of the AG2/PRA cDNA into this vector. Both cDNA inserts contained a stop codon at the 3′ end. The pET constructs were used to transform Escherichia coli BL21(DE3) slyD (40), which was kindly supplied by Ryland Young (Department of Biochemistry and Biophysics, Texas A&M University). This host strain was chosen because it lacks the FK506 binding protein (31), which previous investigators showed to copurify with the E. coli-expressed Ag2/Pra protein during nickel-affinity chromatography (38).
Purification of the recombinant proteins (rAg2/Pra and rPrp2) was conducted as reported previously (47). The pET32b vector used for expression of rAg2/Pra encoded a 109-amino-acid N-terminal thioredoxin fusion peptide (Trx tag; Novagen), which enhanced the solubility of the recombinant protein during the chromatographic isolation procedure. The thioredoxin fusion peptide was removed from the Ni-affinity-purified recombinant protein by proteolysis with biotinylated thrombin (Novagen). The thrombin was then removed with small aliquots of streptavidin-agarose as recommended by the supplier. The endotoxin contents of stock solutions of the two recombinant proteins were determined by use of a Limulus amebocyte lysate kit (QCL-1000; BioWhittaker, Walkersville, MD) as reported previously (32). The stock solutions contained <0.85 endotoxin unit per μg of protein. Confirmation of the purity of rAg2/Pra and rPrp2 was conducted by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and surface-enhanced laser desorption ionization-time-of-flight (SELDI-TOF) mass spectrometry as previously described (23). SELDI-TOF analysis of the chromatographically purified recombinant proteins was performed using an aliphatic reverse-phase chip (H4 protein chip; Ciphergen, Palo Alto, CA) as reported previously (23). Amino acid sequence analysis of the purified recombinant proteins was conducted by high-pressure liquid chromatography (HPLC)-tandem mass spectrometry as previously described (43). The Coomassie blue-stained bands of rAg2/Pra and rPrp2 (31.5 kDa and 14.5 kDa, respectively) were separately excised from the SDS-PAGE gel, subjected to in-gel trypsin digestion, applied to a reverse-phase HPLC column, and introduced into an ion-trap mass spectrometer equipped with a nanospray source (LCQ Deca XP Plus; Finnigan Corp., San Jose, CA). Analysis of the sequence data was conducted as reported previously (43).
Real-time PCR.
Expression levels of AG2/PRA and PRP2 were examined during the parasitic cycle of C. posadasii by quantitative real-time PCR (QRT-PCR) analysis using a LightCycler PCR system (Roche Diagnostics, Indianapolis, IN). Procedures employed for isolation of the parasitic cells at different stages of development, extraction of RNA, and QRT-PCR data analysis were performed as reported previously (43). The sequences of the sense and antisense primers used to measure levels of expression of the AG2/PRA gene were 5′-ATGCAGTTCTCTCACGCTCT-3′ and 5′-TGGTGGGATGTCAATTGGGAC-3′, respectively. This primer pair amplified a 315-bp product from single-stranded cDNA reverse transcribed from parasitic cell-derived RNA preparations. The sequences of the sense and antisense primers used for generation of a 379-bp amplicon of PRP2 were 5′-AACGACGGGTGCGAGAAG-3′ and 5′-ATGATTACTATTTACAGAGT-3, respectively. A 191-bp amplicon of the constitutively expressed glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene of C. posadasii (GenBank accession no. AF288134) was used for normalization of the QRT-PCR assay as previously reported (43). The results of real-time PCR analysis of expression of the genes encoding the two proline-rich proteins during selected stages of the parasitic cycle are presented as ratios of the amount of transcript of each target gene to the amount of GAPDH transcript in each sample (43). The QRT-PCR assays were performed in triplicate, using separately prepared RNA samples.
RT-PCR analysis of gene expression in vivo.
To determine if the AG2/PRA and PRP2 genes were expressed by C. posadasii in vivo, reverse transcription-PCR (RT-PCR) assays were conducted using total RNA isolated from either uninfected lung tissue or pulmonary abscesses infected with C. posadasii as previously described (12). Three C57BL/6 mice (females; 8 weeks old) were challenged by the i.n. route with approximately 70 arthroconidia of C. posadasii and sacrificed 14 days later. Infected and uninfected tissues were dissected from the lungs of each mouse and used as a source of total RNA for separate RT-PCR assays of gene expression. Methods of RNA isolation and RT-PCR analysis were the same as those described previously (12). The same oligonucleotide primers and PCR conditions were used for RT-PCR amplification of the AG2/PRA and PRP2 gene fragments as those described above for the QRT-PCR assays.
IFN-γ ELISPOT assays.
Purified rAg2/Pra or rPrp2 was used to immunize two groups of C57BL/6 mice (four per group) as described above. Two weeks after the second immunization, the spleens of the immunized mice were separately harvested, pooled, macerated, and used to conduct gamma interferon (IFN-γ)-specific enzyme-linked immunospot (ELISPOT) assays as previously described (43, 44). The spleens of a separate control group of mice immunized with adjuvant alone were prepared and examined in the same way. Immune and nonimmune CD4+ T cells were isolated using a CD4+ T-cell isolation kit (Miltenyi Biotec, Inc., Gladbach, Germany). The purity of the T-cell preparations was verified by staining the cells with fluorescein isothiocyanate-conjugated anti-CD4 and anti-CD90 monoclonal antibodies (Miltenyi Biotec), followed by analysis of the labeled cells in an Epics Elite flow cytometer (Beckman Coulter, Inc., Fullerton, CA). To control for nonspecific background in the ELISPOT plates, all wells were corrected by subtracting the number of spots in an identically treated well that contained isolated nonimmune CD4+ cells.
Generation of synthetic peptides used in IFN-γ ELISPOT assays.
Two series of overlapping synthetic peptides covering 100% of the amino acid sequences of Ag2/Pra and Prp2 (Table 1) were produced (Mimotopes, Raleigh, NC). The HPLC-purified peptides were used in IFN-γ ELISPOT assays to evaluate the in vitro responses of immune T cells of C57BL/6 mice immunized with the respective recombinant proteins. The two sets of 20-mer peptides were designed so that contiguous peptide sequences overlapped by 12 amino acids. Individual synthetic peptides supplied by the manufacturer were dissolved in 100% dimethyl sulfoxide (Sigma, St. Louis, MO) to generate peptide stock solutions at a concentration of 1.25 mg/ml. ELISPOT assays were conducted with individual peptides that were diluted with T-cell culture medium (43, 44) to a final concentration of 2.5 μg/ml. The IFN-γ ELISPOT assays were performed as described above.
