Abstract
At least a million people, mainly African children under 5 years old, still die yearly from malaria, and the burden of disease and death has increased. Plasmodium falciparum apical membrane antigen 1 (PfAMA1) is one of the most promising blood-stage malarial vaccine candidates. However, the allelic polymorphism observed in this protein is a potential stumbling block for vaccine development. To overcome the polymorphism- and strain-specific growth inhibition in vitro, we previously showed in a rabbit model that vaccination with a mixture of two allelic forms of PfAMA1 induced parasite growth-inhibitory antisera against both strains of P. falciparum parasites in vitro. In the present study, we have established that, in contrast to a single-allele protein, the antigen mixture elicits primarily antibodies recognizing antigenic determinants common to the two antigens, as judged by an antigen reversal growth inhibition assay (GIA). We also show that a similar reactivity pattern occurs after immunization of mice. By contrast, sera from rhesus monkeys do not distinguish the two alleles when tested by an enzyme-linked immunosorbent assay or by GIA, regardless of whether the immunogen is a single AMA1 protein or the mixture. This is the first report that a malarial vaccine candidate induced different specificities of functional antibodies depending on the animal species immunized. These observations, as well as data available on human immune responses in areas of endemicity, suggest that polymorphism in the AMA1 protein may not be as formidable a problem for vaccine development as anticipated from studies with rabbits and mice.
The malarial parasite remains a scourge on human civilization, and recent data suggest that previous estimates of malaria morbidity and mortality may have significantly underestimated the worldwide burden of this disease. Snow and colleagues estimate that there may be 300 to 500 million clinical cases of malaria annually, a rate approximately twice as high as previous estimates (42). To compound the human toll of this disease, recent macroeconomic analysis by the WHO Commission on Macroeconomics and Health has found that malaria reduces economic growth in sub-Saharan Africa by over 1% per year, with major long-term consequences for the gross national products of the afflicted countries (41). As the burden of disease and death due directly and indirectly to malaria has increased, the need for an effective vaccine has also assumed greater importance (4, 29).
Of the major vaccine candidates directed against blood-stage malaria parasites which are responsible for the pathology associated with this disease, Plasmodium falciparum apical membrane antigen 1 (PfAMA1) is one of the best studied (36). Expressed late in the erythrocytic replication cycle, this 83-kDa membrane protein is initially found in apical organelles called micronemes in the invasive form of the blood-stage parasite known as the merozoite (2, 18). AMA1 is subsequently processed to a 66-kDa form by removal of an N-terminal prosequence and is translocated to the merozoite surface, where additional proteolytic cleavage occurs (24, 25, 34); some AMA1-specific antibodies may interfere with this proteolytic cleavage, thereby inhibiting invasion (11). The AMA1 ectodomain has been described as having three domains based on the disulfide bonding pattern (23), and very recent X-ray crystallographic solutions of AMA1 structures from Plasmodium vivax and P. falciparum should provide a basis for future detailed analysis of structure-function relationships (17, 37). AMA1 appears to play a pivotal role in erythrocyte invasion (44), participating in the attachment and reorientation of the merozoite to the host red cell surface (31). In this context, the AMA1 region closest to the merozoite membrane has been reported to bind to the Kx erythrocyte surface protein (26). Its critical role in merozoite invasion has been supported by its presence in all plasmodial species examined as well as other apicomplexan parasites (10, 21, 47) and by the failure to obtain parasites lacking the AMA1 gene in knockout experiments (44).
Various lines of evidence support AMA1 as a promising blood-stage malarial vaccine candidate: (i) immunization with AMA1 from rodent malarial parasites can protect mice against parasite challenge (1, 3, 33); (ii) immunization of rhesus monkeys with AMA1 from the nonhuman primate parasite Plasmodium knowlesi and immunization of squirrel monkeys with Plasmodium fragile AMA1 can protect the animals against a parasite challenge (5, 9); (iii) P. falciparum AMA1 (PfAMA1) immunization can induce strong protective immunity in Aotus vociferans monkeys, which are highly susceptible to P. falciparum challenge (43); (iv) anti-PfAMA1 antibodies from humans as well as other species can inhibit the invasion of erythrocytes by P. falciparum merozoites in vitro (22, 32); (v) human T-cell proliferative responses to a T-cell epitope of PfAMA1 were associated with a significantly lower risk of parasitemia in a subsequent follow-up in western Kenya (45); and (vi) a prospective study of those living in a coastal region of Kenya found that the presence of antibodies to the full-length ectodomain at the beginning of the transmission season was associated with a reduced risk of subsequent clinical malaria (40).
One of the main concerns related to AMA1 as a vaccine candidate is the fact that it is a polymorphic protein, with numerous amino acid substitutions observed in P. falciparum isolates from various regions of the world. Although PfAMA1 is less polymorphic than other merozoite surface proteins that are also vaccine candidates, such as merozoite surface proteins 1 and 2 (MSP1 and MSP2) (14, 35), there is evidence that the AMA1 polymorphisms are under balancing selection, especially domains I and III (8, 13, 38, 39, 46). It has been assumed that selective pressure by the host immune system is responsible for maintenance of these polymorphisms. In accord with this, several groups have shown that antisera from rabbits directed against PfAMA1 are more effective at in vitro growth inhibition of parasite strains with homologous AMA1 sequences than for strains with heterologous AMA1 sequences (19, 22, 27, 28). It has also been suggested that some of the immune selection pressure operating on AMA1 may function at the level of CD4+ T-cell immunity (40).
Previous studies from our laboratory have explored the specificity of the humoral immune response to AMA1. Antisera from rabbits immunized with a single allelic sequence of PfAMA1 (AMA1-FVO or AMA1-3D7) inhibited in vitro invasion of erythrocytes by a homologous line of P. falciparum parasites better than heterologous parasites (27). By contrast, we showed that immunization of rabbits with a mixture of the same two allelic forms of PfAMA1, designated AMA1-C1, elicited growth-inhibitory antisera with comparable activities against both parental strains of P. falciparum parasites (27). However, the latter finding could be explained either by most of the antibodies being directed against epitopes conserved between the two immunogens or by a mixture of populations of antibodies, some directed primarily against FVO-specific epitopes and others preferentially recognizing 3D7-specific epitopes. Moreover, these patterns of recognition of strain-specific versus conserved antigenic determinants as a function of the immunogen were tested only with rabbits.
In this report, we have expanded our studies of the specificities of the humoral immune responses of rabbits immunized with single versus mixed AMA1 alleles, using both an enzyme-linked immunosorbent assay (ELISA) and a standardized in vitro growth inhibition assay (GIA). We show that rabbits immunized with the PfAMA1-C1 combination primarily produce cross-reactive antibodies to epitopes conserved between the two AMA1 proteins, while single-allele PfAMA1 immunization elicits both cross-reactive and strain-specific antibodies. In addition, we have extended these observations to inbred mice and to rhesus monkeys. While the mice displayed patterns similar to those of the rabbits, rhesus monkeys produced antibodies which did not clearly distinguish these two strains, regardless of whether they were immunized with a single allele or with the AMA1-C1 combination. This species dependence of AMA1 epitope recognition has significant implications for ongoing human clinical trials of AMA1 vaccine candidates and raises questions about the role of amino acid polymorphisms in this protein.
