Abstract
Plasminogen (Plg) binding to the cell surface of Mycoplasma fermentans results in a marked increase in the maximal adherence of the organism to HeLa cells, enhanced Plg activation by the urokinase-type Plg activator, and the induction of the internalization of M. fermentans by eukaryotic host cells (A. Yavlovich, A. Katzenell, M. Tarshis, A. A. Higazi, and S. Rottem, Infect. Immun. 72:5004-5011, 2004). In this study, the M. fermentans Plg binding protein was isolated by affinity chromatography of Triton X-100-solubilized M. fermentans membranes by utilizing a column of a Plg-biotin complex attached to avidin that was eluted with ɛ-aminocaproic acid. The eluted ∼50-kDa protein was identified by mass spectrometric techniques as α-enolase. The possibility that α-enolase, a key cytoplasmatic glycolytic enzyme, resides also on the cell surface of M. fermentans was supported by an immunoblot analysis using polyclonal anti-α-enolase antiserum, which showed that α-enolase was present in a purified M. fermentans membrane preparation, as well as by immunochemical criteria and by immunoelectron microscopy analysis. Our observation that Plg blocked the binding of anti-α-enolase antibodies to a 50-kDa polypeptide band resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of M. fermentans membrane or soluble preparations further supports our notion that mycoplasmal surface α-enolase is a major Plg binding protein of M. fermentans.
Mycoplasmas (class Mollicutes) are wall-less prokaryotes widely distributed in nature. Most mycoplasmas are parasites, exhibiting strict host and tissue specificities, and almost all adhere to the surfaces of eukaryotic cells (18, 19). The adherence of these organisms to host cells is an initial and essential step in tissue colonization and the subsequent development of disease, and adherence-deficient mutants are avirulent (19). The human pathogen Mycoplasma fermentans was isolated from the urogenital tract several decades ago. The interest in this organism has recently increased because of its possible role in the pathogenesis of rheumatoid arthritis (11).
Plasminogen (Plg) is a 92-kDa eukaryotic glycoprotein activated in vivo into the broad-spectrum serine protease plasmin that degrades fibrin and noncollagenous proteins. Plasmin activity results in several physiological and pathophysiological processes, such as fibrinolysis, pericellular proteolysis, tissue penetration of cancer cells, and neuronal cell death (17, 20). Many eukaryotic cells express surface structures that interact with Plg, and specific receptors have been described previously (17). Lysine or lysine analogs such as ɛ-aminocaproic acid (ɛACA) mimic COOH-terminal lysine and thereby inhibit the interaction (20). Recently, it has become evident that Plg is also capable of interacting with a vast number of both gram-positive (14, 15) and gram-negative (21) pathogenic bacteria.
M. fermentans is a typical extracellular microorganism able to adhere to human epithelial cells. Recently, we have shown that M. fermentans binds Plg (23) and that Plg binding markedly increases the adherence of M. fermentans to HeLa cells (24). Furthermore, in the presence of the urokinase-type Plg activator, M. fermentans cells were detected within host cells, suggesting that the ability to bind and activate Plg into plasmin enables M. fermentans to invade host cells (24). Plasmin generated on various bacteria has been shown to degrade mammalian extracellular matrices and, in a few instances, to enhance bacterial metastasis in vitro through reconstituted basement membrane or epithelial cell monolayers (13).
Bacteria expressing Plg receptors on their cell surfaces enhance the activation of Plg by prokaryotic or eukaryotic Plg activators. In essence, Plg receptors and activators turn bacteria into proteolytic organisms by using a host-derived system. In gram-negative bacteria, the filamentous surface appendages fimbriae and flagella form a major group of Plg receptors (21). In gram-positive bacteria, surface-bound enzyme molecules as well as M protein-related structures have been identified as Plg receptors (1). The glycolytic enzymes α-enolase and glyceraldehyde-3-phosphate dehydrogenase are the nonclassical cell surface Plg binding proteins of Streptococcus pneumoniae (2, 3, 16). Plg binds to streptococcal α-enolase through the interaction of the amino-terminal lysine binding domain of Plg with both the carboxy-terminal lysines and the internal motif FYDKERKVYD on the surface-displayed α-enolase (4, 7). In the present study, we have isolated, identified, and described a membrane-bound α-enolase as a key M. fermentans surface protein that mediates Plg binding.