TABLE 1.
Peptide libraries used for immune CD4+ T-cell epitope mapping of two proline-rich proteins of C. posadasii
| Protein and peptide | Amino acid positions | Sequencea |
|---|---|---|
| Ag2/Pra | ||
| 1P1 | 1-20 | MQFSHALIALVAAGLASAQL |
| 1P2 | 9-28 | ALVAAGLASAQLPDIPPCAL |
| 1P3 | 17-36 | SAQLPDIPPCALNCFVEALG |
| 1P4 | 25-44 | PCALNCFVEALGNDGCTRLT |
| 1P5 | 33-52 | EALGNDGCTRLTDFKCHCSK |
| 1P6 | 41-60 | TRLTDFKCHCSKPELPGQIT |
| 1P7 | 49-68 | HCSKPELPGQITPCVEEACP |
| 1P8 | 57-76 | GQITPCVEEACPLDARISVS |
| 1P9 | 65-84 | EACPLDARISVSNIVVDQCS |
| 1P10 | 73-92 | ISVSNIVVDQCSKAGVPIDI |
| 1P11 | 81-100 | DQCSKAGVPIDIPPVDTTAA |
| 1P12 | 89-108 | PIDIPPVDTTAAPEPSETAE |
| 1P13 | 97-116 | TTAAPEPSETAEPTAEPTEE |
| 1P14 | 105-124 | ETAEPTAEPTEEPTAEPTAE |
| 1P15 | 113-132 | PTEEPTAEPTAEPTAEPTHE |
| 1P16 | 121-140 | PTAEPTAEPTHEPTEEPTAV |
| 1P17 | 129-148 | PTHEPTEEPTAVPTGTGGGV |
| 1P18 | 137-156 | PTAVPTGTGGGVPTGTGSFT |
| 1P19 | 145-164 | GGGVPTGTGSFTVTGRPTAS |
| 1P20 | 153-172 | GSFTVTGRPTASTPAEFPGA |
| 1P21 | 161-180 | PTASTPAEFPGAGSNVRASV |
| 1P22 | 169-188 | FPGAGSNVRASVGGIAAALL |
| 1P23 | 175-194 | NVRASVGGIAAALLGLAAYL |
| Prp2 | ||
| 2P1 | 1-20 | MKFSHTLVALAAAGIASAQI |
| 2P2 | 9-28 | ALAAAGIASAQIPNIPPCAL |
| 2P3 | 17-36 | SAQIPNIPPCALMCFIDALG |
| 2P4 | 25-44 | PCALMCFIDALGNDGCEKLT |
| 2P5 | 33-52 | DALGNDGCEKLTDFKCHCAK |
| 2P6 | 41-60 | EKLTDFKCHCAKPELPGKIT |
| 2P7 | 49-68 | HCAKPELPGKITPCVEKACP |
| 2P8 | 57-76 | GKITPCVEKACPNIEARISV |
| 2P9 | 65-84 | KACPNIEARISVSNIVVDQC |
| 2P10 | 73-92 | RISVSNIVVDQCSKAGVPIS |
| 2P11 | 81-100 | VDQCSKAGVPISIPPADTRT |
| 2P12 | 89-108 | VPISIPPADTRTPTQPPSTS |
| 2P13 | 97-116 | DTRTPTQPPSTSPSAPQPTA |
| 2P14 | 105-124 | PSTSPSAPQPTACIPKRRRA |
Bold peptide sequences are those that elicited an in vitro T-cell response in IFN-γ ELISPOT assays, with an arbitrary cutoff value of >40 spots/well.
Nucleotide sequence accession numbers.
The C. posadasii PRP2 nucleotide and deduced protein sequences reported in this paper have been submitted to the GenBank database under accession no. AY102921. The sequences of the CFEM domains of the proline-rich antigens of C. posadasii were deposited in GenBank under the accession numbers listed in Table 2.
TABLE 2.
Sequence comparison of the family of proline-rich proteins of C. posadasii
RESULTS
The reported Ag2/Pra recombinant protein vaccine fails to provide durable protection against pulmonary challenge with Coccidioides.
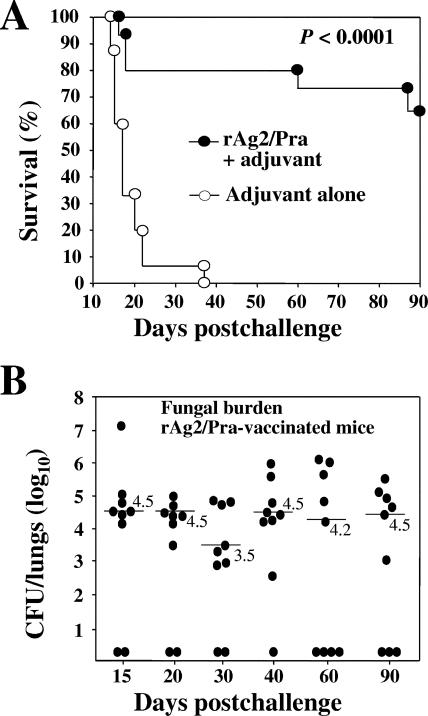
C57BL/6 mice were vaccinated with rAg2/Pra plus the CpG adjuvant preparation and then challenged by the natural i.n. route with a lethal inoculum of viable C. posadasii arthroconidia. Control mice were immunized with the adjuvant preparation alone. The nonvaccinated mice typically began to die at approximately 15 days postchallenge (Fig. 1A), and all showed obvious signs of morbidity by 18 days. On the other hand, the majority of the rAg2/Pra-immunized mice appeared healthy (active, no ruffled fur, and weight gain) during the first 50 days postchallenge. However, during the subsequent 40 days of the experimental period, all survivors began to lose weight, and at least 15% of the animals were lethargic, became moribund, and died. The survival plot in Fig. 1A is representative of three experiments using the same vaccination and challenge protocols. We extended the experimental period to 120 days and recorded additional deaths (>25%) among the group of rAg2/Pra-vaccinated mice (data not shown). Control mice sacrificed at 15 days postchallenge typically showed high numbers of CFU in the lungs (105 to 108) and spleen (103) (43). None of the vaccinated mice had detectable organisms in the spleen at any of the sampling times postchallenge. On the other hand, the results of the fungal burden determinations shown in Fig. 1B indicate that the majority of the rAg2/Pra-vaccinated mice (60 to 90%) retained high numbers of CFU in their lungs (102.7 to 107.2; median range of 103.5 to 104.5) throughout the 90-day period after i.n. challenge. The recombinant Ag2/Pra vaccine appeared to stimulate early protection against pulmonary infection following i.n. challenge with Coccidioides, but this response was not sufficiently robust to contain the pathogen. Based on earlier suggestions that Ag2/Pra is a component of a heterogeneous, polymeric antigen (10) and our past experience that multivalent vaccines provide significantly better protection against coccidioidomycosis than do single recombinant vaccines (42-44), we initiated a search of the C. posadasii database for homologs of the Ag2/Pra protein.