MATERIALS AND METHODS
Antigen preparation.
PfAMA1 was produced and purified as described previously (27). In brief, two synthetic AMA1 genes (based on FVO and 3D7 parasite sequences), codon optimized for expression in Pichia pastoris, were subcloned into the pPIC9K vector and each transformed into P. pastoris. After fermentation, a series of chromatography columns was used for purification and buffer exchange, as follows: a nickel nitrilotriacetic acid Superflow column, a G-25 column, an anion-exchange column, a butyl-Sepharose interaction column, and a Superdex 75 size exclusion column. The characterization of the protein products has been previously reported (27).
Immunizations.
Mouse and rhesus monkey (Macaca mulatta) studies were done in compliance with National Institutes of Health (NIH) guidelines and under the auspices of Animal Care and Use Committee-approved protocols. Rabbit immunization was performed by Spring Valley Laboratories (Frederick, MD).
BALB/c mice (6 to 12 weeks old) were immunized with 1 μg/dose of AMA1-FVO or AMA1-3D7 or an equal-mass mixture of AMA1-FVO and AMA1-3D7 (AMA1-C1). The vaccines were formulated with Alhydrogel (aluminum hydroxide [alum]; HCI Biosector, Frederikssund, Denmark) or Montanide ISA720 (SEPPIC Inc., Fairfield, NJ), and the mice were immunized twice at a 4-week interval and bled on days 0 and 42. Three groups of 10 mice received the alum formulation, and 5 mice were in each ISA720 group.
Three groups of four rabbits were immunized with 25 μg/dose of AMA1-FVO, AMA1-3D7, or AMA1-C1 formulated with Montanide ISA720 twice at a 4-week interval and bled on days 0 and 42. Three groups of five rhesus monkeys were immunized with 25 μg/dose of AMA1-FVO, AMA1-3D7, or AMA1-C1 formulated with Montanide ISA720 on days 0, 28, and 183. They were bled on days 0, 14, 28, 42, 61, 90, 175, 197, 224, 270, and 365.
ELISA.
Ninety-six-well ELISA plates were coated with 100 ng/well of AMA1-FVO or AMA1-3D7 protein at 4°C overnight. After the plates were blocked with 5% skim milk, diluted sera were added to antigen-coated wells in triplicate and incubated for 2 h at room temperature. After extensive washing, the plates were incubated with a species-specific secondary antibody conjugated with alkaline phosphatase for 2 h at room temperature. For mouse samples, 0.1 μg/well of anti-mouse immunoglobulin G (IgG; heavy plus light chains) antibody (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) was used. For rabbit samples, 0.1 μg/well of anti-rabbit IgG (heavy plus light chains) antibody (Kirkegaard & Perry Laboratories, Inc.) was used, and for monkey samples, 0.05 μg/well of anti-monkey IgG(γ) chain antibody (Rockland, Gilbertsville, PA) was used. Bound antibodies were visualized by adding p-nitrophenyl phosphate Sigma 104 substrate (Sigma Chemical Co., St. Louis, MO). The absorbance at 405 nm was read using a SPECTRAmax 340PC microplate reader (Molecular Devices Co., Sunnyvale, CA).
Sera from animals immunized with AMA1-C1 selected for relatively high antibody levels were used to prepare a reference standard for each animal species. After a standard serum pool was prepared, the serum was diluted and stored at −80°C in aliquots until used. For each standard, serially diluted sera were tested and assigned ELISA unit values, defined as the reciprocal of the dilution giving an optical density at 405 nm of 1 for each plate antigen, e.g., AMA1-FVO and AMA1-3D7. For subsequent ELISAs, duplicates of serially diluted standard sera of specific animals were included on each test plate in order to generate a standard curve. The standard curve was used to convert the absorbance values of individual test sera into antibody units (SOFTmax PRO version 3; Molecular Devices Co.).
GIA sample preparation.
Pools of mouse/rabbit/monkey anti-AMA1-FVO, -AMA1-3D7, or -AMA1-C1 sera and pools of normal sera were made. From these pooled sera, anti-AMA1 IgGs and normal IgGs were purified using protein G columns (Pierce, Rockford, IL) according to the manufacturer's instructions. The eluted IgGs were dialyzed against RPMI 1640 and concentrated with centrifugal filter devices (Millipore, Billerica, MA) to a concentration of 4 (mouse), 10 (rabbit), or 20 (monkey) mg/ml and subsequently sterilized with a 0.22-μm filter (Millipore). The purified IgGs were preadsorbed with uninfected human O+ red blood cells (RBCs; 25 μl per 1 ml of the sample) for 1 h to remove anti-human RBC immunoglobulins. For the monkey study, pooled day 0 and individual day 197 monkey sera were also tested by a GIA. To do that, the sera were heat inactivated at 56°C for 20 min and then preadsorbed with uninfected human O+ RBCs. All of the IgGs and sera were aliquoted and frozen at −80°C until needed.
GIA.
The methodology for performing the GIA has been described previously (30). Briefly, test samples, synchronized P. falciparum parasites (late trophozoites and schizonts), and culture medium were applied to 96-well tissue culture plates to achieve a total volume of 100 μl/well and tested in triplicate. The final concentration of the culture was 0.3% ± 0.1% parasitemia, 1% hematocrit in growth medium (RPMI 1640 containing 10% human O+ serum, 25 mM HEPES, 0.4 mM hypoxanthine, 30 mM sodium bicarbonate, and 25 mg/liter of gentamicin).
For the antigen reversal GIA experiments, AMA1 antigens were dialyzed against RPMI 1640. The AMA1 antigens were serially diluted and incubated with anti-AMA1 IgG and incomplete culture medium (total volume, 50 μl/well) for 45 min at room temperature, followed by 15 min of incubation at 37°C in a 96-well tissue culture plate. A parasitized erythrocyte suspension was prepared and added to the plate so that the final concentration of the culture had the same parasitemia and hematocrit levels in growth medium as those in the GIA described above.
The cultures were maintained for 40 to 42 h, and relative parasitemia levels were quantitated by biochemical determination of parasite lactate dehydrogenase.
Percent inhibition of the immune IgG was calculated as 100 − [(A650 of test IgG − A650 of normal RBCs)/(A650 of infected RBCs without any IgG − A650 of normal RBCs) × 100].
Percent inhibition of the monkey antiserum was calculated as 100 − [(A650 of test sera − A650 of normal RBCs)/(A650 of infected RBCs with day 0 monkey sera − A650 of normal RBCs) × 100].
Statistics.
To compare the numbers of ELISA units of a group tested on AMA1-FVO-coated plates and a group tested on 3D7 plates, a Wilcoxon t test was performed. To test the correlation between units of antibody and growth-inhibitory activities, a Spearman rank correlation test was used. All statistical tests were performed by UNISTAT 5.0 (P-STAT Inc., Hopewell, NJ), and probability values less than 0.05 were considered significant. Curve fittings were performed using Sigma Plot (SPSS Inc., Chicago, IL).