MATERIALS AND METHODS
Organisms and growth conditions.
M. fermentans strain PG-18 (kindly provided by S.-C. Lo, Armed Forces Institute of Pathology, Washington, DC) was used throughout the study. The organisms were grown in Chanock medium supplemented with 5% horse serum (9). The cultures were grown for 24 to 48 h at 37°C. Growth was monitored by measuring the absorbance of the culture at 640 nm and by recording pH changes in the growth medium. The organisms were collected by centrifugation at 12,000 × g for 20 min, washed twice, and resuspended in a cold solution of 10 mM Tris-HCl in 250 mM NaCl (pH 7.5; referred to hereinafter as TN buffer). The total protein concentration was determined according to the method of Bradford (6) and adjusted to 0.5 to 1 mg/ml. The number of viable cells was determined by the plating method and presented as the number of CFU. Membrane and soluble-fraction preparations were obtained from intact cells by ultrasonic treatment as described previously (9). The membranes were washed twice and resuspended in TN buffer.
Affinity chromatography and mass spectrometry (MS) analyses.
For affinity chromatography, 2.5 mg of Plg, purified from human plasma as previously described (8), was labeled with EZ-Link maleimide (polyethylene oxide)2-biotin (10 mM) according to the recommendations of the manufacturer (Pierce). The biotin-Plg was incubated with a slurry of avidin acrylic beads (Sigma) for 2 h at room temperature. The beads were then packed in a glass column and washed twice, and the column was equilibrated with phosphate-buffered saline (PBS) containing 1 mM EDTA. Isolated M. fermentans membranes (containing 10 mg of protein) were solubilized in 2% Triton X-100 and applied to the column. The column was washed three to five times with PBS containing 1 mM EDTA and 2% Triton X-100, and proteins bound to the avidin-biotin-Plg complex were eluted with 10 to 50 mM ɛACA. The samples were dialyzed, freeze-dried, redissolved in sample buffer under reducing conditions (62.5 mM Tris-HCl [pH 6.8], 1% sodium dodecyl sulfate [SDS], 20 mM dithiothreitol, 12.5% glycerol, 0.01% bromophenol blue), and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) using a 4 to 20% gel gradient. An analysis of the biotinylated Plg binding proteins showed the presence of a major 50-kDa protein band that could be seen after Coomassie blue staining and four minor protein bands (∼45, 55, 70, and 75 kDa) detected after silver staining.
Matrix-assisted laser desorption ionization-time of flight MS was performed by in-gel trypsin digestion of the Coomassie blue-stained 49-kDa membrane protein that was shown to be the major Plg binding band. Peptides were extracted from the gel with 60% CH3CN in 1% formic acid, and MS was carried out with a Qtof2 system (Micromass, England) using a nanospray attachment (22). The peptide mass data and the tandem mass spectrometry (MS-MS) data were analyzed to determine the amino acid sequence by using the BioLynx package (Micromass, England), and database searches were performed with the Mascot package (Matrix Science, England). Similarity searches using sequences determined via manual analysis were carried out with a BLAST search of the NCBI data bank.
Localization of α-enolase.
A qualitative assessment of α-enolase in cell fractions of M. fermentans was performed by dot immunoblotting. M. fermentans whole-cell extracts and isolated-membrane or soluble-fraction preparations (10 to 100 μg of protein) were immobilized on a nitrocellulose membrane with a Bio-Dot apparatus (Bio-Rad Laboratories). The nitrocellulose membranes were processed by (i) blocking for 1 h at room temperature with skim milk or with PBS containing 1% bovine serum albumin (BSA), (ii) incubation for 16 h at 4°C with a 1:200 dilution of rabbit anti-α-enolase antiserum (kindly provided by S. Hammerschmidt, University of Würzburg, Würzburg, Germany), and (iii) incubation at room temperature for 1 h with horseradish peroxidase-conjugated mouse anti-rabbit immunoglobulin G (IgG; Jackson ImmunoResearch, Inc.). Blots were developed either by using the ECL Western blotting detection reagents (Amersham International Inc.) or by using the o-dianisidine substrate according to the instructions of the manufacturer. To determine the effect of Plg on the binding of rabbit anti-α-enolase antibodies to M. fermentans preparations, polypeptide bands of M. fermentans membrane and soluble fractions were resolved by SDS-PAGE. The polypeptides were transferred onto nitrocellulose membranes by electroblotting using a Hoeffer TE22 electroblotting unit according to the manufacturer's recommendations and treated with rabbit anti-α-enolase antiserum in the presence or absence of Plg (15 μg). The nitrocellulose membranes were processed as described above.