FIG. 1.
(A) Representative evaluation of protective efficacy of rAg2/Pra vaccine plus adjuvant compared to a control preparation that consisted of adjuvant alone. CpG oligodeoxynucleotides were used as the adjuvant, and C57BL/6 mice were challenged by the i.n. route with a potentially lethal inoculum of 50 viable C. posadasii arthroconidia. Immunization was performed subcutaneously. The recombinant protein was administered as two doses (1 μg each). The statistical significance (P value) of the difference between the survival plots for the vaccinated versus nonvaccinated mice is shown. The results are representative of three separate vaccination/survival experiments using the same immunization and challenge protocols. (B) Plots of CFU of C. posadasii detected in dilution plate cultures of lung homogenates of C57BL/6 mice vaccinated with rAg2/Pra plus adjuvant as described for panel A. The animals were sacrificed at different times postchallenge, as indicated. The median CFU values (log10) are indicated by horizontal lines. No statistically significant difference in CFU was observed between the estimates of fungal burden.
Discovery of a proline-rich protein of Coccidioides with structural similarities to Ag2/Pra.
A BLAST search of the translated C. posadasii genomic database, using the Ag2/Pra sequence (GenBank accession no. U32518) as the probe, revealed a 372-bp open reading frame which translates a homolog of the previously reported proline-rich antigen (Fig. 2A). The C. posadasii gene encoding this homolog was designated PRP2. Comparative nucleotide sequence analysis of the 933-bp genomic PCR amplicon and the 372-bp cDNA clone of PRP2 confirmed the presence of two introns, of 62 bp and 64 bp. The AG2/PRA gene has also been reported to contain two introns, of 78 and 101 bp (49). Alignment of the two full-length protein sequences revealed 70% identity and 86% similarity, with the highest homology between amino acids 1 and 98 (Fig. 2A). An 18-residue signal peptide with a cleavage site between A18 and Q19 was predicted for both Prp2 and Ag2/Pra. The signal peptide sequences showed 72% identity and 94% similarity. A conserved, eight-cysteine-containing fungal extracellular membrane (CFEM) domain (30) is located between residues 24 and 83/84 of the two proteins (Fig. 2B). The consensus sequence of this domain is PXC[A/G]X2CX8-12CX1-3[X/T]- DX2-5CXCX9-14CX3-4CX15-16C, where X is any amino acid with the indicated range. Both Prp2 and Ag2/Pra are rich in proline residues (13.7% and 12.9%, respectively). Prp2 contains two TXX′P repeats, in which X is arginine or glutamine and X′ is threonine or proline. This contrasts with the amino acid composition of the repeat sequences of Ag2/Pra, in which X is alanine, histidine, or glutamic acid and X′ is alanine, valine, or glutamic acid (Fig. 2A and B). The threonine-rich region of Prp2 is confined to the sequence between T98 and T115, within which eight O-glycosylation sites are predicted (26). As previously reported (49), the C terminus of Ag2/Pra includes a putative glycosylphosphatidylinositol (GPI) anchor sequence. In contrast, the Prp2 sequence is apparently truncated downstream of the TXX′P repeats and lacks the GPI consensus motif.
FIG. 2.
(A) T-COFFEE alignment (37) of amino acid sequences of two proline-rich proteins of C. posadasii (Ag2/Pra and Prp2). Asterisks indicate amino acid identity, and colons indicate conservative substitutions. (B) Schematic representations of the linear structures of Ag2/Pra and Prp2. The numbers correspond to residues of the protein sequence.
High proline content may influence the conformation of purified recombinant proteins.
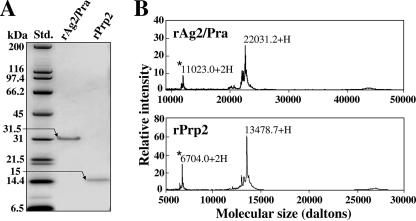
E. coli transformed with the pET32b-AG2/PRA plasmid construct yielded a 31.5-kDa recombinant protein, which appeared as a single Coomassie blue-stained band in an SDS-PAGE gel after nickel-affinity chromatographic purification (Fig. 3A). The predicted molecular size of the translated AG2/PRA gene is 19.4 kDa. We previously demonstrated that the high proline content of Coccidioides proteins typically leads to contradictory data on molecular size based on estimates derived from the deduced protein sequence, on the one hand, and from SDS-PAGE, on the other (23). This anomaly has been suggested to be due to conformational peculiarities of the proteins even after exposure to the reducing conditions of sample preparation. We have shown that SELDI-TOF mass spectrometry is a valuable tool for estimating the accurate molecular sizes of these proteins (23). The result of this analysis of rAg2/Pra shown in Fig. 3B takes into account a 2.7-kDa vector-encoded fusion peptide at the N terminus of the recombinant protein. The estimated molecular size of rAg2/Pra plus the vector peptide by SELDI-TOF is essentially the same as the predicted size. The predicted molecular size of the translated PRP2 gene is 12.9 kDa, while the predicted size of the recombinant Prp2 protein, which includes a three-amino-acid fusion peptide at its N terminus, is 13.3 kDa. As in the case of rAg2/Pra, the estimated molecular size of rPrp2 in the SDS-PAGE gel (approximately 15 kDa) (Fig. 3A) is larger than the molecular size (13.5 kDa) determined by SELDI-TOF mass spectrometry (Fig. 3B). Amino acid sequence analyses of the two purified recombinant proteins were conducted by LC-tandem mass spectrometry of the respective trypsin digests, which yielded two peptide sequences of rAg2/Pra (ISVSNIVVDQCSK and PTASTPAEFPGAGSNVR) and a single peptide sequence of rPrp2 (AGVPISIPPADTR). These sequences matched the respective deduced sequences of the AG2/PRA and PRP2 products.