RESULTS
Characteristics of antibodies elicited by AMA1 vaccination of rabbits.
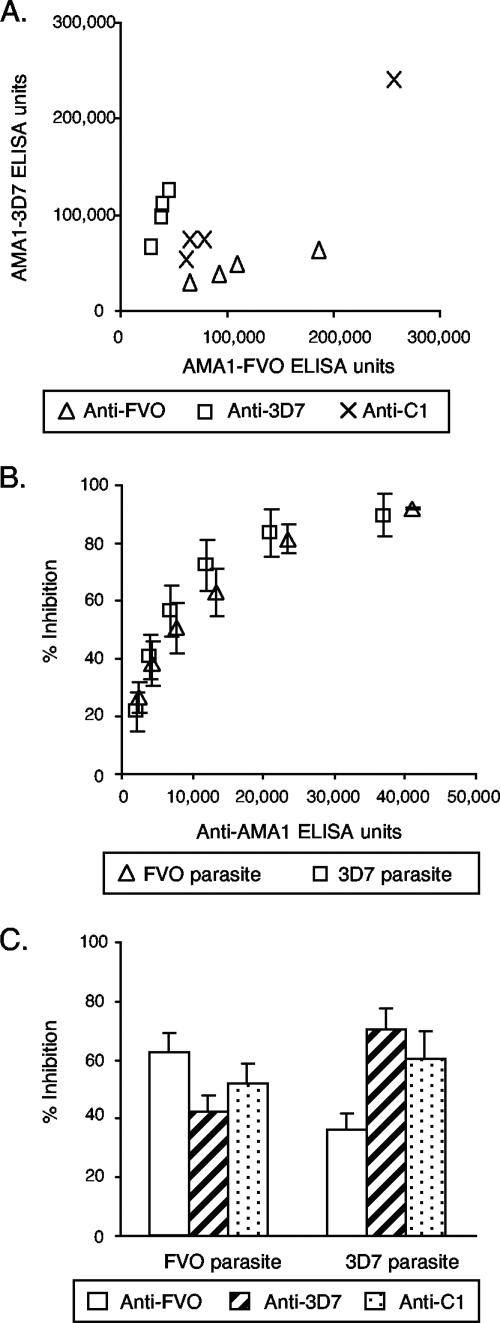
Previous studies from our laboratory had demonstrated that rabbits immunized with either AMA1-FVO or AMA1-3D7 showed higher ELISA activities on the homologous protein than on the heterologous one; by contrast, rabbits immunized with AMA1-C1 showed comparable levels of antibody binding to each of the alleles (27). In addition, there was no significant difference between the levels of antibodies in rabbits immunized with the AMA1-C1 mixture and those in rabbits immunized with the individual AMA1 alleles. The present results confirm those findings (Fig. 1A).
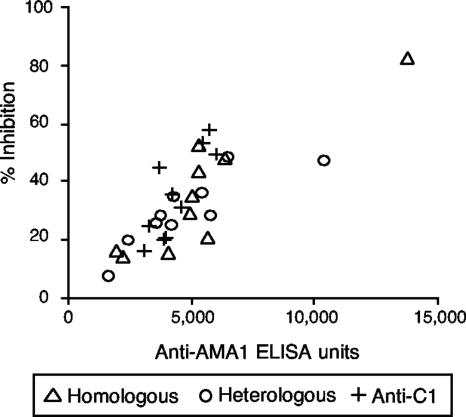
FIG. 1.
Strain specificities of antisera from rabbits immunized with AMA1-FVO, AMA1-3D7, or AMA1-C1 as determined by an ELISA and a GIA. Three groups of four rabbits each were immunized on days 0 and 28 with 25 μg of the three different AMA1 immunogens formulated with Montanide ISA720 and were bled on days 0 and 42. (A) ELISA values for individual animals are plotted. (B) Purified anti-AMA1 IgGs from pooled anti-AMA1-FVO or -3D7 antisera were tested at six different concentrations (1.2 to 2 mg/ml in GIA wells) by a GIA using P. falciparum FVO and 3D7 parasites. Anti-FVO IgG was tested with FVO parasites, and anti-3D7 IgG was tested with 3D7 parasites. The numbers of anti-AMA1-FVO ELISA units against FVO parasites and anti-3D7 ELISA units against 3D7 parasites (x axis) are plotted against percents inhibition (y axis). There is a highly significant correlation between number of ELISA units and percent inhibition (Spearman rank correlation = 0.969, 95% CI = 0.929 to 0.987, P < 0.0001). Percent inhibition values are presented as means ± standard errors from three independent experiments. (C) Purified anti-FVO, -3D7, and -C1 IgGs were diluted with normal rabbit IgG to obtain 10,000 anti-AMA1-FVO ELISA units in individual GIA wells and then mixed with P. falciparum-FVO parasites, and the GIA was completed. Those three IgGs were also diluted to obtain 10,000 anti-AMA1-3D7 ELISA units and tested with 3D7 parasites. Percent inhibition values are presented as means plus standard errors from three independent experiments.
To evaluate the biological activities of the various rabbit antibodies, the parasite growth-inhibitory activities of IgGs isolated from a pool of sera from rabbits immunized with single-allele AMA1 protein were tested against the homologous strain of parasites (Fig. 1B). Six different dilutions of each IgG were tested to establish a relationship between antibody concentration as determined by an ELISA and growth-inhibitory activity. Normal rabbit IgG tested at 2 mg/ml in GIA wells (the same concentration as the highest one for the rabbit immune IgGs tested) showed 5% inhibition for FVO parasites and 4% for 3D7. As shown in Fig. 1B, there is a significant correlation between numbers of ELISA units in GIA wells and percents inhibition of the samples (Spearman rank correlation = 0.969, 95% confidence interval [CI] = 0.929 to 0.987, P < 0.0001), and the relationship follows a hyperbolic curve (r2 = 0.950). Additionally, the two different inhibition curves overlapped.
We have shown that rabbit anti-AMA1 sera had higher growth-inhibitory activities against homologous parasites than against heterologous parasites (27). However, it was not clear whether the difference was caused purely by the quantitative differences between antibodies (e.g., anti-AMA1-FVO antiserum had more ELISA units against FVO than against 3D7) or whether there were any qualitative differences. To unravel the question, the growth-inhibitory activities of the rabbit anti-AMA1 IgGs on heterologous combinations of IgG and parasites were also determined in a standardized GIA. Anti-AMA1-FVO, -AMA1-3D7, and -AMA1-C1 IgGs were diluted with normal rabbit IgG to attain 10,000 anti-AMA1-FVO ELISA units when these three IgGs were tested against P. falciparum FVO parasites by a GIA. For the GIA with 3D7 parasites, the IgGs were diluted in a similar fashion using the 3D7 ELISA values of the IgGs. As shown in Fig. 1C, the homologous combination (e.g., anti-FVO IgG tested against P. falciparum FVO parasites) showed higher inhibitory activity than the heterologous combination (e.g., anti-3D7 IgG against FVO parasites), and anti-C1 IgG showed percent inhibition at a level intermediate between them. When the IgGs were tested at 20,000 ELISA units, the same rank order was found (data not shown). The data showed that single-allele immunization induced fewer ELISA units on plates coated with heterologous AMA1 than on those coated with homologous AMA1, and the antibodies showed less growth-inhibitory activity against heterologous parasites even when they were tested at the same number of ELISA units.
