Abstract
Day-old chicks are very susceptible to infections with Salmonella enterica subspecies. The gut mucosa is the initial site of host invasion and provides the first line of defense against the bacteria. To study the potential of different S. enterica serovars to invade the gut mucosa and trigger an immune response, day-old chicks were infected orally with Salmonella enterica serovar Enteritidis, S. enterica serovar Typhimurium, S. enterica serovar Hadar, or S. enterica serovar Infantis, respectively. The localization of Salmonella organisms in gut mucosa and the number of immune cells in cecum were determined by immunohistochemistry in the period between 4 h and 9 days after infection. Using quantitative real-time reverse transcription (RT)-PCR, mRNA expression of various cytokines, chemokines, and inducible nitric oxide synthase (iNOS) was examined in cecum. As a result, all S. enterica serovars were able to infect epithelial cells and the lamina propria. Notably, serovar Enteritidis showed the highest invasiveness of lamina propria tissue, whereas serovars Typhimurium and Hadar displayed moderate invasiveness and serovar Infantis hardly any invasion capabilities. Only a limited number of bacteria of all serovars were found within intestinal macrophages. Elevated numbers of granulocytes, CD8+ cells, and TCR1+ cells and mRNA expression rates for interleukin 12 (IL-12), IL-18, tumor necrosis factor alpha factor, and iNOS in cecum correlated well with the invasiveness of serovars in the lamina propria. In contrast, changes in numbers of TCR2+ and CD4+ cells and IL-2 mRNA expression seemed to be more dependent on infection of epithelial cells. The data indicate that the capability of Salmonella serovars to enter the cecal mucosa and invade lower regions affects both the level and character of the immune response in tissue.
Salmonella enterica ssp. enterica infection of humans is one of the most important food-borne zoonoses worldwide. In 2005 alone, about 52,000 human cases were reported in Germany (52), but the true figure is assumed to be far higher. While the genus Salmonella is characterized by an extremely high degree of serological diversity, each of the more than 2,500 known Salmonella serovars is potentially capable of producing gastroenteritis, with a few specific representatives being considered to be exceptionally dangerous. Among a number of more or less virulent Salmonella serovars, Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium, followed by Salmonella enterica serovars Infantis, Hadar, and Virchow, are most frequently isolated from humans in European countries (15). The main source of human Salmonella infections is poultry-derived food, mainly eggs and egg products but also chicken meat. Therefore, the primary aim of Salmonella control in poultry is to prevent these organisms from entering the food chain. In addition to efficient hygiene regimes at all stages of production, specific control measures, such as immunization with both live and inactivated Salmonella vaccines against the serovars Enteritidis and Typhimurium, are being used with poultry (16). Nevertheless, human salmonellosis remains a serious public health issue, and therefore, in-depth investigations of basic defense mechanisms against Salmonella in chickens are essential to improve existing vaccines and to develop new, more efficacious vaccines.
Salmonella organisms infect animals as well as humans by the oral route. To induce gastroenteritis, the pathogen must reach the distal ileum and cecum in the first place, where it is set to outcompete the resident microbial flora and must penetrate the mucosal epithelium. For effective elimination of harmful agents in the gastrointestinal tract, a highly specialized lymphoid tissue exists separately in the gut (gut-associated lymphoid tissue [GALT]). An attribute of GALT is the formation of characteristic epithelium covering the lymphoid tissue and containing M cells well equipped for uptake of particular antigens from the lumen. In contrast to the highly organized GALT in mammals, the avian counterpart does not always develop such a specialized epithelium. Indeed, most of the avian gut tissue is separated from the lumen by regular epithelium. Only some unique tissues, such as the cecum tonsils, Peyer's patches, and Meckel's diverticulum, have some parts with the unique epithelium (7, 10). Concerning Salmonella infection in poultry, little is known about how and to what extent Salmonella strains enter the gut mucosa. Only a few studies are focused on the mucosal invasiveness of different Salmonella strains, but experiments either were conducted with ex vivo cell culture systems (33, 42) or permanent cell lines (1, 26) or were gut loop experiments (1). In fact, the mucosal epithelium is not only a physical barrier but also the initiator of the innate immune response to Salmonella. Specialized epithelial cells can produce bactericidal enzymes, and the encounter of bacteria and epithelial cells rapidly stimulates the release of proinflammatory chemokines that attract innate immune cells, such as granulocytes, macrophages, and immature dendritic cells. These cells constitute the first line of defense, are able to engulf and kill pathogens nonspecifically, and trigger a wide range of further immune reactions. An early neutrophil/heterophil and macrophage infiltration in avian gut mucosa has been shown after oral administration of Salmonella strains (63). In vitro studies with avian macrophages additionally showed that avian macrophages are very efficient in bactericidal function against Salmonella (49). Nevertheless, Salmonella organisms are also capable of surviving within macrophages (47, 65). In a case where innate immune mechanisms are not sufficient to prevent bacterial multiplication, adaptive and specific immunity is initiated to completely eradicate the invaders. Adaptive immune mechanisms are divided into cell-mediated immunity, which acts predominantly through effector T cells, and humoral immunity, which operates by means of specific antibodies. In the case of avian Salmonella infection, recent evidence has emerged indicating that humoral immune mechanisms do not play an essential role in clearance of primary infection or in long-term protection (4). Therefore, considerable effort has been undertaken to explore the avian cellular immune defense in different organs, such as the gut, spleen, liver, and blood. Changes in number, distribution, and proliferation of T cells (6, 8) and a delayed-type hypersensitivity response were observed (23). Concerning non-host-adapted serovars, chicks experimentally infected with S. enterica ssp. enterica serovar Enteritidis or Typhimurium were used exclusively in those studies (2, 3, 8, 9, 29, 46, 54). However, there is no in vivo data on the capability of different Salmonella serovars to invade epithelial cells and macrophages in the lower regions of the mucosal lamina propria at present.
Recent progress in cloning of avian cytokine and chemokine genes also promoted further analysis of proinflammatory cytokine and chemokine production during various viral and bacterial infections in chickens (19, 27, 31, 53). Indeed, changing patterns of cytokine mRNA expression have been demonstrated in the course of serovar Typhimurium or serovar Enteritidis infection in avian organs, such as spleen (46, 57) and gut (69), and in avian cells, such as granulocytes (59) and T-cell subsets (9). In mice, Th1 cytokines, which enhance cell-mediated immunity, are crucial for protection against Salmonella infection (35). A protective role for interleukin 1α (IL-1α), tumor necrosis factor alpha, gamma interferon (IFN-γ), IL-12, IL-18, and IL-15 has been demonstrated with a mouse typhoid model (14). On the other hand, IL-4 and IL-10 inhibited the host defense against Salmonella.
The present study was undertaken to characterize serovar Enteritidis, serovar Typhimurium, serovar Hadar, and serovar Infantis as the most commonly isolated, non-host-adapted serovars from both laying hens (16) and broilers (17) with regard to their potential to enter the cecum mucosa, reside in macrophages, and stimulate an immune response. The emergence of immune cells playing a role in the innate and/or adaptive immune response, as well as interleukin, chemokine and inducible nitric oxide synthase (iNOS) mRNA expression, was analyzed after infection of day-old chicks in cecum and compared to Salmonella invasiveness in gut mucosa.
MATERIALS AND METHODS
Chickens.
Specific-pathogen-free White Leghorn chickens were hatched at the facilities of Friedrich-Loeffler-Institute, Jena, Germany, from eggs obtained from Charles River Deutschland GmbH (Extertal, Germany). Experimental and control groups were kept in separate rooms; commercial feed (in powder form without antibiotics or other additives) and drinking water were both available ad libitum. Cleaning and feeding regimens were organized, which prevented cross-contamination effectively throughout the experiment.
Bacterial strains and experimental design.
Newly hatched day-old chicks were infected orally using the following Salmonella serovars (Table 1): serovar Enteritidis 147 (group 1), serovar Typhimurium 9098 (group 2), serovar Hadar 18 (group 3), or serovar Infantis 1326 (group 4). All Salmonella serovars are representatives of the appropriate O groups (B, C1, C2, and D) and tested as good colonizers of the avian gut (40). The Salmonella wild-type strains were inoculated at a dose of 1 × 107 to 2 × 107 CFU per bird at 1 day of age to ensure a high cecum colonization by the strains without inducing severe clinical signs of morbidity (39). Oral administration of 0.1 ml/bird of bacterial suspension was performed by instillation into the crop using a syringe with an attached flexible tube. Another control group (group 5) was neither immunized nor infected. All Salmonella strains used in this study were stored in the Microbank system (PRO-LAB Diagnostics, Ontario, Canada) at −20°C. The bacterial suspensions for infection were cultivated by shaking (18 h at 37°C) in nutrient broth (Sifin, Berlin, Germany). Doses were estimated by measuring extinction at 600 nm against a calibration graph determined for the strains used and were subsequently confirmed by plate counting on nutrient agar (Sifin).
TABLE 1.
Salmonella serovars used in this study
Three birds from each group were sacrificed at 4 h after infection and at 1, 2, 4, 7, and 9 days postinfection (dpi). At each time point, liver and cecum content was collected aseptically for bacteriology. Additionally, ceca were taken from every animal and frozen in liquid nitrogen or stored in RNA-later (Qiagen, Hilden, Germany) until use for immunohistochemistry, confocal microscopy, or quantitative real-time RT-PCR. For flow cytometry, blood and spleen samples of each animal were prepared immediately as described below.
Bacteriology.