FIG. 3.
(A) Molecular sizes of E. coli-expressed recombinant proteins (rAg2/Pra and rPrp2), as estimated by SDS-PAGE. (B) SELDI-TOF mass spectrometry of the identical pair of purified recombinant proteins. The low-intensity peaks (asterisks) represent doubly charged molecules.
AG2/PRA and PRP2 have distinct expression profiles during the parasitic cycle.
For quantitative analysis of gene expression in vitro (Fig. 4A and B), total RNA was isolated from first-generation parasitic cells in the isotropic growth phase (36 h), the segmentation stage (96 h), and the endosporulation phase (132 h) (12). The amount of AG2/PRA transcript increased >2-fold between the isotropic growth phase and the endosporulation stage and showed relatively high levels of expression throughout the parasitic cycle. On the other hand, detection of the PRP2 transcript was essentially restricted to the endosporulation phase of the parasitic cycle. Based on the assumption that the efficiency of QRT-PCR was the same for the two transcripts, it also appeared that the level of expression of AG2/PRA was approximately 10-fold higher than that of the PRP2 gene. To confirm that the genes which encode the two proline-rich proteins were expressed in vivo, RT-PCR was performed, using the same primers employed for the QRT-PCR analyses (Fig. 4C). Template cDNA was derived from total RNA extracted from C. posadasii-infected or uninfected (control) lung tissue. The RT-PCR data indicated that the two genes were expressed in the lungs of mice with coccidioidomycosis at 2 weeks postchallenge. Comparative quantitative analyses of gene expression in vivo were not conducted.
FIG. 4.
(A and B) QRT-PCR plots of AG2/PRA and PRP gene expression in vitro during stages of the parasitic cycle compared to expression levels of the constitutive GAPDH gene. Error bars indicate standard deviations. (C) RT-PCR amplification of AG2/PRA and PRP2 genes expressed in C. posadasii-infected murine lung tissue.
Superior protection against pulmonary challenge was observed with a combined Ag2/Pra-plus-Prp2 recombinant protein vaccine.
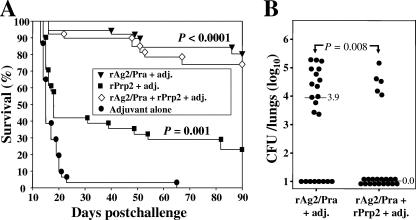
C57BL/6 mice were vaccinated with equal doses of rAg2/Pra and rPrp2 (individually) plus adjuvant and then challenged by the i.n. route (Fig. 5A). A significant difference in percent survival was revealed (80% versus 22.5%, respectively; P = 0.02). On the other hand, the percentage of surviving mice vaccinated with equal doses of the combined antigens at 90 days postchallenge was 73.3%. The results of these vaccination protocols were reproducible, and the data are representative of three separate experiments. In spite of the fact that there was no statistically significant difference in the number of survivors immunized with either rAg2/Pra alone or the combined recombinant proteins (P = 0.78), there was a statistically significant difference in fungal burden between these two groups of mice (Fig. 5B). The percentage of mice with no detectable organisms in the lungs was higher in the case of mice vaccinated with rAg2/Pra plus rPra2 (77.2%) than in the case of mice immunized with rAg2/Pra alone (36.4%). A clear explanation for the presence of high CFU levels in the five mice vaccinated with the combined recombinant protein vaccine is not at hand. We have consistently observed that variation occurs in the levels of residual CFU in the lungs of vaccinated, inbred mice challenged with Coccidioides by the i.n. route (43, 44). Even greater variation in fungal burdens in the lungs of vaccinated mice challenged by the intraperitoneal route has been reported (16, 29, 39). A possible explanation is that the presence of a lung site infection with a high concentration of endosporulating spherules could skew the results of the fungal burden analysis. Because of the variations in fungal burden between identically treated mice, we argue that challenge experiments with large numbers of vaccinated mice are necessary for statistical evaluation of the protective efficacy of experimental vaccines against coccidioidomycosis.
FIG. 5.
(A) Representative evaluation of protective efficacies of single recombinant protein vaccines (rAg2/Pra and rPrp2) and a combined recombinant protein vaccine (rAg2/Pra plus rPrp2). The adjuvant (adj.) used, immunization protocol, and i.n. challenge dose were the same as those described in the legend to Fig. 1A. The statistical significance (P values) of differences between survival plots for the vaccinated versus nonvaccinated mice is shown. (B) Plots of CFU of C. posadasii detected in dilution plate cultures of lung homogenates of C57BL/6 mice vaccinated with either rAg2/Pra plus adjuvant or the combined recombinant proteins (rAg2/Pra plus rPrp2) plus adjuvant and sacrificed at 90 days postchallenge. The median CFU values are shown as described in the legend to Fig. 1B. A statistically significant difference (P value) between the fungal burdens in the lungs of the two groups of vaccinated mice is indicated.
Cellular immunoassays reveal that the recombinant Ag2/Pra and Prp2 proteins have distinct T-cell epitopes.