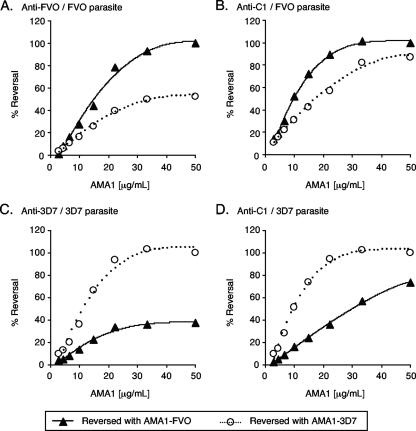
To dissect the fine specificities of the antibodies produced, we evaluated the abilities of AMA1 proteins to reverse growth-inhibitory activity in order to determine whether the combination immunization strategy induced separate populations of antibodies, each of which was specific for a single AMA1 allele, or whether the antibodies induced were primarily directed against conserved epitopes and could recognize both AMA1 alleles. The growth-inhibitory activities of anti-AMA1 IgGs were neutralized by preincubation of the antibodies with various concentrations of either AMA1-FVO or AMA1-3D7 proteins (Fig. 2). The activity of anti-FVO IgG against FVO parasites was completely reversed by AMA1-FVO protein, while the AMA1-3D7 protein reversed only 52% of the activity even at high concentrations of the antigen (Fig. 2A). On the other hand, the activity of anti-C1 IgG against FVO parasites was almost completely reversed by either of the AMA1 proteins at the highest dose tested, although AMA1-FVO protein reversed better than AMA1-3D7 protein at the middle dose (Fig. 2B). The antigen reversal GIA with 3D7 parasites showed that AMA1-FVO protein (50 μg/ml) reversed 38% of anti-3D7 IgG-derived growth inhibitory activity and 74% of anti-C1 IgG-derived activity. The data clearly showed that the AMA1-C1 immunization induced more cross-reactive antibodies than single-allele immunization. Moreover, it is also apparent from these results that the cross-reactive antibodies (e.g., anti-FVO antibodies on 3D7 parasites) and the strain-specific antibodies (e.g., anti-FVO on FVO parasites) were both capable of inhibiting parasite growth in culture. Thus, both conserved and strain-specific epitopes could be targets of growth-inhibitory activity.
FIG. 2.
Fine specificities of rabbit anti-AMA1 antibodies as determined by an antigen reversal GIA. Purified anti-FVO, -3D7, and -C1 IgGs (0.65 mg/ml in GIA wells, which gave 70% ± 10% inhibition) were preincubated with various concentrations of AMA1-FVO or -3D7 proteins prior to mixing with P. falciparum FVO or 3D7 parasites. The percents reversal of growth-inhibitory activity are plotted against the amounts of AMA1 protein used for the antigen reversal. Anti-FVO IgG was tested with FVO parasites (A), anti-3D7 IgG was tested with 3D7 parasites (C), and anti-C1 IgG was tested with both FVO (B) and 3D7 (D) parasites. Data presented are representative of three independent experiments. The line represents regression of the result as determined by use of a hyperbolic equation.
Strain specificity of anti-AMA1 antibodies in mice.
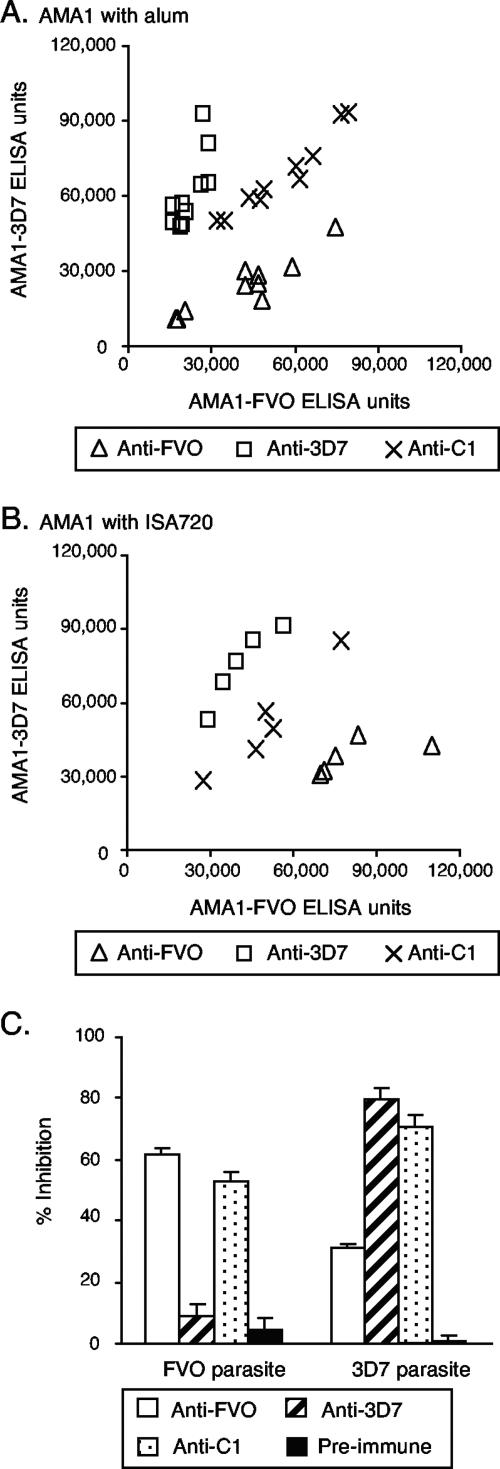
We similarly tested the strain specificities of anti-AMA1 antibodies in mice immunized with AMA1-FVO, AMA1-3D7, or the AMA1-C1 mixture. The immunogens were formulated with Alhydrogel (alum) or Montanide ISA720. Mice were immunized twice at a 1-month interval, and sera obtained 2 weeks after the second injection were tested by an ELISA on plates coated with either AMA1-FVO or AMA1-3D7. Mice immunized with a single form of AMA1 showed significantly higher titers on the homologous protein than on the heterologous protein (Fig. 3A and B). By contrast, when the mixture of the two proteins was used, there was no significant difference in the number of ELISA units tested on either plate. In addition, the levels of antibody were not significantly different for the group of mice that received AMA1-C1 and those that received either of the individual proteins. These observations were true regardless of the adjuvant utilized (alum or Montanide ISA720). We also evaluated the biological activities of the IgGs from pooled mouse preimmune or immune sera by use of a GIA. The GIA was performed with the same concentration of IgG in the well (1 mg/ml), because there were not enough preimmune IgGs to dilute the three immune IgGs to reach the same number of ELISA units. As in the ELISA, the IgG elicited by immunization with a single AMA1 allele showed more inhibitory activity on the homologous parasite than on the heterologous parasite (Fig. 3C). This pattern of strain specificity was similar to that previously seen with rabbits immunized with either the individual proteins or the combination.