A quantitative assessment of the binding of polyclonal rabbit anti-α-enolase antibodies to intact M. fermentans cells was performed by enzyme-linked immunosorbent assay (ELISA) with 96-well MaxiSorb plates (Nunc, Denmark). The ELISA plates coated with M. fermentans (0.1 to 5.0 μg of cell protein/well in TN buffer containing 10 mM CaCl2) were blocked with PBS containing 1% BSA and then treated for 2 h at room temperature with a 1:200 dilution of polyclonal rabbit anti-α-enolase antibodies. The plates were then incubated with mouse anti-rabbit IgG-horseradish peroxidase conjugate for 1 h at 37°C and were developed with the o-dianisidine substrate.
Electron microscopy.
In an attempt to visualize the subcellular localization of α-enolase by transmission electron microscopy (TEM), a preembedding immunogold labeling method was used. M. fermentans cells were grown in Chanock medium (9) to mid-exponential phase and washed twice in TN buffer. The cell protein concentration was adjusted to 200 μg/ml, and aliquots were incubated first for 30 min in a blocking solution containing 5% fetal calf serum in TN buffer and then for 24 h in the cold with a 1:50 dilution of rabbit anti-α-enolase antiserum in TN buffer containing 1% BSA. After several washes in TN buffer, samples were incubated for 3 h at room temperature in goat anti-rabbit IgG coupled to 12-nm colloidal gold particles (Jackson ImmunoResearch Laboratories). The samples were washed twice in TN buffer and then fixed with 2% formaldehyde and 2% glutaraldehyde (in 0.2 M cacodylate buffer, pH 7.4) for 30 min in the cold. Processing of the samples by osmification, dehydration, and embedding in Epon was performed as previously described (10). The samples were then sectioned using an LKB-3 ultramicrotome and observed with a Tecnai 12 electron microscope (Phillips, Eindhoven, The Netherlands) equipped with a MegaView II charge-coupled-device camera.
RESULTS AND DISCUSSION
It has been shown previously that a few Mycoplasma species bind Plg at the cell surface in a lysine-dependent manner (5, 23). Plg binding to M. fermentans markedly increases the adherence of M. fermentans to HeLa cells (23, 24), and it was suggested that in the presence of Plg, M. fermentans adheres to novel sites on the surfaces of HeLa cells (24). Many pathogenic bacteria express Plg receptors on their cell surface, and these receptors may play a role in the dissemination of microorganisms by binding Plg that, when converted to plasmin, can digest extracellular proteins (13). Furthermore, Plg binding and activation into plasmin affects the invasiveness of a variety of microorganisms, including M. fermentans (24). When autoradiograms of ligand blots containing proteins of M. fermentans PG-18 membranes were incubated with 125I-labeled Plg, two polypeptide bands were labeled (23). In an attempt to isolate and characterize the Plg receptors on the cell surface of M. fermentans, solubilized M. fermentans membranes were loaded onto a column containing a Plg-biotin complex attached to avidin-Sephadex. A major protein with a molecular mass of ∼50 kDa was eluted with ɛACA. The sample was found to be digested by trypsin into numerous mass ions that were studied by MS-MS. More than 50% of the ions were found by their MS-MS-generated fragments to correspond with a high degree of confidence to the sequences of α-enolase from various mycoplasmas.