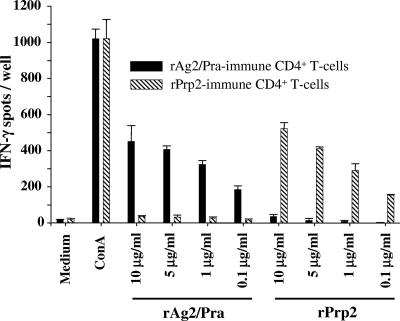
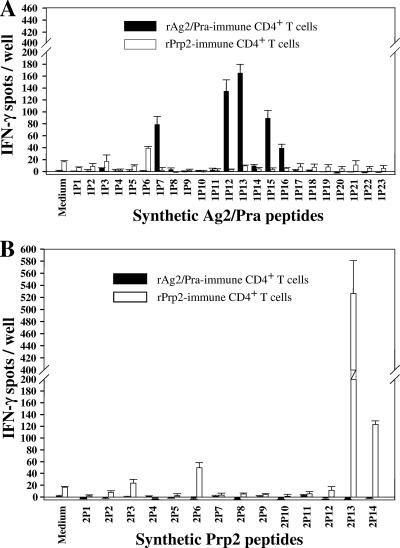
The results of the above vaccination experiments suggested that immunization of mice with each of the two proline-rich proteins in the presence of the CpG adjuvant preparation elicited distinct host immunological responses. To further explore this possibility, we conducted IFN-γ ELISPOT recall assays of immune CD4+ T cells isolated from mice immunized with either rAg2/Pra or rPrp2. The immune T cells were stimulated in vitro with either the homologous or heterologous antigen. The results in Fig. 6 indicate that distinct T-cell epitopes are present in the two recombinant proteins. A linear recall response of immune CD4+ T cells was evident in the presence of decreasing concentrations of the homologous proteins, while little to no in vitro response was detected in the presence of the heterologous antigen at any of the concentrations tested. To further examine this striking difference in immune T-cell responses, we synthesized overlapping 20-mer peptides to map the CD4+ T-cell epitopes of the two proteins. IFN-γ ELISPOT assays were conducted to evaluate the reciprocal response of CD4+ T cells obtained from C57BL/6 mice immunized with either rAg2/Pra or rPrp2 and incubated in vitro with the individual synthetic 20-mer peptides representing each of the full-length proteins (Fig. 7A and B). Peptides 1P6 of Ag2/Pra and 2P6 of Prp2 both showed reactivity with T cells derived from mice immunized with rPrp2 but not with T cells isolated from rAg2/Pra-immunized mice. The amino acid sequences of these two peptides are 80% identical (Table 1). On the other hand, although the sequence of peptide 1P7 of Ag2/Pra is 75% identical to that of the corresponding peptide of Prp2 (2P7), 1P7 stimulated IFN-γ production only in T cells derived from rAg2/Pra-immunized mice. Likewise, peptide 1P12 stimulated T cells isolated only from rAg2/Pra-immunized mice. Peptide 1P13, representing amino acids 97 to 116 of Ag2/Pra, showed the highest recall response of T cells derived from mice immunized with the homologous full-length antigen. The sequences of 1P13 of Ag2/Pra and 2P13 of Prp2 showed essentially no homology, and no reciprocal stimulation of immune T cells was observed. However, peptide 2P13 showed the highest stimulatory response of T cells derived from rPrp2-immunized mice. A common feature of peptides 1P12, 1P13, 1P15, and 1P16 of Ag2/Pra and 2P13 of Prp2 is that they contain one or more TXX′P sequences. A single TXX′P sequence is present in the 1P12 peptide, while the immunostimulatory 1P13, 1P15, and 1P16 peptides of Ag2/Pra contain three, four, and four repeats of this motif, respectively. The major reactive peptide of Prp2 (2P13) contains two TXX′P repeats. However, as previously described, the XX′ residues of the TXX′P motifs of Ag2/Pra and Prp2 are distinct. The immune T-cell-stimulatory 2P14 peptide, which is present at the C terminus of Prp2, does not contain the TXX′P sequence. In summary, the epitope mapping data indicate that with the exception of 1P6 and 2P6, the T-cell epitopes of Ag2/Pra and Prp2 have unique amino acid sequences.
FIG. 6.
Assessment of IFN-γ production by immune CD4+ T cells derived from C57BL/6 mice, conducted by IFN-γ ELISPOT assays. Reciprocal in vitro assays were performed. Recombinant Ag2/Pra- or rPrp2-immune T cells were incubated with either the homologous or heterologous full-length recombinant protein at concentrations of 0.1 to 10 μg/ml of cell culture medium. Isolated CD4+ T cells incubated in medium alone or with concanavalin A (ConA) served as negative and positive controls, respectively. Error bars indicate standard deviations for replicate assays.
FIG. 7.
(A and B) Results of reciprocal IFN-γ assays conducted as described in the legend to Fig. 6, except that the isolated immune CD4+ T cells were incubated with individual purified 20-mer synthetic peptides (2.5 μg/ml) representing the full-length homologous or heterologous vaccine protein. The sequences of contiguous peptides of Ag2/Pra and Prp2 overlapped by 12 amino acids. Error bars indicate standard deviations for replicate assays.
DISCUSSION
As previously discussed, evaluations of the protective efficacies of both the recombinant Ag2/Pra protein and a genetic vaccine with Ag2/PRA against Coccidioides challenge resulted in conflicting data. The results of studies from multiple laboratories on the use of Ag2/Pra as a vaccine against coccidioidomycosis have been reviewed (6). Although a protective response against intraperitoneal infection was reported, the vaccinated mice were not protected against an i.n. challenge. Several factors may have contributed to this contradiction, including the murine strain used, the nature of the adjuvant employed, and the size of the infectious inoculum (41). Although the intraperitoneal route of Coccidioides challenge has been used effectively to screen for antigens which elicit a protective host response against coccidioidomycosis (29), we argue that critical evaluation of candidate vaccines should be conducted with an animal model challenged by the natural i.n. route of infection. Using C57BL/6 mice, which are genetically more resistant to pulmonary infection with Coccidioides than the BALB/c strain (28), we and others have observed approximately 80% survival of rAg2/Pra-vaccinated mice 60 days after i.n. challenge with a lethal inoculum of spores (42). However, in the present study, we revealed that the survivors retained high fungal burdens in the lungs throughout the experimental period postchallenge, and 15% of these vaccinated animals died of overwhelming respiratory infection between 60 and 90 days after i.n. inoculation with Coccidioides. Histopathology of the lungs of the survivors at 90 days revealed large abscesses which contained many parasitic cells (spherules) in various stages of development (not shown). Although the bacterially expressed rAg2/Pra protein vaccine appears to elicit an early protective response to intraperitoneal and pulmonary infection with Coccidioides (29, 41), this response is not sufficiently robust and durable to contain the pathogen or to clear the organism from the majority of infected mice.