FIG. 3.
Reactivity pattern of sera from mice immunized with AMA1-FVO, AMA1-3D7, or AMA1-C1. Mice were immunized with 1 μg of the individual alleles of AMA1 or the combination on days 0 and 28 and bled on days 0 and 42. (A) Groups of 10 mice were immunized with AMA1 formulated on alum. ELISA values for individual mice are plotted. The AMA1-FVO and -3D7 groups show significantly higher titers on the homologous protein than on the heterologous protein (Wilcoxon t test; P = 0.006 for both groups). (B) Groups of five mice were immunized with the same three AMA1 immunogens formulated with Montanide ISA720. (C) Pools of anti-AMA1 sera were prepared from the mice given AMA1 formulated with ISA720, and a pool of preimmune sera was also made. Purified IgGs (1 mg/ml in GIA wells) from these sera were tested by a GIA with P. falciparum FVO or 3D7 parasites. Data are presented as means plus standard errors from three independent experiments. The numbers of ELISA units for anti-FVO, anti-3D7, anti C1, and preimmune IgGs are 28,875, 16,975, 23,700, and 14, respectively, for AMA1-FVO antigen and 13,675, 28,025, 21,150, and 14, respectively, for AMA1-3D7 antigen.
Strain specificity of anti-AMA1 antibodies in rhesus monkeys.
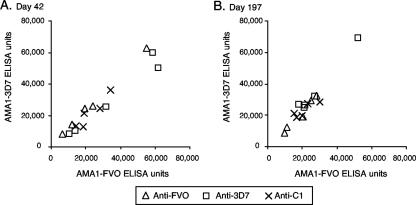
To compare the fine specificities of humoral immune responses to AMA1 in a nonhuman primate, we conducted a similar immunization study with rhesus monkeys. Three groups of five rhesus monkeys each were immunized with AMA1-FVO, AMA1-3D7, or AMA1-C1 formulated with Montanide ISA720 on a day 0, 1-month, and 6-month schedule. In contrast to the previous results, sera from monkeys immunized with a single allelic form of AMA1 showed the same number of ELISA units when tested on either AMA1-FVO- or AMA1-3D7-coated plates (Fig. 4 and Table 1). The ELISA reactivity pattern of sera from animals in the AMA1-C1 group was the same as those in the other two groups. In addition, there was no significant difference in the numbers of ELISA units in any of the three groups when tested on AMA1-FVO- and AMA1-3D7-coated plates using sera obtained at any time point (Wilcoxon t test).
FIG. 4.
ELISA reactivity pattern of sera from rhesus monkeys immunized with either single AMA1 alleles or a combination of AMA1 alleles. Groups of five rhesus monkeys were immunized with 25 μg/dose of AMA1-FVO, AMA1-3D7, or AMA1-C1 formulated with ISA720 on days 0, 28, and 183. They were bled on days 42 and 197. There was no significant difference in ELISA reactivity between the three groups in sera obtained either on day 42 (A) or on day 197 (B).
TABLE 1.
Immunogenicities of AMA1 vaccine in various animal species as judged by ELISAa
| Animal group | No. of ELISA units for indicated immunogen and plate antigen
|
|||||
|---|---|---|---|---|---|---|
| AMA1-FVO
|
AMA1-3D7
|
AMA1-C1
|
||||
| FVO | 3D7 | FVO | 3D7 | FVO | 3D7 | |
| Rabbitb | 105,040 | 42,895 | 37,590 | 96,928 | 94,512 | 90,763 |
| Mouse 1c | 37,173 | 21,897 | 21,806 | 59,946 | 52,664 | 66,485 |
| Mouse 2d | 80,574 | 37,594 | 40,327 | 73,435 | 48,097 | 48,511 |
| Rhesuse | 18,377 | 21,338 | 28,012 | 23,015 | 21,711 | 19,911 |
The geometric means for ELISA units are shown.
Rabbits were immunized as described in the legend to Fig. 1.
Mice were immunized with AMA1 formulated onto alum as described in the legend to Fig. 3.
Mice were immunized with AMA1 formulated with Montanide ISA720 as described in the legend to Fig. 3.
Rhesus monkeys were immunized as described in the legend to Fig. 4. The geometric means for day 42 data are shown.
To determine whether the biological activities of the various rhesus sera paralleled the ELISA results, individual rhesus monkey sera obtained on day 197 were tested by a GIA (Fig. 5). Regardless of the combination of the immunogen used to elicit the anti-AMA1 antiserum and parasite strain, the percent inhibition of the serum was a function of the number of ELISA units in the serum against the particular parasite strain being tested (Spearman rank correlation = 0.785, 95% CI = 0.592 to 0.893, P < 0.0001). Furthermore, the fine specificities of antibodies were assessed by an antigen reversal GIA with purified rhesus anti-AMA1 IgGs. The growth-inhibitory activities of the IgGs from monkeys immunized with single-allele AMA1 protein were reversed to approximately the same degree by either AMA1-FVO or AMA1-3D7 protein, regardless of the immunogen used (Fig. 6). Taken together, these data indicate that in rhesus monkeys there was no difference between single-allele AMA1 immunization and immunization with AMA1-C1 in terms of ELISA units and growth-inhibitory activity in vitro.
FIG. 5.
Cross-reactivities of rhesus anti-AMA1 sera as determined by a GIA. Rhesus monkeys were immunized and bled as described in the legend to Fig. 4. The percents inhibition of the individual day 197 monkey antisera (20 μl/well) were tested against both FVO and 3D7 parasites. The numbers of anti-AMA1-FVO ELISA units against FVO parasites and anti-3D7 ELISA units against 3D7 parasites (x axis) are plotted against percents inhibition determined by the GIA (y axis). There is a significant correlation between number of ELISA units and percent inhibition (Spearman rank correlation = 0.785, 95% CI = 0.592 to 0.893, P < 0.0001).
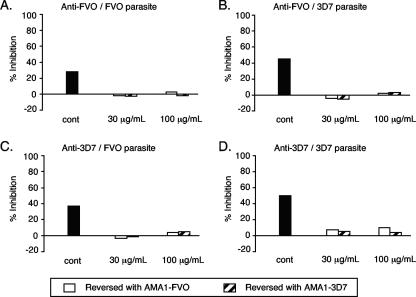
FIG. 6.
Antigen reversal of growth-inhibitory activity from rhesus monkeys immunized with AMA1. Rhesus monkeys were immunized and bled as described in the legend to Fig. 4, and IgG fractions were prepared from pools of day 197 antisera. Purified anti-FVO and -3D7 IgGs (6 mg/ml in GIA wells) were preincubated with various concentrations of AMA1-FVO or -3D7 proteins prior to mixing with P. falciparum FVO or 3D7 parasites. The percents inhibition of growth-inhibitory activity are given for the control (no antigen), 30 μg/ml of AMA1, and 100 μg/ml of AMA1 (used for reversal). Anti-FVO IgG was tested with FVO parasites (A) and 3D7 parasites (B); anti-3D7 IgG was tested with FVO parasites (C) and 3D7 parasites (D). Three data points (for 30 μg/ml of both AMA1 alleles and 100 μg/ml of AMA1-3D7) in panel A are invisible on this scale, because they are too close to zero.