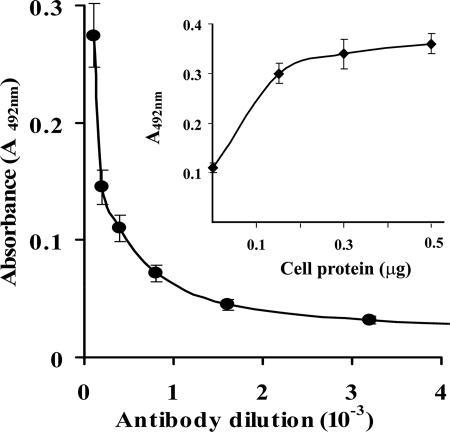
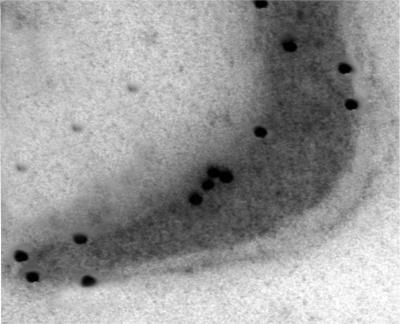
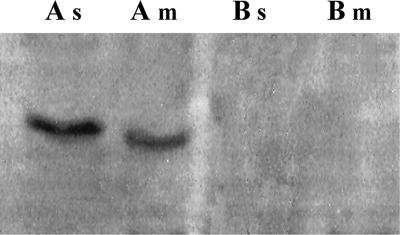
The identification of the glycolytic enzyme α-enolase on the surface of M. fermentans was further supported by ligand blotting immunochemical analyses, ELISA, and electron microscopy. Figure 1 shows that α-enolase could be detected also in preparations of isolated M. fermentans membranes by immunoblot analysis using rabbit anti-α-enolase antibodies. Furthermore, the levels of α-enolase in the membranes were higher than the levels in the soluble fraction. Extensive washing of the membrane preparations with increasing concentrations of NaCl (up to 1 M) in 10 mM Tris buffer (pH 7.5) or with 10 mM EDTA in TN buffer did not affect α-enolase levels (data not shown). A quantitative assessment of the binding of polyclonal rabbit anti-α-enolase antibodies to intact M. fermentans cells by ELISA is presented in Fig. 2. The interaction of the anti-α-enolase antibodies with intact M. fermentans cells was further visualized by TEM analysis of immunogold-stained preparations (Fig. 3) showing the localization of α-enolase on the cell surface of M. fermentans. Figure 4 shows that Plg inhibits the binding of rabbit anti-α-enolase antibodies to a polypeptide band (∼50 kDa) resolved by SDS-PAGE of M. fermentans membrane or soluble preparations, supporting our notion that mycoplasmal surface α-enolase is a major Plg binding protein of M. fermentans. Furthermore, partial competitive inhibition of Plg binding to intact M. fermentans cells was obtained by adding a commercial preparation of α-enolase (Sigma) to the binding assay mixture (data not shown). As the maximal inhibition obtained by adding α-enolase (≥20 mg/well) was only about 50%, it is suggested that more than a single Plg binding protein is present on the surface of M. fermentans.
FIG. 1.
Immunoblot analysis of α-enolase in M. fermentans cell fractions. Intact M. fermentans cells, the soluble fraction, and isolated membranes (10 or 100 μg of protein) were blotted onto nitrocellulose paper and treated with rabbit anti-α-enolase antibodies (1:200). The blots were then treated with horseradish peroxidase-conjugated mouse anti-rabbit IgG and developed with the o-dianisidine substrate as described in Materials and Methods. A, intact cells; B, soluble fraction; C, isolated membranes; D, control without mycoplasmas.
FIG. 2.
Binding of anti-α-enolase antibodies to intact M. fermentans cells. ELISA plates were coated with intact M. fermentans cells (1 μg of cell protein/well). The plate contents were reacted with various dilutions of rabbit anti-α-enolase antibodies, followed by mouse anti-rabbit IgG-horseradish peroxidase conjugate, and results were visualized by the o-dianisidine procedure as described in Materials and Methods. Inset: ELISA of various concentrations of intact M. fermentans cells (up to 0.5 μg of cell protein/well) reacted with a 1:200 dilution of rabbit anti-α-enolase antiserum. Values are means ± standard deviations of results from three independent experiments.
