Abstract
In experimentally infected human volunteers, the cutaneous immune response to Haemophilus ducreyi is orchestrated by serum, polymorphonuclear leukocytes, macrophages, T cells, and myeloid dendritic cells (DC). This response either leads to spontaneous resolution of infection or progresses to pustule formation, which is associated with the failure of phagocytes to ingest the organism and the presence of Th1 and regulatory T cells. In volunteers who are challenged twice, some subjects form at least one pustule twice (PP group), while others have all inoculated sites resolve twice (RR group). Here, we infected PP and RR subjects with H. ducreyi and used microarrays to profile gene expression in infected and wounded skin. The PP and RR groups shared a core response to H. ducreyi. Additional transcripts that signified effective immune function were differentially expressed in RR infected sites, while those that signified a hyperinflammatory, dysregulated response were differentially expressed in PP infected sites. To examine whether DC drove these responses, we profiled gene expression in H. ducreyi-infected and uninfected monocyte-derived DC. Both groups had a common response that was typical of a type 1 DC (DC1) response. RR DC exclusively expressed many additional transcripts indicative of DC1. PP DC exclusively expressed differentially regulated transcripts characteristic of DC1 and regulatory DC. The data suggest that DC from the PP and RR groups respond differentially to H. ducreyi. PP DC may promote a dysregulated T-cell response that contributes to phagocytic failure, while RR DC may promote a Th1 response that facilitates bacterial clearance.
Haemophilus ducreyi causes chancroid, a genital ulcer disease that facilitates acquisition and transmission of human immunodeficiency virus type 1 (13, 67). H. ducreyi enters the host through breaks in the epithelium that occur during intercourse and infects mucosal and stratified squamous epithelium and regional lymph nodes (52). Erythematous papules form at each entry site and evolve into pustules in 2 to 3 days. After a few days to 2 weeks, the pustules ulcerate, and patients typically develop one to four painful ulcers. In the preantibiotic era, chancroid could persist for months and cause giant ulcers, erosion of the infected area, and fibrosis (52, 70). However, spontaneous resolution of ulcers also occurs (27, 70), but its frequency and mechanism are unknown.
To characterize the host response to H. ducreyi, we developed an experimental model in which human volunteers are inoculated with strain 35000HP or its derivatives on the skin of the upper arm via puncture wounds (13, 62). Papules develop within 24 h and either evolve into pustules or spontaneously resolve within 2 to 5 days. Major strengths of the model are that its clinical course and histopathology closely simulate those of natural chancroid (13).
Experimental pustules and natural ulcers contain polymorphonuclear leukocytes (PMN) that form an epidermal abscess and a deep dermal perivascular infiltrate of T cells and macrophages that resembles a loosely formed granuloma (56, 65). The T-cell infiltrate consists predominantly of memory CD4 cells (31). In pustules obtained from subjects infected once, the ratio of myeloid dendritic cells (DC) to plasmacytoid DC is 2.8-fold that in peripheral blood (5a). DC-LAMP- and CD83-positive cells are present in the epidermis and dermis of pustules (36), indicating that DC maturation occurs in response to infection. In pustules, H. ducreyi is surrounded by PMN and macrophages but is not ingested (7), indicating that evasion of phagocytosis is a major mechanism of bacterial survival (3, 35, 50, 78, 84). H. ducreyi also associates with PMN in naturally acquired ulcers (8), suggesting that the interaction between the bacterium and PMN seen in experimental infection is relevant to disease progression. In pustules and natural ulcers, H. ducreyi associates with fibrin (7, 8), suggesting that the bacteria are exposed to plasma that transudates into infected skin. The ability of H. ducreyi to resist the bactericidal activity of serum is another key feature of immune evasion (1, 15, 19, 34, 41).
In the human model, when a subject is inoculated at two or three sites with 35000HP, a pustule may develop at one site while another site(s) resolves (4, 65). Within a subject, the outcomes for sites are not independent; more subjects form pustules at all sites or resolve all sites than have mixed outcomes (63). There is no effect of ethnicity or dose on papule or pustule formation (reference 14 and unpublished data), and the major predictors of pustule formation are gender and host (63). Men and women form papules (become infected) at similar rates, but the odds ratio of men developing pustules is approximately threefold that of women, reflecting the epidemiology of natural chancroid (14). Of 201 volunteers infected once, all sites of infection resolved in 57 (27.4%), while pustules formed at one or more sites in 146 (72.6%). When gender-matched pustule formers and resolvers are reinfected at three sites, 60% of the resolvers and 18% of the pustule formers resolve all inoculated sites (P = 0.029), confirming a host effect on pustule formation (63). This trial allowed us to define two phenotypes in the model, called the PP (subjects who formed at least one pustule in both of two challenges) and RR (subjects in whom all sites resolved in both of two challenges) groups. The mechanism of disease resolution in the RR group is unknown but could be due to the ability of phagocytes or serum to ingest or kill the organism. Interestingly, there are no differences in the abilities of isolated PMN and macrophages derived from the blood of PP and RR subjects to ingest H. ducreyi or in the ability of their sera to kill the organism in vitro (63). These data led us to hypothesize that the microenvironment at the site of infection, determined in part by the bacteria, DC, and T cells, could modulate the function of PMN and macrophages and the success or failure of the host response (63).
Interactions between microbes and DC drive T-cell responses to pathogens (58). Animal models suggest that these interactions may foster T-cell differentiation that promotes either bacterial persistence and abscess formation or clearance and disease resolution (75-77). Type 1 DC (DC1), which present antigen in the context of cytokines such as interleukin-12 (IL-12), IL-18, alpha interferon (IFN-α), and IFN-β and of costimulatory molecules such as CD80 and ICAM-1, promote Th1 responses that facilitate phagocytosis (18, 61, 74). Regulatory DC (DCreg), which present antigen in the context of IL-10 and transforming growth factor β (TGF-β) and express costimulatory molecules such as programmed cell death ligand 1, programmed cell death ligand 2, and Notch, promote the development of regulatory T cells (Treg) (18, 23, 47, 49). Treg suppress Th1 responses and either facilitate organism survival or limit tissue damage by dampening the host response (29, 47, 49). Recently, another class of effector T cells called Th17 cells, which are important in the host defense against extracellular bacteria such as Klebsiella pneumoniae, was described (68). Although it is unclear whether a specific type of DC promotes Th17 development, IL-6 and TGF-β are required for the differentiation and IL-23 for the expansion of Th17 cells in mice (68). In contrast, TGF-β inhibits and IL-1β, IL-6, and IL-23 promote Th17 development in humans (2, 40, 83).
In this study, we examined whether differential DC and T-cell responses to H. ducreyi were associated with the PP and RR phenotypes. To characterize the immune response in tissue, we examined the transcriptional response in papules of PP and RR subjects who were infected a third time. To examine the role of DC in directing the PP and RR responses, we characterized the transcriptional response of blood-derived myeloid DC to H. ducreyi. To test whether cross-reactive T cells primed by commensals were responsible for the PP and RR phenotypes, we characterized proliferative responses of CD4 cells to H. ducreyi and related species of the Pasteurellaceae that colonize humans. We found that the PP and RR groups had divergent transcript responses to the organism in skin and in DC but no evidence that cross-reactive T cells were responsible for the phenotypes. To our knowledge, this is the first report associating transcriptional profiles for DC with the outcome of an infectious disease in humans.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. ducreyi 35000HP is a human-passaged variant of strain 35000 (4). Actinobacillus actinomycetemcomitans (ATCC 29522) was a gift from Dominique Galli, and nontypeable Haemophilus influenzae 1479 was a gift from Timothy Murphy (53). Haemophilus parainfluenzae (ATCC 33392), Haemophilus parahaemolyticus (ATCC 10014), Haemophilus paraphrohaemolyticus (ATCC 29237), and Haemophilus parainfluenzae (“Haemophilus paraphrophilus”) (ATCC 29242) were purchased from the American Type Culture Collection, Manassas, VA. All bacteria were cultured as described previously (25).
Human inoculation experiments.
Eight healthy adult volunteers (seven female and one male; mean age ± standard deviation, 37 ± 12 years), who had been infected twice and either had formed at least one pustule twice or had had the disease resolve at all sites twice, enrolled in the study. Informed consent was obtained from the subjects for human immunodeficiency virus type 1 serology and participation in accordance with the guidelines for human experimentation of the U.S. Department of Health and Human Services and the Institutional Review Board of Indiana University-Purdue University at Indianapolis. One RR subject was excluded due to an underlying illness, one PP subject withdrew from iteration 1 due to a scheduling conflict, and six subjects were infected (Table 1). Procedures for enrollment, exclusion criteria, preparation of the bacteria, inoculation, calculation of the estimated delivered dose, clinical observations, biopsy, and treatment with antibiotics were as described elsewhere (4, 65, 66).
TABLE 1.
PP and RR subjects
| Volunteer no. | Reference for initial infection/second infection | Group | Gendera | Iteration | Estimated delivered dose (CFU) |
|---|---|---|---|---|---|
| 149 | 7/63 | PP | F | 1 | 164 |
| 203 | 25/63 | RR | F | 1 | 164 |
| 216 | 64/63 | RR | F | 1 | 164 |
| 164 | 73/63 | PP | M | 2 | 227 |
| 200 | 25/63 | RR | F | 2 | 227 |
| 243 | 22/this study | PP | F | 3 | 96 |
F, female; M, male.