Using a standard plating method (38), bacterial counts in cecum content and liver were estimated for all birds (three per group and investigation day). For this purpose, homogenized samples were diluted in phosphate-buffered saline, plated on deoxycholate-citrate agar (Sifin) to detect the challenge organisms, and incubated at 37°C for 18 to 24 h. Buffered peptone water was used for preenrichment of the organ homogenates, which were incubated overnight at 37°C and subcultured on the solid agar.
Immunohistochemistry.
For study of Salmonella invasion and cellular influx, frozen sections of all birds were prepared from cecum and stained as described previously (8). Briefly, tissue sections (of 7-μm thickness) were fixed with acetone and subsequently incubated with the appropriate monoclonal antibodies against Salmonella lipopolysaccharide (LPS) (Chemicon, Hofheim, Germany), avian granulocytes (32) (kindly provided by R. Ducatelle, University Gent, Belgium), mononuclear phagocytes (macrophages; CVI 68.1; Institute for Animal Science and Health, Lelystad, The Netherlands), γδ T cells (TCR1), αβ T cells (TCR2), and CD4+ (CT-4), CD8α+ (CT-8), and CD8β+ (EP-42) cells (all from Southern Biotechnology Associates, Eching, Germany), and the secondary goat anti-mouse immunoglobulin (Sigma, Deisenhoven, Germany), as well as the peroxidase-antiperoxidase complex (Sigma). The enzyme-linked antibody was visualized by reaction with 3,3′-diaminobinzidine (Merck, Darmstadt, Germany) and hydrogen peroxide. Sections were counterstained with hematoxylin.
Confocal laser-scanning microscopy.
To study colocalization of macrophages and Salmonella organisms and for quantification of intracellular bacteria by confocal laser-scanning microscopy, acetone-fixed cryostat sections (10-μm thickness) of ceca from all animals (three per group and investigation day) were incubated with primary antibody against Salmonella LPS (immunoglobulin G2a [IgG2a]; Chemicon) and macrophages (CVI 68.1, IgG1; Institute for Animal Science and Health) and detected with the isotype-specific (IgG2a) antibody A-555 (Molecular Probes, Leiden, The Netherlands) and the isotype-specific (IgG1) antibody A-488 (Molecular Probes), respectively. Sections were washed with phosphate-buffered saline three times between each incubation step for 2 min. Finally, the sections were counterstained with 1 μg/ml 4′,6′-diamidino-2-phenylindole (Sigma) and mounted with glycerin gelatin. Colocalization of Salmonella organisms and macrophages was evaluated by using a TCS-SP2 confocal microscope (Leica, Bensheim, Germany) and image analysis. For this purpose, fluorescence image stacks were sequentially registered by the 488-nm (green) and 546-nm (red) channels. A corresponding 4′,6′-diamidino-2-phenylindole wide-field image was subsequently scanned using the Hg lamp for excitation and collected with the photomultiplier tube. To differentiate between epithelium and lamina propria, the images were merged subsequently. At least five images per animal were collected and evaluated.
Image analysis.
Analysis of the number of granulocytes and proportions of immunohistochemically stained areas of immune cells and Salmonella organisms in tissue were performed with each individual bird (three per group and investigation day) by means of the image analysis system analySIS (SIS, Münster, Germany).
For counting of granulocytes, regions of interest (ROI) were drawn interactively, including lamina propria and epithelial lining of cecum. Using computer software, areas were determined and the numbers of granulocytes (characterized by their strong staining and morphology) were counted interactively within ROI and calculated to 1 mm2. At least three ROIs per animal were analyzed.
The changes in percentages of all other immune cell populations were monitored by determining the percentage of positive stained areas in whole-cecum mucosa (epithelial lining and lamina propria). A ROI was drawn (three per animal) and the percentage of stained area determined by using image analysis software.
In the case of Salmonella organisms, analyses were performed for whole-cecum mucosa (epithelium and lamina propria) and separately for the epithelial lining, subepithelial region, and lamina propria basement region. Percentages of Salmonella stained areas of the whole cecum mucosa and epithelium were analyzed by using an image analysis system as described for cell populations (see above). Numbers of Salmonella organisms in subepithelial and basement regions were evaluated individually by using a scoring system (0, no bacteria; 1, very low number; 2, moderate numbers stained; 3, high numbers of bacteria detectable).
For quantification of intracellular salmonellae by colocalization study, a ROI including the lamina propria of each collected confocal image (five per animal) was drawn, the total area of the extracellular salmonellae (green) and intracellular salmonellae (yellow) detected, and the percentage of intracellular salmonellae from all bacteria calculated by using analySIS software.
The number of differently sized Salmonella accumulations (clusters) in 1 mm2 of cecum lamina propria was determined by image analysis of the confocal images by using analySIS software. For this purpose, four classes of Salmonella clusters were defined: 1, single bacterium (0.5 to 3 μm2), 2, small-sized clusters (3 to 10 μm2); 3, medium-sized clusters (10 to 50 μm2); 4, large-sized clusters (50 to 150 μm2).
Additionally, 100 stained cecum macrophages per animal were counted and the percentage of Salmonella-infected cells determined using the confocal images and analySIS software.
Flow cytometry.
For analysis of changes in T-cell composition upon infection, heparinized blood of every animal (three per group and investigation day) was mixed with 3% hetastarch (Sigma Immuno Chemicals, St. Louis, MO) at a ratio of 1:2 and centrifuged at 65 × g for 10 min to allow erythrocytes to sediment. The cells of the supernatant were used for flow-cytometric analysis of blood lymphocyte as described previously (8).
To isolate leukocytes from the spleen, the outer connective tissue was removed and tissue homogenized. After cell detachment by passage through a 20-gauge syringe, lymphocytes were separated by centrifugation using 3% hetastarch according to the procedure for peripheral blood lymphocytes as described previously (8).
Isolated leukocytes (2 × 105) were incubated with fluorescein isothiocyanate (FITC)-labeled monoclonal antibodies (TCR1-FITC and TCR2-FITC; Southern Biotechnology Associates, Eching, Germany) for 30 min in the dark. After the cells were washed, aliquots of 20,000 cells per sample were analyzed using a FACSCalibur flow cytometry system (Becton Dickinson, Heidelberg, Germany) equipped with a 15-mW, 488-nm argon ion laser.
Primer design.
Avian cDNA or mRNA sequences for cytokines, chemokines, iNOS, cytokine-receptor chains, and apoptosis-related proteins were identified using the NCBI home page (http://www.ncbi.nlm.nih.gov/). In cases of the availability only of DNA, the Chicken BLAT Search (http://genome.ucsc.edu/cgi-bin/hgBlat) was used to find exons. Amplification primers were designed by means of the Primer Express 2.0 software (Applied Biosystems, Darmstadt, Germany), with at least one primer of each pair covering an exon-intron boundary. Finally the primers created were checked by using the NCBI BLAST Search (http://www.ncbi.nlm.nih.gov/BLAST/) for specificity.
Each primer pair was tested with a serial dilution of RNA in quantitative real-time RT-PCR to ensure comparable efficiencies of the RT-PCR. The amplification specificity was checked by melting-curve analysis and determination of amplicon size by gel electrophoresis of amplification products in 2% agarose gels.
Quantitative real-time RT-PCR.
To study the dynamics of cytokine production after Salmonella exposure, total RNA was extracted from cecum samples of each individual bird using the RNeasy minikit (Qiagen), following the manufacturer's instructions. Contaminating DNAs were digested using an RNase-free DNase set (Qiagen) and resulting RNAs eluted in 50 μl RNase-free water per 20 mg tissue and stored at −80°C. The resulted mRNAs were evaluated and the quantity and quality determined by spectral analysis (BioPhotometer; Eppendorf, Hamburg, Germany). Only samples with mRNA purity of about 2 (A260/A280 ratio) or above 2 (A260/A230) were used. The quantity of mRNA was adjusted for quantitative RT-PCR and the mRNA expression rates of avian cytokines (IFN-γ, IL-2, IL-12β [p40], IL-18, lipopolysaccharide-induced tumor necrosis factor alpha factor [LITAF]), chemokines (CXCL8b [9E3/CEF4; henceforth named IL-8] and CCL4L1 [henceforth named MIP-1β]) (64), cytokine receptor chains (IL-2Rα and IL-7Rα), iNOS, and apoptosis-related proteins (Fas, Fas ligand, and Bcl-x) determined for every individual bird using the QuantiTect SYBR Green one-step RT-PCR kit (Qiagen) according to the instructions of the manufacturer. Amplification and detection of specific products were performed using Mx3000P real-time PCR equipment (Stratagene, La Jolla, CA) with the following temperature-time profile: one cycle of 50°C for 30 min, 96°C for 15 min, and 45 cycles of 94°C for 30 s, respective annealing temperature for 30 s, followed by 72°C for 30 s. To check the specificities of amplification products, the dissociation-curve mode was used (one cycle at 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s) subsequent to amplification. Primer sequences and annealing temperatures are given in Table 2. The threshold method was used for quantification of the mRNA level (48). Threshold cycle (ΔCT) values were calculated on the basis of the internal standard glyceraldehyde-3-phosphate dehydrogenase. Results were expressed as 2(−ΔΔCT) (n-fold change).
TABLE 2.