FIG. 3.
Visualization of α-enolase by preembedding immunogold staining. α-Enolase was visualized by analyzing ultrathin sections of preembedded M. fermentans preparations treated with rabbit anti-α-enolase antibodies and labeled with goat anti-rabbit IgG coupled to 12-nm colloidal gold particles. The preparations were visualized by TEM as described in Materials and Methods.
FIG. 4.
Effect of Plg on the binding of anti-α-enolase antibodies to M. fermentans cell fractions. Polypeptide bands of M. fermentans membrane (m) and soluble (s) fractions were resolved by SDS-PAGE and analyzed by immunoblotting using rabbit anti-α-enolase antibodies in the absence (A) or presence (B) of 15 μg of Plg/ml. The blots were then treated with horseradish peroxidase-conjugated mouse anti-rabbit IgG and developed with the o-dianisidine substrate as described in Materials and Methods.
The availability of the complete genomic sequence of M. fermentans (A. Strittmatter, The University of Goettingen, personal communications) allowed us to further study the α-enolase of this organism. M. fermentans genomic sequence annotation shows a single copy of an α-enolase sequence. Deducing the amino acid sequence of this enzyme predicts a polypeptide of 454 amino acids, with a calculated molecular mass of the primary translation product of 49,276 Da. A BLAST analysis of the M. fermentans α-enolase protein sequence relative to sequences in the NCBI database was performed. Homologous sequences from other organisms were aligned with the M. fermentans α-enolase sequence by ClustalW multiple-sequence alignment analysis. Like that of S. pneumoniae (3), the α-enolase sequence of M. fermentans lacks a signal sequence or the typical motifs required for membrane anchoring. Nonetheless, the sequence was found to be highly homologous to the α-enolase sequences of a variety of Mycoplasma species as well as that of S. pneumoniae (Table 1) and contains features typical of the Plg binding surface α-enolase described previously (4). These features include a lysine as the C-terminal residue (FYNIK, Plg binding site 1) and a conserved, positively charged lysine-rich internal motif (IYDEKSKKYV, Plg binding site 2). These results support the notion that at least one of the M. fermentans Plg binding proteins is a membrane-associated α-enolase. As in S. pneumoniae, it is likely that the surface α-enolase of M. fermentans binds through lysine-rich binding motifs to the amino-terminal lysine binding Kringle domain of Plg (12), exploiting host properties to the advantage of M. fermentans in tissue invasion.
TABLE 1.
Comparison of α-enolase sequence identities, C termini, and internal putative Plg binding site motifs among various organisms
| Species | % Identity to M. fermentans α-enolase | C-terminal sequence | Sequence of internal motifa | Accession no. |
|---|---|---|---|---|
| M. fermentans | 100 | NIK | IYDEKSKKY | ABW23428 |
| M. agalactiae | 79 | NIK | LYDEKTKKY | CAL59017 |
| M. synoviae | 75 | NLK | LYK--GKYT | AAZ43431 |
| M. mobile | 70 | NLK | LFDKKSKTY | AAT27666 |
| M. capricolum | 58 | NL | LYLEDKKYH | ABC01346 |
| M. pneumoniae | 55 | NIK | FYDDTTKRY | AAB95884 |
| S. pneumoniae | 54 | NLKK | FYDKERKVY | AAK75238 |
Data from reference 4.