DC-H. ducreyi cocultures.
Peripheral blood was obtained from the subjects several months after they were infected a third time. CD14+ peripheral blood monocytes were isolated from peripheral blood mononuclear cells (PBMC) by positive-selection magnetic beads (Miltenyi Biotec, Auburn, CA) and grown in the presence of recombinant human IL-4 (1 ng/ml) and recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) (0.2 ng/ml) (R&D Systems, Minneapolis, MN). After 7 days of culture, the cells were HLA-DR+, CD86+ CD40+, CD3−, CD14−, and CD19−. DC were incubated with nonopsonized H. ducreyi for 90 min at a multiplicity of infection of 20:1, washed to remove nonassociated bacteria, and incubated for an additional 22.5 h. The experiment was done in pairs so that DC from one RR subject were exposed to the same bacterial inoculum as DC from one PP subject. Cells were collected and processed for RNA. Supernatants were collected and analyzed for cytokines using the human Th1/Th2 II cytometric bead array kit per the manufacturer's instructions (BD Biosciences). Cytokine levels were defined as the cytokine concentration in an infected well minus the concentration in the corresponding uninfected well.
Flow cytometry.
Biopsies were processed, and cells were collected and stained, as described previously (32), with the following antibodies conjugated to fluorescein isothiocyanate, allophycocyanin, phycoerythrin, or peridinin chlorophyll a proteins: CD3 (clone SK7), CD14 (MfP9), CD19 (4G7), and CD45 (2D1) (BD Biosciences, San Jose, CA). Monocyte-derived DC were stained with the following antibodies conjugated to fluorescein isothiocyanate, allophycocyanin, phycoerythrin, CyChrome, or peridinin chlorophyll a proteins: CD80 (clone L307.4), CD86 (2331), HLA-DR (646-6), CD40 (5C3), and CD83 (HB15e) (BD Biosciences).
Isolation of RNA.
Skin specimens were placed in RNA Later (QIAGEN, Valencia, CA) for 30 min immediately after biopsy. Tissues were homogenized in lysis buffer using a Mini Bead Beater (Research Products International, Mt. Prospect, IL) with 2.4-mm silica beads, and total RNA was isolated using the RNeasy fibrous tissue minikit (QIAGEN). RNA was prepared from DC using the RNeasy kit (QIAGEN). An additional DNase step was performed on all samples using Ambion's DNA-free kit (Ambion, Austin, TX). RNA integrity was assessed by capillary gel electrophoresis on an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Microarrays.
The same amount (100 ng) of total RNA was used from all samples, which were processed in parallel using premixes to minimize experimental variation. Samples were amplified using the Affymetrix two-cycle protocol. First- and second-strand cDNA syntheses were done using the Affymetrix GeneChip Expression 3′ Amplification two-cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA). The first in vitro transcription used the Ambion MEGAscript T7 high-yield transcription kit (Ambion); the second used the Affymetrix GeneChip Expression 3′ Amplification IVT labeling kit. Biotinylated cRNA was fragmented following standard procedures. Each sample was hybridized to a separate human genome U133 Plus 2 array, which contains approximately 55,000 probe sets corresponding to approximately 39,500 well-substantiated genes, as described previously (46). The arrays were scanned on the Affymetrix GeneChip Scanner 3000 controlled by GCOS software, and data were extracted using the Microarray Software Suite 5 (MAS5) algorithm. Each sample was scaled to a target intensity of 1,000. Probe sets that were absent from more than 66% of the samples in all four groups (PP infected [PPI], PP uninfected [PPU], RR infected [RRI], and RR uninfected [RRU]) were removed before further analysis (45).
After applying the 66% present/absent filter, genome-wide expression data were analyzed using singular value decomposition (SVD), which is also known as principal-component analysis (5). SVD is a mathematical transformation that filters noise and redundant information in expression data, searches for values called eigenvectors that explain the variation of the high-dimensional data, and examines whether there is any separation or clustering in the data. Array samples were plotted in a two-dimensional scatter plot, in which the x or y coordinates represented their projections to two eigenvectors, representing infected versus uninfected samples and PP versus RR subjects.
Array-to-array variation was normalized within either the PP or the RR group, using nonlinear normalization techniques (12). A log-normal distribution was assumed for gene expression data. As both infected and uninfected samples were from either a PP or RR subject, a two-way analysis of variance model that accommodated correlated samples was employed to compare transcript levels under multiple conditions, and P values were calculated with t tests [lm() in R, www.r-project.org]. False-discovery rates (FDR) were calculated for different levels of significance using the following formula: minimum [1, (P value) × (number of transcripts present)/(number of transcripts detected at that P value)].
To examine the biological functions of these transcripts, we utilized websites such as Biocarta (http://www.biocarta.com/) and OMIM (Online Mendelian Inheritance in Man; NCBI). Transcripts were reviewed for polarization of expression of signature genes for different blood cell types (www.affymetrix.com/support/technical/technotes/bloodappendix_technote.pdf), apoptosis pathways, Toll-like receptor (TLR) pathways, cell surface activation markers, cytokines, cytokine receptors, chemokines, and chemokine receptors that may account for different immune responses in the two groups. The two lists were further classified for function using the Gene Ontology categories (http://www.geneontology.org/index.shtml).
The supplemental material includes tables summarizing the immunologically related genes at a ≥2-fold change and a P value of <0.05 for the tissue and DC microarrays. For genes that were detected with multiple probe sets, all values were examined to ensure consistent results, and the probe set with the highest fold change was reported. In the case of discrepant results, we used the results from probe sets corresponding to the 3′ end of the transcript, which are more accurate than those from the 5′ end when RNA is amplified (46). The raw data are available at http://www.ncbi.nlm.nih.gov/geo/ under accession number GSE5574.
Real-time PCR.
IL-10, IL-12, and migration inhibition factor (MIF) were quantified relative to the level of mRNA for RPL35A, which had little sample-to-sample variation in the DC microarrays. The primers and TaqMan probes were designed using Roche's Universal Probe Library Assay Design Center (see Table VI at https://www.roche-applied-science.com). cDNA was made from the same RNA samples used for the microarray experiments with the Roche Transcriptor first-strand cDNA synthesis kit per the manufacturer's instructions. The reaction mixtures consisted of 500 nM of each primer, 5 mM MgCl2, and 200 nM of each probe (LC Fast Start DNA master mix; Roche). The samples were amplified on a Roche LightCycler (94°C for 5 min; 45 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and a final step at 72°C for 7 min). Differences in transcript levels between the groups were compared using the Student t test and the Wilcoxon rank sum test.
Proliferation assays.
PBMC were isolated by Ficoll-Hypaque density gradient centrifugation, frozen, and stored in fetal calf serum with 10% dimethyl sulfoxide. Proliferation assays were performed using the Vybrant CFDA SE cell tracer kit (Molecular Probes) (44, 81). PBMC were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) and incubated with medium alone, 5 μg/ml phytohemagglutinin (PHA), or 5 μg/ml heat-inactivated bacterial cells (25) at 37°C with 5% CO2. Cells were harvested, washed, and stained with propidium iodide and anti-CD4-allophycocyanin (BD Biosciences). Live CD4+ cells were analyzed for expression of CFSE using a FACSCalibur flow cytometer and Cell Quest software (BD Biosciences). The cell division index (CDI) was calculated as (number of CD4+ CFSEdim cells with antigen)/(number of CD4+ CFSEdim cells without antigen). A CDI of greater than 2 is considered a positive response in this assay (44).
RESULTS
Reinfection of PP and RR subjects.
To examine whether subjects who had different clinical outcomes had distinct immune responses at the transcript level to H. ducreyi in tissue, we inoculated PP and RR subjects at three sites with live H. ducreyi 35000HP and at one site with a phosphate-buffered saline control and biopsied all sites 48 h later. We reasoned that by 48 h we could still capture transcripts in RR lesions and compare them to those found in PP lesions and that the infected sites should reflect the response to the bacteria and wounds, while the control site should reflect the response to the wounds. We hypothesized that the PP and RR groups would have different gene expression profiles at their infected sites in response to H. ducreyi.
We infected three subjects in iteration 1 and two subjects in iteration 2 (Table 1). Papules formed at all sites inoculated with live bacteria. We biopsied four sites per subject 48 h after inoculation. RNA was made from the infected site that had the largest diameter of erythema and from the phosphate-buffered saline control site. The biopsies of the remaining two infected sites were pooled and processed for flow cytometry. The mean numbers (PP, 110,000; RR, 92,000) of leukocytes per biopsy obtained from the two groups were not statistically different. The mean percentages of major cell types in the PP and RR lesions were similar for PMN (8.6 versus 10.2), CD14 cells (27.1 versus 36.2), and CD3 cells (63.7 versus 52.7), suggesting that differences in transcript profiles may not merely be due to differences in cell numbers but may reflect qualitative differences in the host response.
Several months after the first five subjects were infected, a sixth subject (no. 243) was infected and formed only one papule. She underwent two biopsies (from the infected site and the wounded site). These biopsies were processed only for RNA.
Differential transcript responses to H. ducreyi in skin are associated with the PP and RR phenotypes.