The native Ag2/Pra protein has been localized to the cell wall of Coccidioides (18) and is highly glycosylated (14). Extraction of the antigenic fraction from formalin-fixed spherules was accomplished by incubation of the cells in 1 N NaOH (9). The resulting alkali-soluble, water-soluble (ASWS) material was characterized as a heterogeneous antigenic complex by 2D-IEP, consisting of Ag2/Pra plus additional macromolecules which comigrated with and appeared to be serologically related to the major precipitate in the electrophoresis gel. The ASWS fraction elicited a delayed-type hypersensitivity response in mice, reacted with patient immunoglobulin M tube precipitin antibody, and protected mice against i.n. challenge with arthroconidia (10). In support of the suggestion that the ASWS fraction contains a complex of Ag2/Pra-related antigens, our BLASTX search of the C. posadasii genomic database revealed seven additional deduced proline-rich proteins, designated Prp2 to -8, which showed structural homology to Ag2/Pra. All but Prp3 are predicted to have a signal peptide, all contain two or more TXX′P repeats, and the C-terminal regions of all but Prp2 and Prp7 include a GPI anchor consensus sequence (not shown). A cysteine-rich region with sequence similarity to the reported CFEM domain (30) was present in each of the newly discovered proline-rich proteins. This polypeptide fragment of the putative Prp family was subjected to CLUSTAL W alignment (45). Preliminary results of the sequence identities and similarities of Prp2-8 with Ag2/Pra are presented in Table 2. Only Prp2 showed high sequence homology to Ag2/Pra. The structural similarity of these two proteins, as well as the prediction that Prp2 has a signal peptide but lacks a GPI anchor, suggested that Prp2 is a secreted antigen and a component of an immunogenic parasitic cell wall complex.
The N-terminal 98-amino-acid sequences of Prp2 and Ag2/Pra show 80% identity. Only six nonconservative amino acid substitutions distinguish these two aligned protein fragments. The cleavage site of the signal peptides is identical (A18/Q19). The conserved CFEM sequence is present in identical locations in Ag2/Pra and Prp2. This domain has been reported to occur in numerous fungal pathogens of plants and animals, is predominantly comprised of hydrophobic residues, and is present in proteins with a signal sequence and either a transmembrane span or a GPI anchor. Both Ag2/Pra and Prp2 lack transmembrane motifs, but Ag2/Pra contains a GPI anchor consensus sequence. The CFEM domain of Ag2/Pra has been reported to contain B-cell epitopes (48). Full-length recombinant Ag2/Pra and PCR-generated Ag2/Pra truncations were expressed in order to map B-cell-reactive epitopes in enzyme-linked immunosorbent and immunoblot assays with sera from infected patients and mice. The prevalence of both linear and conformational B-cell epitopes was predicted to occur between amino acids 19 and 96 of Ag2/Pra. Full-length recombinant Prp2 was recognized in immunoblots by sera from patients with confirmed coccidioidal infection but not by sera from noninfected control patients (not shown). However, to date, there is no evidence that anti-rAg2/Pra antibody plays a role in protective immunity against Coccidioides infection. In fact, high titers of immunoglobulin to rAg2/Pra were detected in sera from patients with chronic and disseminated coccidioidomycosis but not in serum samples from patients with primary, self-limited coccidioidal disease (38). Clinical studies of coccidioidomycosis have traditionally indicated that a rising titer of patient anti-Coccidioides antibody is a poor prognosis of disease outcome (27), although this does not rule out the possibility that protective antibodies exist and play a role in vaccine-induced immunity (34). Downstream of the CFEM domain of Ag2/Pra is an extended threonine-rich region (T97-165), which is truncated in Prp2 (T98-115). Both regions are predicted to contain O-glycosylation sites, but almost three times more exist in Ag2/Pra than in Prp2. These same regions of the two proteins contain TXX′P repeats, with 10 repeats in Ag2/Pra, but only 2 in Prp2. Proline residues can influence polypeptide chain conformation and protein folding, particularly when the prolines occur in repetitive motifs of the protein sequence (4, 33). In support of the occurrence of conformational peculiarities in the bacterially expressed recombinant proteins, we showed that both rAg2/Pra and rPrp2 were impeded in migration during SDS-PAGE separation, even in the presence of reducing agents. The difference in concentrations of proline residues associated with the TXX′P repeats of the two structurally related proteins may also influence their antigenicity (4).
AG2/PRA and PRP2 were distinguished on the basis of their temporal expression and by quantitative analysis of the amounts of gene transcript produced during the parasitic cycle. Expression of PRP2 was essentially restricted to the endosporulation phase, while AG2/PRA was expressed throughout the parasitic cycle. In addition, the amount of PRP2 transcript detected was significantly lower than that of AG2/PRA. The low level of expression of the PRP2 gene may partly account for the relatively poor protection afforded by the recombinant Prp protein vaccine under the immunization conditions employed. We did not evaluate higher vaccination doses of the recombinant Prp2 protein, but rather compared its protective efficacy to that of rAg2/Pra, using an identical immunization protocol. Although vaccination with rAg2/Pra showed a marked increase in the percentage of mice which survived a lethal pulmonary challenge compared to vaccination with the rPrp2 protein, the combined recombinant protein vaccine provided significantly better protection based on long-term survival and pathogen clearance. We suggest that the latter is largely the result of stimulation of a more heterogeneous T-cell repertoire than that induced by the single recombinant protein vaccines. The evidence for this was derived from the results of our IFN-γ ELISPOT assays. We first showed that little to no reciprocal in vitro recall stimulation of rAg2/Pra- and rPrp2-immune CD4+ T cells was evident after their incubation with the heterologous antigen. In spite of the high sequence homology of the two proline-rich proteins, it appeared that the processing of these two antigens by APCs and their presentation to naïve murine T lymphocytes are very different. To further explore this possibility, we repeated the reciprocal ELISPOT assays, using two sets of overlapping peptides which corresponded to the full-length antigens. Only a single peptide of Ag2/Pra (1P6) (Table 1) with 80% sequence identity to the corresponding peptide of Prp2 (2P2) showed cross-reactivity with T lymphocytes obtained from rPrp2-immune mice. Otherwise, the immunostimulatory peptides in each set showed no cross-reactivity. The peptides of Prp2 which elicited the highest T-cell responses were at the C terminus and showed low sequence homology to Ag2/Pra peptides. Three of the immunoreactive peptides of Ag2/Pra were located in the TXX′P repeat domain (1P13, 1P15, and 1P16). This is the region where proline-induced conformational peculiarities are expected to occur, as well as the sites of predicted O-glycosylation of the native antigen. We suggest that both factors may impact antigen processing and presentation by APCs.