Acknowledgments
We are grateful to A. Strittmatter, University of Goettingen, Germany, for making available the nucleotide sequence of the M. fermentans PG-18 genome, to A. Katzenell for excellent technical assistance, and to M. Siman-Tov for critical reading of the manuscript.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Berge, A., and U. Sjobring. 1993. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem. 268:25417-25424. [PubMed] [Google Scholar]
- 2.Bergmann, S., M. Rhode, and S. Hammerschmidt. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72:2416-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, S., D. Wild, O. Diekmann, R. Frank, D. Bracht, G. S. Chhatwal, and S. Hammerschmidt. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49:411-423. [DOI] [PubMed] [Google Scholar]
- 5.Bower, K., S. P. Djordjevic, N. M. Andronicos, and M. Ranson. 2003. Cell surface antigens of Mycoplasma species bovine group 7 bind to and activate plasminogen. Infect. Immun. 71:4823-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Derbise, A., Y. P. Song, S. Parikh, V. A. Fischetti, and V. Pancholi. 2004. Role of C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen binding activities of group A streptococci. Infect. Immun. 72:94-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutsch, D. G., and E. T. Mertz. 1970. Plasminogen: purification from human plasma by affinity chromatography. Science 170:1095-1096. [DOI] [PubMed] [Google Scholar]
- 9.Deutsch, J., M. Salman, and S. Rottem. 1995. An unusual polar lipid from the cell membrane of Mycoplasma fermentans. Eur. J. Biochem. 227:897-902. [DOI] [PubMed] [Google Scholar]
- 10.Feinstein, N., D. Parnas, H. Parnas, J. Dudel, and I. Parnas. 1998. Functional and immunocytochemical identification of glutamate autoreceptors of an NMDA type in crayfish neuromuscular junction. J. Neurophysiol. 80:2893-2899. [DOI] [PubMed] [Google Scholar]
- 11.Gilroy, C. B., A. Keat, and D. Taylor-Robinson. 2001. The prevalence of Mycoplasma fermentans in patients with inflammatory arthritides. Rheumatology (Oxford) 40:1355-1358. [DOI] [PubMed] [Google Scholar]
- 12.Kuusela, P., M. Ullberg, G. Kronvall, T. Tervo, A. Tarkkanen, and O. Saksela. 1992. Surface-associated activation of plasminogen on gram-positive bacteria. Effect of plasmin on the adherence of Staphylococcus aureus. Acta Ophthalmol. Suppl. 70:42-46. [DOI] [PubMed] [Google Scholar]
- 13.Lähteenmäki, K., S. Edelman, and T. K. Korhonen. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 13:79-85. [DOI] [PubMed] [Google Scholar]
- 14.Lähteenmäki, K., P. Kuusela, and T. K. Korhonen. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25:531-552. [DOI] [PubMed] [Google Scholar]
- 15.Lottenberg, R., D. Minning-Wenz, and M. D. P. Boyle. 1994. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 2:20-24. [DOI] [PubMed] [Google Scholar]
- 16.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 17.Plow, E. F., T. Herren, A. Redlitz, L. A. Miles, and J. L. Hoover-Plow. 1995. The cell biology of the plasminogen system. FASEB J. 9:939-945. [DOI] [PubMed] [Google Scholar]
- 18.Rosengarten, R., C. Citti, M. Glew, A. Lischewski, M. Droesse, P. Much, F. Winner, M. Brank, and J. Spergser. 2000. Host-pathogen interactions in mycoplasma pathogenesis: virulence and survival strategies of minimalist prokaryotes. Int. J. Med. Microbiol. 290:15-25. [DOI] [PubMed] [Google Scholar]
- 19.Rottem, S. 2003. Interaction of mycoplasmas with host cells. Physiol. Rev. 83:417-432. [DOI] [PubMed] [Google Scholar]
- 20.Saksela, O., and D. P. Rifkin. 1988. Cell-associated plasminogen activation: regulation and physiological functions. Annu. Rev. Cell Biol. 4:93-126. [DOI] [PubMed] [Google Scholar]
- 21.Ullberg, M., G. Kronvall, I. Karlsson, and B. Wiman. 1990. Receptors for human plasminogen on gram-negative bacteria. Infect. Immun. 8:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilm, M., and M. Mann. 1996. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Yavlovich, A., A. A. Higazi, and S. Rottem. 2001. Plasminogen binding and activation by Mycoplasma fermentans. Infect. Immun. 69:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yavlovich, A., A. Katzenell, M. Tarshis, A. A. Higazi, and S. Rottem. 2004. Mycoplasma fermentans binds to and invades HeLa cells: involvement of plasminogen and urokinase. Infect. Immun. 72:5004-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]