The 12 tissue samples contained intact RNA and were hybridized to separate Affymetrix Human Genome U133 Plus 2 Arrays. SVD showed that the arrays were separated into four distinct groups by two eigenvectors, representing infected versus uninfected samples and PP versus RR subjects (Fig. 1). This analysis suggested that there were distinct global transcriptional responses in infected and uninfected sites and in PP and RR subjects.
FIG. 1.
Plot of SVD of the 12 tissue arrays. The x axis represents the projections of 12 samples onto the eigenvector that depicts the major variation between infected and uninfected samples. The arrow on the x axis points toward higher expression levels in infected samples than in uninfected samples. The y axis represents the projections of 12 samples onto the other eigenvector, which depicts the major variation between PP and RR subjects. The arrow on the y axis points toward higher expression levels in PP samples than in RR samples. Thus, the analysis separates the arrays into four groups that reflect the biology of the PP and RR phenotypes and infected and uninfected sites. Closed circles, RRU; open circles, RRI; closed squares, PPU; open squares, PPI.
We identified transcripts that were up- or downregulated in the infected versus uninfected tissue within each group of subjects with a mixed-model analysis of variance to account for the pairing of an experimental sample and a control sample within each subject. As expected, many transcripts were different, reflecting the infiltration of immune cells at infected sites versus wounded sites. For the comparison between PPI and PPU tissue, there were 9,604 probe sets representing 6,095 unique genes that were differentially regulated at a P value of ≤0.05 with an FDR of 0.14. For the comparison between RRI and RRU tissue, there were 9,284 probe sets representing 5,650 unique genes that were differentially regulated at a P value of ≤0.05 with an FDR of 0.14.
We examined whether the transcripts identified as up- or downregulated in the PPI versus PPU tissues overlapped with those identified in the RRI versus RRU tissues. The tissue RNA was used in its entirety to perform the arrays, and the level of transcript expression could not be validated by quantitative PCR. Thus, we limited our analysis to transcripts with at least twofold difference in expression level at a P value of ≤0.05. Of 4,268 transcripts corresponding to unique genes, 1,543 were found in both groups, while 1,565 were unique to the tissue from the PP group and 1,160 were unique to the tissue from the RR group.
Of the 1,543 common transcripts, we identified 268 that were relevant to immune function and that were differentially regulated in both the PPI versus PPU comparison and the RRI versus RRU comparison (see Table SI in the supplemental material). For the common transcripts, the directions (up- or downregulated) and magnitudes of change were similar in the two groups. These transcripts reflected the infiltration of leukocytes into the infected sites and indicated that the two groups have a core response to H. ducreyi. Upregulated transcripts included markers of antigen presentation and processing, chemokine and chemokine receptors, complement components, cytokines and cytokine receptors, adhesion molecules, IFN-inducible molecules, TLRs, and tumor necrosis factor (TNF)-induced transcripts.
Of the 1,565 transcripts unique to the comparison of the PPI and PPU groups, we identified 52 that were relevant to immune function (see Table SI in the supplemental material). Of the 1,160 transcripts unique to the comparison of the RRI and RRU groups, 58 were relevant to immune function (see Table SI in the supplemental material). Several adhesion molecules, complement and complement-related transcripts, and markers of differentiation, maturation, and activation were upregulated in the RRI sites, while such molecules were downregulated in the PPI sites (Table 2). Several more chemokine and chemokine receptors were upregulated in the RRI sites than in the PPI sites (Table 2). The RRI sites contained downregulated transcripts for markers of tolerance (TGFΒ2 and TGFB1I4) and several prostaglandin synthetases and had upregulated transcripts for CD163 and IL1RN, while the PP sites had upregulated transcripts for several cytokines and cytokine receptors, suggesting that inflammation was resolving at the RR sites and not at the PP sites (21, 51) (Table 2). Taken together, the data are reflective of an effective immune response at the RR sites.
TABLE 2.
Selected transcripts that were differentially regulated in tissue from only one group of volunteers
| Molecule group | Molecule | Regulationa in group:
|
|
|---|---|---|---|
| RR | PP | ||
| Adhesion molecules | ICAM3 | Up | |
| ITGAL | Up | ||
| ITGB7 | Up | ||
| PECAM1 | Up | ||
| CD36 | Down | ||
| ITGB1BP1 | Down | ||
| Chemokines/chemokine | CCL21 | Up | |
| receptors | CCL22 | Up | |
| CCR6 | Up | ||
| CXCL6 | Up | ||
| XCL1 | Up | ||
| XCL2 | Up | ||
| CXCL12 | Down | ||
| CXCL2 | Up | ||
| CXCL5 | Up | ||
| Complement/complement | C1QG | Up | |
| related | C2 | Up | |
| CD59 | Up | ||
| C7 | Down | ||
| Cytokines, receptors, | TNFSF8 | Up | |
| regulators | IL13RA2 | Down | |
| ILF3 | Down | ||
| TGFB2 | Down | ||
| TGFB1I4 | Down | ||
| IL22RA2 | Up | ||
| IL1R1 | Up | ||
| MIF | Up | ||
| TGFBR1 | Up | ||
| TGFBR2 | Up | ||
| TNFRSF12A | Up | ||
| TRAF3 | Up | ||
| IFNGR2 | Down | ||
| Differentiation/maturation/ | CD163 | Up | |
| activation | CD68 | Up | |
| DCNP1 | Up | ||
| HAVCR2 | Up | ||
| ILT7 | Up | ||
| LY75 | Up | ||
| JAM2 | Down | ||
| MAL | Down | ||
| MMD | Down | ||
| Immunomodulatory | IL1RN | Up | |
| LAIR1 | Up | ||
| KLRF1 | Up | ||
| LILRB3 | Up | ||
| Prostaglandins and | PTGES | Up | |
| leukotrienes | ALOX15B | Down | |
| ALOXE3 | Down | ||
| PGDS | Down | ||
| PTGS1 | Down | ||
| LTB4R | Up | ||
Up, gene transcript upregulated >2-fold in infected versus uninfected tissue; down, gene transcript downregulated >2-fold in infected versus uninfected tissue (P < 0.05).
An example of qualitative differences between PP and RR sites is shown in Table 3. Nuclear factor-erythroid 2-related factor 2 (Nrf2) is a transcription factor that regulates the expression of antioxidant genes, whose function results in posttranslational modification of many kinases downstream of TLR4 signaling. Deficiency of Nrf2 leads to a decreased redox state and increased activity of both the MyD88-dependent and MyD88-independent TLR4 pathways (38, 72). In addition, Nrf2 deficiency leads to higher levels of transcript expression in both TLR4 pathways. In models of sepsis and inflammation, Nrf2-deficient mice have higher mortality rates and expression levels of proinflammatory cytokines than wild-type mice (72). We examined the arrays to see if antioxidant synthesis genes and the TLR4 signaling pathways were differentially regulated in the groups. The PPI tissue contained 13 downregulated and 5 upregulated transcripts for glutathione synthesis, while the RRI tissue had 5 downregulated and 4 upregulated transcripts (Table 3). The PPI tissue also contained eight upregulated transcripts and one downregulated transcript in the TLR4 signaling pathways, while the RRI tissue had three upregulated and two downregulated transcripts (Table 3). Taken together, this transcript pattern suggests that the PP response in skin represents a dysregulated, hyperinflammatory state.
TABLE 3.
Oxidative stress response and TLR4 signaling pathway transcripts in infected versus uninfected skin from both groups of volunteers
| Category and gene symbol | Protein | Change in transcript levela in group:
|
|
|---|---|---|---|
| RR | PP | ||
| Oxidative stress response | |||
| CATb | Catalase | — | −2.9 |
| CREB1 | CAMP-responsive element binding protein 1 | 1.7 | 2.2 |
| GPX2 | Glutathione peroxidase 2 | 1.7 | — |
| GSTA1 | Glutathione S-transferase A1 | — | −1.7 |
| GSTA4 | Glutathione S-transferase A4 | −1.9 | −2.7 |
| GSTK1 | Glutathione S-transferase kappa 1 | 2.3 | 1.6 |
| GSTM1b | Glutathione S-transferase M1 | −2.4 | −2.5 |
| GSTM2 | Glutathione S-transferase M2 | −2.4 | −2.7 |
| GSTM3 | Glutathione S-transferase M3 | — | −3.3 |
| GSTM4 | Glutathione S-transferase M4 | — | −2.0 |
| GSTM5 | Glutathione S-transferase M5 | −5.1 | −5.8 |
| GSTO1b | Glutathione S-transferase omega 1 | 3.0 | 2.4 |
| GSTO2 | Glutathione S-transferase omega 2 | −1.8 | −2.0 |
| GSTT1 | Glutathione S-transferase theta 1 | — | −3.0 |
| MGST1b | Microsomal glutathione S-transferase 1 | — | −3.1 |
| MGST3 | Microsomal glutathione S-transferase 3 | — | −2.1 |
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | — | −6.1 |
| TLR4 signaling pathways | |||
| IRAK2 | IL-1 receptor-associated kinase 2 | 3.1 | — |
| IRF3 | IFN regulatory factor 3 | — | 2.1 |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 | −2.1 | — |
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 | — | 2.2 |
| MAP3K7IP3 | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 | — | 2.6 |
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | — | 2.4 |
| MAPK14 | Mitogen-activated protein kinase 14 | −2.0 | — |
| MAPK10 | Mitogen-activated protein kinase 10 | — | −2.4 |
| MAPKAP1 | Mitogen-activated protein kinase-associated protein 1 | — | 1.4 |
| MAPKAPK2 | Mitogen-activated protein kinase-activated protein kinase 2 | — | 1.6 |
| MYD88 | Myeloid differentiation primary response gene 88 | 3.5 | 4.9 |
| TBK1 | TANK-binding kinase 1 | — | 2.1 |
| TICAM2b | TLR adaptor molecule 2 | 2.4 | — |
Values are fold changes in transcript levels between infected and uninfected tissue (P < 0.05). —, not significantly different between infected and uninfected tissue.