DISCUSSION
While AMA1 is a major candidate for a blood-stage vaccine, the issue of polymorphism of AMA1 in various isolates of P. falciparum is potentially a formidable problem for a vaccine based on this protein (6). This problem is compounded by the incomplete information on the biological roles of the various regions of the AMA1 molecule and on the epitopes in those regions which present effective antibody targets. The recent elucidation of the structure of AMA1 from P. vivax (37) and similar analysis of a portion of the PfAMA1 (17) should facilitate future structure-function studies. In the present report, we have expanded our previous observations with rabbits immunized with AMA1, now showing that when rabbits were immunized with a combination of two allelic forms of AMA1, most of the antibodies produced were cross-reactive and were directed against strain-conserved determinants; by contrast, when the rabbits were immunized with a single AMA1 allele, approximately half of the antibodies recognized cross-reactive epitopes and the remainder were allele specific. This general pattern was also the case when mice were immunized with AMA1. Quite a different result was obtained when nonhuman primates were immunized with AMA1. AMA1 immunization of rhesus monkeys primarily elicited cross-reactive antibodies, even when they were immunized with a single allelic variant, and this cross-reactivity was evident not only with an ELISA but also with an in vitro GIA.
The previous findings that rabbit immunization (27) with single AMA1 alleles elicited significant levels of strain-specific antibodies was somewhat surprising in that there is only a 5% difference between the amino acid sequences of the AMA1-FVO and AMA1-3D7 variants. Structural studies indicate that allelic differences are predominantly on the surface of the molecule and would likely be available for antibody binding. These patterns of responsiveness were found regardless of the adjuvant used for immunization (data not shown). Similarly, we found that mice could also recognize these differences, again regardless of the adjuvant utilized (Fig. 3), and that, like rabbits, rodents immunized with the mixture of two alleles of AMA1 produced more cross-reactive antibodies. In addition, the data in Fig. 1C suggest that the rabbit cross-reactive antibodies induced by AMA1-C1 displayed growth-inhibitory activities comparable to or better than cross-reactive antibodies induced by single-allele immunization when tested at the same number of ELISA units. Indeed, when the anti-AMA1-FVO, -AMA1-3D7, and -AMA1-C1 purified IgGs were tested by a GIA with other strains of parasites, such as D10, HB3, and M24, the anti-C1 IgG showed the highest percents inhibition against all of the parasites tested (data not shown), and this result was similar to that previously seen with intact antisera (27).
The GIA studies with purified anti-AMA1 IgGs allowed us to dissect the specificities of antibodies having biological activities against parasites. As shown in Fig. 2, when rabbits were immunized with single-allele AMA1, the parasite-inhibitory activities of the IgGs were nearly completely reversed by the homologous AMA1 protein. By contrast, heterologous AMA1 protein achieved partial reversal, but the reversal curve reached a plateau after the addition of more than 30 μg/ml of AMA1 protein. This indicates that about half of the activity was derived from the anti-AMA1 IgGs that were strain specific, and the other half was from cross-reactive IgGs. This finding corresponds well with the ELISA data, suggesting that approximately half of the activity was strain specific. On the other hand, the antigen reversal growth inhibition with anti-AMA1-C1 IgG showed different patterns. Although homologous protein reversed the growth-inhibitory activity of AMA1-C1 IgG better than heterologous protein at the same dose, the reversal curve did not reach a plateau in the range of the protein concentration tested, and eventually the heterologous protein reversed to a considerable extent (>75%). The results reveal that the AMA1-C1 immunization of rabbits induced primarily cross-reactive antibodies, not a mixture of allele-specific antibodies. Moreover, the fact that anti-AMA1 IgGs from single-allele-immunized rabbits can partially inhibit invasion and growth of heterologous parasites establishes that conserved, cross-reactive epitopes can be targets of invasion-inhibiting antibodies. The additional growth inhibition activities of anti-AMA1 IgGs on homologous parasites show that strain-specific AMA1 epitopes are also potential targets of parasite inhibition. These results support the previous observations of Anders and colleagues that both conserved and variable AMA1 epitopes can be the target of inhibitory antibodies (2, 22, 28).
Surprisingly, rhesus monkeys showed a completely different strain specificity pattern after AMA1 immunization. Even when animals were immunized with single-allele AMA1, the elicited antibodies could bind indistinguishably to both homologous and heterologous AMA1 proteins on ELISA plates, and the antibodies showed cross-reactive biological activities, as judged by a GIA and an antigen reversal GIA. In contrast to the observations with immune rabbit IgG, the GIA reversal data indicated that rhesus anti-AMA1 antibodies could be essentially completely reversed by either AMA1-FVO or AMA1-3D7 protein, regardless of the immunogen that the monkeys received (Fig. 6). Furthermore, Aotus vociferans monkeys immunized with AMA1-FVO produced antibodies which showed nearly the same ELISA reactivities on AMA1-FVO- and AMA1-3D7-coated plates (supplemental figure). Thus, two different nonhuman primate species preferentially recognized conserved antigenic determinants on AMA1.
Some data are available on human antibody responses to PfAMA1. As noted earlier, Anders and colleagues have shown that affinity-purified anti-AMA1 antibodies from those living in an area of malaria endemicity in Papua New Guinea could block parasite invasion in vitro (22). More recently, they examined the allele specificities of plasma samples from Papua New Guinea adults, reporting that antibodies against conserved regions of the molecule were much more common than allele-specific antibodies (7). However, they found some antibodies against polymorphic epitopes, particularly in young children. Polley and colleagues (40) have examined the responses to AMA1-FVO and AMA1-3D7 constructs, as well as subregions of the molecule, in two Kenyan villages. They reported a tight concordance of responses to the FVO and 3D7 ectodomains, suggesting that most antibodies were binding to conserved antigenic determinants. However, they found bias in responses to the two proteins in sera from a number of individuals in the study, suggesting that some people make allele-specific responses. We have also obtained seroepidemiologic data from a longitudinal study of responses to AMA1 in those living in the village of Doneguebougou, Mali (D. J. Diemert, A. Dolo, M. A. Thera, M. Baby, M. Sissoko, M. B. Niambele, B. Kamate, D. A. Diallo, A. Saul, S. Mahanty, O. K. Doumbo, and C. Long, unpublished results). Although we do not know the nature of their previous AMA1 antigenic exposure through infection, these sera recognize AMA1-FVO and AMA1-3D7 proteins in nearly identical fashions, even in the younger age groups. In addition, malaria-naïve U.S. volunteers immunized with AMA1-C1 formulated with Alhydrogel made comparable antibody responses to AMA1-FVO and AMA1-3D7 (30). In that study, it was not determined whether single-allele AMA1 immunization induced cross-reactive antibodies in naïve individuals, as in the case of the nonhuman primates described here.