Primer sequences for real-time RT-PCR
| Avian RNA target and primer description | Sequence (5′-3′) | Annealing temp (°C) | Sequence accession no.b |
|---|---|---|---|
| IFN-γ | |||
| Forward | GCCGCACATCAAACACATATCT | 57 | NM_205149 |
| Reverse | TGAGACTGGCTCCTTTTCCTT | ||
| IL-2 | |||
| Forward | GAGTCTTACGGGTCTAAATC | 53 | NM_204153 |
| Reverse | CACAAAGTTGGTCAGTC | ||
| CXCL8b (IL-8) | |||
| Forward | ATGAACGGCAAGCTTGGAGCT | 57 | NM_205498 |
| Reverse | GCCATAAGTGCCTTTACGATCAG | ||
| IL-12β (p40) | |||
| Forward | TGGTCCACGCTTTGCAGAT | 54 | AJ564201 |
| Reverse | AAGGTTAAGGCGTGGCTTCTTA | ||
| IL-18 | |||
| Forward | TCTGGCAGTGGAATGTACTTCG | 54 | MN_204608 |
| Reverse | CCATTTTCCCATGCTCTTTCTC | ||
| CCL4L1 (MIP-1β) | |||
| Forward | TCCTGCTGCTTCACCTACATCT | 59 | AJ243034 |
| Reverse | ATGAACACAACACCAGCATGAG | ||
| IL-2Rα | |||
| Forward | GCAGATAAATGCCCACGTCTTT | 54 | NM_204596 |
| Reverse | TCAAGGTGTTCCCACTTCGTC | ||
| IL-7Rα | |||
| Forward | AGCATGCTCAGAATGACACGG | 54 | XM_423732 |
| Reverse | TCATCTCCAAAGGTCCCATCA | ||
| Bcl-x | |||
| Forward | GCGAAAGGTCGGATGTGTT | 56 | U26645 |
| Reverse | GCTGGACATTTTCACACCTGA | ||
| Fas | |||
| Forward | AAAGCACTCGGTTTGGAGGT | 54 | AF296875 |
| Reverse | ATGACTCGCAATGTTCACACC | ||
| Fas-L | |||
| Forward | CACTTAACAGGAAACCCCACAC | 54 | AJ890143 |
| Reverse | TTGATCACAAGGCCCTGGT | ||
| LITAF | |||
| Forward | GCTGTTCTATGACCGCCCAGTT | 56 | NM_204267 |
| Reverse | AACAACCAGCTATGCACCCCA | ||
| INOS | |||
| Forward | GAACAGCCAGCTCATCCGATA | 54 | AY648162 |
| Reverse | CCCAAGCTCAATGCACAACTT | ||
| GAPDHa | |||
| Forward | GTCAGCAATGCATCGTGCA | 54 | K01458 |
| Reverse | GGCATGGACAGTGGTCATAAGA |
Glyceraldehyde-3-phosphate dehydrogenase.
NCBI accession numbers.
Statistical analysis.
The data of flow cytometry analysis, quantitative real-time RT-PCR, and image analysis were evaluated statistically. The Student t test for comparison of two independent samples was used to evaluate differences between the groups (each infected group versus the control group). P values of ≤0.05 were considered significant.
Viable counts of bacteria were converted into logarithmic form. For the purpose of statistical analysis, a log10 viable count of <1.47 (limit for direct plate detection) obtained from a sample becoming positive only after enrichment was rated as a log10 value of 1.0. A sample which yielded no Salmonella growth after enrichment was rated as a log10 value of 0. Data were evaluated by multifactorial variance analysis. The factors considered were group and time. P values of ≤0.05 were regarded as statistically significant (Statgraphics Plus software; StatPoint, Inc., Rockville, MD).
RESULTS
Clinical signs after Salmonella infection and bacteriology.
The oral administration of Salmonella wild-type strains of different serovars inoculated at a dose of 1 × 107 to 2 × 107 CFU per bird produced a strong colonization of the intestine and spread to the liver (Table 3) but did not result in morbidity or severe clinical signs. However, signs of intestinal inflammation were observed in some birds in groups challenged with the highly invasive strains serovar Enteritidis 147 and serovar Typhimurium 9098. The different serovars of Salmonella did not reveal relevant differences in their ability to colonize the ceca. Only serovar Enteritidis exhibited slightly but significantly (P ≤ 0.05) reduced counts in cecum content compared with the other serovars at 1 and 2 dpi. Despite their strong capability to colonize the gut, serovar Hadar 18 and serovar Infantis 1326 showed not only a significantly diminished (P ≤ 0.05) but a considerably diminished invasion of the liver at 2, 4, and 7 dpi or on all days postinfection, respectively, compared to serovar Enteritidis and serovar Typhimurium. These differences amounted to 1.5 to 2.0 log10 units, indicating not only their decreased invasion but also their reduced virulence. Although invasion by serovar Enteritidis and serovar Typhimurium of liver tissue was significantly (P ≤ 0.05) different at 7 dpi only, serovar Enteritidis revealed higher counts on all days of examination.
TABLE 3.
Number of Salmonella organisms in liver and cecum content of chickens after oral administration of 2 × 107 CFU/bird on day 1 of age
| Time postinfection | Bacterial count (log10 CFU/g)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Group 1 |
Group 2 |
Group 3 |
Group 4 |
|||||
| Liver | Cecum content | Liver | Cecum content | Liver | Cecum content | Liver | Cecum content | |
| 4 h | 0.5 | 8.5 | 0.7 | 8.7 | 0 | 7.4 | 0 | 8.2 |
| 1 day | 2.3 | 8.7 (c, d) | 2.1 | 9.3 | 2.0 | 9.8 | 0.6 (a) | 10.2 |
| 2 days | 3.2 | 9.1 (d) | 2.9 | 9.5 | 1.7 (a, b) | 9.7 | 0.9 (a, b) | 10.0 |
| 4 days | 3.3 | 9.5 | 2.5 | 9.7 | 1.5 (a, b) | 9.5 | 0.8 (a, b) | 9.3 |
| 7 days | 3.2 | 9.3 | 2.2a | 9.4 | 0.8 (a, b) | 9.0b | 0.3 (a, b) | 9.0 |
| 9 days | 2.5 | 9.2 | 2.1 | 9.1 | 2.1 | 8.8 | 0 (a, b, c) | 8.9 |
Group 1, serovar Enteritidis 147; group 2, serovar Typhimurium 9098; group 3, serovar Hadar 18; group 4, serovar Infantis 1326. Standard error: liver, 0.152; cecum content, 0.071. A parenthetical a, b, c, and/or d indicates a result significantly lower than that for group 1, 2, 3, and/or 4.
Quantification of Salmonella serovars in the cecum wall.
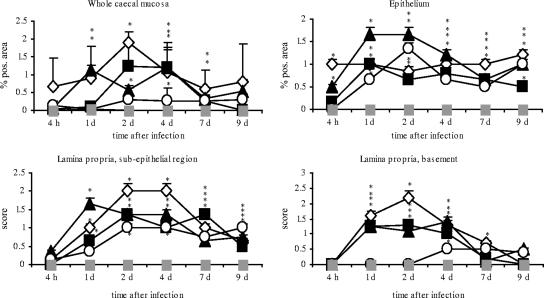
The potential of different Salmonella serovars to invade lower regions of the cecum mucosa was analyzed by using immunohistochemistry and image analysis. All Salmonella serovars were capable of invading the cecum mucosa, but to different degrees (Fig. 1). Serovar Enteritidis, serovar Typhimurium, and serovar Hadar were found in epithelium and in subepithelial and lamina propria basement regions of the cecum. Notably, serovar Enteritidis displayed the best capability of reaching lower zones of the lamina propria. Compared to the other serovars, it was found in the largest quantities in the basement region at 1 and 2 dpi. In contrast, serovar Hadar was most significantly able to infect the epithelium 1, 2, and 4 dpi and serovar Infantis entered solely epithelial cells and regions located directly below the epithelium (subepithelial region).
FIG. 1.
Diagrams showing percentages of Salmonella positive-stained areas in whole avian cecum mucosa and epithelium and the scores for numbers of bacteria in subepithelial and basement regions of cecum lamina propria after infection with different Salmonella serovars. Asterisks indicate a significant difference between the treated and control groups (P ≤ 0.05). ⋄, serovar Enteritidis; ▪, serovar Typhimurium; ▴, serovar Hadar; ○, serovar Infantis; ░⃞, control. d, day; pos., positive.
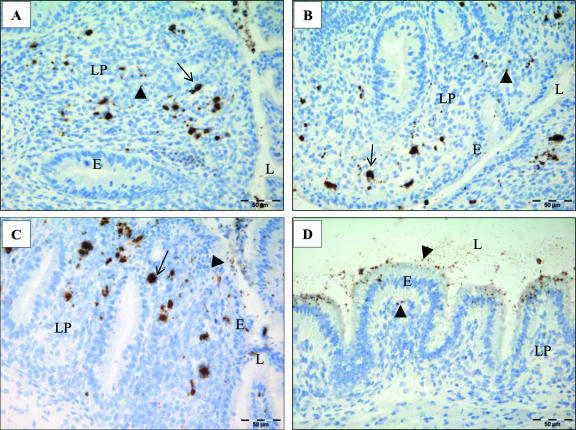
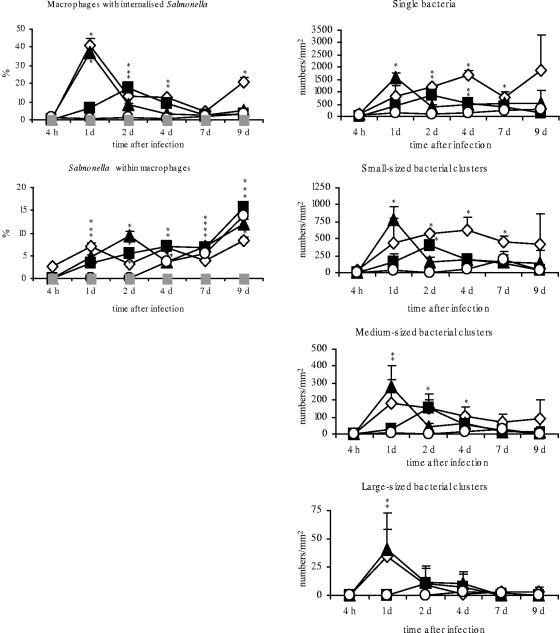
As seen in immunohistochemical staining and analyzed by means of confocal images, the Salmonella serovars presented themselves as both single cells and characteristic bacterial accumulations (see clusters in Fig. 2 and 4). Bacterial clusters were observed exclusively within the lamina propria, whereas single bacteria were seen within the epithelial lining and in the gut lumen but also in the lamina propria. In comparison, serovar Enteritidis, serovar Typhimurium, and serovar Hadar revealed larger accumulations and serovar Infantis showed only small or no accumulations. Bacterial clusters already were seen 1 dpi in serovar Enteritidis-, serovar Hadar-, and serovar Typhimurium-infected animals. While small-sized clusters and single bacteria were found up to 9 dpi for all infected animals, a decline of large-sized clusters was observed for serovar Enteritidis- and serovar Hadar-treated animals between 1 and 4 dpi (Fig. 3).