Transcript detected by multiple probe sets; values shown are from the probe set that had the largest fold change for one of the groups.
Differential DC responses to H. ducreyi are associated with the PP and RR phenotypes.
Myeloid DC are enriched relative to plasmacytoid DC in pustules of subjects infected once (5a). Thus, myeloid DC could orchestrate the differential immune response to the organism in infected tissue. To examine whether DC have differential responses to the bacteria, we derived myeloid DC from PBMC of the PP and RR subjects. After 7 days, DC were incubated with nonopsonized H. ducreyi or in medium alone for 24 h. DC were stained with CD40, CD80, CD83, CD86, and HLA-DR. There was no difference in the percentage or in the mean fluorescence intensity of infected DC that expressed these surface markers compared to uninfected DC that were allowed to mature in the wells for 24 h (data not shown), consistent with previous observations (5a). No differences in expression of these markers were noted between the PP and RR groups.
The 12 DC samples containing intact RNA were hybridized to Affymetrix arrays and analyzed exactly as described for the tissue arrays. At a P value of ≤0.05, for the comparison of RRI and RRU DC, there were 3,317 probe sets representing 2,433 unique genes that were up- or downregulated with an FDR of 0.28. For the comparison of PPI and PPU DC, 3,134 probe sets representing 2,347 unique genes were differentially regulated with an FDR of 0.30.
We examined whether the subset of transcripts that were at least twofold differentially regulated in the RRI versus RRU DC overlapped with the same subset identified in the PPI versus PPU DC. A total of 2,021 transcripts corresponding to unique genes were twofold differentially regulated in both groups, with 464 transcripts unique to the RRI versus RRU DC, 1,107 unique to the PPI versus PPU DC, and 451 common to the two groups.
There were 81 transcripts known to be relevant to immune responses that were common to the infected DC from both groups (see Table SII in the supplemental material). The direction of change was the same for all the common transcripts in the two groups. Key transcripts (Table 4) associated with DC maturation or polarization of T cells that were upregulated in both groups included CD80, ICAM-1, IL-1α, IL-1β, IL-6, IL-8, IL-12p40, IL-15, IL-2Rα, TNF-α, several TNF- and IFN-induced proteins, Jagged-1, and G-CSF. Except for upregulation of programmed cell death ligand 1, the common response to H. ducreyi was generally consistent with a DC1 profile (18).
TABLE 4.
Selected transcripts that were differentially regulated in infected versus uninfected DC derived from both groups of volunteers
| Category and gene symbol | Protein | Change in transcript levela in group:
|
|
|---|---|---|---|
| RR | PP | ||
| DC1 | |||
| CD44b | CD44 antigen (homing function; Indian blood group) | 3.1 | 5.5 |
| CSF3 | Colony-stimulating factor 3 (G-CSF) | 38.0 | 56.6 |
| IL1A | IL-1, alpha | 53.0 | 93.4 |
| IL1Bb | IL-1, beta | 73.9 | 59.2 |
| IL2RA | IL-2 receptor, alpha | 15.8 | 49.4 |
| IL6 | IL-6 (IFN, beta 2) | 247.9 | 692.4 |
| IL8 | IL-8 | 54.5 | 163.6 |
| IL10RA | IL-10 receptor, alpha | 2.2 | 2.3 |
| IL12B | IL-12B (IL-12p40) | 57.0 | 274.8 |
| IL15 | IL-15 | 7.9 | 35.2 |
| JAG1 | Jagged 1 (Alagille syndrome) | 6.4 | 4.0 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 411.2 | 1,021.5 |
| TNFα | TNF (TNF superfamily, member 2) | 14.4 | 51.0 |
| IFN or TNF induced | |||
| GBP1b | Guanylate binding protein 1, IFN inducible, 67 kDa | 17.3 | 28.0 |
| GBP2 | Guanylate binding protein 2, IFN inducible | 9.0 | 20.9 |
| ISG20 | IFN-stimulated gene product, 20 kDa | 30.0 | 372.0 |
| TNFαIP2 | TNF-α-induced protein 2 | 3.6 | 6.4 |
| TNFαIP3b | TNF-α-induced protein 3 | 6.6 | 6.2 |
| TNFαIP6 | TNF-α-induced protein 6 | 51.4 | 98.7 |
| DC maturation | |||
| CD1C | CD1C antigen, c polypeptide | −4.0 | −3.4 |
| CD80 | CD80 antigen (CD28 antigen ligand 1, B7-1 antigen) | 20.1 | 44.9 |
| ICAM-1b | Intercellular adhesion molecule 1 (CD54) | 11.0 | 14.3 |
| DCreg | |||
| PDCD1LG1b | Programmed cell death 1 ligand 1 | 4.2 | 8.4 |
| Signal transduction | |||
| NFKB1 | NF-κB (p105) | 4.4 | 5.0 |
| STAT3 | Signal transducer and activator of transcription 3 | 3.6 | 5.6 |
| TLR/NLRc pathways | |||
| CARD15 | Caspase recruitment domain family, member 15 (NOD2) | 11.8 | 10.5 |
| IRAK3 | IL-1 receptor-associated kinase | 3.6 | 6.7 |
| TLR4b | TLR4 | −2.6 | −3.3 |
| TRAF1b | TNF receptor-associated factor 1 | 8.3 | 9.5 |
Values are fold changes in transcript levels between infected and uninfected tissue (P < 0.05).
Transcript detected by multiple probe sets; values shown are from the probe set that had the largest fold change for one of the groups.
NLR, NOD-like receptor.
There were 74 differentially regulated transcripts unique to infected DC from the PP group (see Table SII in the supplemental material). Key transcripts (Table 5) included many markers of DC1, such as CCR7, CD38, CD58, IL-7, IL-18, LAMP3, and GM-CSF (9, 18, 49, 85). Caspase 1, which processes pro-IL-18, was also upregulated (18). Many IFN-responsive genes were also upregulated. The marked upregulation of GM-CSF and other proinflammatory cytokines suggests that the PP DC could sponsor a hyperinflammatory state. However, the PP DC differentially regulated many transcripts that were markers of DCreg, such as IL-10, CD200, and Notch homolog 2 (17, 18, 37, 48) (Table 5). INDO, encoding IDO, which leads to tryptophan depletion and T-cell death, was upregulated 173-fold. IL-16, which fosters DC maturation, was downregulated, as were CD1D and CD84. SOCS2 and SOCS3, which interfere with TLR 4 signaling and DC function (69), were upregulated. Major histocompatibility complex (MHC) class II DQα, HLA-B, and HLA-F were upregulated, but MHC class II DMβ, MHC class II DMα, and HM13 were downregulated, which could lead to defects in antigen processing and presentation.
TABLE 5.