If humans primarily produce cross-reactive antibodies to AMA1 by natural infection, such a result appears to contradict the evidence that there is a balancing selection for AMA1 polymorphisms, which have been thought to be maintained, at least in part, by selection pressures due to human immune responses (13, 14, 35, 39, 46). Moreover, some direct evidence is available to support the theory that allelic polymorphisms in AMA1 are involved in the susceptibilities of different parasites to antibody-mediated growth inhibition. Healer et al. (19, 20) have constructed transgenic parasites expressing heterologous AMA1 sequences and shown that these alterations are responsible for the reduction in parasite invasion inhibition by specific antisera. However, the antisera used for the GIA in that study were raised in rabbits and the data presented here suggest that these animals are much more likely to recognize strain-specific epitopes on AMA1. Such sera would be less effective in GIA reactions against the heterologous parasites. Whether similar differences among AMA1 transgenic parasites could be observed with human antisera is unknown.
Our observations with nonhuman primates and some of the available human data need to be considered in the context of previous observations that AMA1 polymorphisms are under natural selection. Humans in areas of endemicity are immunized as a result of repeated infection by multiple strains of parasites, not by deliberate immunization. Immunization by infection may alter the specificity of the response, and repeated infection with different parasite strains may favor B-cell clonotypes producing antibodies against conserved determinants; thus, infants and young children may produce more allele-specific antibody responses and selective pressures may primarily act on parasites in these individuals. Also, it is clear that some adults in areas of endemicity produce a larger proportion of allele-specific antibodies (7, 40), and these antibodies may provide selective advantages for some parasites. Previous studies with other malaria proteins (CSP and MSP2) have suggested that T-cell responses could be the source of immune selection pressure (15, 16). While there is no clear evidence for AMA1, Polley and colleagues (40) proposed the possibility of selection pressures on AMA1 domain III by T-cell-mediated immunity. If it is the case, antibody reactivity patterns might not optimally reflect the selection pressures on genetic polymorphisms. Also, selection on AMA1 may not be as significant as that operating on other merozoite molecules. A study of the genotypes of P. falciparum AMA1 and MSP2 using serum samples from people who had multiple infections over 29 months suggested that greater immune selection pressure was acting on MSP2 than on AMA1 (12). Finally, it is also possible that other types of selection pressures besides those resulting from human immune responses may contribute to the maintenance of AMA1 polymorphisms in the field.
To our knowledge, this is the first report that a malarial vaccine candidate induced different specificities of biologically active antibodies depending on the animal species immunized. While it is unclear in this study why rabbits and nonhuman primate models displayed different strain specificity patterns of vaccine-induced anti-AMA1 antibodies, it is important to note that results with animal models may not predict the response patterns of humans to malaria vaccine candidates. Only human trials can address issues of the strain specificity pattern of vaccine-induced anti-AMA1 antibodies. Immunization becomes particularly complex for those living in areas of endemicity with preexisting anti-AMA1 T- and B-cell responses due to previous malaria exposure. An ongoing phase 1 immunization trial of AMA1-C1 in Malian adults will begin to address this issue, but further studies of responses of those in areas of endemicity will be required to gain an understanding of how genetic diversity in AMA1 affects possible protective responses to this important vaccine candidate. However, the present results suggest that if humans preferentially recognize conserved determinants on AMA1, polymorphism may not be a significant problem for vaccine development based on this protein and that inclusion of limited numbers of alleles in a candidate vaccine may suffice to elicit human antibodies that recognize many AMA1 variants.
Supplementary Material
Acknowledgments
We appreciate the help of the NIH Malaria Vaccine Development Branch Fermentation, Protein Purification, and Quality Control groups for AMA1 antigen production and characterization. We particularly acknowledge the contributions of Anthony Stowers and Michael Kennedy. We are also very grateful to Lynn Lambert, Brian Keegan, Josh Reece, and Cheryl Kothe for meticulous execution of animal immunization studies.
We are grateful for the partial support of these studies by the Malaria Vaccine Initiative/PATH. Additional support for these studies came from the intramural program of the National Institute of Allergy and Infectious Diseases/National Institutes of Health.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 8 October 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, L. H., J. M. Hopkins, A. R. Dluzewski, G. Margos, I. T. Williams, M. J. Blackman, C. H. Kocken, A. W. Thomas, and G. H. Mitchell. 2003. Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development. J. Cell Sci. 116:3825-3834. [DOI] [PubMed] [Google Scholar]
- 3.Burns, J. M., Jr., P. R. Flaherty, P. Nanavati, and W. P. Weidanz. 2004. Protection against Plasmodium chabaudi malaria induced by immunization with apical membrane antigen 1 and merozoite surface protein 1 in the absence of gamma interferon or interleukin-4. Infect. Immun. 72:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, R. J., J. R. Sachs, D. J. Wicker, and M. E. Conrad. 1968. Methemoglobinemia provoked by malarial chemoprophylaxis in Vietnam. N. Engl. J. Med. 279:1127-1131. [DOI] [PubMed] [Google Scholar]
- 5.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, J. S. Sullivan, C. L. Morris, and R. F. Anders. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51:711-719. [DOI] [PubMed] [Google Scholar]
- 6.Conway, D. J. 1997. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitol. Today 13:26-29. [DOI] [PubMed] [Google Scholar]
- 7.Cortés, A., M. Mellombo, R. Masciantonio, V. J. Murphy, J. C. Reeder, and R. F. Anders. 2005. Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 73:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortés, A., M. Mellombo, I. Mueller, A. Benet, J. C. Reeder, and R. F. Anders. 2003. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 71:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10:535-552. [DOI] [PubMed] [Google Scholar]
- 10.Donahue, C. G., V. B. Carruthers, S. D. Gilk, and G. E. Ward. 2000. The Toxoplasma homolog of Plasmodium apical membrane antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Mol. Biochem. Parasitol. 111:15-30. [DOI] [PubMed] [Google Scholar]
- 11.Dutta, S., J. D. Haynes, J. K. Moch, A. Barbosa, and D. E. Lanar. 2003. Invasion-inhibitory antibodies inhibit proteolytic processing of apical membrane antigen 1 of Plasmodium falciparum merozoites. Proc. Natl. Acad. Sci. USA 100:12295-12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen, D. P., A. Saul, D. J. Fryauff, J. C. Reeder, and R. L. Coppel. 2002. Alterations in Plasmodium falciparum genotypes during sequential infections suggest the presence of strain specific immunity. Am. J. Trop. Med. Hyg. 67:8-16. [DOI] [PubMed] [Google Scholar]