FIG. 2.
Representative photographs of cecum mucosa after infection of day-old chicks with different Salmonella serovars. Single bacteria are marked with arrowheads and bacterial accumulations with arrows. (A) Serovar Enteritidis infection, 2 dpi: bacteria spread over the whole lamina propria. (B) Serovar Typhimurium infection at 2 dpi: bacteria predominantly located subepithelially. (C) Serovar Hadar infection at 2 dpi: bacteria predominantly located subepithelially. (D) Serovar Infantis infection at 2 dpi: bacteria are predominantly located in epithelial lining.
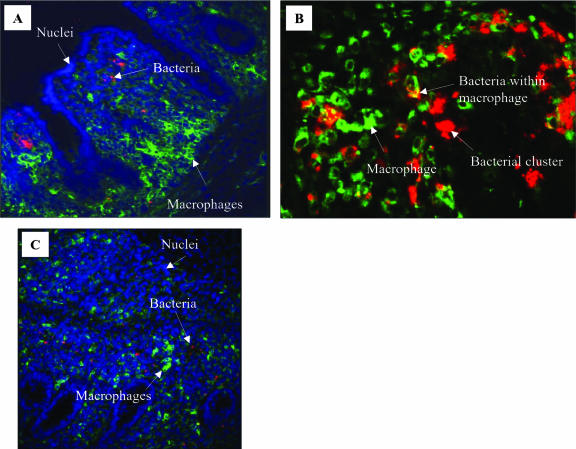
FIG. 4.
Representative confocal laser-scanning micrographs of cecum mucosa of serovar Typhimurium-infected (A) or serovar Enteritidis-infected (B and C) chicks. Red, Salmonella; green, CVI 68.1-positive macrophages; blue, cell nuclei. (A) 1 dpi; different localizations of bacteria (mainly subepithelial) and macrophages (mainly in basement regions) are shown at this time point after infection. Magnification, ×20. (B) 1 dpi; representative region showing colocalization of macrophages and bacteria and Salmonella clusters outside macrophages. Magnification, ×40. (C) 4 dpi; many single bacteria distributed over the whole lamina propria and not colocalized with macrophages. Magnification, ×20.
FIG. 3.
Percentages of Salmonella organisms within CVI 68.1-stained macrophages and Salmonella-positive macrophages and numbers of single bacteria and variably sized bacterial clusters per mm2 after infection of day-old chicks with different Salmonella serovars in cecum mucosa. Asterisks indicate a significant difference between the treated and control groups (P ≤ 0.05). ⋄, serovar Enteritidis; ▪, serovar Typhimurium; ▴, serovar Hadar; ○, serovar Infantis; ░⃞, control. d, day.
Quantification of intracellular salmonellae.
To study colocalization of Salmonella organisms and macrophages, confocal microscopy and image analysis were used. The results showed that most of the bacteria were not associated with macrophages. Only low percentages (between 0 and 16%) of all salmonellae were detected within macrophages (Fig. 3). In serovar Infantis-infected chicks, intracellular bacteria were not seen before 4 dpi.
Salmonella-infected macrophages.
As the colocalization study by confocal laser-scanning microscopy of infected animals revealed, the percentage of macrophages in cecum lamina propria with internalized bacteria ranged between 0.1% (serovar Infantis-infected group, 1 dpi) and 45% (serovar Enteritidis-infected group, 1 dpi) depending on the Salmonella serovar (Fig. 3, % macrophages with internalized Salmonella). The peak number of Salmonella-positive macrophages was seen 1 dpi for serovar Enteritidis- and serovar Hadar-infected chicks and 2 dpi for serovar Typhimurium-treated birds (18% Salmonella-positive macrophages). After infection with serovar Infantis, hardly any colocalization was detected.
Between 2 and 9 dpi, merely about 1 to 3 bacteria were detected in association with macrophages for serovar Enteritidis-, serovar Typhimurium- and serovar Hadar-infected chicks (data not shown). At 1 dpi, the number of intracellular bacteria per macrophage could not be determined because large-scale clustering occurred and a separation into single bacteria was impossible. At this time point, portions of Salmonella clusters were frequently observed as doubly stained for Salmonella and macrophage (Fig. 4).
Quantification of cellular immune response in cecum.
To characterize the potential of different Salmonella serovars to trigger an immune response after invasion of gut mucosa, the occurrence of relevant immune cell subsets in the cecum was investigated by image analysis (Fig. 5).
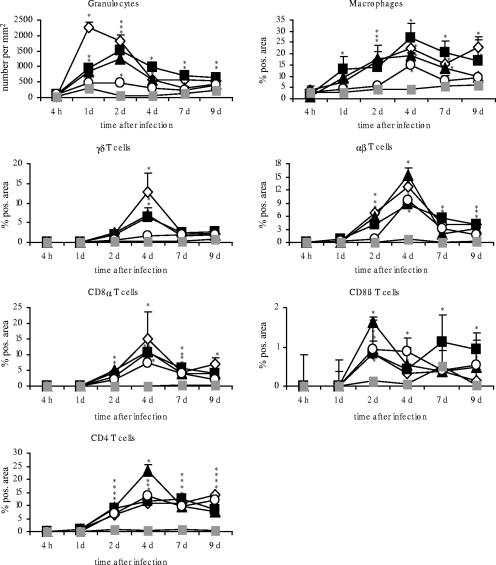
FIG. 5.
Occurrence of immune cells after oral infection of day-old chicks with different Salmonella serovars in cecum. Asterisks indicate a significant difference between the treated and control groups (P ≤ 0.05). ⋄, serovar Enteritidis; ▪, serovar Typhimurium; ▴, serovar Hadar; ○, serovar Infantis; ░⃞, control. d, day; pos., positive.
Already at 1 and 2 dpi, a strong and significant increase of granulocytes was found in serovar Typhimurium-, serovar Hadar-, and particularly in serovar Enteritidis-infected birds (P ≤ 0.05). The elevated number of granulocytes continued up to day 9 after infection. The animal group infected with serovar Infantis also showed a significantly increased number of granulocytes per mm2 in the cecum wall (P ≤ 0.05) but at a very low level compared to the other infected groups. For serovar Enteritidis-, serovar Typhimurium-, and serovar Hadar-infected chicks, granulocytes were predominantly found in the lamina propria and submucosa at 1 dpi. Later on, granulocytes were mostly localized under the epithelial lining (4 dpi), in the epithelium, and within the gut lumen (7 and 9 dpi).
Additionally, a significant increase in the percentage of TCR1+ (γδ) cells was seen at 2, 4, 7, and 9 dpi (P ≤ 0.05) in the cecum mucosa of serovar Enteritidis-, serovar Typhimurium-, and serovar Hadar-infected chicks. The peak was observed at 4 dpi. Starting from 4 dpi, serovar Infantis-infected animals also showed a slight rise of γδ T cells (P ≤ 0.5).
A significant increase in the proportion of CD8α+ cells was detected at 2, 4, 7, and 9 dpi for serovar Enteritidis-, serovar Typhimurium-, serovar Hadar-, and serovar Infantis-infected animals (P ≤ 0.05). The number of CD8α+ cells peaked at 4 dpi in all infected animal groups.
The results concerning the established percentages of granulocytes, γδ T cells, and CD8α+ cells permitted a classification of the Salmonella serovars with regard to their potential to trigger an influx and/or proliferation of these cell populations in the gut mucosa as follows: (i) serovar Enteritidis as a strong mediator, (ii) serovar Typhimurium and serovar Hadar as moderate mediators, and (iii) serovar Infantis as a weak mediator of an immune cell influx.
Interestingly, serovar Infantis-infected animals showed similar changes in the numbers of αβ (TCR2+), CD4+, and CD8α+ T cells in the cecum, as observed with serovar Enteritidis, serovar Hadar, and serovar Typhimurium infection. Numbers of TCR2+ and CD4+ cells were significantly increased after infection in all animal groups (P ≤ 0.05). While the elevated percentages of TCR2+ cells peaked at 4 dpi, CD4+ cells showed a plateau between 2 and 9 dpi in all infected groups (with the exception of serovar Hadar-treated animals at 4 dpi).
Compared to results for controls, the proportions of CD8β+ cells were clearly increased between 2 and 9 days after infection in all infected groups (P ≤ 0.05), but values showed some undulations. CD4+ cells were seen from 2 to 9 dpi in the ceca of all infected animals (P ≤ 0.05). For serovar Hadar-infected chicks, a peak was observed beyond results for the other infected groups at 4 dpi.
Quantification of cellular immune response in peripheral blood and spleen.