Selected transcripts that were differentially regulated in infected versus uninfected DC from only one group of volunteers
| Category and gene symbol | Protein | Change in transcript levela in group:
|
|
|---|---|---|---|
| RR | PP | ||
| DC1 or DC maturation | |||
| CD38 | CD38 antigen (p45) | — | 73.3 |
| CCR7 | Chemokine (C-C motif) receptor 7 | — | 17.1 |
| CD58b | CD58 antigen (lymphocyte function-associated antigen 3) | — | 2.9 |
| LAMP3 | Lysosome-associated membrane protein 3 | — | 14.2 |
| GM-CSF | Colony-stimulating factor 2 (GM-CSF) | — | 50.4 |
| IL1F9 | IL-1 family, member 9 | 3.3 | — |
| IL7 | IL-7 | — | 2.2 |
| IL16 | IL-16 (lymphocyte chemoattractant factor) | — | −4.3 |
| IL18 | IL-18 (IFN-γ-inducing factor) | — | 14.6 |
| CASP1b | Caspase 1 (apoptosis-related cysteine protease) | — | 8.9 |
| IL23α | IL-23, alpha subunit p19 | 3.0 | — |
| IRF4 | IFN regulatory factor 4 | 2.5 | — |
| IRF1 | IFN regulatory factor 1 | 6.8 | — |
| Immunomodulatory | |||
| SOCS2 | Suppressor of cytokine signaling 2 | — | 5.5 |
| SOCS3 | Suppressor of cytokine signaling 3 | — | 30.0 |
| Signal transduction | |||
| STAT4 | Signal transducer and activator of transcription 4 | — | 23.5 |
| STAT1 | Signal transducer and activator of transcription 1 | 2.0 | — |
| Inhibition of maturation | |||
| LILRB1b | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | −2.0 | — |
| DCreg | |||
| CD200 | CD200 antigen | — | 38.8 |
| CD84 | CD84 antigen (leukocyte antigen) | — | −3.0 |
| INDO | Indoleamine-pyrrole 2,3-dioxygenase | — | 173.2 |
| IL10 | IL-10 | — | 7.4 |
| NOTCH2NL | Notch homolog 2 (Drosophila), N-terminal like | — | 2.5 |
| Proinflammatory | |||
| MIF | Macrophage MIF | 2.0 | — |
| TLR pathways | |||
| IRAK1BP1 | IL-1 receptor-associated kinase 1 binding protein 1 | — | 2.6 |
| TBK1 | Tank-binding kinase 1 | — | 2.0 |
| TLR1 | TLR1 | — | 3.5 |
| TRIF | TIR domain containing adaptor inducing IFN-β | — | 2.0 |
| IFN responsive | |||
| G1P3 | IFN, alpha-inducible protein (clone IFI-6-16) | — | 6.6 |
| IFI44 | IFN-induced protein 44 | — | 9.3 |
| IFIH1 | IFN induced with helicase C domain 1 | — | 3.3 |
| IFIT5 | IFN-induced protein with tetratricopeptide repeats 5 | — | 2.8 |
| IFITM1b | IFN-induced transmembrane protein 1 (9-27) | — | 18.4 |
| IFITM2 | IFN-induced transmembrane protein 2 (1-8D) | — | 7.0 |
| IFITM3 | IFN-induced transmembrane protein 3 (1-8U) | — | 10.0 |
| IFI44L | IFN-induced protein 44 | 9.1 | — |
| MNDA | Myeloid cell nuclear differentiation antigen | −2.5 | — |
| Antigen presentation | |||
| CD1D | CD1D antigen, d polypeptide | — | −13.1 |
| HLA-B | MHC, class I, B | — | 2.7 |
| HLA-DQα1 | MHC, class II, DQ alpha 1 | — | 3.0 |
| HLA-DMα | MHC, class II, DM alpha | — | −3.1 |
| HLA-DMβ | MHC, class II, DM beta | — | −11.2 |
| HLA-F | MHC, class I, F | — | 3.1 |
| HM13 | Histocompatibility (minor) 13 | — | −2.1 |
Values are fold changes in transcript levels between infected and uninfected tissue (P < 0.05). —, not significantly different between infected and uninfected tissue.
Transcript detected by multiple probe sets; values shown are from the probe set that had the largest fold change for one of the groups.
There were 28 differentially regulated transcripts unique to the infected DC from the RR group (see Table SII in the supplemental material). Key transcripts (Table 5) included only additional markers of DC1, such as upregulated IL-23αp19, IL-1F9, IRF1, IRF4, and MIF (18, 49, 85). LILRB1 (CD85j), an inhibitory receptor whose activation reduces production of IL-12 and polarizes towards Th2 responses (71), was downregulated.
IL-12p40 was upregulated in DC from both groups, but IL-23αp19 was upregulated only in the RR DC. STAT4 was upregulated in the PP DC, while STAT1 was upregulated in RR DC. This coordinate regulation may reflect programming for subsequent signaling through IL-12/STAT4 and IL-23/STAT1 (6). IL-1β and IL-6, which promote Th17 differentiation in humans, were upregulated in both groups. Since IL-23 also promotes human Th17 development or expansion, RR DC could promote a more profound Th17 response than PP DC. Taken together, the data indicate that while DC from the PP and RR groups share elements of a common response to H. ducreyi, other elements of the DC response are divergent. DC from the PP group express unique transcripts that should promote both Th1 and Treg responses, while DC from the RR group express transcripts should promote a Th1 response and promote expansion of Th17 cells by upregulation of IL-23.
Validation of the DC microarrays by quantitative PCR and cytokine analysis.
RPL35A (a ribosomal protein transcript) was not differentially regulated in either the PP or RR DC. MIF was upregulated in the infected RR DC, IL-10 was upregulated in the infected PP DC, and IL-12p40 was upregulated in both groups. By quantitative PCR, the levels of RPL35A transcripts were not statistically different when infected DC were compared to uninfected DC in both groups. After normalization to RPL35A transcript levels, MIF transcripts were significantly upregulated in the RR DC, IL-10 was significantly upregulated in the PP DC, and IL-12 p40 was significantly upregulated in both groups (all P < 0.01). The directions of the levels of expression of these transcripts were similar to those determined in the array (Table 6), although the magnitude of the fold change was generally higher in real-time PCR than in the array.
TABLE 6.
Comparison of DC transcripts by real-time PCR and microarrays
| Gene | Group | Fold changea by:
|
|
|---|---|---|---|
| Real-time PCR | Microarray | ||
| MIF | RR | 2.1 | 2.0 |
| IL-10 | PP | 47 | 7 |
| IL-12 | RR | 281 | 57 |
| IL-12 | PP | 3,754 | 275 |
Relative to RPL35A.
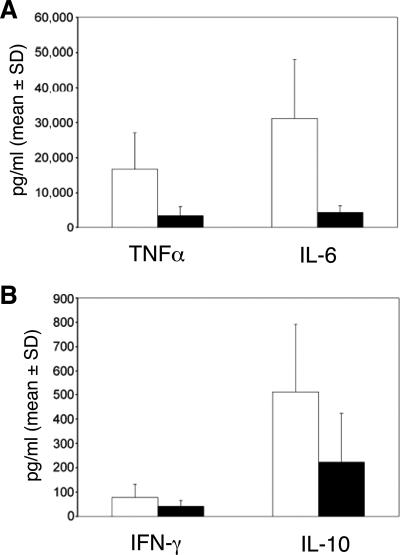
Cytokines were analyzed in the 24-h cell culture supernatants obtained from infected and uninfected DC. Although our sample size lacked sufficient power to determine whether the differences in cytokine levels were significant, the levels of expression of IL-10, TNF-α, and IL-6 (Fig. 2) paralleled the levels of transcript expression for these cytokines expressed by the two groups (Tables 4 and 5). Little IFN-γ was made by both groups, and transcripts for IFN-γ were not differentially regulated in either group. Thus, the cytokines levels produced by the DC were consistent with the relative levels of transcript expression in the microarrays.
FIG. 2.
Cytokine production as measured by cytometric bead array. The culture supernatants from the DC cocultures used for microarray analysis were used for determination of TNF-α and IL-6 (A) and for IFN-γ and IL-10 (B). Values represent the means + standard deviations (SD) of cytokine concentrations in infected wells minus those in uninfected wells from the three persons in each group. White bars, PP group; black bars, RR group.
Proliferative responses to H. ducreyi are not associated with the PP and RR phenotypes.
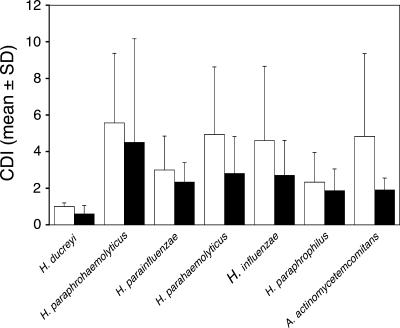
An alternative hypothesis for differential host susceptibility is that the RR group had cross-reactive memory cells that were primed by commensals and provided Th1 responses to H. ducreyi that facilitated clearance, while the PP group lacked such cells. For the five subjects who were inoculated in the first two iterations, PBMCs were obtained prior to the third challenge, 48 h and 7 to 10 days after infection. For all subjects, CD4 cells proliferated in response to PHA, but the mean CDIs for both groups were <1.5 at all three time points for all concentrations (1 to 20 μg/ml) of heat-inactivated H. ducreyi whole cells tested (data not shown). All six subjects were recalled approximately 6 months after the sixth subject was inoculated so that we could measure blastogenic responses to H. ducreyi and related species of the Pasteurellaceae. CD4 cells from the RR and PP groups did not proliferate in response to H. ducreyi (CDI, <1.0), although they did proliferate to various degrees in response to the other bacterial species (Fig. 3). The CDI for PHA was >9 for all samples tested. The concentration of IFN-γ in culture supernatants made in response to H. ducreyi did not differ between the two groups (data not shown). We repeated this experiment for blastogenesis and IFN-γ production in response to H. ducreyi, and the results were identical.
FIG. 3.
Proliferation of CD4 cells in response to various species of the Pasteurellaceae. The data represent the mean CDIs + standard deviations (SD) from the three subjects in each group. White bars, PP group; black bars, RR group.
DISCUSSION
In this study, we showed that the PP and RR subjects who underwent a third challenge shared a common transcript response to H. ducreyi in tissue but also had unique transcript responses, confirming that their immune responses were qualitatively distinct. We defined differentially regulated transcripts within each subject as the difference between the responses at the subject's infected site and wounded site. This approach allowed us to identify the component of the transcript response that was specific to the bacteria and not due to the wounds. The pairing of infected and uninfected samples within each subject removed considerable biological variability and was a strength of the experimental design. A limitation of our tissue array experiment was that our subjects were not gender matched. Although analysis of transcripts in infected tissue is complex in that the transcripts originate from many types of infiltrating cells, such experiments provide meaningful data about infectious processes. In our study, transcripts that were upregulated in the comparison of RRI and RRU skin signified an effective immune response, while transcripts in the comparison of PPI and PPU skin were consistent with a state of immune dysregulation. Similarly, profiling of skin biopsies obtained from patients with tuberculoid and lepromatous leprosy showed that each form of the disease was associated with a unique transcript profile consistent with the Th1 versus Th2 bias that marks each pole of the leprosy spectrum (9).