- 13.Escalante, A. A., H. M. Grebert, S. C. Chaiyaroj, M. Magris, S. Biswas, B. L. Nahlen, and A. A. Lal. 2001. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 113:279-287. [DOI] [PubMed] [Google Scholar]
- 14.Escalante, A. A., A. A. Lal, and F. J. Ayala. 1998. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics 149:189-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franks, S., L. Baton, K. Tetteh, E. Tongren, D. Dewin, B. D. Akanmori, K. A. Koram, L. Ranford-Cartwright, and E. M. Riley. 2003. Genetic diversity and antigenic polymorphism in Plasmodium falciparum: extensive serological cross-reactivity between allelic variants of merozoite surface protein 2. Infect. Immun. 71:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good, M. F., D. Pombo, I. A. Quakyi, E. M. Riley, R. A. Houghten, A. Menon, D. W. Alling, J. A. Berzofsky, and L. H. Miller. 1988. Human T-cell recognition of the circumsporozoite protein of Plasmodium falciparum: immunodominant T-cell domains map to the polymorphic regions of the molecule. Proc. Natl. Acad. Sci. USA 85:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, A., T. Bai, V. Murphy, P. Strike, R. F. Anders, and A. H. Batchelor. 2005. Refolding, purification, and crystallization of apical membrane antigen 1 from Plasmodium falciparum. Protein Expr. Purif. 41:186-198. [DOI] [PubMed] [Google Scholar]
- 18.Healer, J., S. Crawford, S. Ralph, G. McFadden, and A. F. Cowman. 2002. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infect. Immun. 70:5751-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Healer, J., V. Murphy, A. N. Hodder, R. Masciantonio, A. W. Gemmill, R. F. Anders, A. F. Cowman, and A. Batchelor. 2004. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol. Microbiol. 52:159-168. [DOI] [PubMed] [Google Scholar]
- 20.Healer, J., T. Triglia, A. N. Hodder, A. W. Gemmill, and A. F. Cowman. 2005. Functional analysis of Plasmodium falciparum apical membrane antigen 1 utilizing interspecies domains. Infect. Immun. 73:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hehl, A. B., C. Lekutis, M. E. Grigg, P. J. Bradley, J. F. Dubremetz, E. Ortega-Barria, and J. C. Boothroyd. 2000. Toxoplasma gondii homologue of Plasmodium apical membrane antigen 1 is involved in invasion of host cells. Infect. Immun. 68:7078-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 24.Howell, S. A., I. Well, S. L. Fleck, C. Kettleborough, C. R. Collins, and M. J. Blackman. 2003. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J. Biol. Chem. 278:23890-23898. [DOI] [PubMed] [Google Scholar]
- 25.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311-31320. [DOI] [PubMed] [Google Scholar]
- 26.Kato, K., D. C. Mayer, S. Singh, M. Reid, and L. H. Miller. 2005. Domain III of Plasmodium falciparum apical membrane antigen 1 binds to the erythrocyte membrane protein Kx. Proc. Natl. Acad. Sci. USA 102:5552-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy, M. C., J. Wang, Y. Zhang, A. P. Miles, F. Chitsaz, A. Saul, C. A. Long, L. H. Miller, and A. W. Stowers. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 70:6948-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocken, C. H., C. Withers-Martinez, M. A. Dubbeld, A. van der Wel, F. Hackett, A. Valderrama, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 70:4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahanty, S., A. Saul, and L. H. Miller. 2003. Progress in the development of recombinant and synthetic blood-stage malaria vaccines. J. Exp. Biol. 206:3781-3788. [DOI] [PubMed] [Google Scholar]
- 30.Malkin, E. M., D. J. Diemert, J. H. McArthur, J. R. Perreault, A. P. Miles, B. K. Giersing, G. E. Mullen, A. Orcutt, O. Muratova, M. Awkal, H. Zhou, J. Wang, A. Stowers, C. A. Long, S. Mahanty, L. H. Miller, A. Saul, and A. P. Durbin. 2005. Phase 1 clinical trial of apical membrane antigen 1: an asexual blood-stage vaccine for Plasmodium falciparum malaria. Infect. Immun. 73:3677-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell, G. H., A. W. Thomas, G. Margos, A. R. Dluzewski, and L. H. Bannister. 2004. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 72:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair, M., M. G. Hinds, A. M. Coley, A. N. Hodder, M. Foley, R. F. Anders, and R. S. Norton. 2002. Structure of domain III of the blood-stage malaria vaccine candidate, Plasmodium falciparum apical membrane antigen 1 (AMA1). J. Mol. Biol. 322:741-753. [DOI] [PubMed] [Google Scholar]
- 33.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 67:59-68. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira, D. A., V. Udhayakumar, P. Bloland, Y. P. Shi, B. L. Nahlen, A. J. Oloo, W. E. Hawley, and A. A. Lal. 1996. Genetic conservation of the Plasmodium falciparum apical membrane antigen-1 (AMA-1). Mol. Biochem. Parasitol. 76:333-336. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, M. G., V. M. Marshall, J. A. Smythe, P. E. Crewther, A. Lew, A. Silva, R. F. Anders, and D. J. Kemp. 1989. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 9:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizarro, J. C., B. V. Normand, M. L. Chesne-Seck, C. R. Collins, C. Withers-Martinez, F. Hackett, M. J. Blackman, B. W. Faber, E. J. Remarque, C. H. Kocken, A. W. Thomas, and G. A. Bentley. 2005. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science 308:408-411. [DOI] [PubMed] [Google Scholar]
- 38.Polley, S. D., W. Chokejindachai, and D. J. Conway. 2003. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics 165:555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polley, S. D., and D. J. Conway. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polley, S. D., T. Mwangi, C. H. Kocken, A. W. Thomas, S. Dutta, D. E. Lanar, E. Remarque, A. Ross, T. N. Williams, G. Mwambingu, B. Lowe, D. J. Conway, and K. Marsh. 2004. Human antibodies to recombinant protein constructs of Plasmodium falciparum Apical Membrane Antigen 1 (AMA1) and their associations with protection from malaria. Vaccine 23:718-728. [DOI] [PubMed] [Google Scholar]
- 41.Sachs, J., and P. Malaney. 2002. The economic and social burden of malaria. Nature 415:680-685. [DOI] [PubMed] [Google Scholar]
- 42.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434:214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stowers, A. W., M. C. Kennedy, B. P. Keegan, A. Saul, C. A. Long, and L. H. Miller. 2002. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect. Immun. 70:6961-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]
- 45.Udhayakumar, V., S. Kariuki, M. Kolczack, M. Girma, J. M. Roberts, A. J. Oloo, B. L. Nahlen, and A. A. Lal. 2001. Longitudinal study of natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in a holoendemic region of malaria in western Kenya: Asembo Bay Cohort Project VIII. Am. J. Trop. Med. Hyg. 65:100-107. [DOI] [PubMed] [Google Scholar]
- 46.Verra, F., and A. L. Hughes. 2000. Evidence for ancient balanced polymorphism at the Apical Membrane Antigen-1 (AMA-1) locus of Plasmodium falciparum. Mol. Biochem. Parasitol. 105:149-153. [DOI] [PubMed] [Google Scholar]
- 47.Waters, A. P., A. W. Thomas, J. A. Deans, G. H. Mitchell, D. E. Hudson, L. H. Miller, T. F. McCutchan, and S. Cohen. 1990. A merozoite receptor protein from Plasmodium knowlesi is highly conserved and distributed throughout Plasmodium. J. Biol. Chem. 265:17974-17979. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.