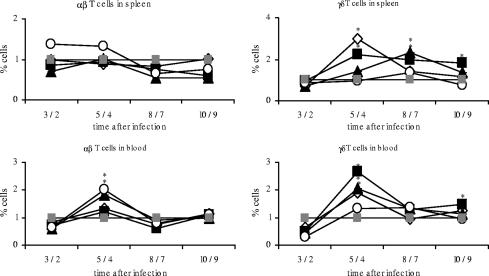
To elucidate to which extent different Salmonella serovars trigger an immune response beyond the local reaction in the gut, the dynamics of T-cells responses in peripheral blood and the spleen was analyzed upon infection (Fig. 6). The resulting data on the proportion of γδ T lymphocytes in blood and spleen tissue replicated the results obtained for the cecum. As with the cecum, the peak of γδ T cells could be observed at 4 dpi. The four Salmonella serovars used induced immune reactions on different levels with hardly any reaction upon serovar Infantis infection and clear responses for serovar Enteritidis-, serovar Hadar-, and serovar Typhimurium-infected chicks (P ≤ 0.05). In contrast, there was a significant increase of αβ T cells at 4 dpi after serovar Infantis infection in peripheral blood and spleen samples (P ≤ 0.05). With the exception of results for serovar Hadar in blood, serovar Enteritidis-, serovar Typhimurium- and serovar Hadar-treated animals did not show any changes in the percentage of αβ T cells in blood and spleen tissue.
FIG. 6.
Relative proportions of alpha/beta and gamma/delta T cells in blood and spleen. Day-old chicks were orally infected with different Salmonella serovars. Results for control animals were set to 1. Asterisks indicate a significant difference between the treated and control groups (P ≤ 0.05). ⋄, serovar Enteritidis; ▪, serovar Typhimurium; ▴, serovar Hadar; ○, serovar Infantis; ░⃞, control. The x axis for each panel shows days postinfection.
Quantification of cytokine, chemokine, apoptosis-related protein, and iNOS mRNA expression in cecum.
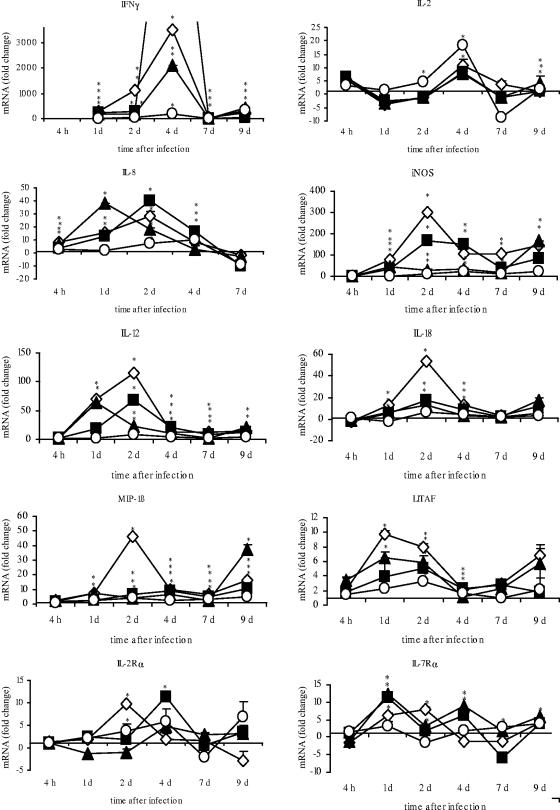
Statistically significantly (P ≤ 0.05) increased mRNA expression of all investigated cytokines, cytokine receptors, chemokines, and iNOS was found in the cecum upon Salmonella infection of day-old chicks. Changes in mRNA expression showed dynamic processes in the course of Salmonella infection (Fig. 7). The standard deviations were always very low and therefore are sometimes not visible in diagrams.
FIG. 7.
Diagrams showing quantifications of different cytokines, chemokines, and iNOS after infection of day-old chicks with different Salmonella serovars. Data are given as n-fold changes in mRNA levels for infected birds relative to levels for noninfected controls. Each value represents the mean for three individual samples. Standard errors are shown as vertical bars. Very small standard deviations are often not visible. Asterisks indicate a significant difference between the treated and control groups (P ≤ 0.05). ⋄, serovar Enteritidis; ▪, serovar Typhimurium; ▴, serovar Hadar; ○, serovar Infantis. d, day.
For interleukins (IL-12 and IL-18), chemokines (MIP-1β and LITAF), and iNOS, changes were most pronounced in serovar Enteritidis-infected animals. After serovar Enteritidis infection, the peak of mRNA expression was established for LITAF at 1 and 2 dpi (up to 10- and 8-fold increase; P ≤ 0.05) and for the others at 2 dpi (for IL-12, up to 116-fold increase; for IL-18, up to 53-fold increase; for iNOS, up to 298-fold increase; for MIP-1β, up to 46-fold increase; P ≤ 0.05). Serovar Infantis-infected chicks showed comparably low increases, with a maximum expression of LITAF mRNA between 1 and 2 dpi (up to two- and threefold increase; P ≤ 0.05), MIP-1β mRNA at 2 dpi (up to fourfold increase; P ≤ 0.05), IL-12 mRNA at 2 dpi (up to eightfold increase; P ≤ 0.05), IL-18 mRNA at 2 and 4 dpi (up to six- and fourfold increase; P ≤ 0.05), and iNOS mRNA at 4 dpi (up to 21-fold increase; P ≤ 0.05). After infection with serovar Typhimurium or serovar Hadar, mRNA expression for iNOS, IL-12, IL-18, MIP-1β, and LITAF was enhanced on different intermediate levels (P ≤ 0.05).
The change of IL-8 mRNA expression was a dynamic process over a period of 4 h to 7 dpi in all animal groups. The level peaked 1 dpi for serovar Hadar-infected animals (up to 38-fold increase; P ≤ 0.05), 2 dpi for serovar Enteritidis-infected animals (up to 28-fold increase; P ≤ 0.05) and serovar Typhimurium-infected animals (up to 40-fold increase; P ≤ 0.05), and 4 dpi for serovar Infantis-infected animals (up to 9-fold increase; P ≤ 0.05).
The mRNA expression of IL-2 was characterized by a significant increase peaking 4 dpi (for serovar Infantis, up to 18-fold increase; for serovar Enteritidis, up to 11-fold increase; for serovar Typhimurium, up to 8-fold increase; for serovar Hadar, up to 7-fold increase; P ≤ 0.05). Notably, the most distinct change in IL-2 mRNA expression was observed 2 and 4 dpi for serovar Infantis-infected chicks (P ≤ 0.05).
Statistically increased (P ≤ 0.05) mRNA expression of IFN-γ was found at all time points (the 4-h time point was not investigated) examined for serovar Enteritidis-, serovar Typhimurium-, serovar Hadar-, and serovar Infantis-infected animals (Table 4) (with the exception of serovar Infantis-infected animals at 7 dpi). Already 1 day after infection with serovar Enteritidis (up to 266-fold increase), serovar Hadar (up to 215-fold increase), serovar Typhimurium (up to 204-fold increase), or serovar Infantis (up to 8-fold change), significantly increased mRNA expression (P ≤ 0.05) was detected (data are not always visible in diagram of Fig. 7). The values peaked at 4 dpi in all animal groups. At days 1 and 2 after infection for chicks treated with serovar Enteritidis (up to 266- and 1,141-fold increases, respectively), at 4 dpi for treatment with serovar Typhimurium (up to 12,105-fold increase), at 7 dpi for treatment with serovar Enteritidis (24-fold increase), and at 9 dpi for treatment with serovar Hadar (up to 430-fold increase), chicks showed the highest values compared to results for the other animal groups. Serovar Infantis-infected birds always showed the lowest values (with the exception of results at 9 dpi).
TABLE 4.
Changes in IFN-γ mRNA expression in cecum samples after infection of day-old chicks with different Salmonella serovars
| Time postinfection (day) | Fold change in IFN-γ mRNA expression after infection witha: |
|||
|---|---|---|---|---|
| SE | STM | SH | SINF | |
| 1 | 266.2 ± 0.9** | 215.7 ± 3.1** | 204.1 ± 0.5** | 8.3 ± 1.6* |
| 2 | 141.5 ± 1.1** | 269.9 ± 1.2** | 178.1 ± 1.8* | 35.3 ± 2.1** |
| 4 | 3500.5 ± 1.4** | 12105.2 ± 1.8** | 2095.8 ± 2.1** | 188.3 ± 2.3** |
| 7 | 24.8 ± 1.7** | 1.33 ± 2.4 | 4.9 ± 0.4** | 0.55 ± 1.2 |
| 9 | 244.7 ± 2.6** | 27.8 ± 2.3* | 430.0 ± 1.1* | 347.7 ± 2.9* |
SE, serovar Enteritidis 147; STM, serovar Typhimurium 9098; SH, serovar Hadar 18; SINF, serovar Infantis 1326. **, significantly different from result for nontreated control animals (P ≤ 0.01); *, significantly different from result for nontreated control animals (P ≤ 0.05).
mRNA expression rates of receptors for IL-2 and IL-7 were changed differentially after Salmonella infection of day-old chicks. While expression of IL-2Rα peaked at 2 dpi after serovar Enteritidis infection (P ≤ 0.05), serovar Typhimurium-, serovar Hadar-, and serovar Infantis-treated animals showed the largest changes at 4 dpi (P ≤ 0.05). The strongest elevated IL-7Rα mRNA expression was seen 1 and 2 dpi in serovar Typhimurium-, and serovar Hadar-, and in serovar Enteritidis-infected animals.