Human skin, the site of H. ducreyi infection, contains Langerhans cells and interstitial DC, which are of the myeloid lineage (33, 42, 58). By flow cytometry, myeloid DC are enriched in experimental pustules (5a). Using myeloid DC differentiated from blood monocytes as surrogates for lesional DC, we also found that DC from both groups shared a core response to H. ducreyi, which was typical of a DC1 response. However, DC from the PP group differentially regulated unique transcripts that should promote both Th1 and Treg responses to H. ducreyi, while those from the RR group differentially regulated unique transcripts that should promote only a Th1 response or allow for expansion of Th17 cells. Our data suggest that differential myeloid DC responses may be the primary mechanism underlying the host responses that result in the PP and RR phenotypes.
T-cell lines isolated from pustules proliferate specifically in response to H. ducreyi, indicating that volunteers who fail to clear the bacteria are sensitized during their initial infection (25). Some of these lines produce IFN-γ and IL-10 but no IL-4 in response to H. ducreyi, suggesting that they contain Th1 and presumptive Treg cells (25). Approximately 15% of subjects who have had pustules biopsied develop hypertrophic scars, demonstrating that an environment that favors excessive collagen synthesis occurs in pustules. TGF-β promotes synthesis of procollagen by fibroblasts and is thought to be important in the pathogenesis of hypertrophic scar formation (54), and TGF-β mRNA is readily amplified from pustules (unpublished data). Immunohistochemistry and flow cytometry reveal that FOXP3-expressing regulatory CD4 cells are also abundant in pustules of persons infected once (unpublished). Taken together, the data suggest that DC in pustule formers prime Th1 and Treg responses that likely promote phagocytic failure in tissue.
CD4 cells in PBMC of PP and RR subjects did not proliferate in response to H. ducreyi before, during, or after a third challenge. We have not yet compared lesional T cells derived from infected PP and RR subjects. We speculate that the lack of proliferation of blood CD4 cells may be due to the development of Treg in the PP group and a low precursor frequency of Th1 cells in the RR group, who clear infection within 2 to 3 days.
The current understanding of responses of antigen-presenting cells (APC) to pathogens is that phylogenetically distinct pathogens such as protozoa, helminths, viruses, and bacteria cause a pathogen-specific transcript response (16, 30). These studies employ APC from normal volunteers and have focused on the common response to the pathogen, not person-to-person variation. Heterogeneous transcript responses to Mycobacterium avium by macrophages derived from healthy volunteers occur (26), which led to the speculation that the heterogeneity could reflect differential donor susceptibility to M. avium. Similarly, variation among donors in the transcript response of PBMC to specific organisms occurs, but the significance of the differences is unclear (11). Had we focused on the differentially regulated transcripts that were shared by our volunteers, we would have concluded that the DC profile to H. ducreyi was a DC1 response. The PP and RR phenotypes allowed us to determine which transcripts were unique to each group and to interpret their biological significance. Our data show that DC from groups of patients who have distinct clinical outcomes when infected with a single pathogen have distinct transcript profiles in response to that pathogen.
Innate recognition of pathogen-associated molecular patterns (PAMPs) is mediated by TLRs. There is high variability in TLR surface expression on APCs derived from different people (79). Genetic polymorphisms in several TLRs are associated with differential human susceptibility to a variety of infectious agents, including staphylococci, Mycobacterium leprae, Neisseria, and Legionella (10, 20, 28, 43, 60). M. leprae produces distinct cytokine responses in tuberculoid and lepromatous leprosy, and these cytokines differentially modulate TLR activation and expression on DC (39). Common polymorphisms in cytokine genes are associated with protection against recurrent Chlamydia trachomatis infection (24, 55, 80). Variability in expression of TLRs, polymorphisms in TLRs, differential modulation of TLR expression or activation, and polymorphisms in cytokine genes could be mechanisms underlying the differential DC responses to H. ducreyi in our model.
Our subjects were normal, healthy adult volunteers who had no history of chronic skin diseases, such as atopic dermatitis, eczema, or psoriasis, or of chronic or recurrent infections. H. ducreyi strains are divided into two classes, which express different oligosaccharides (57), immunotypes or variants of several outer membrane proteins (82), and different proteomes (59). The majority of strains belong to class I, which is represented by 35000HP (82). We do not know if the PP group is more susceptible to infection by class II strains or other bacterial pathogens than the RR group. Similarly, whether the differential DC responses we describe in this study are class I specific or extend to class II strains or other bacterial species is unknown. Nevertheless, to our knowledge this is the first report associating DC transcript profiles with the outcome of an infectious disease in humans.
Future studies will be aimed at determination of the cellular origin of tissue transcripts, comparison of DC and T cells derived from tissues of infected PP and RR subjects, and definition of the mechanism underlying the differential DC responses to H. ducreyi. We will also determine whether myeloid DC derived from PP and RR subjects promote Th1 and Treg responses and Th1 responses, respectively, and whether DC from the two groups have divergent profiles in response to other bacteria. These studies should provide insight into mechanisms underlying differential host susceptibility to infection.
Supplementary Material
Acknowledgments
This work was supported by grants AI31494, AI27863, and AI059384 (to S.M.S.) from the National Institute of Allergy and Infectious Diseases (NIAID). T.L.H. and D.M.J. were supported by NIH grant T32 AI007637 from NIAID. The human challenge trials were supported by grant MO1RR00750 to the GCRC at Indiana University. The microarray experiments were done by the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by the Indiana Genomics Initiative and by The Lilly Endowment, Inc.
All authors have no relevant financial relationships to disclose.
We thank Ron Jerome and Chunxiao Zhu for technical assistance with the arrays; Tom Nutman for advice on analysis of the arrays; Byron Batteiger, Justin Radolf, and Barbara Van Der Pol for criticism of the manuscript; and the volunteers who participated in the trial.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 24 September 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abdullah, M., I. Nepluev, G. Afonina, S. Ram, P. Rice, W. Cade, and C. Elkins. 2005. Killing of dsrA mutants of Haemophilus ducreyi by normal human serum occurs via the classical complement pathway and is initiated by immunoglobulin M binding. Infect. Immun. 73:3431-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acosta-Rodriguez, E. V., G. Napolitani, A. Lanzavecchia, and F. Sallusto. 2007. Interleukins 1β and 6 but not transforming growth factor-β are essential for the differentiation of interleukin 17-producing human T helper cells. Nat. Immunol. 8:942-949. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, H. J., C. Johansson, L. A. Svensson, K. Ahlman, M. Verdrengh, and T. Lagergard. 2002. In vitro and in vivo interactions of Haemophilus ducreyi with host phagocytes. Infect. Immun. 70:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 178:1684-1687. [DOI] [PubMed] [Google Scholar]
- 5.Alter, O., P. O. Brown, and D. Botstein. 2000. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA 97:10101-10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Banks, K. E., T. L. Humphreys, W. Li, B. P. Katz, D. S. Wilkes, and S. M. Spinola. 2007. Haemophilus ducreyi partially activates human myeloid dendritic cells. Infect. Immun. 75:5678-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastos, K. R. B., C. R. F. Marhino, R. Barboza, M. Russo, J. M. Alvarez, and M. R. D'Imperio Lima. 2004. What kind of message does IL-12/IL-23 bring to macrophages and dendritic cells? Microbes Infect. 6:630-636. [DOI] [PubMed] [Google Scholar]
- 7.Bauer, M. E., M. P. Goheen, C. A. Townsend, and S. M. Spinola. 2001. Haemophilus ducreyi associates with phagocytes, collagen, and fibrin and remains extracellular throughout infection of human volunteers. Infect. Immun. 69:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer, M. E., C. A. Townsend, A. R. Ronald, and S. M. Spinola. 2006. Localization of Haemophilus ducreyi in naturally acquired chancroidal ulcers. Microbes Infect. 8:2465-2468. [DOI] [PubMed] [Google Scholar]