Using a bacterial expression system, Peng et al. (39) isolated subunits of Ag2/Pra and tested each as a vaccine against intraperitoneal challenge with Coccidioides in BALB/c mice. They reported that protection by vaccination with full-length Ag2/Pra was almost totally accounted for by the N-terminal subunit (amino acids 1 to 106). This fragment is upstream of the TXX′P repeat domain. In a subsequent report, Jiang et al. (24) showed that the signal peptide of Ag2/Pra (amino acids 1 to 18), administered as either a gene vaccine or a synthetic peptide, induced protective immunity against intraperitoneal challenge but not i.n. challenge in BALB/c mice. We repeated our IFN-γ ELISPOT assays of the same two sets of overlapping peptides of Ag2/Pra and Prp2, using immune CD4+ T cells isolated from BALB/c mice. Our results are in agreement with the conclusion that the N-terminal fragments (amino acids 1 to 106) of both Ag2/Pra and Prp2 induce protective immunity in BALB/c mice (data not shown). C57BL/6 mice are more resistant to coccidioidal infection than the BALB/c strain (28) and express distinct major histocompatibility complex haplotypes (H2-b versus H2-d) (http://jaxmice.jax.org). We anticipated that the T cells from these two inbred strains of mice would respond differently to the same sets of overlapping peptides. Nevertheless, our IFN-γ ELISPOT data (Fig. 7A) suggest that immunostimulatory peptides of Ag2/Pra corresponding to the N-terminal region (i.e., 1P6, 1P7, and 1P12) account for the protective immunity against pulmonary infection reported for C57BL/6 mice vaccinated with the Ag2/Pra N-terminal recombinant subunit (amino acids 1 to 106) (42). However, the majority of these i.n. challenged C57BL/6 mice showed symptoms of morbidity and retained high fungal burdens in the lungs, comparable to the results of our examination of the same murine strain vaccinated with full-length rAg2/Pra.
In summary, we have demonstrated a significant improvement in the level of protective immunity induced by vaccination with the combined rAg2/Pra plus rPrp2 proteins compared to that induced by each of the single recombinant proteins. Results of our cellular immunoassays suggest that the divalent vaccine engaged a broader repertoire of T-cell clones than the monovalent vaccines. It has been suggested that the constituents of such a combined recombinant protein vaccine should be at least additive, if not synergistic (11). We are currently examining the possibility that additional members of the proline-rich protein family of Coccidioides could further enhance protective immunity by their incorporation into a multivalent vaccine against coccidioidomycosis.
Acknowledgments
Support for this study was provided by Public Health Service grant AI19149 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health. The senior author was supported by a supplement to the above grant. Additional support for this project was provided by the Margaret Batts Tobin Foundation, San Antonio, TX.
Editor: A. Casadevall
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awasthi, S. 2007. Dendritic cell-based vaccine against Coccidioides infection. Ann. N. Y. Acad. Sci. 1111:269-274. [DOI] [PubMed] [Google Scholar]
- 3.Barnato, A. E., G. D. Sanders, and D. K. Owens. 2001. Cost-effectiveness of a potential vaccine for Coccidioides immitis. Emerg. Infect. Dis. 7:797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady, L. J., D. G. Cvitkovitch, C. M. Geric, M. N. Addison, J. C. Joyce, P. J. Crowley, and A. S. Bleiweis. 1998. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect. Immun. 66:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall, A., and L. A. Pirofski. 2006. The weapon potential of human pathogenic fungi. Med. Mycol. 44:689-696. [DOI] [PubMed] [Google Scholar]
- 6.Cole, G. T., J. Xue, C. Okeke, E. Tarcha, V. Basrur, R. A. Schaller, R. A. Herr, J.-J. Yu, and C.-Y. Hung. 2004. A vaccine against coccidioidomycosis is justified and attainable. Med. Mycol. 42:189-216. [DOI] [PubMed] [Google Scholar]
- 7.Comrie, A. C. 2005. Climate factors influencing coccidioidomycosis seasonality and outbreaks. Environ. Health Perspect. 113:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordeiro, R. A., R. S. Brilhante, M. F. Rocha, M. A. Fechine, L. M. Camara, Z. P. Camargo, and J. J. Sidrim. 2006. Phenotypic characterization and ecological features of Coccidioides spp. from Northeast Brazil. Med. Mycol. 44:631-639. [DOI] [PubMed] [Google Scholar]
- 9.Cox, R. A., and L. A. Britt. 1985. Antigenic heterogeneity of an alkali-soluble, water-soluble cell wall extract of Coccidioides immitis. Infect. Immun. 50:365-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, R. A., and D. M. Magee. 2004. Coccidioidomycosis: host response and vaccine development. Clin. Microbiol. Rev. 17:804-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutler, J. E., G. S. Deepe, Jr., and B. S. Klein. 2007. Advances in combating fungal diseases: vaccines on the threshold. Nat. Rev. Microbiol. 5:13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado, N., J. Xue, J.-J. Yu, C.-Y. Hung, and G. T. Cole. 2003. A recombinant beta-1,3-glucanosyltransferase homolog of Coccidioides posadasii protects mice against coccidioidomycosis. Infect. Immun. 71:3010-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon, D. M. 2001. Coccidioides immitis as a select agent of bioterrorism. J. Appl. Microbiol. 91:602-605. [DOI] [PubMed] [Google Scholar]
- 14.Dugger, K. O., J. N. Galgiani, N. M. Ampel, S. H. Sun, D. M. Magee, J. Harrison, and J. H. Law. 1991. An immunoreactive apoglycoprotein purified from Coccidioides immitis. Infect. Immun. 59:2245-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dugger, K. O., K. M. Villareal, A. Ngyuen, C. R. Zimmermann, J. H. Law, and J. N. Galgiani. 1996. Cloning and sequence analysis of the cDNA for a protein from Coccidioides immitis with immunogenic potential. Biochem. Biophys. Res. Commun. 218:485-489. [DOI] [PubMed] [Google Scholar]
- 16.Fierer, J., C. Waters, and L. Walls. 2006. Both CD4+ and CD8+ T cells can mediate vaccine-induced protection against Coccidioides immitis infection in mice. J. Infect. Dis. 193:1323-1331. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, M. C., G. L. Koenig, T. J. White, and J. W. Taylor. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73-84. [PubMed] [Google Scholar]
- 18.Galgiani, J. N., S. H. Sun, K. O. Dugger, N. M. Ampel, G. G. Grace, J. Harrison, and M. A. Wieden. 1992. An arthroconidial-spherule antigen of Coccidioides immitis: differential expression during in vitro fungal development and evidence for humoral response in humans after infection or vaccination. Infect. Immun. 60:2627-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guevara-Olvera, L., C.-Y. Hung, J.-J. Yu, and G. T. Cole. 2000. Sequence, expression and functional analysis of the Coccidioides immitis ODC (ornithine decarboxylase) gene. Gene 242:437-448. [DOI] [PubMed] [Google Scholar]
- 20.Hector, R. F., and R. Laniado-Laborin. 2005. Coccidioidomycosis—a fungal disease of the Americas. PLoS Med. 2:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung, C.-Y., K. R. Seshan, J.-J. Yu, R. Schaller, J. Xue, V. Basrur, M. J. Gardner, and G. T. Cole. 2005. Metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect. Immun. 73:6689-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung, C.-Y., J.-J. Yu, P. F. Lehmann, and G. T. Cole. 2001. Cloning and expression of the gene which encodes a tube precipitin antigen and wall-associated β-glucosidase of Coccidioides immitis. Infect. Immun. 69:2211-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, C.-Y., J.-J. Yu, K. R. Seshan, U. Reichard, and G. T. Cole. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, C., D. M. Magee, F. D. Ivey, and R. A. Cox. 2002. Role of signal sequence in vaccine-induced protection against experimental coccidioidomycosis. Infect. Immun. 70:3539-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, C., D. M. Magee, T. N. Quitugua, and R. A. Cox. 1999. Genetic vaccination against Coccidioides immitis: comparison of vaccine efficacy of recombinant antigen 2 and antigen 2 cDNA. Infect. Immun. 67:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153-164. [DOI] [PubMed] [Google Scholar]
- 27.Kirkland, T. N., and G. T. Cole. 2002. Coccidioidomycosis: pathogenesis, immune response and vaccine development, p. 365-399. In R. A. Calderone and L. C. Cihlar (ed.), Fungal pathogenesis: principles and applications. Marcel Dekker, New York, NY.