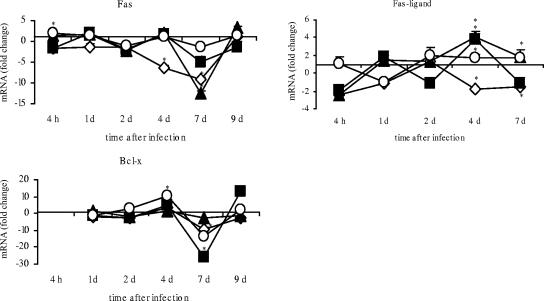
Significantly decreased mRNA expression for Fas was observed at 4 dpi for serovar Enteritidis-infected chicks (P ≤ 0.05) and at 7 dpi (P ≤ 0.05) for all infected animal groups (Fig. 8). Similar results were seen for Bcl-x-mRNA expression. There was significantly decreased expression at 7 dpi in all infected animal groups (P ≤ 0.05). In contrast, Fas ligand expression was heterogenic after infection. At 4 dpi, serovar Enteritidis-infected birds showed significantly decreased expression (P ≤ 0.05) and serovar Hadar-, serovar Typhimurium-, and serovar Infantis-infected chicks showed increased expression (P ≤ 0.05).
FIG. 8.
Quantification of Fas, FasL, and Bcl-x expression after infection of day-old chicks with different Salmonella serovars. Data are given as n-fold changes in mRNA levels for infected birds relative to those for noninfected controls. Each value represents the mean for three individual samples. Standard errors are shown as vertical bars. The very small standard deviations are often not visible. Asterisks indicate a significant difference between the treated and control groups (P ≤ 0.05). ⋄, serovar Enteritidis; ▪, serovar Typhimurium; ▴, serovar Hadar; ○, serovar Infantis. d, day.
DISCUSSION
In the present study, four different Salmonella serovars known to be frequently responsible for human food-borne Salmonella infections were compared regarding their invasiveness and potential to trigger an early immune defense in the avian gut. The Salmonella strains used are good colonizers in the cecum and are representatives of their O groups as tested in several in vivo assays (39, 40). The finding that serovar Enteritidis showed the highest invasiveness and serovar Infantis the lowest value correlated with our data on cellular immune responses and interleukin mRNA expression in the cecum. It has been shown that both the intensity and the character of Salmonella-elicited immune responses depends on the capability of Salmonella serovars to invade the lower regions of the lamina propria.
Various bacterial factors, such as LPS, flagellae, fimbriae, and some outer membrane proteins, have been reported to act as initiators in adhesion to and/or invasion of the epithelium of the alimentary tract by Salmonella organisms. The role of these factors in pathogenesis has remained unclear (21, 42, 66). There are some hints that the O-group antigen may be associated with colonization of the avian gut (13, 60) and of chicken ovaries and the oviduct (21, 22, 50). Additionally, LPS seems to play a role in adherence to avian epithelial cells (60). In mice, the involvement of Salmonella O-group antigens in virulence was demonstrated by the fact that modifications in the LPS side chains result in major virulence changes (44, 45, 61, 62). Whether the LPS O-chain types are responsible for different invasion properties in the present study remains open. However, for colonization of cecum content and epithelium (similar with all serovars), a particular O-group antigen seemed not to be absolutely necessary. Notably, the Salmonella serovars used in this study showed different abilities to invade lower regions of the cecum lamina propria and to trigger particular immune reactions. How far the different O antigens really could be responsible for these various properties must be investigated in further studies. In accordance with the present findings, a very strong invasiveness of serovar Enteritidis compared to that of other serovars had already been shown by results of in vitro studies (1, 42). However, our investigation demonstrated for the first time an in vivo correlation between the invasiveness of Salmonella serovars and the cellular influx, as well as cytokine/chemokine expression in the intestine. The following can be concluded from the present data: (i) serovar Enteritidis is highly invasive and a strong immune stimulator, (ii) serovar Typhimurium and serovar Hadar are moderately invasive and intermediate immune stimulators, and (iii) serovar Infantis is hardly invasive at all and a weak immune stimulator. Notably, serovar Infantis was found to persist in epithelial cells as long as the other serovars examined. Nevertheless, the immune reaction was rather weak upon infection with serovar Infantis.
In vitro studies have shown that interaction between Salmonella and epithelial cells results in a proinflammatory response characterized by the release of several interleukins and chemokines, especially IL-8, the best studied one and the one responsible for specific immune cell attraction (14). Interestingly, Salmonella must access the basolateral surfaces of epithelial cells to initiate the inflammatory response, since an important receptor for bacterial flagellin, Toll-like receptor 5, is located only basolaterally (19, 20). Additionally, macrophage and dendritic cells possess special receptors, such as TLR4, that interact with the LPS- or LPS/LPB-CD14 complex and trigger macrophage activation. This might explain the higher capacity of more-invasive Salmonella serovars to induce granulocyte, macrophage, and γδ-T-cell immigration into the intestinal mucosa, as well as the graded, serovar-dependent mRNA expression of macrophage-specific interleukins in our study. To what extent Salmonella mutants lacking flagella or possessing altered LPS are able to invade cecum mucosa and trigger immune reactions must be investigated in further studies.
While serovar Infantis-infected animals exhibited only a comparably slight increase in numbers of granulocytes, macrophages, and γδ T cells, the TCR2- and CD4-T-cell responses and almost also the CD8-T-cell response were as high as those for serovar Enteritidis-, serovar Typhimurium-, and serovar Hadar-infected birds. A higher CD4 response was observed in our study up to 9 dpi for all infected animals. Similarly, Salmonella-specific CD4-T-cell expansion is rapidly initiated upon infection with serovar Typhimurium in mucosal lymphoid tissue of mice (28, 41, 57, 69). In view of the different invasion capabilities of serovars in the present study, the mechanism of the rapid and similar TCR2- and CD4-T-cell expansions and IL-2 mRNA expression in all animal groups remains unclear. Some models were postulated for the observed rapid T-cell activation in Peyer's patches of mice (51). Beside the possibility that myeloid dendritic cells engulf and process Salmonella antigens and subsequently carry them to T cells, soluble bacterial antigen, such as flagellin, could be excreted into the subepithelial lymph fluid and flow rapidly to the T-cell area without the need for any cellular transport or bacterial dissemination. Indeed, flagellin is the most consistently identified target antigen of Salmonella-specific CD4 T cells (12, 37, 58).
For detection of macrophages in tissue, the CVI 68.1 antibody was used. It is described as a marker for avian monocytes, macrophages, and interdigitating cells, as well as some bursa cells (25). More-detailed studies showed that the antibody identifies a subtype of CD45+, ATPase-positive, vimentin-positive dendritic cells in the avian epidermis (24). In our study, the CVI 68.1-positive cells showed a granular staining pattern within the cytoplasm and a dendritic morphology as seen for tissue macrophages. Whether the antibody detects absolutely all macrophages and cecum dendritic cells must remain an open question. In the present study, most of the salmonellae were not localized in CVI 68.1-positive macrophages but were extracellular in the cecum lamina propria. Thus, the possibility of an extracellular dissemination in gut mucosa and of extracellular virulence mechanisms of Salmonella strains in vivo cannot be ruled out. In mice, escape from infected cells followed by bacterial spread and distribution in the tissues seems to be crucial for the full expression of Salmonella virulence (36). On the other hand, findings of in vitro studies using macrophage cell lines showed strong cellular infection of avian macrophages by salmonellae and the property of reproduction inside the cell (18, 30). In our study, predominantly single bacteria were found in association with macrophages after infection. Similarly, studies with mice showed that high numbers of Salmonella organisms within infected phagocytes were uncommon in vivo (36, 55).
Additionally, size analysis of Salmonella clusters did not completely evince proliferation of bacteria in the lamina propria in the course of our experiment. Otherwise, the single bacteria would be at their maximum number at 1 or 4 dpi and decline afterwards, and big clusters would emerge at the same time. This was not the case, since all cluster sizes exhibited similar dynamic behaviors. Perhaps there is a dynamic process of Salmonella invasion from the gut lumen into the lamina propria and simultaneous destruction in the mucosa. The decline of big clusters and Salmonella-infected macrophages from 1 dpi onward argues for a rapid destruction of bacteria in macrophages. Indeed, the mRNA expression for LITAF and iNOS showed the highest values between 1 and 2 dpi. Additionally, expression of IFN-γ, the most important immune stimulator for macrophages, was significantly increased 1 day after infection.
The observed prominent expression of IFN-γ in parallel with the strong T-cell influx into the cecum lamina propria additionally confirmed the importance of the Th1 immune reaction in Salmonella infection, as shown by other authors (11, 68). IL-12 and IL-18 are interleukins produced by macrophages and promote IFN-γ production by NK cells in mice (34). Whether this is the same in poultry remains unclear. However, the detected peak of IL-12 and IL-18 mRNA at 2 dpi, followed by the highest level of IFN-γ expression at 4 dpi in infected chicks, might be evidence of the same dynamic response in chickens. Interestingly, compared to the serovar Typhimurium group, there is less of an induction of IFN-γ in the serovar Enteritidis group, which shows the highest mRNA expression of IL-12 and IL-18. Perhaps there are additional mechanisms responsible for the level of gene expression. The high n-fold changes in INF-γ expression upon Salmonella infection in the presented study conflict with results found after exposure of older birds to Salmonella (5, 6). In contrast, Withanage et al. (68) showed an up-regulation of IFN-γ mRNA of up to 200-fold after Salmonella challenge. However, the cecum as a representative organ for Salmonella infection in chickens has never been used for comparisons of Salmonella invasiveness.