- 9.Bleharski, J. R., H. Li, C. Meinken, T. G. Graeber, M.-T. Ochoa, M. Yamamura, A. Burdick, E. N. Sarno, M. Wagner, M. Rollinghoff, T. H. Rea, M. Colonna, S. Stenger, B. R. Bloom, D. Eisenberg, and R. L. Modlin. 2003. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science 301:1527-1530. [DOI] [PubMed] [Google Scholar]
- 10.Bochud, P.-Y., T. R. Hawn, and A. Aderem. 2003. Cutting edge: a Toll-like receptor 2 polymorphism that is associated with lepromatous leprosy is unable to mediate mycobacterial signaling. J. Immunol. 170:3451-3454. [DOI] [PubMed] [Google Scholar]
- 11.Boldrick, J. C., A. A. Alizadeh, M. Diehn, S. Dudoit, C. L. Liu, C. E. Belcher, D. Botstein, L. M. Staudt, P. O. Brown, and D. A. Relman. 2002. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc. Natl. Acad. Sci. USA 99:972-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolstad, B. M., R. A. Irizarry, M. Amstrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 13.Bong, C. T. H., M. E. Bauer, and S. M. Spinola. 2002. Haemophilus ducreyi: clinical features, epidemiology, and prospects for disease control. Microbes Infect. 4:1141-1148. [DOI] [PubMed] [Google Scholar]
- 14.Bong, C. T. H., J. Harezlak, B. P. Katz, and S. M. Spinola. 2002. Men are more susceptible to pustule formation than women in the experimental model of Haemophilus ducreyi infection. Sex. Transm. Dis. 29:114-118. [DOI] [PubMed] [Google Scholar]
- 15.Bong, C. T. H., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. A DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 69:1488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaussabel, D., R. T. Semnani, M. A. McDowell, D. Sacks, A. Sher, and T. B. Nutman. 2003. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood 102:672-681. [DOI] [PubMed] [Google Scholar]
- 17.Clark, D. A. 2005. Tolerance signaling molecules. Chem. Immunol. Allergy 89:36-48. [DOI] [PubMed] [Google Scholar]
- 18.de Jong, E. C., H. H. Smits, and M. L. Kapsenberg. 2005. Dendritic cell-mediated T cell polarization. Springer Semin. Immunopathol. 26:289-307. [DOI] [PubMed] [Google Scholar]
- 19.Elkins, C., K. J. Morrow, and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emonts, M., J. A. Hazelzet, R. deGroot, and P. W. M. Hermans. 2003. Host genetic determinants of Neissera meningitidis infections. Lancet 3:565-577. [DOI] [PubMed] [Google Scholar]
- 21.Fabriek, B. O., C. D. Dijkstra, and T. K. van den Berg. 2005. The macrophage scavenger receptor CD163. Immunobiology 210:153-160. [DOI] [PubMed] [Google Scholar]
- 22.Fulcher, R. A., L. E. Cole, D. M. Janowicz, K. L. Toffer, K. R. Fortney, B. P. Katz, P. E. Orndorff, S. M. Spinola, and T. H. Kawula. 2006. Expression of Haemophilus ducreyi collagen binding outer membrane protein NcaA is required for virulence in swine and human challenge models of chancroid. Infect. Immun. 74:2651-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geijtenbeek, T. B. H., S. J. van Vliet, E. A. Koppel, M. Sanchez-Hernandez, C. M. J. E. Vandenbroucke-Grauls, B. Appelmelk, and Y. van Kooyk. 2003. Mycobacteria target DC-sign to suppress dendritic cell function. J. Exp. Med. 197:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisler, W. M., J. Tang, C. Wang, C. M. Wilson, and R. A. Kaslow. 2004. Epidemiological and genetic correlates of incident Chlamydia trachomatis infection in North American adolescents. J. Infect. Dis. 190:1723-1729. [DOI] [PubMed] [Google Scholar]
- 25.Gelfanova, V., T. L. Humphreys, and S. M. Spinola. 2001. Characterization of Haemophilus ducreyi-specific T-cell lines from lesions of experimentally infected human subjects. Infect. Immun. 69:4224-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenwell-Wild, T., N. Vazquez, D. Sim, M. Schito, D. Chatterjee, J. M. Orenstein, and S. M. Wahl. 2002. Mycobacterium avium infection and modulation of human macrophage gene expression. J. Immunol. 169:6286-6297. [DOI] [PubMed] [Google Scholar]
- 27.Hammond, G. W., M. Slutchuk, J. Scatliff, E. Sherman, J. C. Wilt, and A. R. Ronald. 1980. Epidemiologic, clinical, laboratory, and therapeutic features of an urban outbreak of chancroid in North America. Rev. Infect. Dis. 2:867-879. [DOI] [PubMed] [Google Scholar]
- 28.Hawn, T. R., A. Verbon, M. Janer, L. P. Zhao, B. Beutler, and A. Aderem. 2005. Toll-like receptor 4 polymorphisms are associated with resistance to Legionnaires’ disease. Proc. Natl. Acad. Sci. USA 102:2487-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins, S. C., E. C. Lavelle, C. McCann, B. Keogh, E. McNeela, P. Byrne, B. O'Gorman, A. Jarnicki, P. McGuirk, and K. H. G. Mills. 2003. Toll-like receptor 4-mediated innate IL-10 activates antigen-specific regulatory T cells and confers resistance to Bordetella pertussis by inhibiting inflammatory pathology. J. Immunol. 171:3119-3127. [DOI] [PubMed] [Google Scholar]
- 30.Huang, Q., D. Liu, P. Majewski, L. C. Schulte, J. M. Korn, R. A. Young, E. S. Lander, and N. Hacohen. 2001. The plasticity of dendritic cell responses to pathogens and their components. Science 294:870-875. [DOI] [PubMed] [Google Scholar]
- 31.Humphreys, T. L., L. A. Baldridge, S. D. Billings, J. J. Campbell, and S. M. Spinola. 2005. Trafficking pathways and characterization of CD4 and CD8 cells recruited to the skin of humans experimentally infected with Haemophilus ducreyi. Infect. Immun. 73:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphreys, T. L., C. T. Schnizlein-Bick, B. P. Katz, L. A. Baldridge, A. F. Hood, R. A. Hromas, and S. M. Spinola. 2002. Evolution of the cutaneous immune response to experimental Haemophilus ducreyi infection and its relevance to HIV-1 acquisition. J. Immunol. 169:6316-6323. [DOI] [PubMed] [Google Scholar]
- 33.Itano, A. A., and M. K. Jenkins. 2003. Antigen presentation to naive CD4 T cells in the lymph node. Nat. Immunol. 4:733-739. [DOI] [PubMed] [Google Scholar]
- 34.Janowicz, D., I. Leduc, K. R. Fortney, B. P. Katz, C. Elkins, and S. M. Spinola. 2006. A DltA mutant of Haemophilus ducreyi is partially attenuated in its ability to cause pustules in human volunteers. Infect. Immun. 74:1394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janowicz, D. M., K. R. Fortney, B. P. Katz, J. L. Latimer, K. Deng, E. J. Hansen, and S. M. Spinola. 2004. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infect. Immun. 72:4528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janowicz, D. M., K. Tenner-Racz, P. Racz, T. L. Humphreys, C. Schnizlein-Bick, K. R. Fortney, B. Zwickl, B. P. Katz, J. J. Campbell, D. D. Ho, and S. M. Spinola. 2007. Experimental infection with Haemophilus ducreyi in persons who are infected with HIV does not cause local or augment systemic viral replication. J. Infect. Dis. 195:1443-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kluppel, M., and J. L. Wrana. 2005. Turning it up a Notch: cross-talk between TGF beta and Notch signaling. Bioessays 27:115-118. [DOI] [PubMed] [Google Scholar]
- 38.Kolls, J. K. 2006. Oxidative stress in sepsis: a redox redux. J. Clin. Investig. 116:860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krutzik, S. R., M. T. Ochoa, P. A. Sieling, S. Uematsu, Y. W. Ng, A. Legaspi, P. T. Liu, S. T. Cole, P. J. Godowski, Y. Maeda, E. N. Sarno, M. V. Norgard, P. J. Brennan, S. Akira, T. H. Rea, and R. L. Modlin. 2003. Activation and regulation of Toll-like receptors 2 and 1 in human leprosy. Nat. Med. 9:525-532. [DOI] [PubMed] [Google Scholar]
- 40.Laurence, A., and J. O'Shea. 2007. TH-17 differentiation: of mice and men. Nat. Immunol. 8:903-905. [DOI] [PubMed] [Google Scholar]
- 41.Leduc, I., P. Richards, C. Davis, B. Schilling, and C. Elkins. 2004. A novel lectin, DltA, is required for expression of a full serum resistance phenotype in Haemophilus ducreyi. Infect. Immun. 72:3418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, Y.-J. 2001. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell 106:259-262. [DOI] [PubMed] [Google Scholar]
- 43.Lorenz, E., J. P. Mira, K. L. Cornish, N. C. Arbour, and D. A. Schwartz. 2000. A novel polymorphism in the Toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect. Immun. 68:6398-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mannering, S. I., J. S. Morris, K. P. Jensen, A. W. Purcell, M. C. Honeyman, P. M. vanEndert, and L. C. Harrison. 2003. A sensitive method for detecting proliferation of rare autoantigen-specific human T cells. J. Immunol. Methods 283:173-183. [DOI] [PubMed] [Google Scholar]