- 28.Kirkland, T. N., and J. Fierer. 1983. Inbred mouse strains differ in resistance to lethal Coccidioides immitis infection. Infect. Immun. 40:912-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkland, T. N., F. Finley, K. I. Orsborn, and J. N. Galgiani. 1998. Evaluation of the proline-rich antigen of Coccidioides immitis as a vaccine candidate in mice. Infect. Immun. 66:3519-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni, R. D., H. S. Kelkar, and R. A. Dean. 2003. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28:118-121. [DOI] [PubMed] [Google Scholar]
- 31.Leach, M. R., J. W. Zhang, and D. B. Zamble. 2007. The role of complex formation between the Escherichia coli hydrogenase accessory factors HYPB and SlyD. J. Biol. Chem. 282:16177-16182. [DOI] [PubMed] [Google Scholar]
- 32.Li, K., J.-J. Yu, C.-Y. Hung, P. F. Lehmann, and G. T. Cole. 2001. Recombinant urease and urease DNA of Coccidioides immitis elicit an immunoprotective response against coccidioidomycosis in mice. Infect. Immun. 69:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacArthur, M. W., and J. M. Thornton. 1991. Influence of proline residues on protein conformation. J. Mol. Biol. 218:397-412. [DOI] [PubMed] [Google Scholar]
- 34.Magee, D. M., R. L. Friedberg, M. D. Woitaske, S. A. Johnston, and R. A. Cox. 2005. Role of B cells in vaccine-induced immunity against coccidioidomycosis. Infect. Immun. 73:7011-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinnis, M. R., M. B. Smith, and E. Hinson. 2006. Use of the Coccidioides posadasii Δchs5 strain for quality control in the ACCUPROBE culture identification test for Coccidioides immitis. J. Clin. Microbiol. 44:4250-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrow, W. 2006. Holocene coccidioidomycosis: valley fever in early Holocene bison (Bison antiquus). Mycologia 98:669-677. [DOI] [PubMed] [Google Scholar]
- 37.Notredame, C., D. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for multiple sequence alignments. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 38.Orsborn, K. I., and J. N. Galgiani. 1998. Detecting serum antibodies to a purified recombinant proline-rich antigen of Coccidioides immitis in patients with coccidioidomycosis. Clin. Infect. Dis. 27:1475-1478. [DOI] [PubMed] [Google Scholar]
- 39.Peng, T., L. Shubitz, J. Simons, R. Perrill, K. I. Orsborn, and J. N. Galgiani. 2002. Localization within a proline-rich antigen (Ag2/PRA) of protective antigenicity against infection with Coccidioides immitis in mice. Infect. Immun. 70:3330-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roof, W. D., S. M. Horne, K. D. Young, and R. Young. 1994. SlyD, a host gene required for phiX174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J. Biol. Chem. 269:2902-2910. [PubMed] [Google Scholar]
- 41.Shubitz, L., T. Peng, R. Perrill, J. Simons, K. Orsborn, and J. N. Galgiani. 2002. Protection of mice against Coccidioides immitis intranasal infection by vaccination with recombinant antigen 2/PRA. Infect. Immun. 70:3287-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shubitz, L. F., J.-J. Yu, C.-Y. Hung, T. N. Kirkland, T. Peng, R. Perrill, J. Simons, J. Xue, R. A. Herr, G. T. Cole, and J. N. Galgiani. 2006. Improved protection of mice against lethal respiratory infection with Coccidioides posadasii using two recombinant antigens expressed as a single protein. Vaccine 24:5904-5911. [DOI] [PubMed] [Google Scholar]
- 43.Tarcha, E. J., V. Basrur, C.-Y. Hung, M. J. Gardner, and G. T. Cole. 2006. A recombinant aspartyl protease of Coccidioides posadasii induces protection against pulmonary coccidioidomycosis in mice. Infect. Immun. 74:516-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarcha, E. J., V. Basrur, C. Y. Hung, M. J. Gardner, and G. T. Cole. 2006. Multivalent recombinant protein vaccine against coccidioidomycosis. Infect. Immun. 74:5802-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warnock, D. W. 2006. Fungal diseases: an evolving public health challenge. Med. Mycol. 44:697-705. [DOI] [PubMed] [Google Scholar]
- 47.Yu, J.-J., L. Zhang, P. W. Thomas, P. J. Szaniszlo, and G. T. Cole. 1999. Isolation and confirmation of function of the Coccidioides immitis URA5 (orotate phosphoribosyl transferase) gene. Gene 226:233-242. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, Y., V. Tryon, D. M. Magee, and R. A. Cox. 1997. Identification of a Coccidioides immitis antigen 2 domain that expresses B-cell-reactive epitopes. Infect. Immun. 65:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu, Y., C. Yang, D. M. Magee, and R. A. Cox. 1996. Coccidioides immitis antigen 2: analysis of gene and protein. Gene 181:121-125. [DOI] [PubMed] [Google Scholar]