Our study generally demonstrated a rapid and significant mRNA expression of cytokines/chemokines upon Salmonella infection in the cecum. That is in line with observations of other authors (67), who found prompt expression of the chemokines IL-8 and MIP-1β within 12 h after S. enterica serovar Typhimurium infection in the gut of newly hatched chicks. Another study demonstrated an induction of early MIP-1β in spleen and liver tissue but not in intestines (68). Beyond this, we additionally showed rapid mRNA expression of the cytokines IL-12, IL-18, LITAF, and iNOS after Salmonella exposure. Interestingly, the increased expression level of iNOS continued up to 9 dpi, indicating a longer-lasting involvement of macrophages in elimination of the pathogen. With the exception of serovar Infantis, indeed, all Salmonella serovars were still detectable 9 dpi in liver tissue.
The results from the present experiment demonstrate the ability of day-old chicks to express a wide range of cytokines upon oral infection with different Salmonella serovars in the cecum. While the lamina-propria invasion capabilities of the Salmonella strains differed with respect to the serovars, the colonization within epithelial cells was quite the same. Noticeably, the different levels of invasiveness of serovars in the lamina propria correlated with the levels of cytokine production. A Th1-cytokine reaction was observed together with a strong T-cell influx in the cecum. Remarkably, mRNA expression of IL-2 and IL-2Rα and the CD4-T-cell influx seemed to be dependent on the infection of intestinal epithelial cells. Between 2 and 9 dpi, only a limited number of Salmonella organisms were detected inside macrophages, which is in accordance with results from studies with mice (36, 43, 55). The present in vivo experiments can be seen as a step towards a better understanding of possible virulence and persistence mechanisms of different Salmonella serovars and might contribute to the prospective development of high-efficacy vaccines. Future studies should address the identification of more invasion-associated factors to further elucidate pathogenesis and defense mechanisms in Salmonella infection of poultry.
Acknowledgments
This study was supported by the European Union, grant FOOD-CT-2003-505523.
Editor: F. C. Fang
Footnotes
Published ahead of print on 20 August 2007.
REFERENCES
- 1.Aabo, S., J. P. Christensen, M. S. Chadfield, B. Carstensen, J. E. Olsen, and M. Bisgaard. 2002. Quantitative comparison of intestinal invasion of zoonotic serotypes of Salmonella enterica in poultry. Avian Pathol. 31:41-47. [DOI] [PubMed] [Google Scholar]
- 2.Asheg, A., M. Levkut, V. Revajova, Z. Sevcikova, L. Kolodzieyski, and J. Pistl. 2002. T lymphocyte subpopulations and B lymphocyte cells in caecum and spleen of chicks infected with Salmonella enteritidis. Acta Histochem. 104:435-439. [DOI] [PubMed] [Google Scholar]
- 3.Babu, U., M. Scott, M. J. Myers, M. Okamura, D. Gaines, H. F. Yancy, H. Lillehoj, R. A. Heckert, and R. B. Raybourne. 2003. Effects of live attenuated and killed Salmonella vaccine on T-lymphocyte mediated immunity in laying hens. Vet. Immunol. Immunopathol. 91:39-44. [DOI] [PubMed] [Google Scholar]
- 4.Beal, R. K., C. Powers, T. F. Davison, P. A. Barrow, and A. L. Smith. 2006. Clearance of enteric Salmonella enterica serovar Typhimurium in chickens is independent of B-cell function. Infect. Immun. 74:1442-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beal, R. K., C. Powers, P. Wigley, P. A. Barrow, P. Kaiser, and A. L. Smith. 2005. A strong antigen-specific T-cell response is associated with age and genetically dependent resistance to avian enteric salmonellosis. Infect. Immun. 73:7509-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beal, R. K., C. Powers, P. Wigley, P. A. Barrow, and A. L. Smith. 2004. Temporal dynamics of the cellular, humoral and cytokine responses in chickens during primary and secondary infection with Salmonella enterica serovar Typhimurium. Avian Pathol. 33:25-33. [DOI] [PubMed] [Google Scholar]
- 7.Befus, D. A., N. Johnston, G. A. Leslie, and J. Bienenstock. 1980. Gut-associated lymphoid tissue in the chicken. I. Morphology, ontogeny, and some functional charateristics of Peyer's patches. J. Immunol. 125:2626-2632. [PubMed] [Google Scholar]
- 8.Berndt, A., and U. Methner. 2001. Gamma/delta T cell response of chickens after oral administration of attenuated and non-attenuated Salmonella typhimurium strains. Vet. Immunol. Immunopathol. 78:143-161. [DOI] [PubMed] [Google Scholar]
- 9.Berndt, A., J. Pieper, and U. Methner. 2006. Circulating γδ T cells in response to Salmonella enterica serovar enteritidis exposure in chickens. Infect. Immun. 74:3967-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns, R. B., and M. H. Maxwell. 1986. Ultrastructure of Peyer's patches in the domestic fowl and turkey. J. Anat. 147:235-243. [PMC free article] [PubMed] [Google Scholar]
- 11.Cheeseman, J. H., M. G. Kaiser, C. Ciraci, P. Kaiser, and S. J. Lamont. 2007. Breed effect on early cytokine mRNA expression in spleen and cecum of chickens with and without Salmonella enteritidis infection. Dev. Comp. Immunol. 31:52-60. [DOI] [PubMed] [Google Scholar]
- 12.Cookson, B. T., and M. J. Bevan. 1997. Identification of a natural T cell epitope presented by Salmonella-infected macrophages and recognized by T cells from orally immunized mice. J. Immunol. 158:4310-4319. [PubMed] [Google Scholar]
- 13.Craven, S. E., N. A. Cox, J. S. Bailey, N. J. Stern, R. J. Meinersmann, and L. C. Blankenship. 1993. Characterization of Salmonella california and S. typhimurium strains with reduced ability to colonize the intestinal tract of broiler chicks. Avian. Dis. 37:339-348. [PubMed] [Google Scholar]
- 14.Eckmann, L., and M. F. Kagnoff. 2001. Cytokines in host defense against Salmonella. Microbes Infect. 3:1191-1200. [DOI] [PubMed] [Google Scholar]
- 15.European Food Safety Authority. 2004. Opinion of the Scientific Panel on Biological Hazards on the request from the Commission related to the use of vaccines for the control of Salmonella in poultry. EFSA J. 114:1-74. [Google Scholar]
- 16.European Food Safety Authority. 2006. Analysis of the baseline study on the prevalence of Salmonella in laying hen flocks of Gallus gallus. EFSA J. 81:1-71. [Google Scholar]
- 17.European Food Safety Authority. 2007. Report of the task force on zoonoses data collection on the analysis of the baseline survey on the prevalence of Salmonella in broiler flocks of Gallus gallus, in the EU, 2005-2006. EFSA J. 98:1-85. [Google Scholar]
- 18.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gewirtz, A. T., T. A. Navas, S. Lyons, P. J. Godowski, and J. L. Madara. 2001. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167:1882-1885. [DOI] [PubMed] [Google Scholar]
- 20.Gewirtz, A. T., P. O. Simon, Jr., C. K. Schmitt, L. J. Taylor, C. H. Hagedorn, A. D. O'Brien, A. S. Neish, and J. L. Madara. 2001. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J. Clin. Investig. 107:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guard-Petter, J., L. H. Keller, M. M. Rahman, R. W. Carlson, and S. Silvers. 1996. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol. Infect. 117:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guard-Petter, J., C. T. Parker, K. Asokan, and R. W. Carlson. 1999. Clinical and veterinary isolates of Salmonella enterica serovar Enteritidis defective in lipopolysaccharide O-chain polymerization. Appl. Environ. Microbiol. 65:2195-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan, J. O., and R. Curtiss III. 1994. Virulent Salmonella typhimurium-induced lymphocyte depletion and immunosuppression in chickens. Infect. Immun. 62:2027-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igyarto, B. Z., E. Lacko, I. Olah, and A. Magyar. 2006. Characterization of chicken epidermal dendritic cells. Immunology 119:278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeurissen, S. H., E. M. Janse, G. Koch, and G. F. de Boer. 1988. The monoclonal antibody CVI-ChNL-68.1 recognizes cells of the monocyte-macrophage lineage in chickens. Dev. Comp. Immunol. 12:855-864. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser, P., L. Rothwell, E. E. Galyov, P. A. Barrow, J. Burnside, and P. Wigley. 2000. Differential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology 146:3217-3226. [DOI] [PubMed] [Google Scholar]
- 27.Kaiser, P., G. Underwood, and F. Davison. 2003. Differential cytokine responses following Marek's disease virus infection of chickens differing in resistance to Marek's disease. J. Virol. 77:762-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirby, A. C., M. Sundquist, and M. J. Wick. 2004. In vivo compartmentalization of functionally distinct, rapidly responsive antigen-specific T-cell populations in DNA-immunized or Salmonella enterica serovar Typhimurium-infected mice. Infect. Immun. 72:6390-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krajina, T., F. Leithauser, and J. Reimann. 2004. MHC class II-independent CD25+ CD4+ CD8alpha beta+ alpha beta T cells attenuate CD4+ T cell-induced transfer colitis. Eur. J. Immunol. 34:705-714. [DOI] [PubMed] [Google Scholar]