- 45.McClintick, J. N., and H. J. Edenberg. 2006. Effects of filtering by present call on analysis of microarray experiments. BMC Bioinformatics 7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClintick, J. N., R. E. Jerome, C. R. Nicholson, D. W. Crabb, and H. J. Edenberg. 2003. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics 4:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGuirk, P., and K. H. G. Mills. 2002. Pathogen-specific regulatory T cells provoke a shift in the Th1/Th2 paradigm in immunity to infectious diseases. Trends Immunol. 23:450-455. [DOI] [PubMed] [Google Scholar]
- 48.Mellor, A. L., B. Baban, P. Chandler, B. Marshall, K. Jhaver, A. Hansen, P. A. Koni, M. Iwashima, and D. H. Munn. 2003. Induced indoleamine 2,3 dioxygenase expression in dendritic cell subsets suppresses T cell clonal expansion. J. Immunol. 171:1652-1655. [DOI] [PubMed] [Google Scholar]
- 49.Mills, K. H. G. 2004. Regulatory T cells: friend or foe in immunity to infection? Nat. Rev. Immunol. 4:841-855. [DOI] [PubMed] [Google Scholar]
- 50.Mock, J. R., M. Vakevainen, K. Deng, J. L. Latimer, J. A. Young, N. S. van Oers, S. Greenberg, and E. J. Hansen. 2005. Haemophilus ducreyi targets Src family protein tyrosine kinases to inhibit phagocytic signaling. Infect. Immun. 73:7808-7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore, K. W., R. de Waal Malefyt, R. L. Coffman, and A. O'Garra. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683-765. [DOI] [PubMed] [Google Scholar]
- 52.Morse, S. A. 1989. Chancroid and Haemophilus ducreyi. Clin. Microbiol. Rev. 2:137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy, T. F., L. C. Bartos, A. A. Campagnari, M. B. Nelson, and M. A. Apicella. 1986. Antigenic characterization of the P6 protein of nontypable Haemophilus influenzae. Infect. Immun. 54:774-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niessen, F. B., M. P. Andriessen, J. Schalkwijk, L. Visser, and W. Timens. 2001. Keratinocyte-derived growth factors play a role in the formation of hypertrophic scars. J. Pathol. 194:207-216. [DOI] [PubMed] [Google Scholar]
- 55.Opdal, S. H. 2004. IL-10 gene polymorphisms in infectious disease and SIDS. FEMS Immunol. Med. Microbiol. 42:48-52. [DOI] [PubMed] [Google Scholar]
- 56.Palmer, K. L., C. T. Schnizlein-Bick, A. Orazi, K. John, C.-Y. Chen, A. F. Hood, and S. M. Spinola. 1998. The immune response to Haemophilus ducreyi resembles a delayed-type hypersensitivity reaction throughout experimental infection of human subjects. J. Infect. Dis. 178:1688-1697. [DOI] [PubMed] [Google Scholar]
- 57.Post, D. M. B., R. S. Munson, Jr., B. Baker, H. Zhong, J. A. Bozue, and B. W. Gibson. 2007. Identification of genes involved in the expression of atypical lipooligosaccharide structures from a second class of Haemophilus ducreyi. Infect. Immun. 75:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pulendran, B., K. Palucka, and J. Banchereau. 2001. Sensing pathogens and tuning immune responses. Science 293:253-256. [DOI] [PubMed] [Google Scholar]
- 59.Scheffler, N. K., A. M. Falick, S. C. Hall, W. C. Ray, D. M. Post, R. S. Munson, Jr., and B. W. Gibson. 2003. Proteome of Haemophilus ducreyi by 2-D SDS-PAGE and mass spectrometry: strain variation, virulence, and carbohydrate expression. J. Proteome Res. 2:523-533. [DOI] [PubMed] [Google Scholar]
- 60.Smirnova, I., N. Mann, A. Dols, H. H. Derkx, M. L. Hibberd, M. Levin, and B. Beutler. 2003. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc. Natl. Acad. Sci. USA 100:6075-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spellberg, B., and J. E. Edwards, Jr. 2001. Type1/type2 immunity in infectious diseases. Clin. Infect. Dis. 32:76-102. [DOI] [PubMed] [Google Scholar]
- 62.Spinola, S. M., M. E. Bauer, and R. S. Munson, Jr. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 70:1667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spinola, S. M., C. T. H. Bong, A. L. Faber, K. R. Fortney, S. L. Bennett, C. A. Townsend, B. E. Zwickl, S. D. Billings, T. L. Humphreys, M. E. Bauer, and B. P. Katz. 2003. Differences in host susceptibility to disease progression in the human challenge model of Haemophilus ducreyi infection. Infect. Immun. 71:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 71:7178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spinola, S. M., A. Orazi, J. N. Arno, K. Fortney, P. Kotylo, C.-Y. Chen, A. A. Campagnari, and A. F. Hood. 1996. Haemophilus ducreyi elicits a cutaneous infiltrate of CD4 cells during experimental human infection. J. Infect. Dis. 173:394-402. [DOI] [PubMed] [Google Scholar]
- 66.Spinola, S. M., L. M. Wild, M. A. Apicella, A. A. Gaspari, and A. A. Campagnari. 1994. Experimental human infection with Haemophilus ducreyi. J. Infect. Dis. 169:1146-1150. [DOI] [PubMed] [Google Scholar]
- 67.Steen, R. 2001. Eradicating chancroid. Bull. W. H. O. 79:818-826. [PMC free article] [PubMed] [Google Scholar]
- 68.Steinman, L. 2007. A brief history of TH17, the first major revision in the TH1/TH2 hypothesis of T cell-mediated tissue damage. Nat. Med. 13:139-145. [DOI] [PubMed] [Google Scholar]
- 69.Strengell, M., A. Lehtonen, S. Matikainen, and I. Julkunen. 2006. IL-21 enhances SOCS gene expression and inhibits LPS-induced cytokine production in human monocyte-derived dendritic cells. J. Leukoc. Biol. 79:1279-1285. [DOI] [PubMed] [Google Scholar]
- 70.Sullivan, M. 1940. Chancroid. Am. J. Syph. Gon. Vener. Dis. 24:482-521. [Google Scholar]
- 71.Tenca, C., A. Merlo, E. Merck, E. E. M. Bates, D. Saverino, R. Simone, D. Zarcone, G. Trinchieri, C. E. Grossi, and E. Ciccone. 2005. CD85j (leukocyte Ig-like receptor-1/Ig-like transcript 2) inhibits human osteoclast-associated receptor-mediated activation of human dendritic cells. J. Immunol. 174:6757-6763. [DOI] [PubMed] [Google Scholar]
- 72.Thimmulappa, R. K., H. Lee, T. Rangasamy, S. P. Reddy, M. Yamamoto, T. W. Kensler, and S. Biswal. 2006. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Investig. 116:984-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Throm, R. E., and S. M. Spinola. 2001. Transcription of candidate virulence genes of Haemophilus ducreyi during infection of human volunteers. Infect. Immun. 69:1483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 133:133-146. [DOI] [PubMed] [Google Scholar]
- 75.Tzianabos, A. O., A. Chandraker, W. Kalka-Moll, F. Stingele, V. M. Dong, R. W. Finberg, R. Peach, and M. H. Sayegh. 2000. Bacterial pathogens induce abscess formation by CD4+ T-cell activation via the CD28-B7-2 costimulatory pathway. Infect. Immun. 68:6650-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tzianabos, A. O., and D. L. Kasper. 2002. Role of T cells in abscess formation. Curr. Opin. Microbiol. 5:92-96. [DOI] [PubMed] [Google Scholar]
- 77.Tzianabos, A. O., P. R. Russell, A. B. Onderdonk, F. C. Gibson III, C. Cywes, M. Chan, R. W. Finberg, and D. L. Kasper. 1999. IL-2 mediates protection against abscess formation in an experimental model of sepsis. J. Immunol. 163:893-897. [PubMed] [Google Scholar]
- 78.Vakevainen, M., S. Greenberg, and E. J. Hansen. 2003. Inhibition of phagocytosis by Haemophilus ducreyi requires expression of the LspA1 and LspA2 proteins. Infect. Immun. 71:5994-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Visintin, A., A. Mazzoni, J. H. Spitzer, D. H. Wyllie, S. K. Dower, and D. M. Segal. 2001. Regulation of toll-like receptors in human monocytes and dendritic cells. J. Immunol. 166:249-255. [DOI] [PubMed] [Google Scholar]
- 80.Wang, C., J. Tang, W. M. Geisler, P. A. Crowley-Nowick, C. M. Wilson, and R. A. Kaslow. 2005. Human leukocyte antigen and cytokine gene variants as predictors of recurrent Chlamydia trachomatis infection in high-risk adolescents. J. Infect. Dis. 191:1084-1092. [DOI] [PubMed] [Google Scholar]
- 81.Wells, A. D., H. Gudmundsdottir, and L. A. Turka. 1997. Following the fate of individual T cells throughout activation and clonal expansion. Signals from T cell receptor and CD28 differentially regulate the induction and duration of a proliferative response. J. Clin. Investig. 100:3173-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White, C. D., I. Leduc, C. Jeter, C. Harris, and C. Elkins. 2005. Haemophilus ducreyi outer membrane determinants, including DsrA, define two clonal populations. Infect. Immun. 73:2387-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilson, N. J., K. Boniface, J. R. Chan, B. S. McKenzie, W. M. Blumenschein, J. D. Mattson, B. Basham, K. Smith, T. Chen, F. Morel, J. C. Lecron, R. A. Kastelein, D. J. Cua, T. K. McClanahan, E. P. Bowman, and R. de Waal Malefyt. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8:950-957. [DOI] [PubMed] [Google Scholar]
- 84.Wood, G. E., S. M. Dutro, and P. A. Totten. 2001. Haemophilus ducreyi inhibits phagocytosis by U-937 cells, a human macrophage-like cell line. Infect. Immun. 69:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zou, G. M., and Y. K. Tam. 2002. Cytokines in the generation and maturation of dendritic cells: recent advances. Eur. Cytokine Netw. 13:186-199. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.