- 30.Kramer, J., A. H. Visscher, J. A. Wagenaar, and S. H. Jeurissen. 2003. Entry and survival of Salmonella enterica serotype Enteritidis PT4 in chicken macrophage and lymphocyte cell lines. Vet. Microbiol. 91:147-155. [DOI] [PubMed] [Google Scholar]
- 31.Lowenthal, J. W., B. Lambrecht, T. P. van den Berg, M. E. Andrew, A. D. Strom, and A. G. Bean. 2000. Avian cytokines—the natural approach to therapeutics. Dev. Comp. Immunol. 24:355-365. [DOI] [PubMed] [Google Scholar]
- 32.Mandi, Y., T. Veromaa, K. Baranji, A. Miczak, I. Beladi, and P. Toivanen. 1987. Granulocyte-specific monoclonal antibody inhibiting cytotoxicity reactions in the chicken. Immunobiology 174:292-299. [DOI] [PubMed] [Google Scholar]
- 33.Martin, G. D., H. Chart, E. J. Threlfall, E. Morgan, J. M. Lodge, N. L. Brown, and J. Stephen. 2000. Invasiveness of Salmonella serotypes Typhimurium and Enteritidis of human gastro-enteritic origin for rabbit ileum: role of LPS, plasmids and host factors. J. Med. Microbiol. 49:1011-1021. [DOI] [PubMed] [Google Scholar]
- 34.Mastroeni, P., S. Clare, S. Khan, J. A. Harrison, C. E. Hormaeche, H. Okamura, M. Kurimoto, and G. Dougan. 1999. Interleukin 18 contributes to host resistance and gamma interferon production in mice infected with virulent Salmonella typhimurium. Infect. Immun. 67:478-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mastroeni, P., J. A. Harrison, J. H. Robinson, S. Clare, S. Khan, D. J. Maskell, G. Dougan, and C. E. Hormaeche. 1998. Interleukin-12 is required for control of the growth of attenuated aromatic-compound-dependent salmonellae in BALB/c mice: role of gamma interferon and macrophage activation. Infect. Immun. 66:4767-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mastroeni, P., and M. Sheppard. 2004. Salmonella infections in the mouse model: host resistance factors and in vivo dynamics of bacterial spread and distribution in the tissues. Microbes Infect. 6:398-405. [DOI] [PubMed] [Google Scholar]
- 37.McSorley, S. J., B. T. Cookson, and M. K. Jenkins. 2000. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 164:986-993. [DOI] [PubMed] [Google Scholar]
- 38.Methner, U., S. al-Shabibi, and H. Meyer. 1995. Experimental oral infection of specific pathogen-free laying hens and cocks with Salmonella enteritidis strains. Zentbl. Vet. Med. B 42:459-469. [DOI] [PubMed] [Google Scholar]
- 39.Methner, U., P. A. Barrow, D. Gregorova, and I. Rychlik. 2004. Intestinal colonisation-inhibition and virulence of Salmonella phoP, rpoS and ompC deletion mutants in chickens. Vet. Microbiol. 98:37-43. [DOI] [PubMed] [Google Scholar]
- 40.Methner, U., A. Berndt, and A. Haase. 2006. Exploitation of intestinal colonisation inhibition between Salmonella organisms in vaccines for poultry—potential and limitations, p. 585-586. In Proceedings of the International Symposium “Salmonella and Salmonellosis,” Saint-Malo, France.
- 41.Mittrucker, H. W., A. Kohler, and S. H. Kaufmann. 2002. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar Typhimurium infection. Infect. Immun. 70:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizumoto, N., K. Sasai, H. Tani, and E. Baba. 2005. Specific adhesion and invasion of Salmonella Enteritidis in the vagina of laying hens. Vet. Microbiol. 111:99-105. [DOI] [PubMed] [Google Scholar]
- 43.Monack, D. M., D. M. Bouley, and S. Falkow. 2004. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J. Exp. Med. 199:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 45.Murray, G. L., S. R. Attridge, and R. Morona. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 188:2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamura, M., H. S. Lillehoj, R. B. Raybourne, U. S. Babu, and R. A. Heckert. 2004. Cell-mediated immune responses to a killed Salmonella enteritidis vaccine: lymphocyte proliferation, T-cell changes and interleukin-6 (IL-6), IL-1, IL-2, and IFN-gamma production. Comp. Immunol. Microbiol. Infect. Dis. 27:255-272. [DOI] [PubMed] [Google Scholar]
- 47.Okamura, M., H. S. Lillehoj, R. B. Raybourne, U. S. Babu, R. A. Heckert, H. Tani, K. Sasai, E. Baba, and E. P. Lillehoj. 2005. Differential responses of macrophages to Salmonella enterica serovars Enteritidis and Typhimurium. Vet. Immunol. Immunopathol. 107:327-335. [DOI] [PubMed] [Google Scholar]
- 48.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qureshi, M. A., C. L. Heggen, and I. Hussain. 2000. Avian macrophage: effector functions in health and disease. Dev. Comp. Immunol. 24:103-119. [DOI] [PubMed] [Google Scholar]
- 50.Rahman, M. M., J. Guard-Petter, and R. W. Carlson. 1997. A virulent isolate of Salmonella enteritidis produces a Salmonella typhi-like lipopolysaccharide. J. Bacteriol. 179:2126-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravindran, R., and S. J. McSorley. 2005. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology 114:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert-Koch-Institut. 2006. Epidemiol. Bull. 13:99-107. [Google Scholar]
- 53.Rothwell, L., J. R. Young, R. Zoorob, C. A. Whittaker, P. Hesketh, A. Archer, A. L. Smith, and P. Kaiser. 2004. Cloning and characterization of chicken IL-10 and its role in the immune response to Eimeria maxima. J. Immunol. 173:2675-2682. [DOI] [PubMed] [Google Scholar]
- 54.Sasai, K., M. Aita, H. S. Lillehoj, T. Miyamoto, T. Fukata, and E. Baba. 2000. Dynamics of lymphocyte subpopulation changes in the cecal tonsils of chickens infected with Salmonella enteritidis. Vet. Microbiol. 74:345-351. [DOI] [PubMed] [Google Scholar]
- 55.Sheppard, M., C. Webb, F. Heath, V. Mallows, R. Emilianus, D. Maskell, and P. Mastroeni. 2003. Dynamics of bacterial growth and distribution within the liver during Salmonella infection. Cell Microbiol. 5:593-600. [DOI] [PubMed] [Google Scholar]
- 56.Sijben, J. W., K. C. Klasing, J. W. Schrama, H. K. Parmentier, J. J. van der Poel, H. F. Savelkoul, and P. Kaiser. 2003. Early in vivo cytokine genes expression in chickens after challenge with Salmonella typhimurium lipopolysaccharide and modulation by dietary n-3 polyunsaturated fatty acids. Dev. Comp. Immunol. 27:611-619. [DOI] [PubMed] [Google Scholar]
- 57.Srinivasan, A., J. Foley, and S. J. McSorley. 2004. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated Salmonella causes interclonal competition. J. Immunol. 172:6884-6893. [DOI] [PubMed] [Google Scholar]
- 58.Strindelius, L., L. Degling Wikingsson, and I. Sjoholm. 2002. Extracellular antigens from Salmonella enteritidis induce effective immune response in mice after oral vaccination. Infect. Immun. 70:1434-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swaggerty, C. L., M. H. Kogut, P. J. Ferro, L. Rothwell, I. Y. Pevzner, and P. Kaiser. 2004. Differential cytokine mRNA expression in heterophils isolated from Salmonella-resistant and -susceptible chickens. Immunology 113:139-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner, A. K., M. A. Lovell, S. D. Hulme, L. Zhang-Barber, and P. A. Barrow. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valtonen, M. V. 1977. Role of phagocytosis in mouse virulence of Salmonella typhimurium recombinants with O antigen 6,7 or 4,12. Infect. Immun. 18:574-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valtonen, M. V., M. Plosila, V. V. Valtonen, and P. H. Makela. 1975. Effect of the quality of the lipopolysaccharide on mouse virulence of Salmonella enteritidis. Infect. Immun. 12:828-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Immerseel, F., J. De Buck, I. De Smet, J. Mast, F. Haesebrouck, and R. Ducatelle. 2002. The effect of vaccination with a Salmonella enteritidis aroA mutant on early cellular responses in caecal lamina propria of newly-hatched chickens. Vaccine 20:3034-3041. [DOI] [PubMed] [Google Scholar]
- 64.Wang, J., D. L. Adelson, A. Yilmaz, S.-H. Sze, Y. Jin, and J. J. Zhu. 2005. Genomic organization, annotation, and ligand-receptor inferences of chicken chemokines and chemokine receptor genes based on comparative genomics. BMC Genomics 6:45-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wick, M. J. 2004. Living in the danger zone: innate immunity to Salmonella. Curr. Opin. Microbiol. 7:51-57. [DOI] [PubMed] [Google Scholar]
- 66.Wilson, R. L., J. Elthon, S. Clegg, and B. D. Jones. 2000. Salmonella enterica serovars Gallinarum and Pullorum expressing Salmonella enterica serovar Typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect. Immun. 68:4782-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Withanage, G. S., P. Kaiser, P. Wigley, C. Powers, P. Mastroeni, H. Brooks, P. Barrow, A. Smith, D. Maskell, and I. McConnell. 2004. Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar Typhimurium. Infect. Immun. 72:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Withanage, G. S., P. Wigley, P. Kaiser, P. Mastroeni, H. Brooks, C. Powers, R. Beal, P. Barrow, D. Maskell, and I. McConnell. 2005. Cytokine and chemokine responses associated with clearance of a primary Salmonella enterica serovar Typhimurium infection in the chicken and in protective immunity to rechallenge. Infect. Immun. 73:5173-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yrlid, U., M. Svensson, A. Hakansson, B. J. Chambers, H. G. Ljunggren, and M. J. Wick. 2001. In vivo activation of dendritic cells and T cells during Salmonella enterica serovar Typhimurium infection. Infect. Immun. 69:5726-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]